Abstract
Proteases, including intracellular proteases, play roles at many different stages of malignant progression. Our focus here is cathepsin B, a lysosomal cysteine cathepsin. High levels of cathepsin B are found in a wide variety of human cancers, levels that often induce secretion and association of cathepsin B with the tumor cell membrane. In experimental models, such as transgenic models of murine pancreatic and mammary carcinomas, causal roles for cathepsin B have been demonstrated in initiation, growth/tumor cell proliferation, angiogenesis, invasion, and metastasis. Tumor growth in transgenic models is promoted by cathepsin B in tumor-associated cells, for example, tumor-associated macrophages, as well as in tumor cells. In transgenic models, the absence of cathepsin B has been associated with enhanced apoptosis, yet cathepsin B also has been shown to contribute to apoptosis. Cathepsin B is part of a proteolytic pathway identified in xenograft models of human glioma; targeting only cathepsin B in these tumors is less effective than targeting cathepsin B in combination with other proteases or protease receptors. Understanding the mechanisms responsible for increased expression of cathepsin B in tumors and association of cathepsin B with tumor cell membranes is needed to determine whether targeting cathepsin B could be of therapeutic benefit.
Keywords: Cancer, Cathepsin B, Cysteine proteases
1 Introduction
Proteases perform essential functions in such processes as ovulation [1,2], fertilization [3], bone remodeling [4], cell migration [5–8], inflammation [9–12], angiogenesis [13,14], and apoptosis [15–18]. Proteases not only perform nonspecific roles, such as hydrolysis of dietary proteins by pancreatic proteases [19], but also act as processing enzymes that can perform highly selective and limited cleavage of substrates as seen for such proteases as calpains [20]. Changes in the expression patterns of proteases underlie numerous human pathological processes, including arthritis [21–23], neurodegenerative disorders [24, 25], inflammatory processes [9], and cardiovascular diseases [26]. Critical functions for proteases in a wide variety of cancers have been identified as evidenced by the more than 7000 reviews on this topic to date. There are approximately 600 proteases in the human genome [27] that are classified as aspartic, cysteine, metallo, serine, or threonine.
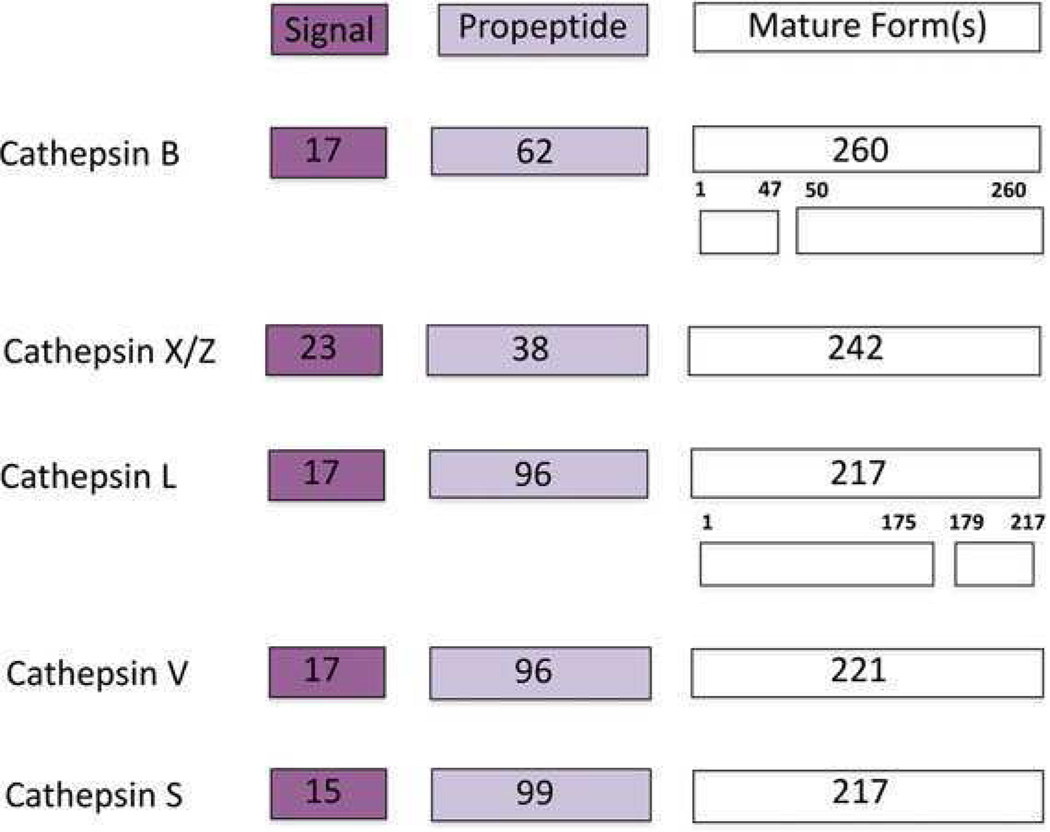
Cathepsin B (EC 3.4.22.1), a lysosomal cysteine protease that is structurally similar to the papaya enzyme papain [28], is one of 11 human cysteine cathepsins (B, C, F, H, L, K, O, S, V, W, X/Z). All the members of the family have been identified in the human genome and characterized molecularly and structurally (for review, see [29]). Cathepsin B is expressed constitutively and linked to general protein turnover in lysosomes. Cathepsin B is synthesized on the rough endoplasmic reticulum (RER) as a preproenzyme of 339 amino acids [30, 31] with a signal peptide of 17 amino acids (Fig. 1). The signal peptide directs the protein into the lumen of RER where the signal peptide is removed and an inactive 43/46 kDa precursor form, procathepsin B, is formed. Procathepsin B is then transported through the RER to the Golgi apparatus where it is glycosylated at two asparagine residues by mannose-containing oligosaccharides with phosphorylated mannose residues. The phosphorylated protein binds to mannose-6-phosphate receptors in the trans-Golgi network and is transported to lysosomes via transport vesicles. The propeptide functions as an inhibitor as well as to stabilize the enzyme. In the acidic environment of lysosomes, procathepsin B can undergo autocatalytic activation as a result of proteolytic cleavage and dissociation of the propeptide, leading to formation of active cathepsin B. Alternatively, cathepsin B can be activated by cathepsin D, an aspartic protease [32], and the serine proteases cathepsin G, urokinase-type plasminogen activator (uPAR), tissue-type plasminogen activator, and elastase [33, 34]. The removal of the propeptide along with six amino acid residues from the C terminus generates a 31 kDa mature single chain form of cathepsin B. A proteolytic cleavage between residues 47 and 50 and excision of the dipeptide generates the double chain form consisting of a heavy chain of 25 kDa and a light chain of 5 kDa (Fig. 1) [30, 31, 35, 36].
Figure 1.
Schematic of cathepsin B protein depicting the signal sequence, propeptide, and single chain and double chain forms. Cathepsin B is compared with other cysteine cathepsins that are implicated in cancer progression. Number of amino acids for each domain is indicated in boxes: cathepsin B [31], cathepsin X/Z [144], cathepsin L [145], cathepsin V [146], cathepsin S [147].
Structurally, cathepsin B is a bilobal protein with cysteine, histidine, and aspartic acid forming active site of the enzyme at the interface between the two lobes [37]. Cathepsin B can function as an endopeptidase, cleaving internal peptide bonds, as well as an exopeptidase (carboxydipeptidase activity) [37, 38]. This dual activity is due to the presence of an occluding loop that interferes with access of substrates to the active site [39]. At acidic pH, the occluding loop partially blocks the active site of the molecule preventing large substrates from entering the active site, yet allowing access of synthetic substrates or the carboxy terminus of proteins. Thus, cathepsin B has carboxydipeptidase activity at acidic pH. At neutral pH, the loop is displaced and no longer blocks the active site, therefore large substrates can enter the active site and cathepsin B can function as an endopeptidase [39,40].
