Abstract
A growing number of functions are emerging for RNAi in the nucleus, in addition to well-characterized roles in post-transcriptional gene silencing in the cytoplasm. Epigenetic modifications directed by small RNA have been shown to cause transcriptional repression in plants, fungi, and animals. Additionally growing evidence indicates that RNAi regulates transcription through interaction with transcriptional machinery. Nuclear small RNAs include small interfering RNA (siRNA) and PIWI-interacting RNA (piRNA) and are implicated in nuclear processes such as transposon regulation, heterochromatin formation, developmental gene regulation and genome stability.
Introduction
Since the discovery that double stranded RNA (dsRNA) can robustly silence genes in C. elegans and plants, RNAi has become a new paradigm for understanding gene regulation. The mechanism is well conserved across model organisms and utilizes short antisense RNA to inhibit translation, or to degrade cytoplasmic mRNA by post-transcriptional gene silencing (PTGS). PTGS protects against viral infection, prevents transposon mobilization, and regulates endogenous genes. Three classes of small RNA can regulate genes by targeting transcripts in the cytoplasm. These are miRNA that are hairpin-derived with imperfect complementarity to target transcripts and cause translational repression, small interfering RNA (siRNA) with perfect complementarity to targets and cause transcript degradation, and piwi RNA (piRNA) which target transposon transcripts in animal germlines. Traditionally the term RNAi has been used to describe siRNA pathways, however the mechanistic details of diverse small RNA pathways are converging, so in this review we use RNAi as an umbrella term to describe silencing that is dependent on small RNA.
In plants and fungi, RNAi pathways in the nucleus can repress target genes at the transcriptional level by guiding epigenetic modification of chromatin, for example via histone and DNA methyltransferases. At first these pathways were thought to be absent from metazoans, but recently, a parallel mechanism has been found in the germline. These findings have revealed a conserved nuclear role for RNAi in transcriptional gene silencing (TGS), and because it occurs in the germline, TGS can lead to transgenerational inheritance in absence of the initiating RNA, but dependent on endogenously produced small RNA. Such epigenetic inheritance is familiar in plants, but only recently described in metazoans.
In this review we cover the extremely broad range of nuclear RNAi pathways that have been discovered across organisms, so that readers can appreciate the conservation between these pathways, while being aware of important differences. As such we can only scratch the surface of pathways in each organism, however readers are directed to other articles for an in-depth analysis when appropriate. We concentrate on the siRNA and piRNA small RNA pathways because their nuclear roles are best understood.
To begin we outline the biogenesis of small RNA, focusing on the subcellular localization of these processes. Next, a mechanistic understanding of nuclear RNAi is described in model systems where it has been elucidated. Nuclear small RNA function in the germlines of a broad range of organisms where mechanistic details have not fully emerged, so we discuss the important biological significance of their roles, including in transposon regulation, epigenetic inheritance, and developmental gene regulation, and suggest parallels that can be drawn to well understood mechanistic models. Finally we end forward thinking by exploring the newly developing relationship between nuclear RNAi, genome maintenance, and DNA repair.
Biogenesis of Nuclear Small RNA
The siRNA and piRNA pathways differ in the source of the primary RNA that elicits a response and the mechanism by which small RNA is subsequently produced and amplified. The Argonaute family of proteins are the effectors of RNAi and this family consists of two subclades: Ago proteins which are ubiquitously expressed and bind miRNA and siRNA, and Piwi proteins which were originally discovered in the germline and bind piRNA1–3. Table 1 summarizes the characteristics of the nuclear siRNA and piRNA pathways for some of the organisms described in this review, highlighting the nuclear effector, size class, and proteins involved in biogenesis.
Table 1.
Overview of small RNA classes and their functions within the nucleus.
