ABSTRACT
G protein-coupled receptors (GPCRs) are transmembrane receptors that relay signals from the external environment inside the cell, allowing an organism to adapt to its surroundings. They are known to detect a vast array of ligands, including sugars, amino acids, pheromone peptides, nitrogen sources, oxylipins, and light. Despite their prevalence in fungal genomes, very little is known about the functions of filamentous fungal GPCRs. Here we present the first full-genome assessment of fungal GPCRs through characterization of null mutants of all 15 GPCRs encoded by the aflatoxin-producing fungus Aspergillus flavus. All strains were assessed for growth, development, ability to produce aflatoxin, and response to carbon sources, nitrogen sources, stress agents, and lipids. Most GPCR mutants were aberrant in one or more response processes, possibly indicative of cross talk in downstream signaling pathways. Interestingly, the biological defects of the mutants did not correspond with assignment to established GPCR classes; this is likely due to the paucity of data for characterized fungal GPCRs. Many of the GPCR transcripts were differentially regulated under various conditions as well. The data presented here provide an extensive overview of the full set of GPCRs encoded by A. flavus and provide a framework for analysis in other fungal species.
IMPORTANCE
Aspergillus flavus is an opportunistic pathogen of crops and animals, including humans, and it produces a carcinogenic toxin called aflatoxin. Because of this, A. flavus accounts for food shortages and economic losses in addition to sickness and death. Effective means of combating this pathogen are needed to mitigate its deleterious effects. G protein-coupled receptors (GPCRs) are often used as therapeutic targets due to their signal specificity, and it is estimated that half of all drugs target GPCRs. In fungi such as A. flavus, GPCRs are likely necessary for sensing the changes in the environment, including food sources, developmental signals, stress agents, and signals from other organisms. Therefore, elucidating their functions in A. flavus could identify ideal receptors against which to develop antagonists.
INTRODUCTION
Aspergillus flavus is a soilborne fungus that acts as a saprophyte, decomposing dead organic matter. A. flavus is also an opportunistic pathogen of both plants and animals. It is capable of infecting a range of crops, both pre- and postharvest, including corn, peanuts, and cottonseed, where it produces the carcinogenic mycotoxin aflatoxin (AF). Ingestion of low doses of AF can lead to hepatocellular carcinoma, while high doses are toxic and lethal. The loss of crops infected with this fungus leads to food shortages and economic losses in the billion-dollar range in the United States alone (1). Humans and animals can also be infected by A. flavus via the inhalation of spores, and it is the second leading cause of aspergillosis after Aspergillus fumigatus. It is commonly associated with manifestations of aspergillosis in the skin, oral mucosa, and subcutaneous tissues (2). In order to thrive in such vastly different environments, the fungus must be able to compete with other organisms for nutrition and defend itself against attacks from other microbes and the plant and animal hosts it invades. Therefore, the ability to sense and respond to the various environments in which it lives is critical for A. flavus’ survival.
G protein-coupled receptors (GPCRs) are transmembrane (TM) proteins that detect external signals and transmit that information inside the cell to elicit some type of response. They contain 7 TM helices that are joined by internal and external loops. The N terminus resides outside the cell, while the C terminus is inside. GPCRs bind heterotrimeric G proteins, which are composed of an α, β, and γ subunit, and the α subunit is bound to GTP or GDP and is capable of GTP hydrolysis. Upon activation by their cognate ligands, GPCRs undergo a conformational change. This triggers the exchange of GTP for GDP on the Gα subunit, resulting in dissociation of the G protein. Both the Gα subunit and the Gβγ dimer can initiate downstream signaling pathways. Additional proteins that impact this reaction are regulators of G protein signaling (RGS), which quench the signal by accelerating GTPase activity of the Gα subunit. When GTP is hydrolyzed to GDP, the G protein reassociates in its inactive heterotrimeric state. Signaling is also terminated by internalization of the GPCR, which is regulated by phosphorylation and ubiquitination of the GPCR C terminus, triggering endocytosis and degradation of the receptor (reviewed in reference 3).
Both the Gα and Gβγ subunits can act as effectors of downstream signaling cascades. These have been well studied in the model yeast Saccharomyces cerevisiae. Sugars stimulate a cAMP-protein kinase A (PKA) pathway in which Gα activates adenylate cyclase, which converts ATP to cAMP. The increased concentration of cAMP results in the release of inhibitory subunits from PKA, thereby activating the enzyme. PKA then phosphorylates downstream proteins to impact the cellular response. Following yeast pheromone sensing, the Gβγ dimer acts as the effector for the mitogen-activated protein kinase (MAPK) pathway, which consists of a kinase cascade that regulates mating (4). Components of both the cAMP-PKA and MAPK pathways have been identified in the aspergilli and found to regulate germination, sporulation, mycotoxin production, and stress tolerance (reviewed in references 5 and 6).
Based on their potential importance to fungal growth and survival, GPCRs have been the subject of numerous bioinformatics studies. As a result, the entire set of GPCRs encoded by various fungi has been predicted for Aspergillus nidulans, Aspergillus fumigatus, Aspergillus oryzae, Magnaporthe grisea, Cryptococcus neoformans, Neurospora crassa, Verticillium spp., and Trichoderma spp. (7–12), with numbers ranging from 7 (C. neoformans) to 22 (Verticillium dahliae), in contrast to the three GPCRs in the S. cerevisiae genome. However, the functions of almost all of these GPCRs remain unknown. In this study, we identified the putative GPCRs encoded by A. flavus and carried out functional studies to investigate the roles of these GPCRs in growth, development, response to various carbon and nitrogen sources, stress agents, and fatty acid-derived signals. From this, we can begin to classify A. flavus’ GPCRs by their biological functions. We found that many receptors appear to be involved in multiple processes, highlighting the cross talk that occurs between different signal transduction pathways.
RESULTS
A. flavus encodes 15 putative GPCRs.
We began our search for A. flavus GPCRs by examining the set of 16 GPCRs reported for A. nidulans (7). In an effort to focus on the canonical GPCRs, our study did not include the PTH11-like membrane-spanning proteins, of which A. nidulans is predicted to encode at least 25 (7, 8). A BLAST search of the amino acid sequences of each of the 16 A. nidulans GPCRs against the A. flavus genome yielded 77 total hits, 57 of which were unique. GPCRs pass the membrane seven times, so two protein topology prediction tools, TMHMM 2.0 (13, 14) and TopPred 1.10 (15, 16), were used to determine which of these 57 proteins were likely GPCRs. Twenty-nine of the fifty-seven sequences were not predicted to encode any transmembrane (TM) domains, so they were eliminated. Of the remaining 28 proteins, 13 were predicted to have 7 TM domains by at least one of the prediction tools. Two additional proteins that were homologous to A. nidulans GprF and GprO were predicted to have fewer than 7 TM domains, but since their A. nidulans orthologs were predicted to have 7 TM domains, they were also included with the rest of the A. flavus GPCRs, bringing the total to 15 (Table 1).
TABLE 1 .
Overview of A. flavus GPCRs
| Gene | Gene ID (AFLA_x) | No. of amino acids | Class | Conserved domain (note) | No. of TM domainsa |
|---|---|---|---|---|---|
| gprA | 060740 | 374 | I | STE2 GPCR (S. cerevisiae pheromone receptor) | 6/7 |
| gprB | 061620 | 465 | II | STE3 GPCR (S. cerevisiae pheromone receptor) | 7/7 |
| gprC | 074150 | 444 | III | Git3; Git3_C (S. pombe glucose receptor) | 7/6 |
| gprD | 135680 | 415 | III | Git3; Git3_C (S. pombe glucose receptor) | 7/7 |
| gprF | 006880 | 300 | IV | PQ loop repeat (S. pombe nitrogen sensor) | 4/5 |
| gprG | 067770 | 426 | IV | PQ loop repeat (S. pombe nitrogen sensor) | 7/7 |
| gprH | 006920 | 428 | V | Secretin family (signal through cAMP pathways) | 7/7 |
| gprJ | 127870 | 322 | IV | PQ loop repeat (S. pombe nitrogen sensor) | 7/7 |
| gprK | 009790 | 560 | VI | RGS domain (regulator of G protein signaling) | 7/7 |
| gprM | 075000 | 490 | VII | [No conserved domains] | 7/7 |
| gprO | 032130 | 282 | VIII | Hemolysin III related (broad range of ligands) | 6/7 |
| gprP | 088190 | 502 | VIII | Hemolysin III related (broad range of ligands) | 7/7 |
| gprR | 023070 | 523 | VI | RGS domain (regulator of G protein signaling) | 7/7 |
| gprS | 006320 | 266 | IV | PQ loop repeat (S. pombe nitrogen sensor) | 7/7 |
| nopA | 117970 | 312 | IX | Bacteriorhodopsin-like (photoreactive) | 7/7 |
The first number is predicted by TMHMM; the second is predicted by TopPred.
We also compared the A. flavus genome to the reported A. oryzae GPCRs (7) since these two species are believed to be the same species, with A. oryzae constituting a domesticated clade (17). Similar to what was reported for A. oryzae, no orthologs for A. nidulans GprE, GprI, or GprN were found in the A. flavus genome. A novel A. oryzae GPCR not found in A. nidulans was identified and named GprQ. Its A. flavus ortholog (AFLA_132040; 99% identity, with an E value of 7e−115) had only four (TMHMM) or three (TopPred) predicted TM domains. Since A. oryzae GprQ was reported to encode only five predicted TM domains, the A. flavus homolog was not included in the set of GPCRs.
The BLAST search also identified two GPCRs that were not previously identified in the aspergilli, GprR and GprS. Like GprK, GprR harbors an RGS (regulator of G protein signaling) domain at the C-terminal end. This type of GPCR was first discovered in Arabidopsis thaliana and called A. thaliana RGS1 (AtRGS1). AtRGS1 regulates cell proliferation as well as sensitivity to sugars via its functional RGS domain. Unlike canonical GPCRs, it does not trigger the exchange of GTP for GDP on a G protein. Upon sensing glucose, AtRGS1 interacts with the constitutively active Gα subunit A. thaliana GPA1 (AtGPA1), resulting in hydrolysis of GTP and subsequent deactivation of the G protein (18–20). GPCR-RGS hybrids have since been found in several species of filamentous ascomycete fungi in addition to the aspergilli, including N. crassa, M. grisea, Fusarium graminearum, and several Trichoderma species (7, 10, 12). GprS contains a PQ loop repeat, grouping it with the class IV receptors GprF, GprG, and GprJ. The function of the PQ loop repeat domain is unknown, although this class contains the Schizosaccharomyces pombe nitrogen starvation sensor Stm1 (7).
