Abstract
Objective
To determine whether the mRNA concentrations of inflammation response genes in isolated adipocytes and in cultured preadipocytes are related to adipocyte size and in vivo insulin action in obese individuals.
Design
Cross-sectional inpatient study.
Subjects
Obese Pima Indians with normal glucose tolerance.
Measurements
Adipocyte diameter (by microscope technique; n=29), expression of candidate genes (by quantitative real-time PCR) in freshly isolated adipocytes (monocyte chemoattractant protein [MCP] 1 and MCP2, macrophage inflammatory protein [MIP] 1α, MIP1β and MIP2, macrophage migration inhibitory factor [MIF], tumor necrosis factor alpha, interleukin [IL] 6 and IL8; n=22) and cultured preadipocytes (MCP1, MIP1α, MIF, IL6 and matrix metalloproteinase 2; n=33) from subcutaneous abdominal adipose tissue (by aspiration biopsy, n=34), body fat by dual-energy X-ray absorptiometry, glucose tolerance by 75-gram oral glucose tolerance test, and insulin action by euglycemic-hyperinsulinemic clamp (insulin infusion rate 40 mU/m2.min)(all n=34).
Results
MIF was the only gene whose expression in both freshly isolated adipocytes and cultured preadipocytes was positively associated with adipocytes diameter and negatively associated with peripheral and hepatic insulin action (all P<0.05). In multivariate analysis, the association between adipocyte MIF mRNA concentrations and adipocytes diameter was independent of percent body fat (P=0.03), whereas adipocyte MIF mRNA concentrations but not adipocytes diameter independently predicted peripheral insulin action. The mRNA expression concentrations of MIF gene in adipocytes were not associated with plasma concentrations of MIF, but were negatively associated with plasma adiponectin concentrations (P=0.004). In multivariate analysis, adipocyte MIF RNA concentrations (P=0.03) but not plasma adiponectin concentrations (P=0.4) remained a significant predictor of insulin action.
Conclusions
Increased expression of MIF gene in adipose cells may be an important link between obesity characterized by enlarged adipocytes and insulin resistance in normal glucose tolerant people.
Keywords: adipocytes, preadipocytes, MIF, insulin action, inflammation, adiponectin
Introduction
Obesity and insulin resistance are associated with chronic sub-clinical inflammation (1;2). More than a decade ago it became evident that adipose tissue itself may play an active role in the obesity-related inflammation via its production of proinflammatory cytokines (3;4). Further studies have shown that inflammation-related genes are the most prominently expressed genes in white adipose tissue in animal models of obesity (5–7). Moreover, increased expression of inflammatory cytokines in adipose tissue in these animals precedes hyperinsulinemia and overt hyperglycemia (6) indicating that increased adipose tissue inflammation may have a pathophysiological role in metabolic complications of obesity (8).
More detailed studies of different cell types within adipose tissue have shown that many of those cytokines are predominantly produced by cells of non-adipose lineage (9;10), including macrophages (11), that increasingly infiltrate adipose tissue of obese individuals (5;6). However, inflammation-related genes are still amongst the most overexpressed genes in cells of adipose lineage such as freshly isolated matured adipocytes (12) and cultured preadipocytes (13) from obese individuals. It has been hypothesized that under positive energy balance, as adipose cells increase in size, they start to produce molecules with chemoattracting and activating effects on monocytes/macrophages (8). In fact, macrophage content in human subcutaneous adipose tissue is positively related to adipocyte size (5) and large adipocytes produce higher concentrations of chemokines than small ones (14).
It has been shown that subjects with enlarged subcutaneous adipocytes are, on average, more insulin resistant and at higher risk for developing diabetes mellitus than those with a similar degree of adiposity but relatively smaller adipocytes (15;16). Given that many individuals who are very obese are still relatively insulin sensitive (17;18), we hypothesized that increased expression of inflammation-related genes in subcutaneous abdominal adipocytes in obese subjects would be more specifically related to adipocyte size and predict in vivo insulin action. Candidate genes were mainly selected from genes known to be expressed at higher levels in adipocytes and/or cultured preadipocytes of obese Pima Indians compared to lean controls (12;13). In adipocytes, we determined the expression levels of: monocyte chemoattractant protein (MCP) 1, macrophage inflammatory protein (MIP) 1α, MIP 1β, MIP 2α, macrophage migration inhibitory factor (MIF), tumor necrosis factor alpha (TNFα) and interleukin-8 (IL8). In addition, we included interleukin 6 (IL6) because of its possible role in the development of insulin resistance and MCP 2 because of its potential role in attracting macrophages into adipose tissue. In cultured preadipocytes we studied the expression levels of MCP1, MIP1α, MIF, IL6 and matrix metalloproteinase 2 (MMP2).
Methods
Subjects in this study were at least half Pima (or closely related Tohono O’Odham) Indians from the Gila River Indian Community. All subjects were between 18 and 45 years of age, obese (BMI≥30 kg/m2), with normal plasma fasting and 2-h glucose concentration values according to a 75-gram oral glucose tolerance test (OGTT, World Health Organization 1999 criteria; <6.1 and <7.8 mmol/l, respectively), and nonsmokers at the time of the study. Acute and chronic diseases were excluded on the basis of medical history, physical examination, and routine laboratory tests. Subjects with evidence of serious medical conditions (including autoimmune, cerebrovascular, and ischemic heart disease) or taking any medications, including contraceptives, were excluded. The protocol was approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases, and all subjects provided written informed consent before participation. To minimize changes in glucose and adipose metabolism resulting from ovarian hormonal effects, the female subjects were studied during the follicular phase (days 0 through 14) of the menstrual cycle.
All subjects were admitted to the National Institutes of Health Clinical Research Unit in Phoenix, Arizona, and were placed on a weight-maintaining diet (containing 50% of calories as carbohydrates, 30% as fat, and 20% as protein) for 2–3 days before clinical testing. Body composition was measured by dual-energy X-ray absorptiometry using a total body scanner (DPX-L; Lunar Radiation, Madison, WI). At least 3 days after admission and after a 12-h overnight fast, subjects underwent OGTT to exclude impaired glucose tolerance or diabetes. Plasma glucose concentrations were determined by the glucose oxidase method (Beckman Instruments, Fullerton, CA) and plasma insulin concentrations by an automated immunoassay (Access; Beckman Instruments). Plasma MIF was measured using a human specific sandwich enzyme-linked immunosorbent assay (ELISA) from R&D Systems (Minneapolis, MN). Serum samples were diluted 1:10 for analysis and all samples were run in duplicate in the same assay. Total plasma adiponectin concentration was determined using a validated sandwich ELISA employing an adiponectin-specific antibody (intra-assay and interassay coefficients of variation 3.3% and 7.4%, respectively). Luminex bead array (LINCOplex, Linco Research Inc., St. Charles, MO) multianalyte detection system was used to determine serum concentrations of 13 cytokines (IL-1β, IL-2 , IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12, IL-13, IFN-γ, GM-CSF, and TNF-α).