| Small RNA Class | Size | Nuclear Effector | Biogenesis Factors | Genomic Origin | Nuclear Function |
|---|---|---|---|---|---|
| S. pombe | |||||
| siRNA | 21–23nt | Ago1 | Dicer: Dcr1 RdRP: Rdp1 |
Heterochromatic repeat regions | Centromere function Heterochromatin formation and spreading RNA Pol II regulation |
| Arabidopsis | |||||
| siRNA | 24 nt | AGO4, AGO6, AGO9 | Dicer: DCL3 RdRP: RDR2 |
Heterochromatic repetitive regions enriched for transposons and retroelements | Systemic RNA dependent DNA methylation (RdDM) Reproductive strategy (meiosis/apospory) |
| C. elegans | |||||
| siRNA (22G) | 22 nt | NRDE-3 | Primary siRNA biogenesis: ERGO-1 Dicer: DCR-1 RdRP: RRF-3 (Primary), RRF-1 (Secondary) |
Disperse genomic loci | Systemic heterochromatin formation RNA Pol II regulation |
| piRNA (21U) | 21 nt | WAGO9, WAGO10 with 22G for TGS. | PRG1 loaded with 21U for 22G production. | Two clusters on Chromosome IV | Heritable heterochromatin formation |
| Drosophila | |||||
| siRNA | ~ 22 nt | Ago2 | Dicer: Dcr2 | Transposons, repetitive elements, convergent transcription units, and structured loci. | Heterochromatin formation Chromosome segregation RNA Pol II regulation |
| piRNA | 23 to 29 | Piwi | Ping-pong: Piwi, Aub (primary) Ago3 (secondary) Zucchini |
Primary: heterochromatic piRNA clusters antisense to transposons, maternally deposited piRNA Secondary: sense transcripts of active transposons |
Chromosome segregation Heterochromatin formation Telomere homeostasis |
| Mouse | |||||
| siRNA | 21 to 22 nt | Ago2 | Dicer: Dcr1 | Dispersed naturally occurring dsRNA, pseudo-genes | Speculative |
| piRNA | 24 to 31 nt | Miwi2 | Ping-pong cycle: Mili (primary) Miwi2 (secondary) Maelstrom |
Primary: sense transcripts of active transposons Secondary: antisense transcripts |
De Novo DNA methylation |
siRNA Biogenesis
Double stranded RNA (dsRNA) is thought to be the trigger for most if not all siRNA biogenesis and can be generated by several means (Figure 1). Once dsRNA is available the biogenesis of siRNA requires action of the RNAse III like Dicer family of enzymes. Dicer cleaves dsRNA into 20–25nt siRNA duplexes with 2nt 3′ OH overhangs4 and 5′ monophosphates4,5. Dicer independent mechanisms of siRNA production have also been proposed in Neurospora6, S. pombe7 and C. elegans8. The cellular location in which dsRNA processing occurs has implications for how siRNA biogenesis and nuclear effects are regulated. In S. pombe transcription, processing, RdRP amplification, and Ago mediated target cleavage are all intimately linked in the nucleus (figure 1a)4,9–12. In animals siRNA processing was originally thought to occur in the cytoplasm3 however recent studies in Drosophila have shown that DCR2 is found predominantly in the nucleus challenging this view13. This is in contrast to C. elegans where in depth studies have validated the cytoplasmic processing of many siRNA pathways14 (figure 1b).
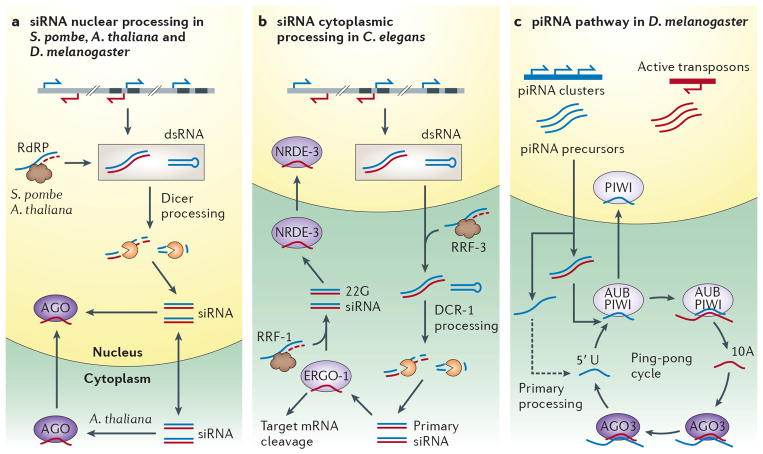
Figure 1. Generalized pathways depicting the biogenesis of nuclear small RNA.