2 Regulation of cathepsin B
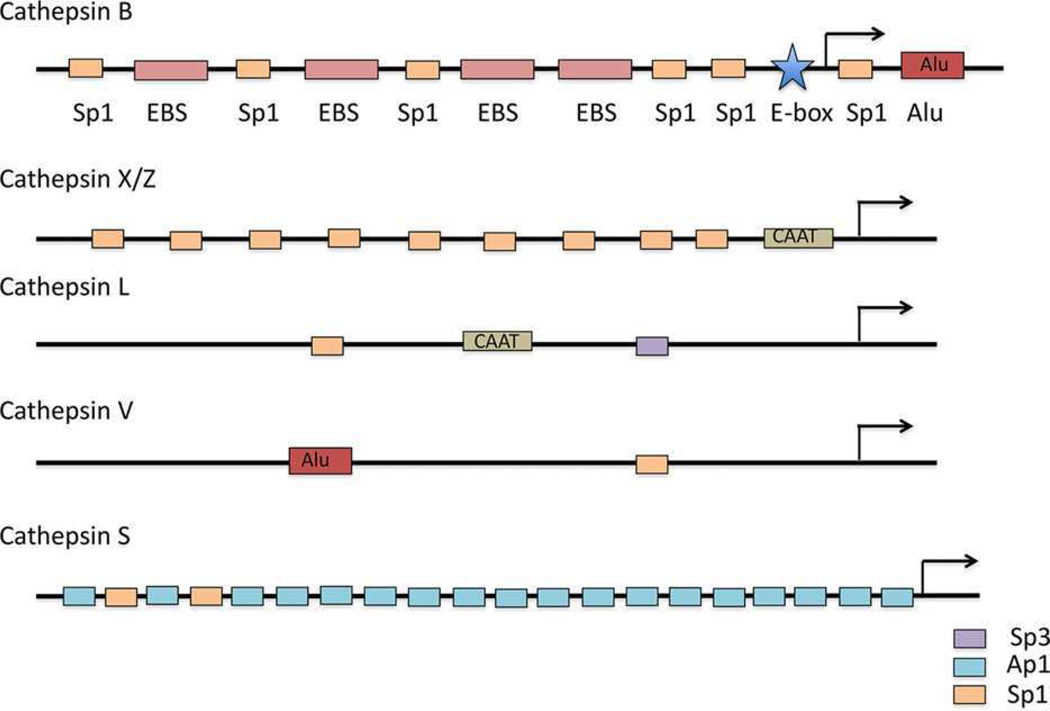
Cathepsin B is regulated at multiple levels from transcription through posttranslational processing and trafficking to activation and inhibition. High levels of expression of cathepsin B at both transcriptional and protein levels have been observed in cancers, for example, esophageal [41], gastric [42], prostate [43], glioblastoma [44, 45], breast [46]. Cathepsin B, a single copy gene located on chromosome 8p22, spans about 27 kilo-bases in the genome [47]. In Barrett’s esophagus, a premalignant lesion, there is amplification of cathepsin B as a result of its location within a novel amplicon [41]. Multiple promoters for the gene have been identified, including in glioma [47]. Upstream of exon 1 is a 2.2 kilobase promoter region that is rich in GC content, yet lacks the characteristic TATA and CAAT boxes. This region has six specificity protein 1 (Sp1) binding sites, four E-twenty six (Ets) binding sites, and one enhancer box (E-box) that regulate transcription (Fig. 2) [45]. Ets1, Sp1, and Sp3 have been shown to activate transcription of cathepsin B [45]. Elevated levels of cathepsin B in cancers thus correlate with Sp1 and Sp3 proteins being high in cancer cells and tissues [48]. Ets1 is a proto-oncogene that enhances invasiveness of carcinoma cells and endothelial cells and is predictive of poor prognosis [49]. In breast cancer, Etsl is overexpressed [50], is elevated in drug resistant cell lines [51], and regulates matrix metalloproteinases (MMPs) that are associated with an invasive phenotype [52]. The E-box in the cathepsin B promoter region is indispensable for promoter activity [53,54]. Upstream stimulatory factors (USFs) increase or repress expression of cathepsin B by binding to the E-box [53, 54]. Intriguingly, USFs are stress-responsive transcription factors [55] and can modulate hypoxia-inducible factor mediated transcription [56], which has been linked to metastasis of breast cancers [57]. In the case of cathepsin B, expression is increased by binding of USF1 and USF2 to the E-box and repressed by binding of USF2c, an alternatively spliced form of USF2 [54].
Figure 2.
Diagram of cathepsin B promoter. The promoter of cathepsin B is compared to promoters of other cysteine cathepsins that are implicated in cancer progression. These promoters are GC rich, do not have a well-defined transcription start site (lack a TATA-box), and have multiple transcription factor binding sites: cathepsin B [60, 148], cathepsin X/Z [149], cathepsin L [150], cathepsin V [151], cathepsin S [147]. Bakhshi et al. [152] and Seth et al. [153] have described two alternative cathepsin L promoters with several putative transcription factor binding sites, but here we depict three sites that Jean et al. [150] confirmed to interact with transcription factors. EBS, Ets binding site.
Posttranscriptionally, cathepsin B is regulated by alternative splicing. Thirty-five splice variants and 27 protein products have been identified (Ensembl). Two 5’ splice variants of cathepsin B are found commonly in pathologies, such as cancer and osteoarthritis [58]. One mRNA lacks exon 2 (i.e., splice variant CB(−2)) and encodes full-length enzyme, but is translated twice as efficiently as the unspliced transcript [59]. This may be due to exon 2 deriving from an Alu element [60] as Alu sequences have been shown to affect gene expression [61]. The second common mRNA is lacking exons 2 and 3 (i.e., splice variant CB(−2,3)). Exon 3 encodes the signal peptide and the first 34 of the 62 amino acids of the propeptide that includes the glycosylation site for binding to mannose phosphate receptors. Therefore, translation of CB(−2,3) produces a truncated form of cathepsin B that does not traffic to the endosomal-lysosomal compartment [59]. Mehtani et al. [62] demonstrated that in vitro translation of CB(−2,3) results in a product that can be folded into an active protease. On the other hand, Baici et al. have shown that CB(−2,3) cannot be properly folded in situ and thus truncated cathepsin B is catalytically inactive [58, 63]. Truncated cathepsin B is targeted to mitochondria [64] at which location the enzyme is involved in cell death [58]. Alternative splicing of cathepsin B to transcripts lacking exon 2 or exons 2 and 3 in cancer and osteoarthritis is likely related to the increased expression of cathepsin B protein in these pathologies as well as to the changes in cathepsin B forms and in its trafficking, both intracellularly and pericellularly, in these pathologies.
In cancers, cathepsin B is found in lysosomes that are perinuclear as well as in vesicles throughout the cytoplasm and at the cell periphery [65, 66]. Subcellular fractionation of tumors established that cathepsin B sediments with both other lysosomal enzymes and plasma membrane markers [67]. Cathepsin B has been localized to the surface of oncogene transformed cells, for example, MCF10AneoT breast epithelial cells [66], and of a variety of cancer cells [68–70]. Activity-based probes for cysteine cathepsins label tumor cell surface cathepsin B have shown that in cathepsin B deficient tumor cells another cysteine cathepsin, that is, cathepsin X, replaces cathepsin B on the tumor cell surface [71]. One of the mechanisms responsible for association of cathepsin B with the tumor cell surface is binding of cathepsin B to the light chain of the annexin II heterotetramer, which localizes to caveolae in the tumor cell membrane [72–74]. Proteases and receptors of the plasminogen cascade also bind to the annexin II heterotetramer, suggesting a role for the annexin II heterotetramer in the induction of proteolytic cascades. Both procathepsin B and active cathepsin B are secreted from tumor cells [75–78]. The secretion of cathepsin B can occur through shedding of membrane vesicles, microvesicles, or exosomes [79, 80]. For further discussion of potential mechanisms for tumor cell surface binding of cathepsin B, see Mohamed and Sloane [81].
The final step of regulation of cathepsin B is at the level of activity through endogenous cysteine protease inhibitors, the cystatins (for reviews, see [82–86]). Cystatins are subdivided into three families as follows: cystatin 1, 2, and 3 [86]. The type 1 and 2 cystatins are single chain polypeptides. Type 1 cystatins, that is, cystatins A and B (also called stefins), are approximately 100 amino acids and are found mainly in the cytoplasm. Type 2 cystatins, for example, cystatins C, D, E/M and S, are approximately 120 amino acid residues and are secreted. Type 3 cystatins are multidomain, high molecular mass proteins (60–120 kDa), such as kininogens. Although cystatin A (stefin A) can inhibit cathepsin B, it is less effective against cathepsin B than against cathepsins L, V, and S due to the presence of the occluding loop in cathepsin B [87]. Cathepsin B is also inhibited by cystatin B (stefin B) [85] and cystatin C [87]. Low mRNA and protein levels of cystatin B are detected in atypical as compared to benign meningioma along with high protein levels of cathepsin B [88]. Changes in the expression of cystatins A and E/M have also been observed in advanced stage prostate and breast cancers, respectively [89–91]. These examples indicate that changes in expression of endogenous inhibitors in malignancy may affect activity of cathepsin B.