Insulin action was assessed at physiological insulin concentrations during a hyperinsulinemic-euglycemic glucose clamp (19). Briefly, after an overnight fast a primed (30 µCi) continuous [0.3 µCi/min] 3-[3H] glucose infusion was started to determine endogenous glucose production (EGP). Two hours after starting the isotope infusion, a primed continuous intravenous insulin infusion was administered for 100 min at a constant rate of 40 mU·m−2 body surface area·min−1. Blood samples for measurement of 3-[3H] glucose specific activity were collected at the end of the basal period and every 10 min during the final 40 min of insulin infusion. Under basal conditions, EGP was calculated as the 3-[3H] glucose infusion rate divided by the steady-state plasma 3-[3H] glucose specific activity. During the insulin clamp, EGP was calculated from Steele's equation (20) and was used as surrogate for insulin sensitivity in the liver. The rate of total insulin-stimulated glucose disposal was calculated for the last 40 min of the insulin infusion and was corrected for the rate of EGP. Individual variation in plasma glucose and insulin concentrations during the clamp were taken into account in the calculation of the insulin-mediated glucose disposal (M) (19;21). All measurements derived from the clamp were normalized to estimated metabolic body size (EMBS, or fat-free mass + 17.7 kg) to account for the fact that the intercept of the relationship between fat-free mass and resting metabolic rate is not zero (22).
Fat biopsies were obtained after a 12-h overnight fast, between 0830 and 1000 AM. Subcutaneous abdominal adipose tissue was removed from the periumbilical region by percutaneous needle biopsy under local anesthesia (lidocaine 1%). The biopsy specimen was placed on a sterile nylon mesh, rinsed with sterile 0.9% NaCl solution and cleaned of visible connective tissue and blood vessels in Hank's Buffered Saline Solution (HBSS) supplemented with 5.5 mM glucose.
Isolation of the adipocytes and stromal vascular fraction containing preadipocytes (and other cells) was performed according to a previously described method (12;13). Briefly, adipose tissue was digested in HBSS buffer containing 5.5 mM glucose, 5% fatty acid free BSA (Introgen/Serologicals, Norcross, GA ) and 3.3 mg/mL type I collagenase (Worthington Biochemical Corp., Lakewood, NJ) for 30 min in a 37°C water bath. The digestion mixture was passed through a sterile 230 µm stainless steel tissue sieve (Thermo EC, Holbrook, NY), and the adipocytes were allowed to float by gravity and collected for adipocyte size measurement and RNA isolation.
The preadipocyte-containing infranatant was collected into a separate tube and washed several times with HBSS. The stromal vascular fraction cells (hereafter referred to as preadipocytes) were resuspended in standard medium consisting of Medium 199 (Life Technologies, Grand Island, NY, USA) supplemented with 1 µg/ml amphotericin B, 100 U/ml penicillin G sodium, 100 µg/ml streptomycin sulphate, 2 mmol/l Glutamax-1 and 10% heat-inactivated FBS (Life Technologies). The cell suspension was strained through a sterile 25-µm stainless steel tissue sieve (Thermo EC, Holbrook, NY, USA). The filtrate was transferred to a T75 culture flask and maintained in an incubator at 37°C in 5% CO2. Cells were allowed to attach and, the next day, floating red blood cells were removed by aspiration and the culture medium was replenished. At sub-confluency, the cultured cells were trypsinised and plated at a concentration of approximately 1.5×106 cells/15-cm dish for RNA extraction, which was carried out approximately 14 days after the biopsy date. Media was changed every 2–3 days throughout the culturing period. Preadipocyte culturing was completed in 33 of 34 samples obtained.
RNA from adipocytes and cultured preadipocytes was extracted using RNeasy Mini Kit from Qiagen (Valencia, CA). During the extraction, RNA was treated with RNAse-free DNAse (Qiagen) according to the manufacturer's instruction. RNA extracts were stored at −70°C until shipped on dry ice for analyses to Phoenix VA Health Care System (adipocytes) and Pennington Biomedical Research Center (preadipocytes).
Twenty-two of the 34 volunteers had sufficient amount of adipocyte RNA for the assay. The RNA was used to synthesize cDNA using iScript cDNA Synthesis Kit (BioRad, Hercules, CA). Quantitative real-time PCR (QRT-PCR) was carried out using SYBR Green chemistry (iQ SYBR Green Supermix; BioRad) and analyzed by an iCycler iQ Real-Time Detection System (BioRad) following the manufacturer’s recommendation. Successful cDNA synthesis was verified by PCR amplification of β2-microglobulin transcript using forward primer 5′ - TGT CTT TCA GCA AGG ACT GGT C - 3′ and reverse primer 5′- TGA TGC TGC TTA CAT GTC TCG AT - 3′. Quantification of candidate genes mRNA concentrations was carried out using gene specific primers (see Suppl. 1 for primer sequences). Each sample was run in duplicate and the mean value was used to calculate transcript level. A standard curve for each primer-probe set was generated by serial dilution of cDNA from a healthy subject done in triplicate. Reactions without template were included as negative controls. The reaction was carried out at 95°C for 3 min and followed by 40 cycles of amplification (94°C for 20 s, 60°C for 40 s). The reaction was further subjected to 66 cycles of 0.5°C increments (10s each) beginning at 62°C for melting curve analysis to confirm the specificity of amplification products. The mRNA concentrations of candidate genes were normalized to the geometric mean of those for human cyclophilin, TATA box binding protein and 18s ribosomal RNA. For the preadipocyte gene expression work, QRT-PCR reactions were performed as previously described (23) on an ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA). The mRNA concentrations of candidate genes were normalized to those for human cyclophilin. Each sample was run in duplicate and mean values were used for the expression of each gene (primer sequences in Suppl. 1). Preadipocyte gene expression analysis was performed on samples from 33 volunteers.