Based on their domain structures, A. flavus’ 15 putative GPCRs were assigned to nine different classes. These classes have been previously described (7, 21), and a few notes on each class are included in Table 1. The amino acid sequences for all 15 GPCRs, as well as the sequences for GPCRs from A. nidulans, A. fumigatus, A. oryzae, F. graminearum, M. grisea, N. crassa, Trichoderma reesei, and V. dahliae, were aligned using the Clustal Omega software tool (22), and a phylogenetic tree was generated using the Phylogeny.fr software tool (23) (see Fig. S1 in the supplemental material). From this analysis, it was apparent that the A. flavus GPCRs GprF, GprH, GprS, and GprR are not orthologous to any of A. oryzae’s predicted GPCRs, despite the high degree of genetic similarity between the two fungi. Interestingly, of the two GPCRs that were not previously found in the aspergilli, GprS was most closely related to two GPCRs from V. dahliae, while GprR was unique among the fungi analyzed here.
In order to verify the coding sequences of the A. flavus GPCRs, 13 of the 15 GPCRs were amplified, cloned, and sequenced from A. flavus mRNA. cDNA for gprJ and gprM could not be amplified. Two GPCRs, gprF and gprO, appeared to be misannotated. In both cases, the actual sequence improved the proteins’ alignments with their A. nidulans orthologs, and for GprO, it changed the number of predicted TM domains from six to seven (TopPred), with the updated prediction reported in Table 1. GprF was only predicted to encode four (TMHMM) or five (TopPred) TM domains, but since A. nidulans GprF was predicted to encode seven TM domains, it was included with the set of A. flavus GPCRs.
Individual GPCRs deleted in A. flavus.
All 15 of the GPCRs were individually disrupted in the CA14 ΔpyrG Δku70 strain of A. flavus by replacing the GPCR-encoding gene with pyrG. Deletion mutants were confirmed by PCR (data not shown) and Southern blotting (see Fig. S2 in the supplemental material), and confirmed isolates were checked for marker gene effects by growing on media with and without uridine and uracil (UU) (see >Fig. S3 to S5; also data not shown) (24). The ΔgprG, ΔgprK, ΔgprM, and ΔgprR mutants all exhibited growth defects that were at least partially remediated by supplementation of UU.
To investigate the impact of UU supplementation on a phenotypic assay, a subset of mutants was examined in greater detail. First, gprR was disrupted in strain NRRL3357 of A. flavus. Like the CA14 ΔgprR mutant, the NRRL3357 ΔgprR mutant was severely restricted in growth in the absence of UU, demonstrating that the need for pyrimidine supplementation was conserved in different genetic backgrounds (see Fig. S3 in the supplemental material). Strains were then exposed to two different stressors: alkaline pH (pH 8) and cell wall stress (via Congo red), with and without UU. The wild types for both the CA14 and NRRL3357 backgrounds were inhibited to similar degrees regardless of UU supplementation, whereas the addition of UU was required to observe stress-induced inhibition for both ΔgprR mutants (see Fig. S3). Multiple independent isolates of a mutant in the same genetic background were similarly examined. The ΔgprM mutant had shown a marker gene effect for all three confirmed transformants, and without UU, the effects of the stresses were more dramatic (see Fig. S4). As a final test of the effects of UU supplementation, multiple isolates of two mutants that had not shown marker gene effects, the ΔgprD and ΔgprH mutants, were analyzed. Independent isolates of both mutations showed similar patterns of inhibition regardless of UU supplementation (see Fig. S5). The impacts of alkaline pH and cell wall stress were quantified for all of the isolates of the ΔgprR, ΔgprM, ΔgprD, and ΔgprH mutants, revealing that multiple isolates of the same mutant behaved similarly (see Table S1). Therefore, a single isolate for each GPCR disruption was chosen, and UU was included in the media for all subsequent experiments.
GPCRs important in basic A. flavus development and aflatoxin synthesis.
Various fungal developmental processes have previously been linked to G protein signaling. To elucidate which GPCRs may be involved in mediating these signals, we measured the germination rate, AF biosynthesis, sporulation under both light and dark conditions, and production of spores and sclerotia at high and low cell densities, described below.
(i) Germination.
Germination studies in A. nidulans have revealed several components of G protein signaling to be important in that process. The heterotrimeric G protein GanB-SfaD-GpgA activates a cAMP pathway in response to glucose to drive germination, and this is abrogated by RGS RgsA (25–27). There is also evidence of a small GTPase, RasA, controlling germination independently of cAMP (25, 28). The only GPCRs implicated in germination thus far are A. nidulans GprD (21) and A. fumigatus GprC and GprD (29). Therefore, germination of the entire set of A. flavus Δgpr mutants was monitored from 4 to 9 h postinoculation. During this time period, the wild type went from 1% to 77% germinated spores (data not shown). The germination rates of the Δgpr mutants were measured, and a heat map was generated with the percentage of mutant germination versus wild-type germination for each time point (Table 2). Aside from a slightly enhanced germination rate at 6 h by the ΔgprO mutant, the rest of the mutants were either equivalent or impaired in germination compared to the wild type. The most severely deficient mutants were the ΔgprA and ΔgprK strains, with <50% of the wild-type germination rate at nearly all time points measured. Many of the other mutants, including the ΔgprB, ΔgprC, ΔgprH, ΔgprJ, ΔgprP, ΔgprR, and ΔgprS strains, were impaired in germination as well.
TABLE 2 .
Developmental phenotypes of A. flavus Δgpr mutantsa
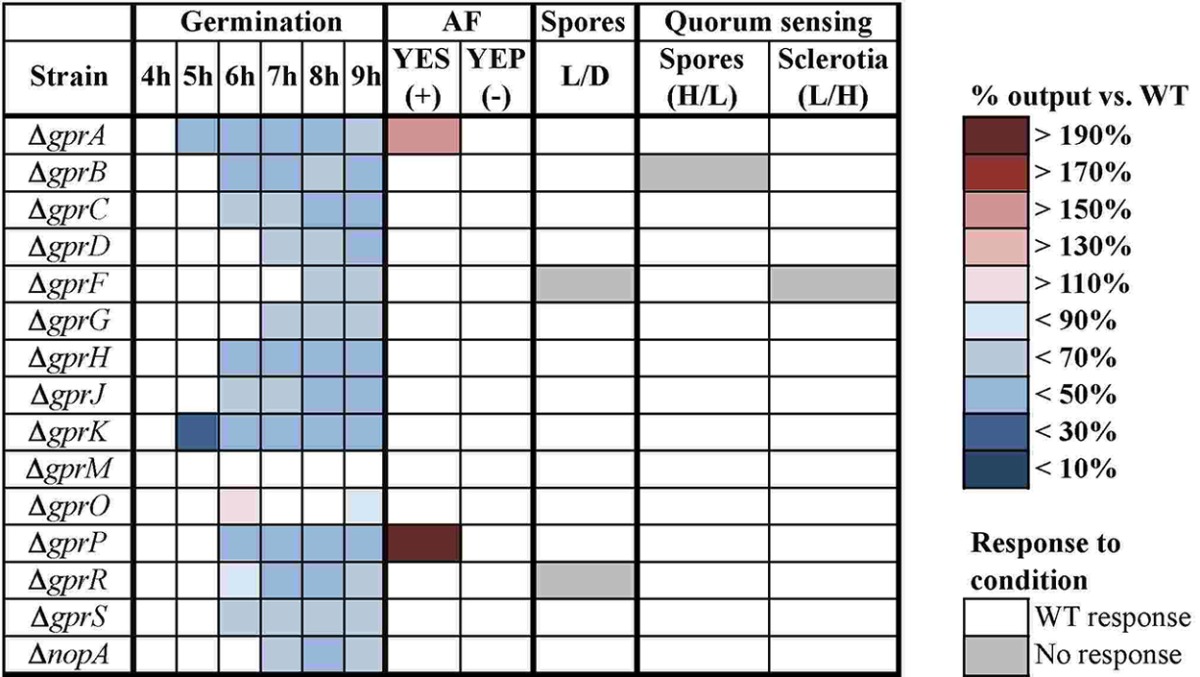
GPCR mutants were assayed for germination rate at 4 to 9 h postinoculation and for aflatoxin (AF) production on AF-promoting (YES) and AF-repressing (YEP) media. For both assays, shaded squares represent data points that were significantly different from those for the wild type (WT). The color of each box indicates the amount of germinated spores or AF produced as a percentage of WT germination or AF produced. The ratios of spores produced under light and dark conditions (L/D), as well as spores and sclerotia produced by high-density (H) and low-density (L) inocula, were determined, and any data point that did not exhibit the same pattern as the WT is shaded gray. For all four experiments, statistical significance was determined by a Student t test, with P < 0.05. Deletions are shown in the “Strain” column.
(ii) Light-induced sporulation.
Rhodopsin is well characterized as a receptor of light, and A. flavus encodes one GPCR, NopA, that belongs to the class IX bacteriorhodopsin-like GPCR family. Since light is an inducer of sporulation, the fungi were exposed to continuous light or continuous dark, and their spores were counted. The wild type and most of the mutants produced significantly more spores in the light versus production in the dark. Two mutants, the ΔgprF and ΔgprR mutants, showed no significant difference between the two conditions. Notably, the ΔnopA mutant exhibited the wild-type response to light (Table 2).
(iii) Quorum sensing.
A. flavus undergoes a quorum-sensing-mediated developmental shift in which at low inoculum density, it produces many sclerotia and very few spores. The opposite is observed at high inoculum density (30). Because an exogenous signal mediates this phenomenon, GPCRs are plausible targets for relaying the signal. In fact, two GPCRs in the A. flavus strain NRRL3357, encoded by gprC and gprD, have already been implicated as quorum-sensing receptors based on their inability to respond to a high-density extract (31). Surprisingly, all 15 Δgpr mutants produced more sclerotia and fewer spores at high density than the wild type (data not shown). However, the trend of decreasing sclerotia and increasing spore production as inoculum density increased was still observed for most of the mutants, with the exceptions of the ΔgprB and ΔgprF mutants There was no significant difference in the amount of spores produced at high and low densities for the ΔgprB mutant, nor was there a significant difference in sclerotia production at the two densities for the ΔgprF mutant (Table 2).
(iv) Aflatoxin.
G protein signaling has been shown to regulate synthesis of AF and its precursor, sterigmatocystin (ST). The Gα subunit FadA and its RGS FlbA are part of an adenylate cyclase/cAMP/protein kinase A (PKA) (PkaA) pathway that controls ST production in A. nidulans via transcriptional and posttranscriptional regulation of aflR (32–34). The orthologous pathway regulates AF biosynthesis in Aspergillus parasiticus (35) and A. flavus (J. W. Bok and N. P. Keller, unpublished). Nothing is known about which GPCR(s) might be initiating this pathway, so the panel of Δgpr mutants was tested for their ability to make AF. AF was extracted and measured following growth on AF-inducing and AF-repressing media (YES [2% yeast extract and 6% sucrose, pH 5.8] and YEP [2% yeast extract and 6% peptone, pH 5.8], respectively). No mutant gained the ability to produce detectable AF on YEP. However, on YES medium, the ΔgprA mutant made ~1.5× more AF than the wild type, and the ΔgprP mutant made ~2.3× more AF than the wild type (Table 2). None of the other mutants were affected in their ability to produce AF.