3 Cathepsin B and malignancy
Cathepsin B as a cysteine cathepsin often linked to cancer progression is considered a potential therapeutic target (for review, see [29,35,81,92–99]). Elevated levels of cathepsin B have been observed by both genomic and proteomic analyses (for reviews, see [93, 100]). In a proteome analysis, Ebert et al. [42] found that cathepsin B is overexpressed in 60% of gastric cancer patients. Elevated cathepsin B serum levels were present in advanced stage patients as compared to healthy individuals. Proteomic studies of breast, thyroid, and colorectal cancers also found increases in cathepsin B protein [101–103].
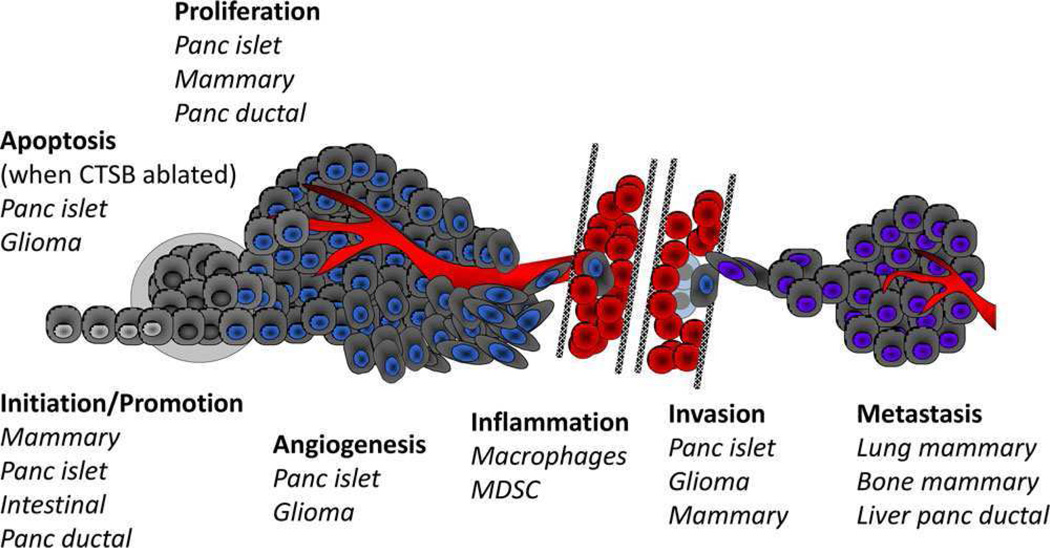
The finding that the expression of cathepsin B is high in many different tumors led to the hypothesis that this enzyme plays a causal role in progression of these tumors. Supporting this hypothesis are studies in which expression of cathepsin B has been downregulated by antisense, siRNA and shRNA technologies, studies that verified roles for cathepsin B at various stages of tumor development (Fig. 3). In brain tumors, Rao and his colleagues have performed a comprehensive analysis of the roles played by cathepsin B, downregulating cathepsin B alone or in combination with members of other proteolytic pathways (for reviews, see [92, 98, 99]). Antisense downregulation of cathepsin B has also been shown to reduce motility and invasion of osteosarcoma cells [104] and shRNA downregulation of cathepsin B to reduce degradation of type I collagen in vitro and bone metastasis in vivo in a murine mammary carcinoma model [105]. Downregulation of cathepsin B in conjunction with downregulation of other proteases/protease receptors has shown that cathepsin B is part of a proteolytic network that involves MMPs and the plasminogen activator cascade. In meningioma, down-regulation of cathepsin B and MMP9 reduces growth in vivo and reduces growth, invasion, angiogenesis, and regulation of downstream kinase signaling pathways in vitro [106], whereas downregulation of cathepsin B and uPAR reduces transforming growth factor β1 (TGF) induced signaling and invasion [107]. In glioma, inhibition of α3β1 integrin and the tetraspanin CD151 occurs when cathepsin B and uPAR are downregulated [108]. Other pathways implicated in cancer progression have also been shown to be linked to decreased expression of cathepsin B, for example, downregulation of secreted protein acidic and rich in cysteine [109], annexin A2 [110], β1-integrin [111], caveolin [73]. In myeloid tumor cells, increases in cathepsin B expression that are mediated by TGF-β1 enhance their carcinogenic potential [112]. Overexpressing cathepsin B and uPAR in meningioma cells in which the two proteases had been knocked down rescues phosphorylation of focal adhesion kinase and angiogenesis [113]. Similarly, overexpression of cathepsin B in mammary cancer cells increases their invasiveness in vitro and tumor progression in vivo [114, 115]. These studies thus establish that overexpression of cathepsin B promotes a malignant phenotype.
Figure 3.
Cancers and stages of initiation and progression in which cathepsin B has been demonstrated by in vivo experimentation to play a causal role. See text for further details; names of cancers are shown italics; cartoon is adapted from [97].
In vivo models have been used to establish causal roles for cathepsin B at multiple points during the development of tumors, that is, initiation, proliferation, angiogenesis, invasion, inflammation, apoptosis, and metastasis. Joyce and colleagues observed that, when cathepsin B knockout mice are crossed with a transgenic model for pancreatic islet cell carcinogenesis, there is a reduction in tumor initiation, proliferation, angiogenesis, and invasion [116]. In this model, genetic ablation of cathepsin B results in an increase in apoptosis [116]. In a model for squamous cell carcinoma, cathepsin B did not promote tumor development; however, another cysteine cathepsin, that is, cathepsin C, did [117]. Thus, there appear to be tissue specific roles for cysteine cathepsins, including cathepsin B, in promoting malignancy. In similar studies in which cathepsin B knockout mice are crossed with a transgenic model for mammary carcinoma, Peters, Reinheckel, and colleagues [71, 118] demonstrated a role for cathepsin B in initiation, proliferation, invasion, and metastasis, including a role for macrophage cathepsin B in metastasis of the mammary carcinomas to the lungs. Cathepsin B does not have any effect on tumor necrosis factor (TNF) mediated apoptosis in this transgenic mammary tumor model [118]. However, in in vitro systems selective damage to lysosomal membranes (using lysosomotropic reagents like L-leucyl-L-leucine methyl ester) induces release of lysosomal proteases, including cathepsin B. This leads to cathepsin B mediated cleavage of proapoptotic Bid to tBid and release of cytochrome c from mitochondria [119]. Cathepsin B has also been reported to cleave antiapoptotic proteins, such as Bcl-2, Bcl-xL, and Bak, thereby inactivating them [120]. Cathepsin B thus plays a role in TNF-triggered and caspase-initiated apoptotic cascades [121, 122] as well as in TNF-related apoptosis-inducing ligand mediated apoptosis [123]. Spesetal. [124] have however recently reported that cysteine cathepsins, while involved, are not critical for TNF-related apoptosis-inducing ligand mediated apoptosis. These studies do not support those in vivo studies demonstrating that cathepsin B loss results in increased apoptosis and would be consistent with cathepsin B playing antagonistic roles in apoptosis, roles that will need to be defined for each cell type and tissue and that may change with malignant progression.
In order to evaluate whether high levels of cathepsin B alter malignant progression, mice overexpressing human cathepsin B have been crossed with the mouse mammary tumor virus polyoma middle T antigen transgenic mouse model of mammary carcinoma [115]. Mammary tumors in the double transgenic mice exhibit a 20-fold increase in cathepsin B activity and grow faster [115]. Lung metastases in these mice are larger and more frequent, supporting a tumor-promoting role for cathepsin B [115]. Using a double transgenic model in which human cathepsin B is overexpressed in either the mammary tumor cells, the stromal cells, or both cell types, Bengsch et al. [114] demonstrated that promotion of tumor progression is due to overexpression of cathepsin B in the tumor cells. In contrast, overexpression of cathepsin B in macrophages creates a microenvironment that is permissive for tumor growth, but does not directly affect the invasive phenotype [114]. Another example of a paracrine action of cathepsin B has been shown for invasion of esophageal epithelial cells. This invasion is mediated as a result of activation of fibroblasts by TGF-β1 [125], which is activated by cathepsin B [126]. Thus, cathepsin B is playing a critical role in crosstalk between tumor cells and other cells in the tumor microenvironment, in some cases resulting in increased invasion and in others being permissive for tumor growth.