An aliquot of the adipocyte-suspension was used for cell size measurement by microscope technique. Briefly, packed adipocytes were mixed at a ratio of 1:2 with Medium 199 (Life Technologies, Grand Island, NY) containing 1% heat-inactivated FBS (Life Technologies), 1% BSA (Introgen/Serologicals), and 50 nM adenosine (Sigma-Aldrich, St. Louis, MO) on a chambered slide covered with a cover slip. Representative pictures of the adipocytes were taken using a Polaroid Microcam (Polaroid) on an inverted microscope (Eclipse TE200), and the adipocyte cell perimeters on scanned images were measured using Scion Image (Scion, Frederick, MD) by three independent readers. The inter-reader variability was 4.7%. The mean cell diameter from samples was calculated from the mean cell perimeter of the three readings. As recommended (24), only samples with at least 100 sized cells (n=29) were included in further analyses.
Statistical analyses were performed using the software of the SAS Institute (Cary, NC). Each gene mRNA concentration was normalized by taking the residuals after linear regression to the reference gene(s). Depending on the data distribution, Student-t-or Mann-Whitney test was used for comparison between males and females, and Pearson-r or Spearman-ρ correlation was used to test for simple or partial correlations between the variables. General linear regression analysis was used to test for independent determinants of the outcomes after adjusting for confounders. Non-normally distributed data were logarithmically transformed to approximate normal distribution before being analyzed in the multivariate analysis. P-values less than 0.05 were considered to be statistically significant.
Results
Anthropometric and metabolic characteristics of the study population are presented in Table 1. Women had on average higher percentage of body fat (BF, P<0.0001) and larger adipocytes (P =0.0002) compared to men (Table 1). Therefore gender was considered as a confounder in all analyses involving BF and adipocyte diameter. Metabolic characteristics of the subgroup of subjects who had available gene expression data in isolated adipocytes were similar. The expression levels of the candidate genes were not significantly different between men and women (data not shown).
Table 1.
Anthropometric and metabolic characteristics of the study population
| Variable | All | Men | Women |
|---|---|---|---|
| N | 34 | 18 | 16 |
| Age (years) | 24 (23, 32) | 24 (23, 28) | 25 (20, 34) |
| BMI (kg.m−2) | 36 (5) | 36 (4) | 36 (5) |
| Body fat (%) | 36 (5) | 33 (3) | 39 (4)c |
| Adipocyte diameter (µm)a | 67 (9) | 62 (8) | 73 (5)b |
| Fasting plasma glucose (mmol.L−1) | 4.93 (0.50) | 4.90 (0.53) | 4.95 (0.48) |
| 2h plasma glucose (mmol.L−1) | 6.25 (5.17, 7.0) | 6.25 (4.39, 7.0) | 6.31 (5.44, 6.94) |
| Fasting plasma insulin (µU.mL−1) | 11 (6) | 11 (7) | 11 (5) |
| 2-h plasma insulin (µU.mL−1) | 58 (27, 84) | 43 (17, 146) | 63 (40, 83) |
| M (µmol.kgEMBS−1.min−1) | 14.3 (12.7, 17.4) | 14 (12.4, 17.4) | 14.7 (12.8, 17.5) |
| EGPfast (µmol.kgEMBS−1.min−1) | 9.8 (2.5) | 9.4 (1.7) | 10.2 (1.1) |
| EGPins (µmol.kgEMBS−1.min−1) | 1.7 (0, 3.5) | 1.7 (0, 3.2) | 1.7 (1.4, 3.8) |
Expressed as means (SD) or median (lower quartile, upper quartile); data availability
29 (13F)
P< 0.001
P< 0.0001
sex comparison; M, insulin-mediated glucose disposal; EGP, endogenous glucose production; EMBS, estimated metabolic body size (fat-free mass + 17.7)
The expression levels of macrophage migration inhibitory factor (MIF) gene in cultured preadipocyte was the only one of the adipocyte or preadipocyte genes studied that was significantly associated (positively) with adiposity (BF, P=0.01, Table 2). A positive correlation was found between mean adipocyte diameter and expression levels of MIF in both adipocytes and preadipocytes (both P=0.02, Table 2 and Figure 1). On the other hand, adipocyte diameter was negatively associated with the expression levels of macrophage inflammatory protein 1a (MIP1a) in preadipocytes (P=0.02, Table 2). After adjustment for sex and BF, only the MIF expression in adipocytes was independently related to adipocyte diameter (P=0.03), while neither adipocyte diameter nor BF were significant predictors of preadipocyte MIF mRNA concentrations (P=0.09 and P=0.08 respectively).
Table 2.
Correlation of clinical characteristics with the expression levels of adipocyte and preadipocyte candidate genes.
| Gene | BFb | ADb | FPG | 2hPGa | FPI | 2hPI | Ma | EGPfast | EGPAins | |
|---|---|---|---|---|---|---|---|---|---|---|
| Adipocytes: | MCP1 | −0.22 | −0.24 | 0.17 | 0.15 | −0.14 | −0.11 | −0.11 | −0.27 | 0.1 |
| n=22 | MCP2 | −0.19 | −0.26 | 0.22 | 0.26 | −0.15 | −0.04 | −0.19 | −0.38 | 0.16 |
| MIF | 0.14 | 0.53c | 0.43c | 0.49c | 0.59d | 0.72e | −0.50c | 0.21 | 0.75f | |
| MIP1α | −0.20 | −0.27 | 0.15 | −0.02 | −0.24 | 0.05 | 0.14 | 0.02 | 0.15 | |
| MIP1β | −0.16 | −0.31 | 0.17 | 0.02 | −0.21 | 0.04 | 0.10 | −0.05 | 0.18 | |
| MIP2 | −0.32 | −0.43 | 0.05 | 0.07 | −0.35 | −0.09 | 0.09 | −0.20 | 0.12 | |
| IL6 | −0.20 | −0.41 | 0.05 | 0.11 | −0.19 | −0.09 | −0.06 | −0.35 | 0.09 | |
| IL8 | −0.35 | −0.39 | 0.03 | −0.08 | −0.41 | −0.17 | 0.16 | −0.23 | −0.07 | |
| TNFα | −0.25 | −0.18 | 0.24 | 0.05 | −0.07 | 0.24 | 0.13 | −0.10 | 0.20 | |
| Preadipocytes: | MCP1 | 0.09 | −0.15 | 0.02 | 0.06 | −0.14 | 0.10 | 0.02 | −0.15 | 0.02 |
| n=13 | MIF | 0.44c | 0.44c | 0.53d | 0.34c | 0.50d | 0.38c | −0.40c | 0.07 | 0.41c |
| MIP1α | 0.04 | −0.46c | 0.01 | 0.13 | −0.29 | −0.20 | −0.01 | −0.20 | 0.03 | |
| IL6 | −0.12 | 0.24 | −0.04 | 0.10 | 0.002 | −0.02 | 0.09 | −0.18 | −0.08 | |
| MMP2 | 0.08 | 0.24 | −0.18 | 0.17 | 0.18 | 0.19 | −0.14 | −0.13 | 0.13 |
Pearson or aSpearman, simple or bpartial (sex) correlation coefficients
P< 0.05
P< 0.01
P<0.001
P<0.0001
AD, adipocyte diameter; BF, body fat; FPG, fasting glucose; 2hPG, 2-hour glucose; FPI, fasting insulin; M, glucose disposal; EGP, endogenous glucose production; MCP, monocyte chemoattractant protein; MIP, macrophage inflammatory protein; MIF, macrophage migration inhibitory factor; TNF, tumor necrosis factor; IL, interleukin; MMP, matrix metalloproteinase
Figure 1.