GPCRs impact growth on various carbon and nitrogen sources.
In addition to developmental cues, GPCRs are able to detect the presence or absence of important nutrients. GprC and GprD belong to class III, which is the group that also contains the sugar receptors S. cerevisiae Gpr1 and S. pombe Git3. Gpr1 senses glucose and sucrose (with a stronger affinity for sucrose) and transmits the signal through a G protein/cAMP/PKA pathway (36–38). Other Gpr1 homologs are also carbon source sensors, such as S. pombe Git3 (reviewed in reference 39) and N. crassa GPR-4 (40), although the role of C. albicans Gpr1 in carbon sensing is ambiguous (41, 42). While a carbon-sensing GPCR has not been identified in the aspergilli, it has been shown that carbon sensing is mediated by the heterotrimeric G protein GanB-SfaD-GpgA in A. nidulans (27). To identify A. flavus GPCRs involved in carbon sensing, strains were grown on a variety of carbon and nitrogen sources. These included glucose, galactose, xylose, sucrose, and corn oil for carbon sources. Corn oil is mainly composed of C18 and C16 fatty acids (43). The ΔgprA, ΔgprC, ΔgprJ, ΔgprK, and ΔgprR mutants were impaired in growth on several of the carbon sources tested (Table 3). The ΔgprR mutant was restricted in growth on every carbon source tested except galactose and corn oil.
TABLE 3 .
Growth of A. flavus Δgpr mutants on various carbon and nitrogen sourcesa
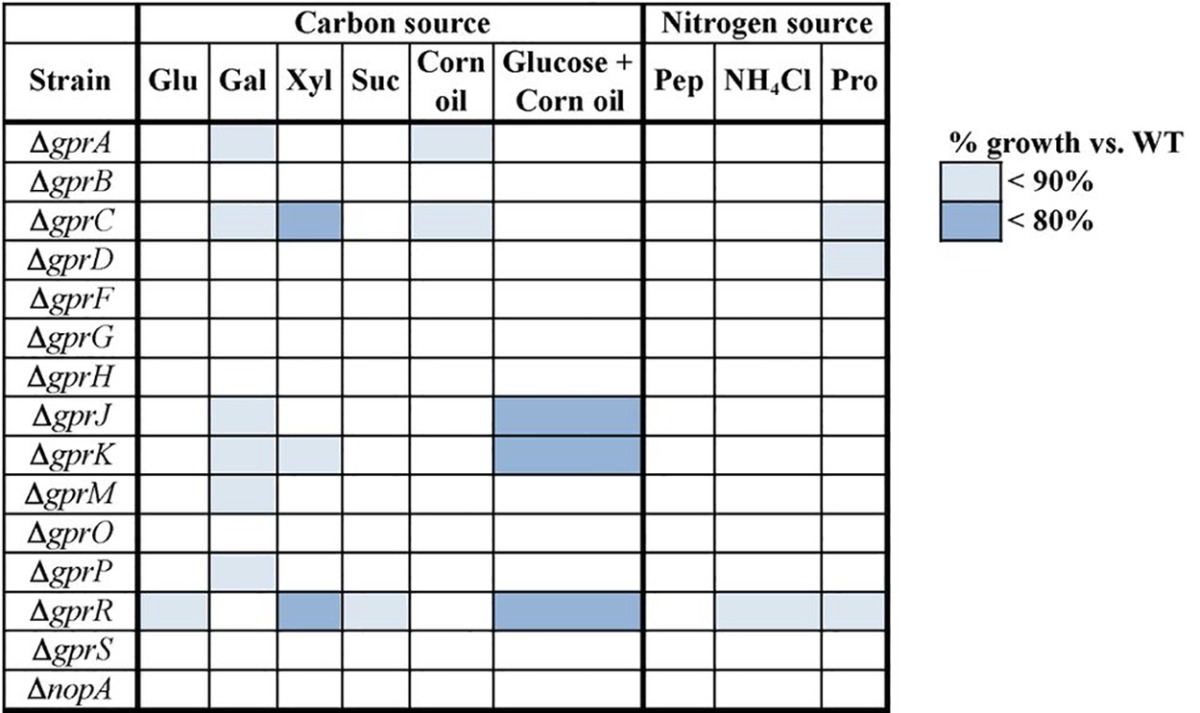
Strains were grown on a variety of media with different sources of carbon and nitrogen. The radial growth was measured, and mutants were compared to the WT on the same medium. The carbon sources were glucose (Glu), galactose (Gal), xylose (Xyl), sucrose (Suc), corn oil, and corn oil with glucose. The nitrogen sources were peptone (Pep), ammonium chloride, and proline (Pro). Shaded boxes indicate data points that were significantly different from those for the WT, with the color representing the degree of growth inhibition compared to the WT. Significance was determined by a Student t test, with P < 0.05. Deletions are shown in the “Strain” column.
Class IV receptors are grouped together based on their relatedness to the S. pombe nitrogen starvation sensor Stm1, and A. flavus has four receptors in this group. Nitrogen starvation results in cell cycle arrest in S. pombe, which allows the yeast to remain viable either until nutrition becomes available or mating partners are present. stm1 is expressed at low, moderate, and high levels on peptone, ammonium, and proline, respectively. Deletion of stm1 causes premature cell cycle arrest under nitrogen starvation, while overexpression of stm1 represses vegetative growth under nitrogen-replete conditions (44). Therefore, the mutants were tested for their ability to grow on peptone, ammonium chloride, and proline. While no mutant was restricted on peptone, mutants carrying both ΔgprC and ΔgprD deletions—the class III receptors—were inhibited on proline. The ΔgprR mutant was impaired on ammonium chloride and proline (Table 3).
GPCRs contribute to fungal stress responses.
Fungi transmit the myriad stress signals they encounter through several conserved, interconnected signal transduction pathways. Multiple stimuli trigger each pathway, and the pathways themselves engage in cross talk with one another. For example, the cell wall integrity (CWI) MAPK pathway is triggered in response not only to cell wall stress but to osmoregulatory and oxidative stresses as well, among others (reviewed in reference 45). In addition to the CWI pathway, osmotic stress is also managed by another MAPK pathway, the high-osmolarity glycerol (HOG) pathway (reviewed in reference 46). pH tolerance is maintained by the Pal/Rim alkaline response pathway. It transmits its signal via PalH, a 7-TM-domain protein that interacts with an arrestin protein to initiate endocytosis, ultimately activating the PacC transcription factor (reviewed in reference 47). Besides their overlapping roles in stress tolerance, what these pathways have in common is the prevalence of G protein signaling components. To examine whether any of A. flavus’ GPCRs mediate stress responses, the mutants were exposed to a variety of stressors that inhibit the radial growth of the wild type. The percent inhibition of growth in the presence versus absence of the stressor was compared between the wild type and each mutant, and the changes in inhibition between wild type and the mutants are presented as a heat map (Table 4).
TABLE 4 .
Stress responses of A. flavus Δgpr mutantsa
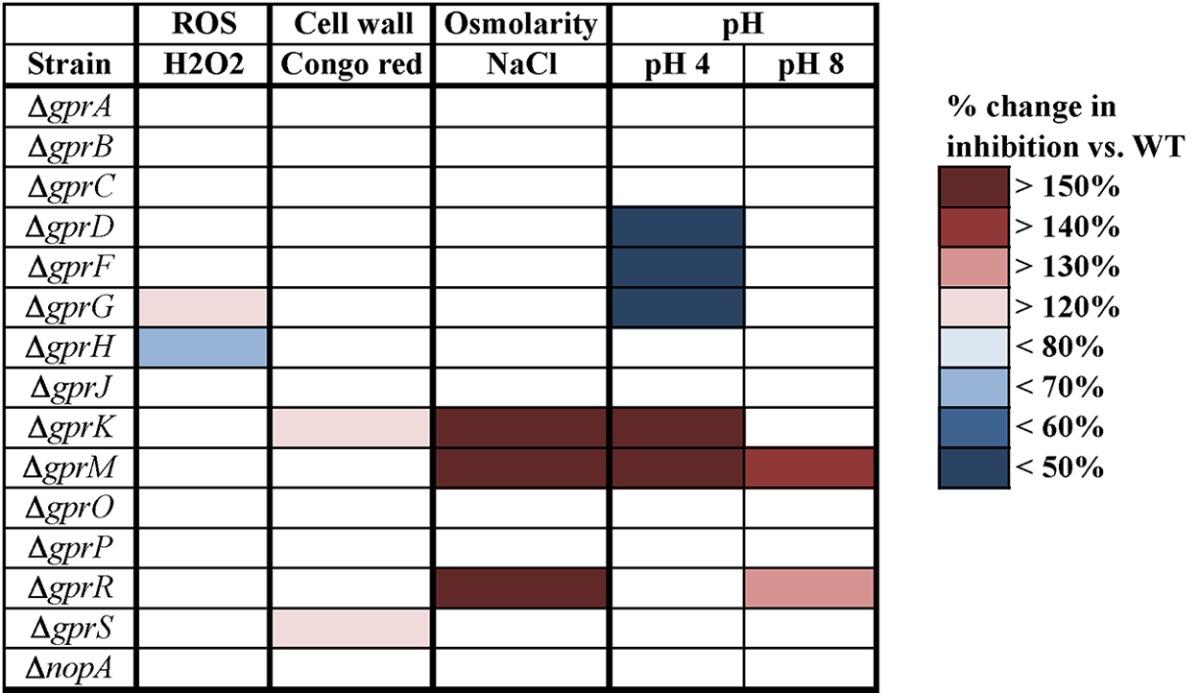
Strains were exposed to a variety of stresses, including reactive oxygen species (ROS), cell wall stress, osmolarity stress, and high and low pHs. Percent inhibition of growth under stress versus growth on control medium (GMM) was measured. Shaded boxes indicate data points in which the percent inhibition of the mutant differed significantly from that of the WT, and the degree of the difference is indicated by the different colors. Darker red tones indicate mutants with greater sensitivity to the stressor, while darker blue tones denote the opposite. Significance was determined by a Student t test, with P < 0.05. Deletions are shown in the “Strain” column.