There is an extensive literature indicating a role for proteolytic pathways and the ability of one protease to compensate for the loss of another, including striking evidence in transgenic models of compensation for the loss of cathepsin B. In a transgenic mouse model of pancreatic ductal adenocarcinoma, cathepsin B deficiency delays progression of premalignant lesions and pancreatic carcinoma [127]. In these, tumors cathepsin L is upregulated, thus compensating for the loss of cathepsin B. In the mouse mammary tumor virus polyoma middle T antigen mammary carcinoma model, there is compensation not just for a loss of cathepsin B, but specifically for the loss of cathepsin B on the tumor cell surface. Cathepsin X is redistributed to the tumor cell surface of cathepsin B deficient mammary carcinoma cells [71]. Cathepsin X in contrast to cathepsin B is exclusively a carboxypeptidase and thus these findings would be consistent with tumor cell surface cathepsin B functioning as a carboxypeptidase. The ability of cathepsin X to compensate for loss of cathepsin B has also been observed in premalignant intestinal polyps that form in mice with a truncated adenomatous polyposis coli gene [128]. Knockdown of cathepsin B reduces polyposis and infiltration of myeloid-derived suppressor cells (MDSCs), yet increases expression of cathepsin X. An important role for cathepsin B in MDSC also has been observed in two mouse models of pancreatic neuroendocrine carcinogenesis [129]. In these tumors ablation of MMP-9 results in homing of cathepsin B expressing MDSC to the invasive front of the tumors. Cathepsin B in MSDC is thus compensating for the absence of MMP-9 in tumor cells, indicating that proteases of different classes can compensate for one another and that a protease in one cell type can compensate for a protease in another cell type, an observation that emphasizes the critical role played by the tumor microenvironment.
A noncellular aspect of the tumor microenvironment is the generation of an acidic microenvironment around solid tumors [130–133]. This is relevant to cathepsin B as this “lysosomal enzyme” is optimally active at slightly acidic pH [36]. Furthermore, secretion of cathepsin B from malignant cells can be increased by exposing the cells to a slightly acidic extracellular pH [69, 134]. In collaborative studies with the Gillies laboratory, we have shown that tumor invasion occurs in regions of low pH and established a role for cathepsin B in the acid-mediated invasion [135]. We have recently demonstrated that increased degradation of the basement membrane protein type-IV collagen at acidic pH is mediated by cathepsin B [137]. The pattern of pericellular degradation of type-IV collagen observed is consistent with an active role for tumor cell surface cathepsin B, perhaps cathepsin B bound to the light chain of the annexin II heterotetramer in caveolae and on invadopodia [136]. Based on our studies in breast cancer and building on those of Cardelli and colleagues on prostate cancer [137], we propose a model wherein extracellular acidosis is transduced via acid-stimulated ion channels into a signal impinging on the phosphoinositide 3-kinase pathway, microtubules, and RhoA to promote the secretion of cathepsin B at the acidic and invasive edges of tumors, thus, enhancing matrix degradation and tumor invasion.
4 Conclusion
We do not yet have a thorough understanding of all the functions of cathepsin B, including all the functions of this highly promiscuous protease [36] in cancer. Targeting this enzyme may thus reveal yet unknown roles for cathepsin B in cancer and may result in unexpected findings that reflect the specificity of the inhibitors for cathepsin B or compensatory mechanisms, such as elevation in levels/membrane association of cathepsin X as discussed above [71, 128]. Preclinical studies in mouse tumor models have shown that targeting cathepsin B, along with other cysteine cathepsins, with broad-spectrum inhibitors can be efficacious. This is the case in pancreatic islet tumors [138, 139], yet is not true in mammary tumors as a result of tissue differences in inhibitor bioavailability [140]. On the other hand, clinical use of broad-spectrum inhibitors for cysteine cathepsins is likely to encounter the same problems that were found in clinical trials of MMP inhibitors, that is, that some proteases protect against tumor development [97]. In the case of cysteine cathepsins, cathepsin L has been shown to be protective in two mouse models of skin carcinogenesis, one transgenic and one chemically induced [141, 142]. A highly selective inhibitor of cathepsin B, CA074, has been shown to reduce metastasis of a syngeneic mouse mammary tumor to bone [105]. Nonetheless, given the compensatory changes observed in other cysteine cathepsins in mice deficient in cathepsin B [71, 118, 127], use of an inhibitor that is highly selective for cathepsin B may not prove to be effective over the extended time frame needed for treatment in the clinic. Studies by Rao and colleagues on brain tumors [15, 106–108, 113] suggest that cathepsin B is part of a proteolytic cascade that involves proteases of several classes and thus that targeting cathepsin B as well as other protease pathways, for example, the plasminogen cascade through uPAR, is needed. Another type of combination therapy that has been shown to have efficacy is the use of chemotherapeutics and broad-spectrum cysteine cathepsin inhibitors, which greatly reduces both growth and invasion of murine pancreatic islet tumors [138]. Whether cathepsin B will prove to be a druggable target requires further information as we do not yet know whether cathepsin B is an appropriate therapeutic target in a given tumor type or for an individual patient or when during the course of tumor progression that anticathepsin B therapies might prove most effective. We do not know whether the functions of cathepsin B are dynamic and change during the course of malignant progression; whether the cathepsin B playing critical roles in malignant progression comes from tumor cells, tumor-associated cells, or both, factors that need to be considered in designing therapeutic strategies; the mechanisms by which cathepsin B is affected by interactions of the tumor with both its cellular and noncellular microenvironment; etc. Elevated expression of cathepsin B can be documented at the transcript and protein levels in many tumors, as described here. This elevated expression does not, however, necessarily translate into elevated activity as cathepsin B is synthesized as an inactive proenzyme that requires activation. In addition, as noted, there are changes in tumors in the expression of endogenous cysteine protease inhibitors that impact cathepsin B activity. Of note is that elevated activities of cathepsin B at the membrane of tumor cells can be used in activating prodrugs to treat tumors. This has been shown recently in a preclinical study of the efficacy of a cathepsin B cleavable doxorubicin prodrug against hepatocellular carcinoma, in which the prodrug reduced metastasis and exhibited less toxicity than doxorubicin [143]. Clearly further studies defining the multiple and temporal roles of cathepsin B in cancer as well as its cellular localization and interactions with other pathways that contribute to malignancy are needed if cathepsin is to be targeted therapeutically.
Acknowledgments
This work was supported inpart by U.S. Public Health Service Grants: R01 CA56586 and R01 CA131990.
Abbreviations
- E-box
enhancer box
- Ets
E-twenty six
- MDSC
myeloid-derived suppressor cells
- MMP
matrix metalloproteinase
- RER
rough endoplasmic reticulum
- Sp
specificity protein
- TGF
transforming growth factor
- TNF
tumor necrosis factor
- uPAR
urokinase plasminogen activator receptor
- USF
upstream stimulatory factors
Footnotes
The authors have declared no conflict of interest.