a | siRNA processing takes place in the nucleus in S. pombe and Drosophila and the nucleoulus in Arabidopsis. dsRNA can be produced by convergent transcription, complementary transcripts, structured loci, or by RdRP activity in Arabidopsis and S. pombe. Dicer proteins generate siRNA that is loaded into an Argnoaute protein. In Arabidopsis siRNA are transported to the cytoplasm where Argonaute is loaded and then imported into the nucleus. b | In C. elegans siRNA processing occurs in the cytoplasm in a two-step fashion. Primary trigger dsRNA arises from nuclear transcription or the RdRP activity of RRF-3, which acts on transcripts in the cytoplasm. Primary processing by DCR-1 produces primary 26 nt siRNA which are loaded into ERGO-1. Loaded ERGO-1 can both facilitate PTGS in the cytoplasm and with RRF-1 generate secondary 22G siRNA siRNA. Secondary 22G siRNA is loaded into the nuclear Argonaute NRDE-3 in the cytoplasm that is then transported into the nucleus. c | piRNA biogenesis via the ping-pong cycle in the Drosophila female germline. Primary precursor piRNA antisense to active transposons (blue) is transcribed from heterochromatic piRNA clusters, and sense mRNA from active transposons (pink). In the cytoplasm primary processing generates antisense piRNA from primary precursor that is then loaded into Aub or Piwi and cleaves sense transposon mRNA to produce sense piRNA. Additional antisense piRNA is produced by Ago3 mediated cleavage of antisense primary piRNA transcripts, completing the cycle. Only loaded Piwi is imported into the nucleus.
Once generated the siRNA duplexes are loaded into an appropriate effector Argonaute protein. The subcellular location where Argonaute loading takes place is not yet fully understood across model organisms. In Arabidopsis nuclear AGO4 loading is cytoplasmic and mediated by the heat shock protein HSP90, after which it is then imported into the nucleus15. A requirement for HSP90 in Ago loading has also been observed in Drosophila however where this process occurs is not known16. Like Arabidopsis, siRNA processing is nuclear in S. pombe, however it is not known where Ago1 loading occurs. The C. elegans nuclear Argonaute NRDE-3 is imported into the nucleus only when loaded with secondary siRNA that is produced in the cytoplasm17. If cytoplasmic loading of Argonaute proteins is conserved across species this would have important implications for the regulation of nuclear RNAi.
piRNA biogenesis
The biogenesis of piRNA primarily occurs via a process known as the ping-pong cycle (Figure 1c) initially described in the Drosophila germline18,19. First, piRNA genomic clusters are transcribed to produce primary piRNA precursors. In the cytoplasm an unknown mechanism processes primary piRNA precursors into short 23–29nt antisense piRNA with a strong 5′ uridine bias. These short ssRNA are loaded into the Piwi family Argonaute proteins Aub and Piwi. In the cytoplasm, the loaded Aub/Piwi then targets mRNA of active transposons for cleavage; this produces sense piRNA, which have a strong adenine bias at position 10. The sense piRNA is loaded into the Piwi family member Ago3, which then directs cleavage of primary piRNA precursors and the subsequent production of more antisense piRNA, completing the ping-pong cycle19. In the female germline Aub protein is restricted to the cytoplasm whereas Piwi is predominantly nuclear, indicating that Aub plays a larger role in the ping-pong cycle20. The nuclear localization of Piwi is lost in Ago3 mutants, suggesting that once loaded with piRNA produced by the ping-pong cycle Piwi is imported into the nucleus20. A less understood ping-pong independent piRNA biogenesis pathway operates in the somatic follicle cells that surround the female oocytes that is Piwi-dependent and Aub/Ago3 independent (figure 4c)20,21.
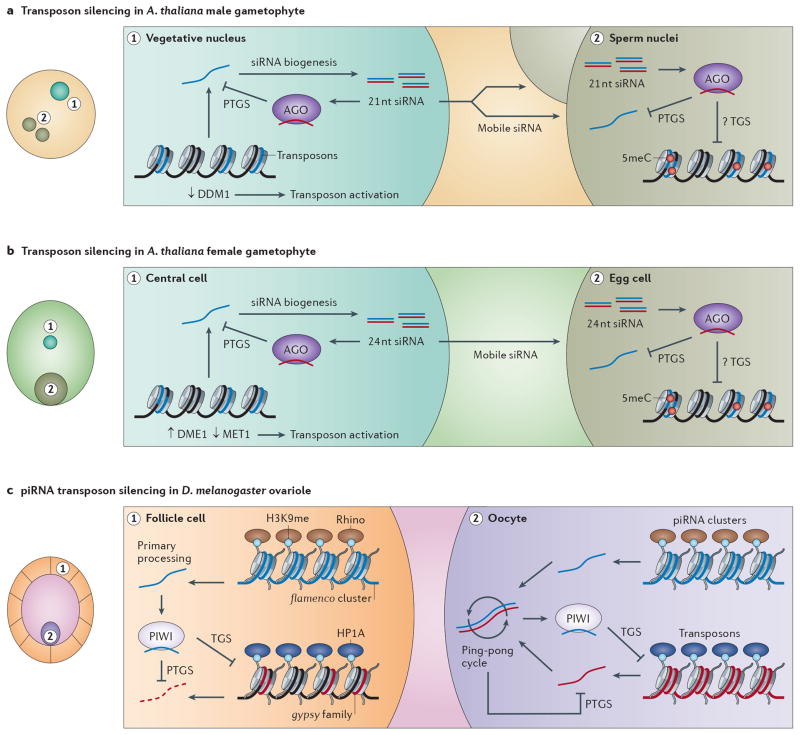
Figure 4. RNAi mediated transposon silencing in the germline.