Hydrogen peroxide was used at a concentration of 3 mM to assay oxidative stress. Loss of gprH rendered the fungus more resistant to hydrogen peroxide, while the ΔgprG mutant was more sensitive. Cell wall stress was created by growing the fungi on Congo red, which weakens the cell wall by binding to growing chitin chains, preventing their attachment to other cell wall components (48, 49). The ΔgprK, and ΔgprS mutants were both more sensitive to Congo red. The impact of osmotic pressure was measured with 1 M sodium chloride. Three mutants, carrying the ΔgprK, ΔgprM, and ΔgprR deletions, were more sensitive than the wild type to the hyperosmotic condition. Finally, the strains were grown on media buffered to pH 4 and pH 8. The ΔgprM mutant was more sensitive to both conditions, while the ΔgprK and ΔgprR mutants were more sensitive to acidic and alkaline pHs, respectively. Alternatively, the ΔgprD, ΔgprF, and ΔgprG mutants were more resistant to acidic pH.
GPCRs are involved in response to lipids and oxylipins.
It has long been speculated that oxylipins are detected by fungal GPCRs, since this is their mode of perception in mammalian cells (reviewed in references 3, 5, and 50). To address this, strains were exposed to the oxylipins methyl jasmonate (MeJA) and 13-hydroperoxy-octadecadienoic acid [13(S)-HpODE] and to the fatty acid linoleic acid. Any mutant that deviated from the wild-type response to these chemicals is indicated in Table 5 with a shaded box.
TABLE 5 .
A. flavus Δgpr mutant responses to fatty acids and oxylipinsa
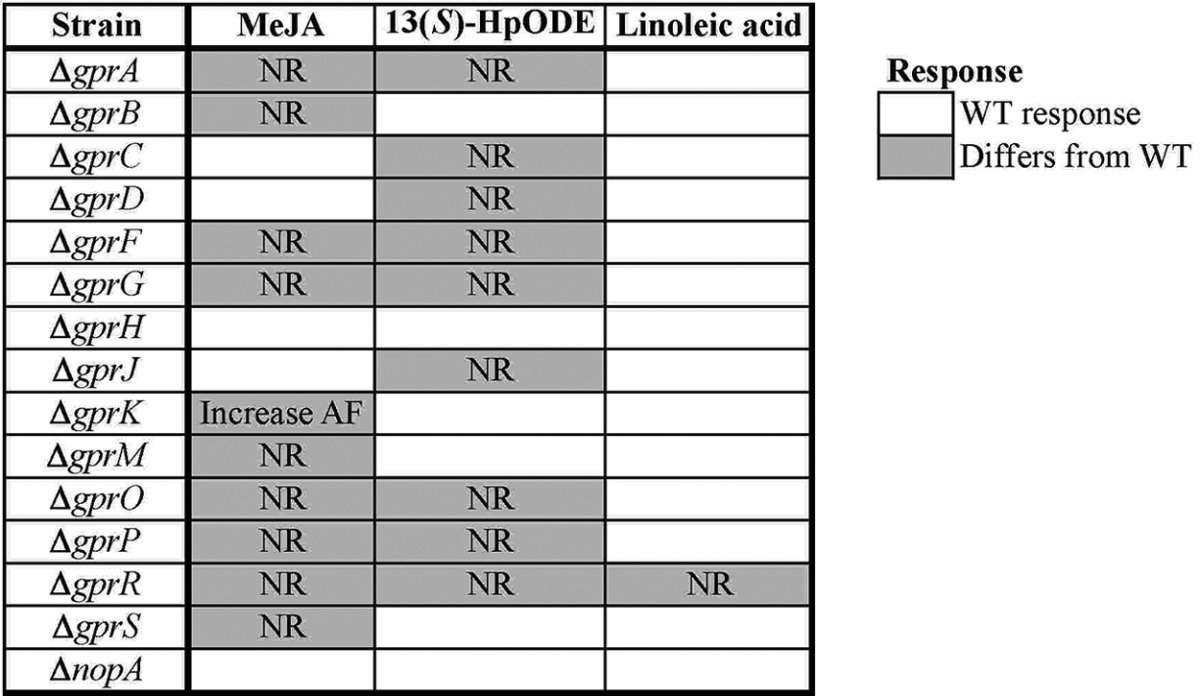
Strains were incubated with or without MeJA, a repressor of AF biosynthesis, and AF was extracted and measured by HPLC. Strains were also exposed to disks soaked with the sporulation inducers 13(S)-HpODE and linoleic acid or the negative control ethanol (EtOH), and spores surrounding the disks were counted. Shaded boxes represent data points that did not exhibit the WT response, and the mutant response is indicated (“NR” means “no response”). Statistical significance was determined by a Student’s t test with P < 0.05. Deletions are shown in the “Strain” column.
The effect of MeJA on AF production varies depending on the experimental setup and concentration of MeJA used (51). Under certain conditions, MeJA has been shown to repress AF production by A. flavus (52), but under others, it has enhanced AF synthesis by A. parasiticus (53). For this study, an AF-repressive protocol was followed (51) in which cultures were exposed to 10−4 M MeJA, and AF was measured after 3 days. AF repression was observed for the wild type and five of the Δgpr mutants: the ΔgprC, ΔgprD, ΔgprH, ΔgprJ, and ΔnopA mutants. The rest of the mutants showed no change in AF production except for the ΔgprK mutant, which actually made more AF when exposed to MeJA (Table 5). A lack of response to MeJA cannot be attributed to a basic biosynthetic defect because, as previously shown, none of the mutants were impaired in AF biosynthesis on YES media (Table 2).
13(S)-HpODE, as well as its precursor fatty acid, linoleic acid, has been shown to induce sporulation in A. flavus (54). Stimulation of conidiation by 13(S)-HpODE was lost for the ΔgprA, ΔgprC, ΔgprD, ΔgprF, ΔgprG, ΔgprJ, ΔgprO, ΔgprP, and ΔgprR mutants. To test whether this represented a general sporulation defect or a specific lack of response to 13(S)-HpODE, sporulation was also monitored in response to linoleic acid. All of the mutants except the ΔgprR mutant responded as did the wild type to linoleic acid, indicating that the lack of response to 13(S)-HpODE was specific for all of the above mutants except the ΔgprR mutant (Table 5).
GPCRs are differentially expressed under disparate conditions.
To further understand potential roles of A. flavus GPCRs, their expression levels under different conditions from various published studies were investigated. The results are compiled in Table 6, which revealed several GPCRs to be differentially expressed on maize versus liquid culture mycelia, and these GPCRs do not overlap with those that are differentially expressed on maize versus wheat (55). The corn kernel germ is mainly composed of lipids, while the endosperm is predominantly starch, and A. flavus preferentially grows in the lipid-rich portions of the kernel (56). Interestingly, four GPCRs, encoded by gprC, gprH, gprM, and gprS, were differentially expressed on the germ versus the endosperm (55). Two separate studies compared genomic expression from mycelial growth versus sclerotial production, though the methods used were quite different between the two studies (55, 57). The strains of A. flavus, media, and growth conditions (i.e., liquid versus solid culture) that were used varied, as did the methods for measuring gene expression (i.e., microarray versus transcriptome sequencing [RNA-seq]). Both studies showed that many of A. flavus’ GPCRs were differentially expressed depending on whether the fungus was growing as mycelia or producing sclerotia, although the expression patterns differed greatly between the two studies. 5-Azacytidine (5-AC) is an inhibitor of AF production and sporulation (58), but application of 5-AC to cultures of A. flavus did not impact expression of any of its GPCRs (59).
TABLE 6 .
Expression of GPCR-encoding genes under various conditionsa
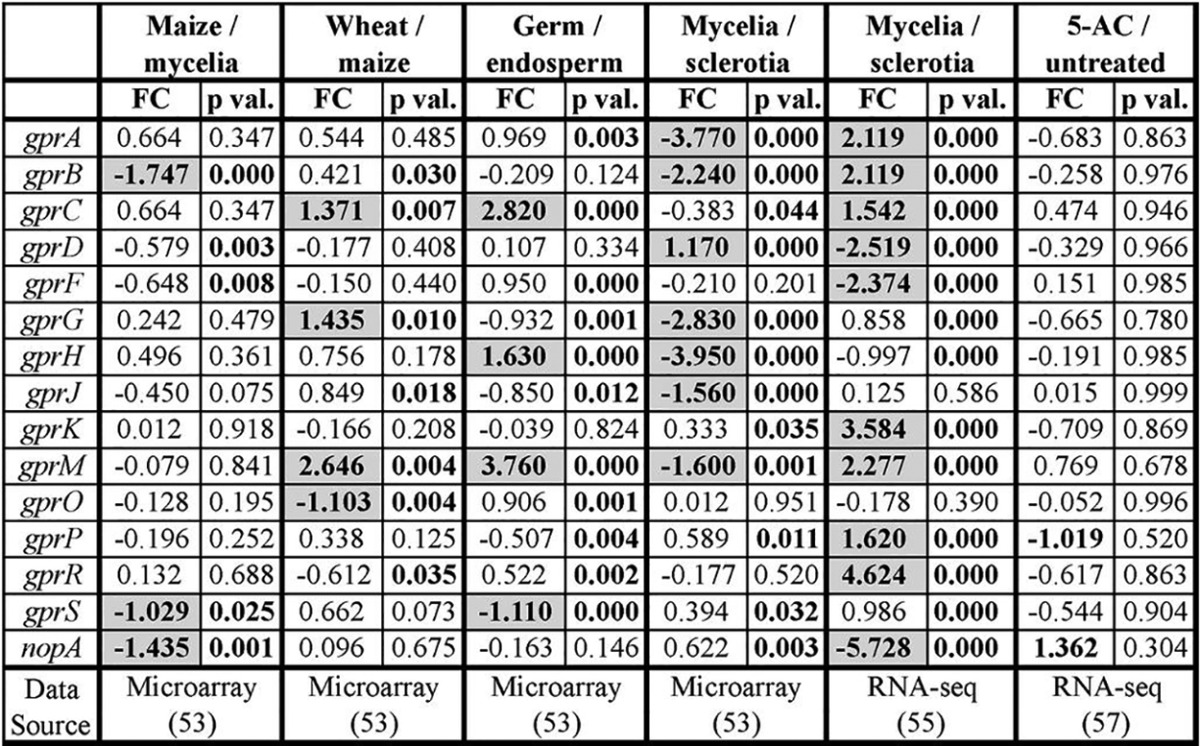
Expression data from several different studies were mined for expression levels of the GPCRs under various conditions. For each data set, the log2-fold change (FC) value and P value (p val.) are reported. The first four data sets contain microarray data, while the last two contain RNA-seq data. The first data set (“Maize/mycelia”) reports the FC in expression from A. flavus grown as mycelia in liquid culture to A. flavus grown on maize. The second (“Wheat/maize”) reports the FC in expression from A. flavus growing on maize to A. flavus growing on wheat. The third (“Germ/endosperm”) reports the FC in expression from A. flavus growing on the corn kernel endosperm to growth on the kernel germ. The fifth and sixth data sets (“Mycelia/sclerotia”) report the FC in expression from A. flavus growing as mycelia to producing sclerotia. The final data set reports the FC in expression from untreated A. flavus cultures to those exposed to 5-AC.