References
- 1.Liu YX, Liu XM, Nin LF, Shi L, et al. Serine protease and ovarian paracrine factors in regulation of ovulation. Front. Biosci. 2013;18:650–664. doi: 10.2741/4128. [DOI] [PubMed] [Google Scholar]
- 2.Barbolina MV, Stack MS. Membrane type 1-matrix metalloproteinase: substrate diversity in pericellular proteolysis. Semin. Cell Dev. Biol. 2008;19:24–33. doi: 10.1016/j.semcdb.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo R, Bugge TH. Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu. Rev. Cell Dev. Biol. 2011;27:213–235. doi: 10.1146/annurev-cellbio-092910-154247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georges S, Ruiz Velasco C, Trichet V, Fortun Y, et al. Proteases and bone remodelling. Cytokine Growth Factor Rev. 2009;20:29–41. doi: 10.1016/j.cytogfr.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Staun-Ram E, Miller A. Cathepsins (S and B) and their inhibitor Cystatin C in immune cells: modulation by interferon-beta and role played in cell migration. J. Neuroimmunol. 2011;232:200–206. doi: 10.1016/j.jneuroim.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer A, Nethe M, Hordijk RL. Ubiquitin links to cytoskeletal dynamics, cell adhesion and migration. Biochem. J. 2012;442:13–25. doi: 10.1042/BJ20111815. [DOI] [PubMed] [Google Scholar]
- 8.Wolf K, Friedl P. Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol. 2011;27:736–744. doi: 10.1016/j.tcb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 9.van Kempen LC, de Visser KE, Coussens LM. Inflammation, proteases and cancer. Eur. J. Cancer. 2006;42:728–734. doi: 10.1016/j.ejca.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Broder C, Becker-Pauly C. The metalloproteases meprin alpha and meprin beta: unique enzymes in inflammation, neurodegeneration, cancer and fibrosis. Biochem. J. 2013;450:253–264. doi: 10.1042/BJ20121751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heymann MC, Rosen-Wolff A. Contribution of the inflammasomes to autoinflammatory diseases and recent mouse models as research tools. Clin. Immunol. 2013;747:175–184. doi: 10.1016/j.clim.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Reichel CA, Kanse SM, Krombach F. At the interface of fibrinolysis and inflammation: the role of urokinase-type plasminogen activator in the leukocyte extravasation cascade. Trends Cardiovasc. Med. 2012;22:192–196. doi: 10.1016/j.tcm.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Breuss JM, Uhrin P. VEGF-initiated angiogenesis and the uPA/uPAR system. Cell Adhes. Migr. 2012;6:535–615. doi: 10.4161/cam.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deryugina EI, Quigley JR. Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: contrasting, overlapping and compensatory functions. Biochim. Biophys. Acta. 2010;1803:103–120. doi: 10.1016/j.bbamcr.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malla RR, Gopinath S, Gondi CS, Alapati K, et al. uPAR and cathepsin B downregulation induces apoptosis by targeting calcineurin A to BAD via Bcl-2 in glioma. J Neurooncol. 2012;707:69–80. doi: 10.1007/s11060-011-0727-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Stoka V, Turk B, Turk V. Lysosomal cysteine proteases: structural features and their role in apoptosis. IUBMB Life. 2005;57:347–353. doi: 10.1080/15216540500154920. [DOI] [PubMed] [Google Scholar]
- 17.Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanova S, Repnik U, Bojic L, Petelin A, et al. Lysosomes in apoptosis. Methods Enzymol. 2008;442:183–199. doi: 10.1016/S0076-6879(08)01409-2. [DOI] [PubMed] [Google Scholar]
- 19.Beck IT. The role of pancreatic enzymes in digestion. Am. J. Clin. Nutr. 1973;26:311–325. doi: 10.1093/ajcn/26.3.311. [DOI] [PubMed] [Google Scholar]
- 20.Campbell RL, Davies PL. Structure-function relationships in calpains. Biochem. J. 2012;447:335–351. doi: 10.1042/BJ20120921. [DOI] [PubMed] [Google Scholar]
- 21.Krane SM. Elucidation of the potential roles of matrix metalloproteinases in skeletal biology. Arthritis Res. Ther. 2003;5:2–4. doi: 10.1186/ar600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Troeberg L, Nagase H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta. 2012;1824:133–145. doi: 10.1016/j.bbapap.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baici A, Lang A, Zwicky R, Muntener K. Cathepsin B in osteoarthritis: uncontrolled proteolysis in the wrong place. Semin. Arthritis Rheum. 2005;34:24–28. doi: 10.1016/j.semarthrit.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Hill JW, Rosenberg GA. Multiple roles of metalloproteinases in neurological disorders. Prog. Mol. Biol. Transl. Sci. 2011;99:241–263. doi: 10.1016/B978-0-12-385504-6.00006-3. [DOI] [PubMed] [Google Scholar]
- 25.Todi SV, Paulson HL. Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011;34:370–382. doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luttun A, Dewerchin M, Collen D, Carmeliet P. The role of proteinases in angiogenesis, heart development, restenosis, atherosclerosis, myocardial ischemia, and stroke: insights from genetic studies. Curr. Atheroscler. Rep. 2000;2:407–116. doi: 10.1007/s11883-000-0079-z. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Otin C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 28.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turk V, Stoka V, Vasiljeva O, Renko M, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirschke H, Barrett AJ, Rawlings ND. Proteinases 1: lysosomal cysteine proteinases. Protein Profile. 1995;2:1581–1643. [PubMed] [Google Scholar]
- 31.Mort JS, Buttle DJ. Cathepsin B. Int. J. Biochem. Cell Biol. 1997;29:715–720. doi: 10.1016/s1357-2725(96)00152-5. [DOI] [PubMed] [Google Scholar]
- 32.van der Stappen JW, Williams AC, Maciewicz RA, Paraskeva C. Activation of cathepsin B, secreted by a colorectal cancer cell line requires low pH and is mediated by cathepsin D. Int. J. Cancer. 1996;67:547–554. doi: 10.1002/(SICI)1097-0215(19960807)67:4<547::AID-IJC14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Dalet-Fumeron V, Guinec N, Pagano M. In vitro activation of pro-cathepsin B by three serine proteinases: leucocyte elastase, cathepsin G, and the urokinase-type plasminogen activator. FEBS Lett. 1993;332:251–254. doi: 10.1016/0014-5793(93)80643-9. [DOI] [PubMed] [Google Scholar]
- 34.Dalet-Fumeron V, Boudjennah L, Pagano M. Competition between plasminogen and procathepsin B as a probe to demonstrate the in vitro activation of procathepsin B by the tissue plasminogen activator. Arch. Biochem. Biophys. 1996;335:351–357. doi: 10.1006/abbi.1996.0516. [DOI] [PubMed] [Google Scholar]
- 35.Frosch BA, Berquin I, Emmert-Buck MR, Moin K, et al. Molecular regulation, membrane association and secretion of tumor cathepsin B. APMIS. 1999;707:28–37. doi: 10.1111/j.1699-0463.1999.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 36.Cavallo-Medved D, Moin K, Sloane B, Cathepsin B. UCSD Nat. Mol. Pages. 2011 [PMC free article] [PubMed] [Google Scholar]
- 37.Musil D, Zucic D, Turk D, Engh RA, et al. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 1991;10:2321–2330. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keppler D, Sloane BF. Cathepsin B: multiple enzyme forms from a single gene and their relation to cancer. Enzyme Protein. 1996;49:94–105. doi: 10.1159/000468619. [DOI] [PubMed] [Google Scholar]
- 39.Illy C, Quraishi O, Wang J, Purisima E, et al. Role of the occluding loop in cathepsin B activity. J. Biol. Chem. 1997;272:1197–1202. doi: 10.1074/jbc.272.2.1197. [DOI] [PubMed] [Google Scholar]
- 40.Quraishi O, Nagler DK, Fox T, Sivaraman J, et al. The occluding loop in cathepsin B defines the pH dependence of inhibition by its propeptide. Biochemistry. 1999;38:5017–5023. doi: 10.1021/bi981950o. [DOI] [PubMed] [Google Scholar]
- 41.Hughes SJ, Glover TW, Zhu XX, Kuick R, et al. A novel amplicon at 8p22–23 results in overexpression of cathepsin B in esophageal adenocarcinoma. Proc. Natl. Acad. Sci. USA. 1998;95:12410–12415. doi: 10.1073/pnas.95.21.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ebert MR, Kruger S, Fogeron ML, Lamer S, et al. Overexpression of cathepsin B in gastric cancer identified by proteome analysis. Proteomics. 2005;5:1693–1704. doi: 10.1002/pmic.200401030. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez PL, Farre X, Nadal A, Fernandez E, et al. Expression of cathepsins B and S in the progression of prostate carcinoma. Int. J. Cancer. 2001;95:51–55. doi: 10.1002/1097-0215(20010120)95:1<51::aid-ijc1009>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 44.Rempel SA, Rosenblum ML, Mikkelsen T, Yan RS, et al. Cathepsin B expression and localization in glioma progression and invasion. Cancer Res. 1994;54:6027–6031. [PubMed] [Google Scholar]