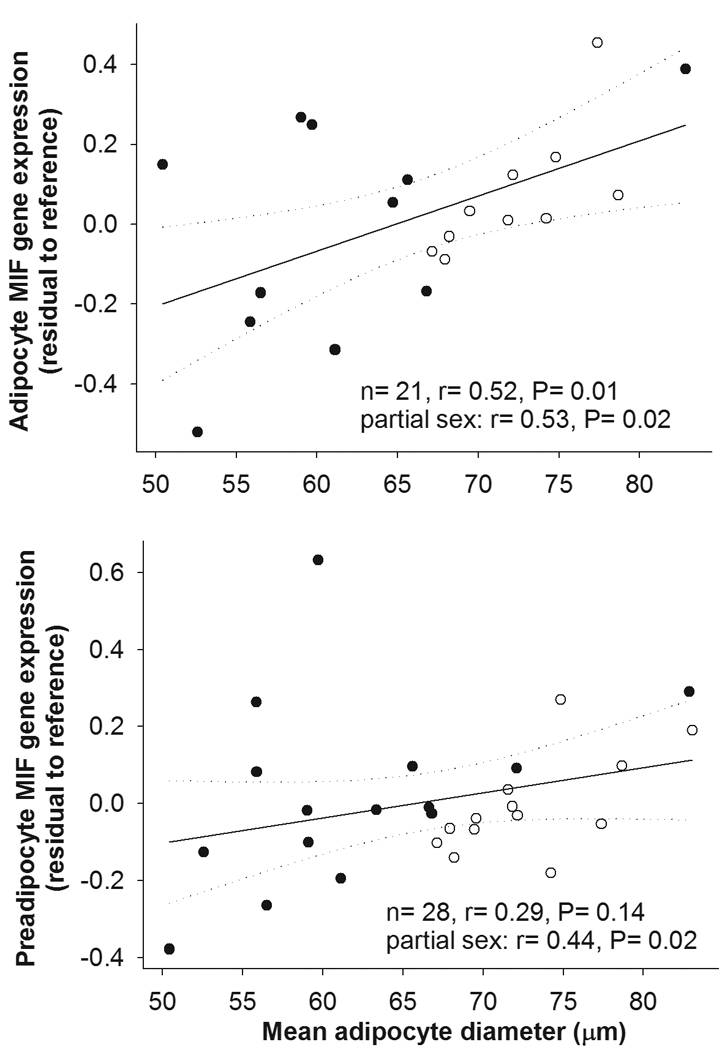
Correlation of the mRNA concentrations of MIF in freshly isolated adipocytes and cultured preadipocytes from subcutaneous abdominal adipose tissue with mean adipocyte diameter. Symbols: close circles – males, open circles – females.
As shown in Table 2, MIF was the only gene whose increased mRNA expression in isolated adipocytes was significantly associated with high fasting and 2-hour plasma concentrations of glucose (P<0.05 and P=0.02, respectively) and insulin (P=0.004 and P=0.0002, respectively), and with reduced insulin action at the periphery (M, P=0.02) and liver (EGPins, P<0.0001, Figure 2). Similarly, increased expression level of MIF mRNA in cultured preadipocytes was the only significant correlate of high fasting and 2-hour plasma concentrations of glucose (P=0.002 and P=0.02, respectively) and insulin (P=0.003 and P=0.03, respectively) (Table 2). M and EGPins was positively correlated with MIF mRNA concentrations in cultured preadipocytes as well (P=0.02 and P=0.01 respectively; Table 2, Figure 2).
Figure 2.
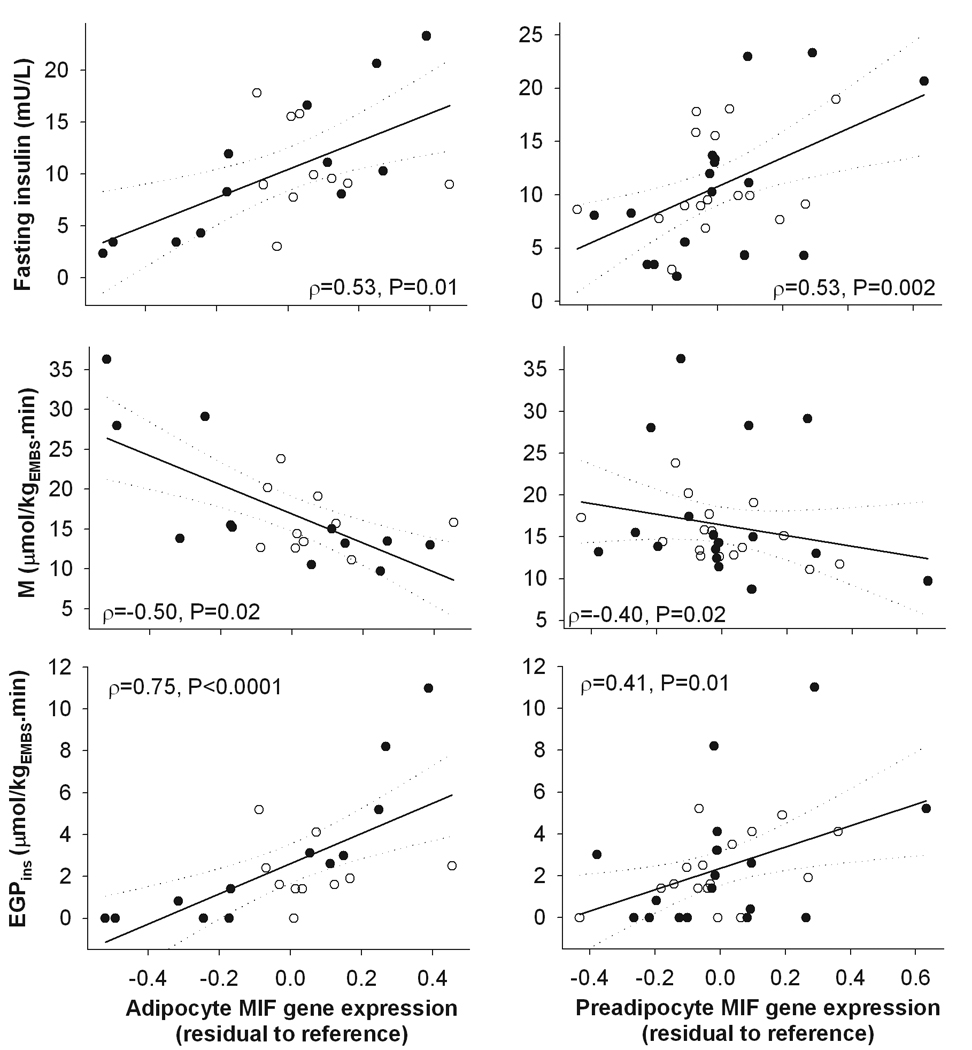
Spearman correlation between the expression levels of MIF gene in freshly isolated adipocytes or cultured preadipocytes from subcutaneous abdominal adipose tissue and fasting plasma insulin, insulin-mediated glucose disposal (M), and endogenous glucose production (EGP) during the clamp. Symbols: close circles – males, open circles – females.