DISCUSSION
Several works have bioinformatically identified the GPCRs encoded by various fungi. These include A. nidulans, A. fumigatus, A. oryzae, M. grisea, C. neoformans, N. crassa, Verticillium spp., and Trichoderma spp. (7–12). Functions have been assigned for a few of these GPCRs. However, this is the first study to identify functions for a fungus’ entire set of GPCRs, providing leads on most of the GPCRs that can be pursued further in future studies.
An attempt was made to identify the activating ligands of A. flavus’ GPCRs through heterologous expression in S. cerevisiae. Despite testing more than 60 potential ligands, no ligand-receptor pair was elucidated (data not shown). Individually deleting each GPCR, however, led to an abundance of findings that highlight the complexity of signal transduction networks. Of the 15 deletion mutants, all but two (the ΔgprO and ΔnopA mutants) exhibited aberrancies in multiple processes. This was not unexpected, since there is overlap in many of the pathways that drive various aspects of fungal development. For example, germination in A. nidulans is triggered by a carbon source (28), so without the receptor(s) that detects that signal, the fungus would likely be impaired in both germination and growth on various carbons sources, as was the case with the ΔgprA, ΔgprC, ΔgprJ, ΔgprK, ΔgprP, and ΔgprR mutants.
Stress pathways are also interconnected among themselves and with other developmental pathways, making it difficult to decipher which GPCRs are tied to which responses. Nitrogen starvation, for example, creates a stressful environment for cells. Chung et al. (44) found that the nitrogen starvation sensor Stm1 was important for growth on different nitrogen sources and that it also appeared to stimulate a stress response pathway. These are just a couple examples of cross talk between different signaling and development pathways, though many more such instances exist, creating a domino effect when a component of one pathway is disrupted.
Table 7 lists each GPCR’s predicted function based on its sequence alignment as well as the various processes it may be involved in based on the functional studies reported here. Interestingly, none of the predicted functions (Table 1) were validated by the experimental results as presented here, which aligns with the paucity of data on the functions of filamentous fungus GPCRs. In the case of the yeast pheromone receptor Ste2 and Ste3 homologs GprA and GprB, a role in mating was not evaluated in this study. However, in A. nidulans, gprA and gprB are required for fruiting-body formation during self-fertilization (60). A. flavus is a heterothallic fungus, and it undergoes sexual reproduction when the appropriate mating type is present (61). Therefore, GprA and GprB are likely to be important for detecting the presence of mating partners and triggering sexual reproduction.
TABLE 7 .
Potential roles of A. flavus GPCRs
| Strain description | Predicted role(s) | Observed role(s) |
|---|---|---|
| ΔgprA | Mating | Germination; AF repression; carbon source sensing; oxylipin sensing |
| ΔgprB | Mating | Germination; quorum sensing; MeJA sensing |
| ΔgprC | Glucose sensing | Germination; carbon and nitrogen sensing; 13(S)-HpODE sensing |
| ΔgprD | Glucose sensing | Nitrogen sensing; ROS, cell wall, acidic pH stress response; 13(S)-HpODE sensing |
| ΔgprF | Nitrogen sensing | Light sensing; quorum sensing; acidic pH stress response; oxylipin sensing |
| ΔgprG | Nitrogen sensing | ROS and acidic pH stress responses; oxylipin sensing |
| ΔgprH | Methionine sensing | Germination; ROS stress response |
| ΔgprJ | Nitrogen sensing | Germination; carbon sensing; 13(S)-HpODE sensing |
| ΔgprK | Unknown | Germination; carbon sensing; cell wall, osmotic, and acidic stress response, MeJA sensing |
| ΔgprM | Unknown | Carbon sensing; osmotic and pH stress responses; MeJA sensing |
| ΔgprO | Unknown | Oxylipin sensing |
| ΔgprP | Unknown | Germination; AF repression; carbon sensing; oxylipin sensing |
| ΔgprR | Unknown | Germination; light sensing; carbon and nitrogen sensing; osmotic and alkaline pH stress responses; lipid and oxylipin sensing |
| ΔgprS | Nitrogen sensing | Germination; cell wall stress response; MeJA sensing |
| ΔnopA | Light sensing | Unknown |
GprC and GprD were predicted to be carbon source sensors based on their homology with the S. pombe glucose receptor Git3. However, only GprC displayed any growth defects on different carbon sources, suggesting that despite their high degree of sequence similarity, GprC and GprD have distinct roles. In addition to GprC, several other putative GPCRs may be involved in carbon source sensing. Deletion of gprA, gprJ, gprK, and gprR also resulted in impaired growth on multiple carbon sources. It was shown in S. cerevisiae that its carbon sensor Gpr1 was partially redundant with Ras2, a small GTPase, such that a Δgpr1 mutant grew normally but a Δgpr1 Δras2 strain had a severe growth defect (36). Perhaps several of A. flavus’ receptors have overlapping functions in carbon sensing with each other and/or with other proteins, such as small GTPases, which would explain why all of the single GPCR mutants were still viable. It is also possible that several of the GPCRs work as heterodimers, as is found in other systems (62), and thus their full role in fungal development will be uncovered only in multiple deletion strains.
Class IV GPCRs are S. pombe Stm1 homologs and are therefore predicted to have a role in nitrogen sensing. A. flavus’ four class IV GPCRs are GprF, GprG, GprJ, and GprS. However, none of these mutants showed any deviation from the wild-type phenotype on the three nitrogen sources tested. Instead, the ΔgprC, ΔgprD, and ΔgprR mutants were restricted on proline, with the ΔgprR mutant also showing restriction on ammonium chloride. This is reminiscent of the S. pombe Δstm1 mutant showing premature cell cycle arrest on proline, a poor nitrogen source (44). Further experimentation is required to definitively classify these receptors as nitrogen sensors, though it is interesting that GprC and GprD may have overlapping roles in nitrogen sensing and divergent roles in carbon sensing.
Not much is known about class V receptors. They are predicted to be cAMP sensors, though there is no evidence that this occurs in Aspergillus. A. flavus encodes only one class V receptor, GprH. It is homologous to C. neoformans Gpr4, which is a methionine sensor. The Gpr4 protein is internalized in response to methionine, and a Δgpr4 mutant does not produce a spike in cAMP in the presence of methionine as is seen in the wild type. The Δgpr4 mutant is defective in capsule production and mating (9). Heterologous expression of A. flavus GprH in yeast revealed it to be active in the absence of any added ligand (data not shown), although efforts to determine whether this was due to methionine in the media were inconclusive.
NopA belongs to the bacteriorhodopsin-like class of GPCRs (class IX). Rhodopsins consist of an opsin protein and a retinal chromophore and act as light-responsive GPCRs or proton pumps (reviewed in reference 63). In spite of the wealth of knowledge about rhodopsins, the function of many opsins, including NopA, remains enigmatic. The first opsin identified in fungi was N. crassa Nop-1 (64), which binds retinal (65). It has no obvious function, though its expression is induced during conidiation, which is a light-responsive process (66). The Fusarium fujikuroi Nop-1 ortholog also lacks an obvious function, although it appears to play a role in the regulation of retinal biosynthesis (67).
Based on its sequence, A. flavus NopA is an opsin-related protein instead of a rhodopsin, meaning that it lacks a critical lysine residue required for binding retinal. Opsin-related proteins are found throughout the fungal kingdom, though nothing is known about what cofactors, if any, they bind to and what their functions may be (68). Other than a slight germination defect, the ΔnopA mutant did not appear different from the wild type in any of the development, growth, stress, or lipid response assays described here. Either NopA is functionally redundant with another receptor, or else it has a role that was not addressed by these experiments.
Five GPCRs from three different classes (classes VI, VII, and VIII) had no predicted functions. GprK and GprR both belong to class VI, characterized by hybrid GPCR-RGS proteins. Although the ΔgprK and ΔgprR mutants shared some phenotypes, the ΔgprR mutant was distinct for its defective growth/response in nearly every assay performed. This suggests that GprR might regulate a process that is required for growth, and without it, the mutant is impaired in its response to many other stimuli. GprO and GprP are both class VII receptors. They may share overlapping functions in oxylipin sensing, but GprP clearly has additional roles in germination and AF production.
In this study, the Δgpr mutants were evaluated for their basic growth and development, growth on a variety of carbon and nitrogen sources, stress responses, and responses to lipids and oxylipins. An area that was not explored but is likely to be extremely important for A. flavus is the roles of its GPCRs during interaction with other organisms. A. flavus is naturally found in the soil, where it encounters many different microbes, and it is also capable of infecting plants and humans. These interactions are mediated by signaling between organisms, and it is quite possible that some of these signals are transduced via A. flavus’ GPCRs.
GPCRs are an attractive antifungal target for their lock-and-key specificity. In fact, approximately half of all drugs target GPCRs (69). Finding antagonists for GPCRs that are required for fungal invasion of a host, for example, could be a very effective way of combating infection. No single GPCR was identified here as being absolutely required for growth in culture, but perhaps certain receptors are required to grow or produce toxins in a host and/or function as heterodimers. By unraveling the functions of A. flavus’ GPCRs, new therapeutic targets may emerge for reducing the impact of this pathogen.
MATERIALS AND METHODS
Culture conditions.
Strains (see Table S2 in the supplemental material) were grown on glucose minimal medium (GMM) (33) with ammonium salts instead of nitrate salts unless otherwise mentioned. In some cases, 5 mM uridine and 5 mM uracil were added, and this is denoted as “plus UU.” For genomic DNA (gDNA) extraction, strains were grown on liquid GMM (NH4+) with 0.5% yeast extract added.
Strain construction.
Strain genotypes and sources are summarized in Table S2 in the supplemental material. All primers used for strain construction are listed in Table S3. GPCRs were disrupted using homologous recombination to replace each gene with pyrG in the parental strain CA14Δku70ΔpyrG (70). Table S3 indicates which primers were used to amplify the GPCR 5′ and 3′ flanks. The pyrG marker gene was amplified from A. fumigatus genomic DNA using primers 61 and 62. In all cases, the 5′ flank forward primer and 3′ flank reverse primer were used to amplify the entire double-joint cassette. CA14Δku70ΔpyrG was also complemented to prototrophy to generate an isogenic “wild type” for comparison with the other mutants. To achieve this, A. fumigatus pyrG was targeted to the nkuA locus. Primers 63 and 64 and primers 65 and 66 were used to amplify the nkuA 5′ and 3′ flanks, respectively. The entire construct was amplified with primers 63 and 66.