- 45.Yan S, Berquin IM, Troen BR, Sloane BF. Transcription of human cathepsin B is mediated by Sp1 and Ets family factors in glioma. DNA Cell Biol. 2000;79:79–91. doi: 10.1089/104454900314591. [DOI] [PubMed] [Google Scholar]
- 46.Kos J, Werle B, Lah T, Brunner N. Cysteine proteinases and their inhibitors in extracellular fluids: markers for diagnosis and prognosis in cancer. Int. J. Biol. Markers. 2000;15:84–89. doi: 10.1177/172460080001500116. [DOI] [PubMed] [Google Scholar]
- 47.Berquin IM, Cao L, Fong D, Sloane BF. Identification of two new exons and multiple transcription start points in the 5’-untranslated region of the human cathepsin-B-encoding gene. Gene. 1995;759:143–149. doi: 10.1016/0378-1119(95)00072-e. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann. Anat. 2010;792:275–283. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Dittmer J. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2003;2:29–49. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buggy Y, Maguire TM, McGreal G, McDermott E, et al. Overexpression of the Ets-1 transcription factor in human breast cancer. Br. J. Cancer. 2004;97:1308–1315. doi: 10.1038/sj.bjc.6602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kars MD, Iseri OD, Gunduz U. Drug resistant breast cancer cells overexpress ETS1 gene. Biomed. Pharmacother. 2010;64:458–462. doi: 10.1016/j.biopha.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Dittmer J, Vetter M, Blumenthal SG, Lindemann RK, et al. Importance of etsl proto-oncogene for breast cancer progression. Zentralbl. Gynakol. 2004;726:269–271. doi: 10.1055/s-2004-822758. [DOI] [PubMed] [Google Scholar]
- 53.Yan S, Jane DT, Dufresne MJ, Sloane BR. Transcription of cathepsin B in glioma cells: regulation by an E-box adjacent to the transcription initiation site. Biol. Chem. 2003;384:1421–1427. doi: 10.1515/BC.2003.157. [DOI] [PubMed] [Google Scholar]
- 54.Yan S, Sloane BR. Isolation of a novel USR2 isoform: repressor of cathepsin B expression. Gene. 2004;337:199–206. doi: 10.1016/j.gene.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 56.Hu J, Stiehl DR, Setzer C, Wichmann D, et al. Interaction of HIF and USF signaling pathways in human genes flanked by hypoxia-response elements and E-box palindromes. Mol. Cancer Res. 2011;9:1520–1536. doi: 10.1158/1541-7786.MCR-11-0090. [DOI] [PubMed] [Google Scholar]
- 57.Semenza GL. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends Mol. Med. 2012;18:534–543. doi: 10.1016/j.molmed.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baici A, Muntener K, Willimann A, Zwicky R. Regulation of human cathepsin B by alternative mRNA splicing: homeostasis, fatal errors and cell death. Biol. Chem. 2006;387:1017–1021. doi: 10.1515/BC.2006.125. [DOI] [PubMed] [Google Scholar]
- 59.Gong Q, Chan SJ, Bajkowski AS, Steiner DR, et al. Characterization of the cathepsin B gene and multiple mRNAs in human tissues: evidence for alternative splicing of cathepsin B pre-mRNA. DNA Cell Biol. 1993;12:299–309. doi: 10.1089/dna.1993.12.299. [DOI] [PubMed] [Google Scholar]
- 60.Berquin IM, Ahram M, Sloane BR. Exon 2 of human cathepsin B derives from an Alu element. FEBS Lett. 1997;479:121–123. doi: 10.1016/s0014-5793(97)01445-2. [DOI] [PubMed] [Google Scholar]
- 61.Mighell AJ, Markham AR, Robinson PA. Alu sequences. FEBS Lett. 1997;477:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 62.Mehtani S, Gong Q, Panella J, Subbiah S, et al. In vivo expression of an alternatively spliced human tumor message that encodes a truncated form of cathepsin B. Subcellular distribution of the truncated enzyme in COS cells. J. Biol. Chem. 1998;273:13236–13244. doi: 10.1074/jbc.273.21.13236. [DOI] [PubMed] [Google Scholar]
- 63.Muntener K, Willimann A, Zwicky R, Svoboda B, et al. Folding competence of N-terminally truncated forms of human procathepsin. B. J. Biol. Chem. 2005;280:11973–11980. doi: 10.1074/jbc.M413052200. [DOI] [PubMed] [Google Scholar]
- 64.Muntener K, Zwicky R, Csucs G, Rohrer J, et al. Exon skipping of cathepsin B: mitochondrial targeting of a lysosomal peptidase provokes cell death. J. Biol. Chem. 2004;279:41012–11017. doi: 10.1074/jbc.M405333200. [DOI] [PubMed] [Google Scholar]
- 65.Sloane BR, Moin K, Sameni M, Tait LR, et al. Membrane association of cathepsin B can be induced by transfection of human breast epithelial cells with c-Ha-ras oncogene. J. Cell Sci. 1994;707(Pt 2):373–384. doi: 10.1242/jcs.107.2.373. [DOI] [PubMed] [Google Scholar]
- 66.Sameni M, Elliott E, Ziegler G, Rortgens PH, et al. Cathepsin B and D are localized at the surface of human breast cancer cells. Pathol. Oncol. Res. 1995;7:43–53. doi: 10.1007/BF02893583. [DOI] [PubMed] [Google Scholar]
- 67.Rozhin J, Robinson D, Stevens MA, Lah TT, et al. Properties of a plasma membrane-associated cathepsin B-like cysteine proteinase in metastatic B16 melanoma variants. Cancer Res. 1987;47:6620–6628. [PubMed] [Google Scholar]
- 68.Demchik LL, Sameni M, Nelson K, Mikkelsen T, et al. Cathepsin B and glioma invasion. Int. J. Dev. Neurosci. 1999;77:483–494. doi: 10.1016/s0736-5748(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 69.Rozhin J, Sameni M, Ziegler G, Sloane BR. Pericellular pH affects distribution and secretion of cathepsin B in malignant cells. Cancer Res. 1994;54:6517–6525. [PubMed] [Google Scholar]
- 70.Yan Z, Deng X, Chen M, Xu Y, et al. Oncogenic c-Ki-ras but not oncogenic c-Ha-ras up-regulates CEA expression and disrupts basolateral polarity in colon epithelial cells. J. Biol. Chem. 1997;272:27902–27907. doi: 10.1074/jbc.272.44.27902. [DOI] [PubMed] [Google Scholar]
- 71.Vasiljeva O, Papazoglou A, Kruger A, Brodoefel H, et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 72.Cavallo-Medved D, Dosescu J, Linebaugh BE, Sameni M, et al. Mutant K-ras regulates cathepsin B localization on the surface of human colorectal carcinoma cells. Neoplasia. 2003;5:507–519. doi: 10.1016/s1476-5586(03)80035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cavallo-Medved D, Mai J, Dosescu J, Sameni M, et al. Caveolin-1 mediates the expression and localization of cathepsin B, pro-urokinase plasminogen activator and their cell-surface receptors in human colorectal carcinoma cells. J. Cell Sci. 2005;77S:1493–1503. doi: 10.1242/jcs.02278. [DOI] [PubMed] [Google Scholar]
- 74.Victor BC, Anbalagan A, Mohamed MM, Sloane BR, et al. Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion. Breast Cancer Res. 2011;13:R115–R128. doi: 10.1186/bcr3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Recklies AD, Tiltman KJ, Stoker TA, Poole AR. Secretion of proteinases from malignant and nonmalignant human breast tissue. Cancer Res. 1980;40:550–556. [PubMed] [Google Scholar]
- 76.Koblinski JE, Dosescu J, Sameni M, Moin K, et al. Interaction of human breast fibroblasts with collagen I increases secretion of procathepsin B. J. Biol. Chem. 2002;277:32220–32227. doi: 10.1074/jbc.M204708200. [DOI] [PubMed] [Google Scholar]
- 77.Linebaugh BE, Sameni M, Day NA, Sloane BR, et al. Exocytosis of active cathepsin B enzyme activity at pH 7.0, inhibition and molecular mass. Eur. J. Biochem. FEBS. 1999;264:100–109. doi: 10.1046/j.1432-1327.1999.00582.x. [DOI] [PubMed] [Google Scholar]
- 78.Klose A, Wilbrand-Hennes A, Zigrino R, Weber E, et al. Contact of high-invasive, but not low-invasive, melanoma cells to native collagen I induces the release of mature cathepsin B. Int. J. Cancer. 2006;118:2735–2743. doi: 10.1002/ijc.21700. [DOI] [PubMed] [Google Scholar]
- 79.Cavanaugh PG, Sloane BR, Bajkowski AS, Gasic GJ, et al. Involvement of a cathepsin B-like cysteine proteinase in platelet aggregation induced by tumor cells and their shed membrane vesicles. Clin. Exp. Metastasis. 1983;7:297–307. doi: 10.1007/BF00121192. [DOI] [PubMed] [Google Scholar]