Sex-adjusted adipocyte diameter was significantly associated with M (log10, β=-0.009, SE=0.004, P=0.02) before but not after (P=0.9) further adjustment for MIF gene expression level in adipocytes. The expression of MIF in adipocytes was the only significant predictor of M in this model (β=-0.36, SE=0.14, P=0.02). The effect of preadipocyte MIF gene expression on M disappeared (P=0.9) after adjustment for adipocyte diameter. EGPins was not associated with adipocyte diameter (P=0.7)
Finally, we tested whether differences in circulatory factors may explain the relationships between the expression of MIF gene in adipose cells and insulin action. Plasma concentration of MIF neither correlated with the mRNA expression levels of MIF gene in adipocytes or preadipocytes (Figure 3) nor was a significant predictor of M (ρ=0.19, P=0.3) or EGPins (ρ=-0.07, P=0.7). Adiponectin was the only adipokine whose circulatory concentration was significantly associated with both the expression levels of MIF gene in adipocytes (but not in preadipocytes, Figure 3) and insulin action (M, ρ=0.46, P=0.006; EGPins, ρ=-0.57, P=0.0004). In a multivariate model, adipocyte MIF expression (β=-0.32, SE=0.13, P=0.03) but not plasma adiponectin (P=0.4) predicted M, while neither of the two variables predicted EGPins (P=0.6 and P=0.15 respectively).
Figure 3.
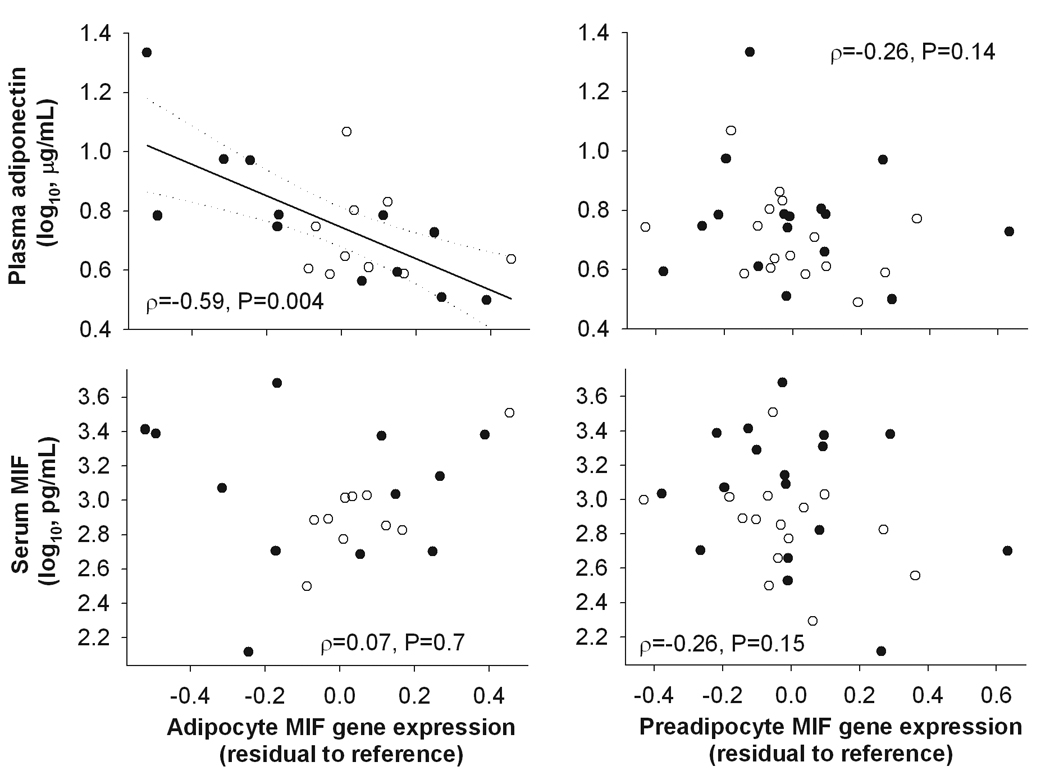
Spearman correlations between the expression levels of MIF gene in freshly isolated adipocytes or cultured preadipocytes from subcutaneous abdominal adipose tissue and fasting serum MIF and plasma adiponectin concentrations. Symbols: close circles – males, open circles – females.
Discussion
The results of the present study demonstrate that the mean size of adipocytes from abdominal subcutaneous adipose tissue was positively associated with the expression levels of the gene for macrophage migration inhibitory factor (MIF) in both freshly isolated adipocytes and cultured preadipocytes of subcutaneous adipose tissue from obese Pima Indians. In addition, increased levels of MIF mRNA from both cell types were associated with higher fasting and 2-hour plasma glucose and insulin concentrations, and reduced peripheral and hepatic insulin action.