All of the above constructs were transformed individually into parental strain CA14Δku70ΔpyrG (70), with the exception of ΔgprR, which was also transformed into strain 3357.5 (71). Transformation of the fungus was carried out as described previously (31). At least 12 independent isolates were screened by PCR, followed by Southern analysis to confirm PCR-positive mutants. The same primers that were used to amplify the full double-joint cassette were used to amplify Southern probes from wild-type gDNA. Confirmed mutants were grown with and without UU to check for marker gene effects (24), and a single isolate of each mutant was selected for subsequent analyses.
Bioinformatic characterization of GPCRs.
The 15 putative A. flavus GPCRs were identified by a BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search of the A. nidulans GPCRs (7) against the A. flavus genome. The topology prediction software programs TMHMM 2.0 (13, 14) and TopPred 1.10 (15, 16) were used to predict the number of TM domains for all unique hits. GPCRs were aligned using the software program Clustal Omega (22), and a phylogenetic tree was generated using Phylogeny.fr (23).
Fungal development assays. (i) Germination assay.
Germination was assayed using a 48-well plate, with 400 µl of GMM(NH4+) plus UU plus 0.1% yeast extract containing 106 spores/ml dispensed per well (three replicates per strain). The plate was incubated at 29°C, and germination was measured at 4, 5, 6, 7, 8, and 9 h postinoculation. The plate was imaged on an Eclipse TS100 inverted microscope (Nikon) with an EOS Rebel T3 camera (Canon) fitted into the eyepiece. The Cell Counter tool in the ImageJ program (72) was used to count the number of germinated spores per 100 spores. A spore was counted as germinated when the germ tube extended for at least the length of the spore.
(ii) Aflatoxin analysis.
Strains were grown on 10-cm-diameter plates containing 25 ml YES (2% yeast extract and 6% sucrose, pH 5.8) or YEP (2% yeast extract and 6% peptone, pH 5.8) with uridine and uracil (plus UU) and 1.5% agar. Spores (103) in 1 µl of 0.1% Tween 20 were point inoculated onto the center of each plate. Three replicates per strain were placed in the dark at 30°C for 5 days. A 15-mm-diameter core was punched from each plate and homogenized in 3 ml 0.01% Tween 20.
To extract AF, 3 ml of ethyl acetate was added to each tube, and the tubes were shaken vigorously and spun at 3,000 rpm for 10 min. The organic layer was removed, dried down, and resuspended in 1 ml 50:40:10 water-methanol-acetonitrile. Samples were filtered through an Acrodisc syringe filter with a nylon membrane (0.45 µm; Pall Corporation) and run on a PerkinElmer Flexar instrument equipped with a Zorbax Eclipse XDB C18 column (150 by 4.6 mm, 5 μm, 100 Å; Agilent). The column was equilibrated in the running solvent (50:40:10 water-methanol-acetonitrile), and 20 µl was injected and run isocratically for 11 min with 100% running solvent at a flow rate of 0.8 ml/min. AF was detected by a fluorescent detector with an emission wavelength of 455 nm and excitation wavelength of 365 nm.
(iii) Impact of light and dark on sporulation
.Strains were grown on 10-cm plates containing 10 ml GMM(NH4+) plus UU (15 g/liter agar). Five hundred spores in 1 µl 0.1% Tween 20 were point inoculated into the center of each plate, and plates were incubated for 3 days at 30°C in either constant light or constant dark. There were three replicates per strain per condition. Following incubation, a 15-mm core was punched from the center of each plate and homogenized in 3 ml 0.1% Tween 20. A 100-µl aliquot was removed and diluted, and spores were enumerated using a hemocytometer.
(iv) Density dependence assay.
The impact of GPCRs on density dependence was assayed with the method of Horowitz Brown et al. (30) with some modifications. A 35.3 mM concentration of glutamine (Gln) was used as a nitrogen source since NH4+ does not support the normal density-dependent development pattern. Ten-centimeter plates containing 10 ml GMM(Gln) plus UU plus 2% sorbitol (15 g/liter agar) were overlaid with 103 (low density) or 107 (high density) spores in 3 ml of the base medium containing 7.5 g/liter agar. Plates were incubated in the dark at 30°C for 6 days. Following incubation, three replicates were processed for sclerotia production, and the other three replicates were processed for spore production.
To measure sclerotia, plates were sprayed with 70% ethanol to wash off conidia. The sclerotia were scraped from the plates, frozen in liquid nitrogen, and lyophilized. The dry weight was recorded. To measure spore production, three 15-mm cores were punched from each plate and homogenized in 3 ml 0.1% Tween 20. A 100-µl aliquot was removed, diluted, and counted on a hemocytometer.
Growth on alternative carbon and nitrogen sources.
To assess the effects of a variety of carbon sources, strains were grown on a variety of media, the base being MM(NH4+) plus UU (15 g/liter agar) as described above. The various carbon sources were added at the following concentrations: glucose, galactose, and xylose, 1%; sucrose, 0.95%. To test alternative nitrogen sources, GMM plus UU (15 g/liter agar) with nitrogen-free salts added was the base medium. Three grams per liter peptone or 8.13 g/liter proline was added, and these media were compared to GMM(NH4+) plus UU (15 g/liter agar). Forty milliliters of medium was dispensed into square 10-cm by 10-cm plates and inoculated with 500 spores of each strain in 1 µl 0.1% Tween 20. Plates were incubated at 30°C for 3 days with three replicate plates per condition.
Fungal stress assays.
Several different media were used to assess the roles of GPCRs in stress responses. The base for these media was GMM(NH4+) plus UU (15 g/liter agar). The following stressors were added to the media after autoclaving: hydrogen peroxide (3 mM), Congo red (200 µg/ml), and sodium chloride (1 M). Media were also buffered to two different pHs before autoclaving: pH 4 (100 mM NaH2PO4 and 100 mM NaCl) and pH 8 (100 mM Na2HPO4). The pH stress media contained 30 g/liter agar, so they were compared to GMM(NH4+) plus UU with 30 g/liter agar. Forty milliliters of medium was dispensed into square 10-cm by 10-cm plates and inoculated with 500 spores of each strain in 1 µl 0.1% Tween 20. Plates were incubated at 30°C for 3 days with three replicate plates per condition.
Fatty acid assays. (i) Impact of methyl jasmonate on aflatoxin production.
To test the impact of methyl jasmonate (MeJA) on the ability of the GPCR mutants to synthesize aflatoxin, the method of Meimaroglou et al. (51) was followed with some modifications. One hundred spores were inoculated into 10-cm plates containing 10 ml YES (2% yeast extract and 6% sucrose, pH 5.8) plus UU liquid medium with or without 0.1 mM MeJA (Sigma-Aldrich). Three replicates per condition per strain were incubated in the dark at 30°C for 3 days. The contents of each plate were transferred to a 50-ml conical tube and homogenized.
To extract aflatoxin, 5 ml ethyl acetate was added to each tube for 15 min with periodic shaking. The mixtures were spun at 3,000 rpm for 5 min, and 3 ml of the organic layer was transferred to a glass vial, dried down, and resuspended in 1 ml 50:40:10 water-methanol-acetonitrile. They were analyzed by high-performance liquid chromatography (HPLC) as described above.
(ii) Impact of fatty acids on sporulation.
Disk assays were carried out to measure the impact of linoleic acid and 13(S)-HpODE (Cayman Chemical) on sporulation, following the method of Calvo et al. (54) with some modifications. Ten-centimeter plates containing 10 ml YGT (0.5% yeast extract, 2% glucose, and 1 ml/liter trace elements) plus UU with 1.5% agar were overlaid with 104 spores per plate in 3 ml YGT plus UU with 0.75% agar. One-centimeter filter disks (Whatman, grade 1) were soaked with either 1 mg linoleic acid in 8 µl ethanol, 1 µg 13(S)-HpODE in 8 µl ethanol, or 8 µl ethanol by itself. The disks were placed at the center of the plates (one disk per plate), and the plates were incubated at 30°C for 3 days (linoleic acid plates) or 4 days [13(S)-HpODE] along with ethanol control plates at each time point.
Following incubation, an 11-mm core containing the disk was removed and discarded. A 23-mm ring was punched out around the removed portion and homogenized in 3 ml Tween H2O. A 100-µl aliquot was removed and diluted, and spores were counted on a hemocytometer.
Expression analysis.
Publicly available RNA-seq and microarray expression data for A. flavus under various conditions was accessed from the publication itself or from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), depending on where the data were deposited. For the GEO data, the GEO2R Web tool was used to compare different samples within an experiment. The log2-fold change (FC) values and P values for A. flavus’ 15 GPCRs were collected from the various data sets.
Statistical analysis.
Statistically significant differences were determined by using an unpaired Student’s t test with a two-tailed distribution and a P value of <0.05.