- 80.Giusti I, D’Ascenzo S, Millimaggi D, Taraboletti G, et al. Cathepsin B mediates the pH-dependent proinvasive activity of tumor-shed microvesicles. Neoplasia. 2008;10:481–488. doi: 10.1593/neo.08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mohamed MM, Sloane BR. Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 82.Turk V, Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-c. [DOI] [PubMed] [Google Scholar]
- 83.Sloane BR. Cathepsin B and cystatins: evidence for a role in cancer progression. Semin. Cancer Biol. 1990;7:137–152. [PubMed] [Google Scholar]
- 84.Sloane BR, Moin K, Krepela E, Rozhin J. Cathepsin B and its endogenous inhibitors: the role in tumor malignancy. Cancer Metastasis Rev. 1990;9:333–352. doi: 10.1007/BF00049523. [DOI] [PubMed] [Google Scholar]
- 85.Turk B, Turk V, Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol. Chem. 1997;378:141–150. [PubMed] [Google Scholar]
- 86.Abrahamson M, Alvarez-Rernandez M, Nathanson CM. Cystatins. Biochem. Soc. Symp. 2003;70:179–199. doi: 10.1042/bss0700179. [DOI] [PubMed] [Google Scholar]
- 87.Pavlova A, Krupa JC, Mort JS, Abrahamson M, et al. Cystatin inhibition of cathepsin B requires dislocation of the proteinase occluding loop. Demonstration by release of loop anchoring through mutation of his 110. FEBS Lett. 2000;487:156–160. doi: 10.1016/s0014-5793(00)02337-1. [DOI] [PubMed] [Google Scholar]
- 88.Trinkaus M, Vranic A, Dolenc VV, Lah TT. Cathepsins B and L and their inhibitors stefin B and cystatin C as markers for malignant progression of benign meningiomas. Int. J. Biol. Markers. 2005;20:50–59. doi: 10.1177/172460080502000108. [DOI] [PubMed] [Google Scholar]
- 89.Sinha AA, Quast BJ, Korkowski JC, Wilson MJ, et al. The relationship of cathepsin B and stefin A mRNA localization identifies a potentially aggressive variant of human prostate cancer within a Gleason histologic score. Anticancer Res. 1999;19:2821–2829. [PubMed] [Google Scholar]
- 90.Sinha AA, Quast BJ, Wilson MJ, Rernandes ET, et al. Ratio of cathepsin B to stefin A identifies heterogeneity within Gleason histologic scores for human prostate cancer. Prostate. 2001;48:274–284. doi: 10.1002/pros.1107. [DOI] [PubMed] [Google Scholar]
- 91.Shridhar R, Zhang J, Song J, Booth BA, et al. Cystatin M suppresses the malignant pheno-type of human MDA-MB-435S cells. Oncogene. 2004;23:2206–2215. doi: 10.1038/sj.onc.1207340. [DOI] [PubMed] [Google Scholar]
- 92.Gondi CS, Rao JS. Cathepsin B as a cancer target. Expert Opin. Ther. Targets. 2013;17:281–291. doi: 10.1517/14728222.2013.740461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jedeszko C, Sloane BR. Cysteine cathepsins in human cancer. Biol. Chem. 2004;385:1017–1027. doi: 10.1515/BC.2004.132. [DOI] [PubMed] [Google Scholar]
- 94.Kuester D, Lippert H, Roessner A, Krueger S. The cathepsin family and their role in colorectal cancer. Pathol. Res. Pract. 2008;204:491–500. doi: 10.1016/j.prp.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 95.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;27:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol. Sci. 2008;29:22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 97.Sloane BR, List K, Ringleton B, Matrisian L. Proteases in cancer – significance for invasion and metastasis. In: Brix K, Stoecker W, editors. Proteases—Structure and Function. New York: Springer-Verlag; in press. [Google Scholar]
- 98.Lakka SS, Gondi CS, Rao JS. Proteases and glioma angiogenesis. Brain Pathol. 2005;15:327–341. doi: 10.1111/j.1750-3639.2005.tb00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat. Rev. Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 100.Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clin. Chim. Acta. 2000;291:113–135. doi: 10.1016/s0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 101.McKerrow JH, Bhargava V, Hansell E, Huling S, et al. A functional proteomics screen of proteases in colorectal carcinoma. Mol. Med. 2000;6:450–160. [PMC free article] [PubMed] [Google Scholar]
- 102.Srisomsap C, Subhasitanont P, Otto A, Mueller EC, et al. Detection of cathepsin B up-regulation in neoplastic thyroid tissues by proteomic analysis. Proteomics. 2002;2:706–712. doi: 10.1002/1615-9861(200206)2:6<706::AID-PROT706>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 103.Wulfkuhle JD, Sgroi DC, Krutzsch H, McLean K, et al. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 2002;62:6740–6749. [PubMed] [Google Scholar]
- 104.Krueger S, Haeckel C, Buehling R, Roessner A. Inhibitory effects of antisense cathepsin B cDNA transfection on invasion and motility in a human osteosarcoma cell line. Cancer Res. 1999;59:6010–6014. [PubMed] [Google Scholar]
- 105.Withana NR, Blum G, Sameni M, Slaney C, et al. Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res. 2012;72:1199–1209. doi: 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tummalapalli P, Spomar D, Gondi CS, Olivero WC, et al. RNAi-mediated abrogation of cathepsin B and MMP-9 gene expression in a malignant meningioma cell line leads to decreased tumor growth, invasion and angiogenesis. Int. J. Oncol. 2007;31:1039–1050. [PMC free article] [PubMed] [Google Scholar]
- 107.Gogineni VR, Gupta R, Nalla AK, Velpula KK, et al. uPAR and cathepsin B shRNA impedes TGR-beta1-driven proliferation and invasion of meningioma cells in a XIAP-dependent pathway. Cell death Dis. 2012;3:e439. doi: 10.1038/cddis.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rao Malla R, Gopinath S, Alapati K, Gorantla B, et al. Knockdown of cathepsin B and uPAR inhibits CD151 and alpha3beta1 integrin-mediated cell adhesion and invasion in glioma. Mol. Carcinog. 2013;52:777–790. doi: 10.1002/mc.21915. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109.Girotti MR, Rernandez M, Lopez JA, Camafeita E, et al. SPARC promotes cathepsin B-mediated melanoma invasiveness through a collagen l/alpha2beta1 integrin axis. J. Invest. Dermatol. 2011;737:2438–2447. doi: 10.1038/jid.2011.239. [DOI] [PubMed] [Google Scholar]
- 110.Wang YX, Lv H, Li ZX, Li C, et al. Effect of shRNA mediated down-regulation of Annexin A2 on biological behavior of human lung adencarcinoma cells A549. Pathol. Oncol. Res. 2012;18:183–190. doi: 10.1007/s12253-011-9427-2. [DOI] [PubMed] [Google Scholar]
- 111.Sameni M, Dosescu J, Yamada KM, Sloane BF, et al. Functional live-cell imaging demonstrates that beta1-integrin promotes type IV collagen degradation by breast and prostate cancer cells. Mol. Imaging. 2008;7:199–213. [PMC free article] [PubMed] [Google Scholar]
- 112.Reisenauer A, Eickelberg O, Wille A, Heimburg A, et al. Increased carcinogenic potential of myeloid tumor cells induced by aberrant TGR-beta1-signaling and upregulation of cathepsin B. Biol. Chem. 2007;388:639–650. doi: 10.1515/BC.2007.072. [DOI] [PubMed] [Google Scholar]
- 113.Gupta R, Nalla AK, Gogineni VR, Chetty C, et al. uPAR/cathepsin B overexpression reverse angiogenesis by rescuing RAK phosphorylation in uPAR/cathepsin B down regulated meningioma. PloS One. 2011;6:e17123. doi: 10.1371/journal.pone.0017123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Bengsch R, Buck A, Gunther SC, Seiz JR, et al. Cell type-dependent pathogenic functions of overexpressed human cathepsin B in murine breast cancer progression. Oncogene. 2013 doi: 10.1038/onc.2013.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sevenich L, Werner R, Gajda M, Schurigt U, et al. Transgenic expression of human cathepsin B promotes progression and metastasis of polyoma-middle-T-induced breast cancer in mice. Oncogene. 2011;30:54–64. doi: 10.1038/onc.2010.387. [DOI] [PubMed] [Google Scholar]
- 116.Gocheva V, Zeng W, Ke D, Klimstra D, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ruffell B, Affara NI, Cottone L, Junankar S, et al. Cathepsin C is a tissue-specific regulator of squamous carcinogenesis. Genes Dev. 2013;27:2086–2098. doi: 10.1101/gad.224899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vasiljeva O, Korovin M, Gajda M, Brodoefel H, et al. Reduced tumour cell proliferation and delayed development of high-grade mammary carcinomas in cathepsin B-deficient mice. Oncogene. 2008;27:4191–4199. doi: 10.1038/onc.2008.59. [DOI] [PubMed] [Google Scholar]