Adipocytes produce proteins that impact distant tissues, such as liver, skeletal muscle and brain, as well as proteins that affect either neighboring adipocytes or other local cell types within adipose tissue (25). Translational and proteomic studies have shown that adipocytes produce factors that attract macrophages into adipose tissue (12;26). The production of these signals may change with the expansion of adipocytes, both in size and number, and the relatively activated macrophages may in turn produce cytokines that can modify the functions of adipose cells and have significant impact on metabolic processes in distant organs (8). In fact, animal data show that increased inflammation in adipose tissue precedes the development of obesity-induced insulin resistance (6).
MIF is a widely expressed protein with potent pro-inflammatory action (27). In adipose cells the secretion of MIF increases while MIF mRNA concentrations are relatively constant throughout the differentiation process (26). The release of MIF protein from human subcutaneous and omental adipocytes positively correlate with the donors’ body mass index (26). In our study in Pima Indians, the expression levels of MIF in preadipocytes positively correlated with different measures of whole body and regional obesity, while MIF expression in adipocytes correlated only with the adipocyte size. This difference may be due to the smaller number of subjects with data on adipocyte MIF mRNA concentrations (type II error).
Increased concentrations of MIF in circulation have been reported in subjects with T2DM and impaired glucose tolerance (28;29). Higher MIF concentrations in circulation also predicted increased risk of T2DM in Caucasian women (30). A previous study showed that Pima Indians, a population with high propensities for obesity and T2DM, had higher circulating MIF concentrations compared to Caucasians, and MIF concentrations appeared to be increased in non-diabetic Pima Indians with relatively impaired peripheral insulin action (31). The lack of significant association between plasma concentrations of MIF and measures of obesity or insulin action in our study may be explained by the rather narrow range of adiposity and insulin sensitivity in our subjects. In addition, our data indicate that adipocytes or preadipocytes may not be a major source of MIF in the circulation and that the association between MIF from adipose cells and insulin action may be of an autocrine and/or paracrine nature.
Studies in immune cells indicate several putative mechanisms of autocrine/paracrine effects of MIF, including the activation of extracellular signal-regulated kinase 1 (ERK1)/ERK2 - members of the family of mitogen-activated protein kinases (MAPKs), up-regulation of toll like receptor 4 (TLR4) or inhibition of p53 mediated apoptosis (27). ERK activation in adipocytes inhibits secretion of the insulin-sensitizing hormone adiponectin (32). In our study, the expression level of MIF in adipocytes was negatively associated with plasma adiponectin concentration. It could be speculated that local MIF in adipose tissue may inhibit adiponectin secretion, which in turn may result in impaired insulin action in the skeletal muscle and the liver. However, this was not supported by the multivariate model where MIF expression but not circulating adiponectin was a significant predictor of insulin action. Alternatively, MIF may activate TLR4, which stimulates the activity of IκB/NFκB pathway. Activation of this pro-inflammatory pathway can affect insulin signaling via increased release of proinflammatory cytokines (2). Conversely, genetic or immunologic inhibition of MIF in mice prevented TNF-α induced insulin resistance, while recombinant MIF inhibited insulin signaling in adipocytes (33). Alternatively, elevated MIF expression in adipocytes may go along with elevated MIF expression in other cell types, such as resident macrophages within skeletal muscle or Kupfer cells in the liver, thereby directly affecting peripheral and hepatic insulin sensitivity. Embryonic fibroblasts from mice with genetic MIF deficiency show increased expression of pro-adipogenic genes (33). Increased adipogenic capacity of the precursor cells may lead to relatively more small, newly differentiated adipocytes compared to older, lipid-laden large fat cells (34). In fact, adipocyte size was inversely associated with differentiation capacity of cultured preadipocytes in a previous study from our group (35). Lower proportion of newly differentiated adipocytes may also explain reduced adiponectin production and secretion (36). However, it must be noted that knock-down of MIF gene inhibited differentiation and decreased expression of adiponectin mRNA in murine 3T3-L1 preadipocytes (37), which may represent species-specific differences. Adipocyte hypertrophy may also be a consequence of antiapoptotic action of MIF via the p53 pathway (27). According to this scenario adipocyte enlargement and insulin resistance may represent two independent characteristics related to MIF overproduction from adipocytes.
Plasma glucose and insulin levels were positively associated with the expression levels of MIF gene in adipocytes and preadipocytes in our study. Past experiments in 3T3L1 adipocyte line have shown that that the expression of MIF could be stimulated by glucose and insulin, indicating that increased expression of MIF may be a consequence of metabolic dysregulation in obesity (38). However, this was not confirmed in a recent study in isolated mature human adipocytes (26). Furthermore, the association between plasma glucose or insulin concentration and expression of MIF was still present in the cultured preadipocytes, i.e. cells that underwent multiple division cycles after being separated from the in vivo environment for at least two weeks. This indicates that increased expression of MIF may represent an intrinsic characteristic of adipose cell lineage of obese individuals with relatively enlarged adipocytes and impaired insulin action.
There are some potential caveats in interpreting the results of this study. First, our study had an exploratory character because of limited availability of biological material. Therefore we cannot exclude the possibility that some associations were significant by chance (type 1 error) as may be indicated by multiple comparison tests (suppl. 2). However, consistent results of MIF expression between two different cell types and laboratory settings, i.e. adipocytes and cultured preadipocytes, may counteract such an explanation. Inflammation-related genes can also be expressed by macrophages in adipose tissue. Previous experiment from our lab indicate that although macrophages may be present in the preadipocyte cultures, they were not likely to be a major contaminant (13). Collagenase digestion of adipose tissue during isolation could potentially stimulate transcription of inflammation-related genes in adipocytes (39). Although this may have affected the gene expression levels in our study, all our samples were subjected to the same digestion procedure that likely affected all samples similarly. Furthermore, among the measured genes transcripts, the mRNA concentrations of MIF may be the least affected by the digestion procedure because of its constitutive expression pattern (27). We cannot exclude that the average cell size might have been underestimated in subjects with the largest fat cells due to their increased propensity for disruption/lysis when treated by collagenase but if anything, this would likely have led to an underestimation of the relationship between adipocyte size and MIF expression (40). Also, it must be noted that hepatic insulin action was overestimated in over 40% of the study group with complete suppression of EGP due to both insulin dose used and dilution effect of unlabelled glucose (41). Therefore some caution is warranted when interpreting the results on hepatic insulin sensitivity.