SUPPLEMENTAL MATERIAL
Alignment of fungal GPCRs. GPCR protein sequences were aligned using Clustal Omega, and a phylogenetic tree was generated using Phylogeny.fr. A. flavus GPCRs are colored red. Download
Confirmation of disruption of GPCR-encoding genes. Disruption of each GPCR was confirmed by Southern blotting. For all blots, strain TKJA13.1 was used as the wild type, though for several blots, the probe did not hybridize to the wild-type DNA. The expected band sizes are as follows: gprA, WT = 8.5, 2.8 kb; Δ = 7.8, 2.8, 1.6 kb; gprB, WT = 10.4, 4.0 kb; Δ = 8.9, 4.0, 1.8 kb; gprC, WT = 5.3, 2.1 kb; Δ = 4.9, 2.1, 1.1 kb; gprD, WT = 6.1, 2.3 kb; Δ = 5.8, 2.3, 1.1 kb; gprF, WT = 4.0 kb; Δ = 2.9, 2.2 kb; gprG, WT = 10.3 kb; Δ = 9.0, 3.3, 2.3 kb; gprH, WT = 6.1 kb; Δ = 4.9, 1.9 kb; gprJ, WT = 7.4 kb; Δ = 5.2, 2.8 kb; gprK, WT = 7.1 kb; Δ =5.3, 2.2 kb; gprM, WT = 6.9 kb; Δ = 5.1, 2.2 kb; gprO, WT = 6.6 kb; Δ = 4.3, 3.3 kb; gprP, WT = 5.8 kb; Δ = 3.9, 2.4 kb; gprR, WT = 5.6 kb; Δ = 4.1, 2.4 kb; gprS, WT = 6.7 kb; Δ = 5.9, 1.8 kb; nopA, WT = 7.6 kb; Δ = 5.7, 2.9 kb. Download
Impact of uridine and uracil on ΔgprR strain stress responses. (A) ΔgprR mutants in two different genetic backgrounds (CA14 and NRRL3357) were compared on neutral pH (pH 6.5) versus alkaline pH (pH 8) with and without supplementation of uridine and uracil (UU). After 3 days, radial growth was measured. (B) The same strains were also compared on glucose minimal medium (GMM) versus GMM amended with the cell wall stressor Congo red, both with and without UU. After 3 days, radial growth was measured. In both panels A and B, the percent growth inhibition caused by the stressor was calculated, and a two-tailed Student t test (P < 0.05) was carried out to assess differences between percent inhibition with and without UU supplementation. Differences that were not significant are denoted “n.s.,” and significant differences are marked with an asterisk (*, P < 0.05; **, P < 0.01). Download
Impact of uridine and uracil on the stress responses of three independent ΔgprM transformants. (A) Three independent ΔgprM transformants in the CA14 background were compared to the wild type (WT) on neutral pH (pH 6.5) versus alkaline pH (pH 8) with and without supplementation of uridine and uracil (UU). After 3 days, radial growth was measured. (B) The same strains were also compared on glucose minimal medium (GMM) versus GMM amended with the cell wall stressor Congo red, both with and without UU. After 3 days, radial growth was measured. In both panels A and B, the percent growth inhibition caused by the stressor was calculated, and a two-tailed Student’s t test (P < 0.05) was carried out to assess differences between percent inhibition with and without UU supplementation. Differences that were not significant are denoted “n.s.,” and significant differences are marked with an asterisk (P < 0.05). Download
Impact of uridine and uracil on the stress responses of mutants that do not exhibit a marker gene effect. (A) Independent ΔgprD and ΔgprH transformants in the CA14 background were compared to the wild type (WT) on neutral pH (pH 6.5) versus alkaline pH (pH 8) with and without supplementation of uridine and uracil (UU). After 3 days, radial growth was measured. (B) The same strains were also compared on glucose minimal medium (GMM) versus GMM amended with the cell wall stressor Congo red, both with and without UU. After 3 days, radial growth was measured. In both panels A and B, the percent growth inhibition caused by the stressor was calculated, and a two-tailed Student’s t test (P < 0.05) was carried out to assess differences between percent inhibition with and without UU supplementation. Differences that were not significant are denoted “n.s.,” and significant differences are marked with an asterisk (*, P < 0.05; **, P < 0.01). Download
Stress responses of multiple isolates of a subset of Δgpr mutants; strains were exposed to alkaline pH (pH 8) and cell wall stress (Congo red); percent inhibition of growth under stress versus growth on control medium (GMM) was measured; shaded boxes indicate data points in which the percent inhibition of the mutant differed significantly from that of the WT, as determined by a two-tailed Student’s t test (P < 0.05), and the degree of the difference is indicated by the different colors; darker red tones indicate mutants with greater sensitivity to the stressor, while darker blue tones denote the opposite.
All strains used in this study and their genotypes
Oligonucleotide primers used for strain construction and confirmation
ACKNOWLEDGMENTS
This work was supported by NSF grant IOS-0965649 (to N.P.K. and K.J.A.).
Footnotes
Citation Affeldt KJ, Carrig J, Amare M, Keller NP. 2014. Global survey of canonical Aspergillus flavus G protein-coupled receptors. mBio 5(5):e01501-14. doi:10.1128/mBio.01501-14.
REFERENCES
- 1. Robens J, Cardwell K. 2003. The costs of mycotoxin management to the USA: management of aflatoxins in the United States. J. Toxicol. Toxin Rev. 22:139–152. 10.1081/TXR-120024089 [DOI] [Google Scholar]
- 2. Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. 2007. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153:1677–1692. 10.1099/mic.0.2007/007641-0 [DOI] [PubMed] [Google Scholar]
- 3. Xue C, Hsueh YP, Heitman J. 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol. Rev. 32:1010–1032. 10.1111/j.1574-6976.2008.00131.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Versele M, Lemaire K, Thevelein JM. 2001. Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2:574–579. 10.1093/embo-reports/kve132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brodhagen M, Keller NP. 2006. Signalling pathways connecting mycotoxin production and sporulation. Mol. Plant Pathol. 7:285–301. 10.1111/j.1364-3703.2006.00338.x [DOI] [PubMed] [Google Scholar]
- 6. Ma D, Li R. 2013. Current understanding of HOG-MAPK pathway in Aspergillus fumigatus. Mycopathologia 175:13–23. 10.1007/s11046-012-9600-5 [DOI] [PubMed] [Google Scholar]
- 7. Lafon A, Han KH, Seo JA, Yu JH, d’Enfert C. 2006. G-protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet. Biol. 43:490–502. 10.1016/j.fgb.2006.02.001 [DOI] [PubMed] [Google Scholar]
- 8. Kulkarni RD, Thon MR, Pan H, Dean RA. 2005. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 6:R24. 10.1186/gb-2005-6-3-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xue C, Bahn YS, Cox GM, Heitman J. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667–679. 10.1091/mbc.E05-07-0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. 2007. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61:423–452. 10.1146/annurev.micro.61.080706.093432 [DOI] [PubMed] [Google Scholar]
- 11. Zheng H, Zhou L, Dou T, Han X, Cai Y, Zhan X, Tang C, Huang J, Wu Q. 2010. Genome-wide prediction of G protein-coupled receptors in Verticillium spp. Fungal Biol 114:359–368. 10.1016/j.funbio.2010.02.008 [DOI] [PubMed] [Google Scholar]
- 12. Gruber S, Omann M, Zeilinger S. 2013. Comparative analysis of the repertoire of G protein-coupled receptors of three species of the fungal genus Trichoderma. BMC Microbiol. 13:108. 10.1186/1471-2180-13-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–182 [PubMed] [Google Scholar]
- 14. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 15. Von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494. 10.1016/0022-2836(92)90934-C [DOI] [PubMed] [Google Scholar]
- 16. Claros MG, von Heijne G. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685–686 [DOI] [PubMed] [Google Scholar]
- 17. Kurtzman CP, Smiley MJ, Robnett CJ, Wicklow DT. 1986. DNA relatedness among wild and domesticated species in the Aspergillus flavus group. Mycologia 78:955–959. 10.2307/3807436 [DOI] [Google Scholar]
- 18. Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. 2003. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301:1728–1731. 10.1126/science.1087790 [DOI] [PubMed] [Google Scholar]
- 19. Chen JG, Jones AM. 2004. AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 389:338–350. 10.1016/S0076-6879(04)89020-7 [DOI] [PubMed] [Google Scholar]
- 20. Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. 2007. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc. Natl. Acad. Sci. U. S. A. 104:17317–17322. 10.1073/pnas.0704751104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han KH, Seo JA, Yu JH. 2004. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol. Microbiol. 51:1333–1345. 10.1111/j.1365-2958.2003.03940.x [DOI] [PubMed] [Google Scholar]
- 22. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Palmer JM, Mallaredy S, Perry DW, Sanchez JF, Theisen JM, Szewczyk E, Oakley BR, Wang CC, Keller NP, Mirabito PM. 2010. Telomere position effect is regulated by heterochromatin-associated proteins and NkuA in Aspergillus nidulans. Microbiology 156:3522–3531. 10.1099/mic.0.039255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fillinger S, Chaveroche MK, Shimizu K, Keller N, d’Enfert C. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001–1016. 10.1046/j.1365-2958.2002.02933.x [DOI] [PubMed] [Google Scholar]
- 26. Han KH, Seo JA, Yu JH. 2004. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Galpha) signalling. Mol. Microbiol. 53:529–540. 10.1111/j.1365-2958.2004.04163.x [DOI] [PubMed] [Google Scholar]
- 27. Lafon A, Seo JA, Han KH, Yu JH, d’Enfert C. 2005. The heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171:71–80. 10.1534/genetics.105.040584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osherov N, May G. 2000. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155:647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gehrke A, Heinekamp T, Jacobsen ID, Brakhage AA. 2010. Heptahelical receptors GprC and GprD of Aspergillus fumigatus are essential regulators of colony growth, hyphal morphogenesis, and virulence. Appl. Environ. Microbiol. 76:3989–3998. 10.1128/AEM.00052-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horowitz Brown S, Zarnowski R, Sharpee WC, Keller NP. 2008. Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl. Environ. Microbiol. 74:5674–5685. 10.1128/AEM.00565-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Affeldt KJ, Brodhagen M, Keller NP. 2012. Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins (Basel) 4:695–717. 10.3390/toxins4090695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hicks JK, Yu JH, Keller NP, Adams TH. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 16:4916–4923. 10.1093/emboj/16.16.4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimizu K, Keller NP. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shimizu K, Hicks JK, Huang TP, Keller NP. 2003. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics 165:1095–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roze LV, Beaudry RM, Keller NP, Linz JE. 2004. Regulation of aflatoxin synthesis by FadA/cAMP/protein kinase A signaling in Aspergillus parasiticus. Mycopathologia 158:219–232. 10.1023/B:MYCO.0000041841.71648.6e [DOI] [PubMed] [Google Scholar]
- 36. Xue Y, Batlle M, Hirsch JP. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 17:1996–2007. 10.1093/emboj/17.7.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, Winderickx J, de Winde JH, Thevelein JM. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002–1012. 10.1046/j.1365-2958.1999.01413.x [DOI] [PubMed] [Google Scholar]
- 38. Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16:293–299. 10.1016/j.molcel.2004.10.004 [DOI] [PubMed] [Google Scholar]