- 119.Cirman T, Oresic K, Mazovec GD, Turk V, et al. Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J. Biol. Chem. 2004;279:3578–3587. doi: 10.1074/jbc.M308347200. [DOI] [PubMed] [Google Scholar]
- 120.Droga-Mazovec G, Bojic L, Petelin A, Ivanova S, et al. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J. Biol. Chem. 2008;283:19140–19150. doi: 10.1074/jbc.M802513200. [DOI] [PubMed] [Google Scholar]
- 121.Foghsgaard L, Wissing D, Mauch D, Lademann U, et al. Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 2001;753:999–1010. doi: 10.1083/jcb.153.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mathiasen IS, Hansen CM, Foghsgaard L, Jaattela M. Sensitization to TNF-induced apoptosis by 1,25-dihydroxy vitamin D(3) involves up-regulation of the TNF receptor 1 and cathepsin B. Int. J. Cancer. 2001;93:224–231. doi: 10.1002/ijc.1325. [DOI] [PubMed] [Google Scholar]
- 123.Guicciardi ME, Bronk SF, Werneburg NW, Gores GJ. cFLIPL prevents TRAIL-induced apoptosis of hepatocellular carcinoma cells by inhibiting the lysosomal pathway of apoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G1337–G1346. doi: 10.1152/ajpgi.00497.2006. [DOI] [PubMed] [Google Scholar]
- 124.Spes A, Sobotic B, Turk V, Turk B. Cysteine cathepsins are not critical for TRAIL- and CD95-induced apoptosis in several human cancer cell lines. Biol. Chem. 2012;393:1417–1431. doi: 10.1515/hsz-2012-0213. [DOI] [PubMed] [Google Scholar]
- 125.Andl CD, McCowan KM, Allison GL, Rustgi AK. Cathepsin B is the driving force of esophageal cell invasion in a fibroblast-dependent manner. Neoplasia. 2010;72:485–498. doi: 10.1593/neo.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guo M, Mathieu PA, Linebaugh B, Sloane BF, et al. Phorbol ester activation of a proteolytic cascade capable of activating latent transforming growth factor-betaL a process initiated by the exocytosis of cathepsin B. J. Biol. Chem. 2002;277:14829–14837. doi: 10.1074/jbc.M108180200. [DOI] [PubMed] [Google Scholar]
- 127.Gopinathan A, Denicola GM, Frese KK, Cook N, et al. Cathepsin B promotes the progression of pancreatic ductal adenocarcinoma in mice. Gut. 2012;61:877–884. doi: 10.1136/gutjnl-2011-300850. [DOI] [PubMed] [Google Scholar]
- 128.Gounaris E, Tung CH, Restaino C, Maehr R, et al. Live imaging of cysteine-cathepsin activity reveals dynamics of focal inflammation, angiogenesis, and polyp growth. PloS One. 2008;3:e2916. doi: 10.1371/journal.pone.0002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shchors K, Nozawa H, Xu J, Rostker F, et al. Increased invasiveness of MMP-9-deficient tumors in two mouse models of neuroendocrine tumorigenesis. Oncogene. 2013;32:502–513. doi: 10.1038/onc.2012.60. [DOI] [PubMed] [Google Scholar]
- 130.Kato Y, Ozawa S, Miyamoto C, Maehata Y, et al. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013;73:89–96. doi: 10.1186/1475-2867-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat. Rev. Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 132.Wike-Hooley JL, Haveman J, Reinhold HS. The relevance of tumour pH to the treatment of malignant disease. Radiother. Oncol. 1984;2:343–366. doi: 10.1016/s0167-8140(84)80077-8. [DOI] [PubMed] [Google Scholar]
- 133.Gillies RJ, Raghunand N, Karczmar GS, Bhujwalla ZM. MRI of the tumor microenvironment. J. Magn. Reson. Imaging. 2002;16:430–450. doi: 10.1002/jmri.10181. [DOI] [PubMed] [Google Scholar]
- 134.Glunde K, Guggino SE, Solaiyappan M, Pathak AR, et al. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 2003;5:533–545. doi: 10.1016/s1476-5586(03)80037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Estrella V, Chen T, Lloyd M, Wojtkowiak J, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73:1524–1535. doi: 10.1158/0008-5472.CAN-12-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rothberg JM, Sameni M, Moin K, Sloane BF. Live-cell imaging of tumor proteolysis: impact of cellular and non-cellular microenvironment. Biochim. Biophys. Acta. 2012;7324:123–132. doi: 10.1016/j.bbapap.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Steffan JJ, Snider JL, Skalli O, Welbourne T, et al. Na+/H +exchangers and RhoA regulate acidic extracellular pH-induced lysosome trafficking in prostate cancer cells. Traffic. 2009;10:737–753. doi: 10.1111/j.1600-0854.2009.00904.x. [DOI] [PubMed] [Google Scholar]
- 138.Bell-McGuinn KM, Garfall AL, Bogyo M, Hanahan D, et al. Inhibition of cysteine cathepsin protease activity enhances chemotherapy regimens by decreasing tumor growth and invasiveness in a mouse model of multistage cancer. Cancer Res. 2007;67:7378–7385. doi: 10.1158/0008-5472.CAN-07-0602. [DOI] [PubMed] [Google Scholar]
- 139.Elie BT, Gocheva V, Shree T, Dalrymple SA, et al. Identification and pre-clinical testing of a reversible cathepsin protease inhibitor reveals anti-tumor efficacy in a pancreatic cancer model. Biochimie. 2010;92:1618–1624. doi: 10.1016/j.biochi.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schurigt U, Sevenich L, Vannier C, Gajda M, et al. Trial of the cysteine cathepsin inhibitor JPM-OEt on early and advanced mammary cancer stages in the MMTV-PyMT-transgenic mouse model. Biol. Chem. 2008;389:1067–1074. doi: 10.1515/BC.2008.115. [DOI] [PubMed] [Google Scholar]
- 141.Dennemarker J, Lohmuller T, Mayerle J, Tacke M, et al. Deficiency for the cysteine protease cathepsin L promotes tumor progression in mouse epidermis. Oncogene. 2010;29:1611–1621. doi: 10.1038/onc.2009.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Benavides F, Perez C, Blando J, Contreras O, et al. Protective role of cathepsin L in mouse skin carcinogenesis. Mol. Carcinog. 2011;51:352–361. doi: 10.1002/mc.20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang Q, Zhong YJ, Yuan JR, Shao LH, et al. Targeting therapy of hepatocellular carcinoma with doxorubicin prodrug PDOX increases anti-metastatic effect and reduces toxicity: a preclinical study. J. Transl. Med. 2013;77:192. doi: 10.1186/1479-5876-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nagler DK, Menard R. Human cathepsin X: a novel cysteine protease of the papain family with a very short proregion and unique insertions. FEBS Lett. 1998;434:135–139. doi: 10.1016/s0014-5793(98)00964-8. [DOI] [PubMed] [Google Scholar]
- 145.Ritonja A, Popovic T, Kotnik M, Machleidt W, et al. Amino acid sequences of the human kidney cathepsins H and L. FEBS Lett. 1988;228:341–345. doi: 10.1016/0014-5793(88)80028-0. [DOI] [PubMed] [Google Scholar]
- 146.Bromme D, Li Z, Barnes M, Mehler E. Human cathepsin V functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization, and chromosomal localization. Biochemistry. 1999;38:2377–2385. doi: 10.1021/bi982175f. [DOI] [PubMed] [Google Scholar]
- 147.Wiederanders B, Bromme D, Kirschke H, von Figura K, et al. Phylogenetic conservation of cysteine proteinases. Cloning and expression of a cDNA coding for human cathepsin S. J. Biol. Chem. 1992;267:13708–13713. [PubMed] [Google Scholar]
- 148.Yan S, Sloane BR. Molecular regulation of human cathepsin B: implication in pathologies. Biol. Chem. 2003;384:845–854. doi: 10.1515/BC.2003.095. [DOI] [PubMed] [Google Scholar]
- 149.Deussing J, von Olshausen I, Peters C. Murine and human cathepsin Z: cDNA-cloning, characterization of the genes and chromosomal localization. Biochim. Biophys. Acta. 2000;7497:93–106. doi: 10.1016/s0167-4781(00)00021-x. [DOI] [PubMed] [Google Scholar]
- 150.Jean D, Guillaume N, Rrade R. Characterization of human cathepsin L promoter and identification of binding sites for NF-Y, Sp1 and Sp3 that are essential for its activity. Biochem. J. 2002;367:173–184. doi: 10.1042/0264-6021:3610173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Itoh R, Kawamoto S, Adachi W, Kinoshita S, et al. Genomic organization and chromosomal localization of the human cathepsin L2 gene. DNA Res. 1999;6:137–140. doi: 10.1093/dnares/6.2.137. [DOI] [PubMed] [Google Scholar]
- 152.Bakhshi R, Goel A, Seth R, Chhikara R, et al. Cloning and characterization of human cathepsin L promoter. Gene. 2001;275:93–101. doi: 10.1016/s0378-1119(01)00650-3. [DOI] [PubMed] [Google Scholar]
- 153.Seth R, Mahajan VS, Chauhan SS. Transcription of human cathepsin L mRNA species hCATL B from a novel alternative promoter in the first intron of its gene. Gene. 2003;327:83–91. doi: 10.1016/s0378-1119(03)00838-2. [DOI] [PubMed] [Google Scholar]