In conclusion, our data show that increased expression of MIF gene in cells of adipocyte lineage is associated with obesity characterized by enlarged subcutaneous abdominal adipocytes, reduced circulatory adiponectin, and impaired insulin action on glucose uptake and production. Although these results indicate a compelling role for this abundant and potent pro-inflammatory cytokine in the development of metabolic complications of obesity, further studies are necessary to elucidate mechanisms underlying and the clinical risks related to these associations.
Supplementary Material
Acknowledgments
This work was funded by the intramural research program of the NIDDK/NIH/DHHS. We gratefully acknowledge Thomas Brookshire, PA and nursing and dietary staffs of the NIH metabolic unit for the care of the volunteers, and Emma Rousseau and Shannon Parrington for excellent technical assistance. We are grateful to the members and leaders of the Gila River Indian Community for their continuing co-operation in our studies. We thank Dr. Susanne Votruba for helpful comments on this manuscript.
Reference List
- 1.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444/7121:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 2.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;1167:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259/5091:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 4.Mohamed-Ali V, Goodrick S, Rawesh A, Katz DR, Miles JM, Yudkin JS, Klein S, Coppack SW. Subcutaneous Adipose Tissue Releases Interleukin-6, But Not Tumor Necrosis Factor-{alpha}, in Vivo. Journal of Clinical Endocrinology Metabolism. 1997;82/12:4196–4200. doi: 10.1210/jcem.82.12.4450. [DOI] [PubMed] [Google Scholar]
- 5.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112/12:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112/12:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moraes RC, Blondet A, Birkenkamp-Demtroeder K, Tirard J, Orntoft TF, Gertler A, Durand P, Naville D, Begeot M. Study of the Alteration of Gene Expression in Adipose Tissue of Diet-Induced Obese Mice by Microarray and Reverse Transcription-Polymerase Chain Reaction Analyses. Endocrinology. 2003;144/11:4773–4782. doi: 10.1210/en.2003-0456. [DOI] [PubMed] [Google Scholar]
- 8.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003;112/12:1785–1788. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fain JN, Madan AK, Hiler ML, Cheema P, Bahouth SW. Comparison of the Release of Adipokines by Adipose Tissue, Adipose Tissue Matrix, and Adipocytes from Visceral and Subcutaneous Abdominal Adipose Tissues of Obese Humans. Endocrinology. 2004;145/5:2273–2282. doi: 10.1210/en.2003-1336. [DOI] [PubMed] [Google Scholar]
- 10.Clement K, Viguerie N, Poitou C, Carette C, Pelloux V, Curat CA, Sicard A, Rome S, Benis A, Zucker JD, Vidal H, Laville M, Barsh GS, Basdevant A, Stich V, Cancello R, Langin D. Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J. 2004;18/14:1657–1669. doi: 10.1096/fj.04-2204com. [DOI] [PubMed] [Google Scholar]
- 11.Curat C, Wegner V, Sengen+¿s C, Miranville A, Tonus C, Busse R, Bouloumi+¬ A. Macrophages in human visceral adipose tissue: increased accumulation in obesity and a source of resistin and visfatin. Diabetologia. 2006;49/4:744–747. doi: 10.1007/s00125-006-0173-z. [DOI] [PubMed] [Google Scholar]
- 12.Lee YH, Nair S, Rousseau E, Allison DB, Page GP, Tataranni PA, Bogardus C, Permana PA. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48/9:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair S, Lee YH, Rousseau E, Cam M, Tataranni PA, Baier LJ, Bogardus C, Permana PA. Increased expression of inflammation-related genes in cultured preadipocytes/stromal vascular cells from obese compared with non-obese Pima Indians. Diabetologia. 2005;48/9:1784–1788. doi: 10.1007/s00125-005-1868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skurk T, Alberti-Huber C, Herder C, Hauner H. Relationship between Adipocyte Size and Adipokine Expression and Secretion. J Clin Endocrinol Metab. 2007;92/3:1023–1033. doi: 10.1210/jc.2006-1055. [DOI] [PubMed] [Google Scholar]
- 15.Weyer C, Foley JE, Bogardus C, Tataranni PA, Pratley RE. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia. 2000;43/12:1498–1506. doi: 10.1007/s001250051560. [DOI] [PubMed] [Google Scholar]
- 16.Larson-Meyer DE, Heilbronn LK, Redman LM, Newcomer BR, Frisard MI, Anton S, Smith SR, Alfonso A, Ravussin E the Pennington CALERIE Team. Effect of Calorie Restriction With or Without Exercise on Insulin Sensitivity, {beta}-Cell Function, Fat Cell Size, and Ectopic Lipid in Overweight Subjects. Diabetes Care. 2006;29/6:1337–1344. doi: 10.2337/dc05-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abate N, Garg A, Peshock RM, Stray-Gundersen J, Grundy SM. Relationships of generalized and regional adiposity to insulin sensitivity in men. J Clin Invest. 1995;96/1:88–98. doi: 10.1172/JCI118083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogardus C, Lillioja S, Mott DM, Hollenbeck C, Reaven G. Relationship between degree of obesity and in vivo insulin action in man. Am J Physiol. 1985;248/3 Pt 1:E286–E291. doi: 10.1152/ajpendo.1985.248.3.E286. [DOI] [PubMed] [Google Scholar]
- 19.Lillioja S, Mott DM, Howard BV, Bennett PH, Yki-Jarvinen H, Freymond D, Nyomba BL, Zurlo F, Swinburn B, Bogardus C. Impaired glucose tolerance as a disorder of insulin action. Longitudinal and cross-sectional studies in Pima Indians. N Engl J Med. 1988;318/19:1217–1225. doi: 10.1056/NEJM198805123181901. [DOI] [PubMed] [Google Scholar]
- 20.Steele R. Influences of glucose loading and of injected insulin on hepatic glucose output. Ann N Y Acad Sci. 1959;82:420–430. doi: 10.1111/j.1749-6632.1959.tb44923.x. [DOI] [PubMed] [Google Scholar]