- 39. Hoffman CS. 2005. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33:257–260. 10.1042/BST0330257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li L, Borkovich KA. 2006. GPR-4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa. Eukaryot. Cell 5:1287–1300. 10.1128/EC.00109-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miwa T, Takagi Y, Shinozaki M, Yun CW, Schell WA, Perfect JR, Kumagai H, Tamaki H. 2004. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3:919–931. 10.1128/EC.3.4.919-931.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maidan MM, De Rop L, Serneels J, Exler S, Rupp S, Tournu H, Thevelein JM, Van Dijck P. 2005. The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16:1971–1986. 10.1091/mbc.E04-09-0780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strocchi A. 1982. Fatty acid composition and triglyceride structure of corn oil, hydrogenated corn oil, and corn oil margarine. J. Food Sci. 47:36–39. 10.1111/j.1365-2621.1982.tb11021.x [DOI] [Google Scholar]
- 44. Chung KS, Won M, Lee SB, Jang YJ, Hoe KL, Kim DU, Lee JW, Kim KW, Yoo HS. 2001. Isolation of a novel gene from Schizosaccharomyces pombe: stm1+ encoding a seven-transmembrane loop protein that may couple with the heterotrimeric Gα2 protein, Gpa2. J. Biol. Chem. 276:40190–40201. 10.1074/jbc.M100341200 [DOI] [PubMed] [Google Scholar]
- 45. Fuchs BB, Mylonakis E. 2009. Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8:1616–1625. 10.1128/EC.00193-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duran R, Cary JW, Calvo AM. 2010. Role of the osmotic stress regulatory pathway in morphogenesis and secondary metabolism in filamentous fungi. Toxins (Basel) 2:367–381. 10.3390/toxins2040367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Selvig K, Alspaugh JA. 2011. pH response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology 39:249–256. 10.5941/MYCO.2011.39.4.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roncero C, Durán A. 1985. Effect of calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ram AF, Klis FM. 2006. Identification of fungal cell wall mutants using susceptibility assays based on calcofluor white and Congo red. Nat. Protoc. 1:2253–2256. 10.1038/nprot.2006.397 [DOI] [PubMed] [Google Scholar]
- 50. Christensen SA, Kolomiets MV. 2011. The lipid language of plant-fungal interactions. Fungal Genet. Biol. 48:4–14. 10.1016/j.fgb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 51. Meimaroglou DM, Galanopoulou D, Markaki P. 2009. Study of the effect of methyl jasmonate concentration on aflatoxin biosynthesis by Aspergillus parasiticus in yeast extract sucrose medium. Int. J. Microbiol. 2009:842626. 10.1155/2009/842626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goodrich-Tanrikulu M, Mahoney NE, Rodriguez SB. 1995. The plant growth regulator methyl jasmonate inhibits aflatoxin production by Aspergillus flavus. Microbiology 141:2831–2837 [DOI] [PubMed] [Google Scholar]
- 53. Vergopoulou S, Galanopoulou D, Markaki P. 2001. Methyl jasmonate stimulates aflatoxin B1 biosynthesis by Aspergillus parasiticus. J. Agric. Food Chem. 49:3494–3498. 10.1021/jf010074+ [DOI] [PubMed] [Google Scholar]
- 54. Calvo AM, Hinze LL, Gardner HW, Keller NP. 1999. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 65:3668–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, Horowitz-Brown S, Woloshuk CP, Yu J, Keller NP, Payne GA. 2010. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 11:213–226. 10.1111/j.1364-3703.2009.00594.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keller N, Butchko R, Sarr B, Phillips T. 1994. A visual pattern of mycotoxin production in maize kernels by Aspergillus spp. Phytopathology 84:483–488. 10.1094/Phyto-84-483 [DOI] [Google Scholar]
- 57. Wu X, Zhou B, Yin C, Guo Y, Lin Y, Pan L, Wang B. 2014. Characterization of natural antisense transcript, sclerotia development and secondary metabolism by strand-specific RNA sequencing of Aspergillus flavus. PLoS One 9:e97814. 10.1371/journal.pone.0097814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin JQ, Zhao XX, Wang CC, Xie Y, Li GH, He ZM. 2013. 5-Azacytidine inhibits aflatoxin biosynthesis in Aspergillus flavus. Ann. Microbiol. 63:763–769. 10.1007/s13213-012-0531-7 [DOI] [Google Scholar]
- 59. Lin JQ, Zhao XX, Zhi QQ, Zhao M, He ZM. 2013. Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet. Biol. 56:78–86. 10.1016/j.fgb.2013.04.007 [DOI] [PubMed] [Google Scholar]
- 60. Seo JA, Han KH, Yu JH. 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53:1611–1623. 10.1111/j.1365-2958.2004.04232.x [DOI] [PubMed] [Google Scholar]
- 61. Horn BW, Moore GG, Carbone I. 2009. Sexual reproduction in Aspergillus flavus. Mycologia 101:423–429. 10.3852/09-011 [DOI] [PubMed] [Google Scholar]
- 62. Borroto-Escuela DO, Brito I, Romero-Fernandez W, Di Palma M, Oflijan J, Skieterska K, Duchou J, Van Craenenbroeck K, Suárez-Boomgaard D, Rivera A, Guidolin D, Agnati LF, Fuxe K, Borroto-Escuela DO, Brito I, Romero-Fernandez W, Di Palma M, Oflijan J, Skieterska K, Duchou J, Van Craenenbroeck K, Suárez-Boomgaard D, Rivera A, Guidolin D, Agnati LF, Fuxe K. 2014. The G protein-coupled receptor heterodimer network (GPCR-HetNet) and its hub components. Int. J. Mol. Sci. 15:8570–8590. 10.3390/ijms15058570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spudich JL. 1998. Variations on a molecular switch: transport and sensory signalling by archaeal rhodopsins. Mol. Microbiol. 28:1051–1058. 10.1046/j.1365-2958.1998.00859.x [DOI] [PubMed] [Google Scholar]
- 64. Bieszke JA, Braun EL, Bean LE, Kang S, Natvig DO, Borkovich KA. 1999. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. U. S. A. 96:8034–8039. 10.1073/pnas.96.14.8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. 1999. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry (Mosc.) 38:14138–14145 [DOI] [PubMed] [Google Scholar]
- 66. Bieszke JA, Li L, Borkovich KA. 2007. The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Curr. Genet. 52:149–157. 10.1007/s00294-007-0148-8 [DOI] [PubMed] [Google Scholar]
- 67. Estrada AF, Avalos J. 2009. Regulation and targeted mutation of opsA, coding for the NOP-1 opsin orthologue in Fusarium fujikuroi. J. Mol. Biol. 387:59–73. 10.1016/j.jmb.2009.01.057 [DOI] [PubMed] [Google Scholar]
- 68. Brown LS. 2004. Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem. Photobiol. Sci. 3:555–565. 10.1039/b315527g [DOI] [PubMed] [Google Scholar]
- 69. Drews J. 2000. Drug discovery: a historical perspective. Science 287:1960–1964. 10.1126/science.287.5460.1960 [DOI] [PubMed] [Google Scholar]
- 70. Chang PK, Scharfenstein LL, Wei Q, Bhatnagar D. 2010. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 81:240–246. 10.1016/j.mimet.2010.03.010 [DOI] [PubMed] [Google Scholar]
- 71. He ZM, Price MS, OBrian GR, Georgianna DR, Payne GA. 2007. Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiol. 7:104. 10.1186/1471-2180-7-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of fungal GPCRs. GPCR protein sequences were aligned using Clustal Omega, and a phylogenetic tree was generated using Phylogeny.fr. A. flavus GPCRs are colored red. Download
Confirmation of disruption of GPCR-encoding genes. Disruption of each GPCR was confirmed by Southern blotting. For all blots, strain TKJA13.1 was used as the wild type, though for several blots, the probe did not hybridize to the wild-type DNA. The expected band sizes are as follows: gprA, WT = 8.5, 2.8 kb; Δ = 7.8, 2.8, 1.6 kb; gprB, WT = 10.4, 4.0 kb; Δ = 8.9, 4.0, 1.8 kb; gprC, WT = 5.3, 2.1 kb; Δ = 4.9, 2.1, 1.1 kb; gprD, WT = 6.1, 2.3 kb; Δ = 5.8, 2.3, 1.1 kb; gprF, WT = 4.0 kb; Δ = 2.9, 2.2 kb; gprG, WT = 10.3 kb; Δ = 9.0, 3.3, 2.3 kb; gprH, WT = 6.1 kb; Δ = 4.9, 1.9 kb; gprJ, WT = 7.4 kb; Δ = 5.2, 2.8 kb; gprK, WT = 7.1 kb; Δ =5.3, 2.2 kb; gprM, WT = 6.9 kb; Δ = 5.1, 2.2 kb; gprO, WT = 6.6 kb; Δ = 4.3, 3.3 kb; gprP, WT = 5.8 kb; Δ = 3.9, 2.4 kb; gprR, WT = 5.6 kb; Δ = 4.1, 2.4 kb; gprS, WT = 6.7 kb; Δ = 5.9, 1.8 kb; nopA, WT = 7.6 kb; Δ = 5.7, 2.9 kb. Download
Impact of uridine and uracil on ΔgprR strain stress responses. (A) ΔgprR mutants in two different genetic backgrounds (CA14 and NRRL3357) were compared on neutral pH (pH 6.5) versus alkaline pH (pH 8) with and without supplementation of uridine and uracil (UU). After 3 days, radial growth was measured. (B) The same strains were also compared on glucose minimal medium (GMM) versus GMM amended with the cell wall stressor Congo red, both with and without UU. After 3 days, radial growth was measured. In both panels A and B, the percent growth inhibition caused by the stressor was calculated, and a two-tailed Student t test (P < 0.05) was carried out to assess differences between percent inhibition with and without UU supplementation. Differences that were not significant are denoted “n.s.,” and significant differences are marked with an asterisk (*, P < 0.05; **, P < 0.01). Download
Impact of uridine and uracil on the stress responses of three independent ΔgprM transformants. (A) Three independent ΔgprM transformants in the CA14 background were compared to the wild type (WT) on neutral pH (pH 6.5) versus alkaline pH (pH 8) with and without supplementation of uridine and uracil (UU). After 3 days, radial growth was measured. (B) The same strains were also compared on glucose minimal medium (GMM) versus GMM amended with the cell wall stressor Congo red, both with and without UU. After 3 days, radial growth was measured. In both panels A and B, the percent growth inhibition caused by the stressor was calculated, and a two-tailed Student’s t test (P < 0.05) was carried out to assess differences between percent inhibition with and without UU supplementation. Differences that were not significant are denoted “n.s.,” and significant differences are marked with an asterisk (P < 0.05). Download
Impact of uridine and uracil on the stress responses of mutants that do not exhibit a marker gene effect. (A) Independent ΔgprD and ΔgprH transformants in the CA14 background were compared to the wild type (WT) on neutral pH (pH 6.5) versus alkaline pH (pH 8) with and without supplementation of uridine and uracil (UU). After 3 days, radial growth was measured. (B) The same strains were also compared on glucose minimal medium (GMM) versus GMM amended with the cell wall stressor Congo red, both with and without UU. After 3 days, radial growth was measured. In both panels A and B, the percent growth inhibition caused by the stressor was calculated, and a two-tailed Student’s t test (P < 0.05) was carried out to assess differences between percent inhibition with and without UU supplementation. Differences that were not significant are denoted “n.s.,” and significant differences are marked with an asterisk (*, P < 0.05; **, P < 0.01). Download
Stress responses of multiple isolates of a subset of Δgpr mutants; strains were exposed to alkaline pH (pH 8) and cell wall stress (Congo red); percent inhibition of growth under stress versus growth on control medium (GMM) was measured; shaded boxes indicate data points in which the percent inhibition of the mutant differed significantly from that of the WT, as determined by a two-tailed Student’s t test (P < 0.05), and the degree of the difference is indicated by the different colors; darker red tones indicate mutants with greater sensitivity to the stressor, while darker blue tones denote the opposite.
All strains used in this study and their genotypes
Oligonucleotide primers used for strain construction and confirmation