- 21.Best JD, Taborsky GJ, Halter JB, Porte D. Glucose disposal is not proportional to plasma glucose level in man. Diabetes. 1981;30/10:847–850. doi: 10.2337/diab.30.10.847. [DOI] [PubMed] [Google Scholar]
- 22.Lillioja S, Bogardus C. Obesity and insulin resistance: lessons learned from the Pima Indians. Diabetes Metab Rev. 1988;4/5:517–540. doi: 10.1002/dmr.5610040508. [DOI] [PubMed] [Google Scholar]
- 23.Dubois SG, Heilbronn LK, Smith SR, Albu JB, Kelley DE, Ravussin E. Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. Obesity (Silver Spring) 2006;14/9:1543–1552. doi: 10.1038/oby.2006.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith U, Sjostrom L, Bjornstorp P. Comparison of two methods for determining human adipose cell size. J Lipid Res. 1972;13/6:822–824. [PubMed] [Google Scholar]
- 25.Trujillo ME, Scherer PE. Adipose Tissue-Derived Factors: Impact on Health and Disease. Endocr Rev. 2006 doi: 10.1210/er.2006-0033. er. [DOI] [PubMed] [Google Scholar]
- 26.Skurk T, Herder C, Kraft I, Muller-Scholze S, Hauner H, Kolb H. Production and Release of Macrophage Migration Inhibitory Factor from Human Adipocytes. Endocrinology. 2005;146/3:1006–1011. doi: 10.1210/en.2004-0924. [DOI] [PubMed] [Google Scholar]
- 27.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3/10:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yabunaka N, Nishihira J, Mizue Y, Tsuji M, Kumagai M, Ohtsuka Y, Imamura M, Asaka M. Elevated serum content of macrophage migration inhibitory factor in patients with type 2 diabetes. Diabetes Care. 2000;23/2:256–258. doi: 10.2337/diacare.23.2.256. [DOI] [PubMed] [Google Scholar]
- 29.Herder C, Kolb H, Koenig W, Haastert B, Muller-Scholze S, Rathmann W, Holle R, Thorand B, Wichmann HE. Association of Systemic Concentrations of Macrophage Migration Inhibitory Factor With Impaired Glucose Tolerance and Type 2 Diabetes: Results from the Cooperative Health Research in the Region of Augsburg, Survey 4 (KORA S4) Diabetes Care. 2006;29/2:368–371. doi: 10.2337/diacare.29.02.06.dc05-1474. [DOI] [PubMed] [Google Scholar]
- 30.Herder C, Klopp N, Baumert J, Muller M, Khuseyinova N, Meisinger C, Martin S, Illig T, Koenig W, Thorand B. Effect of macrophage migration inhibitory factor (MIF) gene variants and MIF serum concentrations on the risk of type 2 diabetes: results from the MONICA/KORA Augsburg Case-Cohort Study, 1984–2002. Diabetologia. 2007 doi: 10.1007/s00125-007-0800-3. [DOI] [PubMed] [Google Scholar]
- 31.Vozarova B, Stefan N, Hanson R, Lindsay RS, Bogardus C, Tataranni PA, Metz C, Bucala R. Plasma concentrations of macrophage migration inhibitory factor are elevated in Pima Indians compared to Caucasians and are associated with insulin resistance. Diabetologia. 2002;45/12:1739–1741. doi: 10.1007/s00125-002-0896-4. [DOI] [PubMed] [Google Scholar]
- 32.Juan CC, Chuang TY, Chang CL, Huang SW, Ho LT. Endothelin-1 Regulates Adiponectin Gene Expression and Secretion in 3T3-L1 Adipocytes via Distinct Signaling Pathways. Endocrinology. 2007;148/4:1835–1842. doi: 10.1210/en.2006-0654. [DOI] [PubMed] [Google Scholar]
- 33.Atsumi T, Cho YR, Leng L, McDonald C, Yu T, Danton C, Hong EG, Mitchell RA, Metz C, Niwa H, Takeuchi J, Onodera S, Umino T, Yoshioka N, Koike T, Kim JK, Bucala R. The Proinflammatory Cytokine Macrophage Migration Inhibitory Factor Regulates Glucose Metabolism during Systemic Inflammation. J Immunol. 2007;179/8:5399–5406. doi: 10.4049/jimmunol.179.8.5399. [DOI] [PubMed] [Google Scholar]
- 34.Danforth E., Jr Failure of adipocyte differentiation causes type II diabetes mellitus? Nat Genet. 2000;26/1:13. doi: 10.1038/79111. [DOI] [PubMed] [Google Scholar]
- 35.Permana PA, Nair S, Lee YH, Luczy-Bachman G, Vozarova de Court, Tataranni PA. Subcutaneous abdominal preadipocyte differentiation in vitro inversely correlates with central obesity. Am J Physiol Endocrinol Metab. 2004;286/6:E958–E962. doi: 10.1152/ajpendo.00544.2003. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Jansson PA, Nagaev I, Jack MM, Carvalho E, Sunnerhagen KS, Cam MC, Cushman SW, Smith U. Evidence of impaired adipogenesis in insulin resistance. Biochem Biophys Res Commun. 2004;317/4:1045–1051. doi: 10.1016/j.bbrc.2004.03.152. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda D, Sakaue S, Kamigaki M, Ohira H, Itoh N, Ohtsuka Y, Tsujino I, Nishimura M. Knockdown of Macrophage Migration Inhibitory Factor Disrupts Adipogenesis in 3T3-L1 Cells. Endocrinology. 2008;149/12:6037–6042. doi: 10.1210/en.2008-0158. [DOI] [PubMed] [Google Scholar]
- 38.Sakaue S, Nishihira J, Hirokawa J, Yoshimura H, Honda T, Aoki K, Tagami S, Kawakami Y. Regulation of macrophage migration inhibitory factor (MIF) expression by glucose and insulin in adipocytes in vitro. Molecular Medicine. 1999;5/6:361–371. [PMC free article] [PubMed] [Google Scholar]
- 39.Ruan H, Zarnowski MJ, Cushman SW, Lodish HF. Standard Isolation of Primary Adipose Cells from Mouse Epididymal Fat Pads Induces Inflammatory Mediators and Down-regulates Adipocyte Genes. J Biol Chem. 2003;278/48:47585–47593. doi: 10.1074/jbc.M305257200. [DOI] [PubMed] [Google Scholar]
- 40.Hirsch J, Gallian E. Methods for the determination of adipose cell size in man and animals. J Lipid Res. 1968;9/1:110–119. [PubMed] [Google Scholar]
- 41.Hother-Nielsen O, Henriksen JE, Holst JJ, Beck-Nielsen H. Effects of insulin on glucose turnover rates in vivo: Isotope dilution versus constant specific activity technique. Metabolism. 1996;45/1:82–91. doi: 10.1016/s0026-0495(96)90204-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