a | In the supportive vegetative nucleus of the Arabidopsis male gametophyte ddm1 expression is repressed which leads to the loss of cytosine methylation and reveals transposons. Transposons are processed into 21nt siRNA that are mobile and can direct PTGS in the sperm nuclei. They may also impact transposons transcriptionally by directing or inhibiting epigenetic modification. Red lollipops represent 5meC. b | The supportive central cell of the Arabidopsis female gametophyte reveals transposons for transcription by downregulating the maintenance DNA methyltransferase MET1 and expressing the DNA glycosylase DEMETER causing a loss of cytosine methylation. This activates the RdDM pathway and produces 24nt siRNA that may be transported to the egg cell to enforce transcriptional silencing through AGO9. Red lollipops represent 5meC. c | In the Drosophila ovariole the flamenco cluster is expressed in somatic follicle cells, and generates piRNA independently of the ping-pong cycle. Loaded Piwi silences the gypsy family of retrotransposons which could otherwise form infectious particles. In oocytes and surrounding nurse cells all piRNA clusters are expressed and the primary transcripts enter the ping-pong cycle to produce piRNA. Active transposons are post-transcriptionally silenced, and nuclear Piwi promotes transcriptional silencing via H3K9 methylation, and HP1a localization. The HP1a homolog Rhino binds to heterochromatic piRNA clusters in place of HP1a and promotes transcription.
Stability and turnover play an important role in the regulation of both the siRNA and piRNA pathways. Methylation of small RNA is a major determinant of their stability. Both piRNA and siRNA are 2′-O-methylated by the enzyme Hen1 across organisms (for extensive review see Ji and Chen, 2012)22. This methylation protects small RNA from both 3′ uridylation and 3′ truncation, which cause small RNA degradation and turnover. The specificity of Hen1 could therefor contribute to cell-type specific small RNA profiles, and thus determine targets of RNAi, however such a mechanism has yet been uncovered.
Mechanisms of Nuclear RNAi
Transcriptional Gene Silencing (TGS) was the first function of nuclear RNAi to be discovered, and refers to the process by which RNAi can reduce transcription by guiding localized heterochromatin formation at target genomic loci. A question that arises from this mechanism is how sequence specific targeting of chromatin modifications is achieved? As in the cytoplasm the substrate for nuclear RNAi has been shown to be RNA molecules, but these must be in close proximity to the locus they arose from so that epigenetic modification can be specific. This has lead to a model of co-transcriptional gene silencing (CTGS), whereby nuclear small RNA target nascent RNA molecules from RNA polymerases, and the effector complexes themselves interact with and regulate transcriptional machinery. The two examples of nuclear RNAi described here in mechanistic detail reveal that positive feedback loops are involved in chromatin modification. The nuclear RNAi complexes themselves are both attracted to repressive epigenetics marks, and deposit them, creating robust silencing at target loci.
Nuclear RNAi at Heterochromatic Loci
Nuclear RNAi in S. pombe: TGS
A role for RNAi in TGS was identified in S. pombe where it is required for the formation of constitutive heterochromatin at pericentromeres. They are highly enriched for H3K9 methylation (H3K9me) and are composed of varying numbers of repeat units that are bi-directionally transcribed to form dsRNA that is then processed by Dcr1 into siRNA23. The RNA dependent RNA polymerase complex (RDRC) interacts with both Dcr14 and Ago124 to produce dsRNA and siRNA from Ago1 targeted transcripts and amplify the siRNA response. siRNA are loaded into Ago1, the principle member of the RNA Induced Transcriptional Silencing Complex (RITSC), and guide the RITSC to nascent pericentromeric ncRNA transcripts (Figure 2). The chromodomain protein Chp1 is also a member of the RITSC and contributes to its localization to heterochromatin by binding H3K9me25. Once the RITSC is localized to repeat loci it facilitates H3K9 methylation by recruiting the cryptic loci regulator complex (CLRC) which contains Clr4, the sole H3K9 methyltransferase in S. pombe26. Interestingly the catalytic slicing activity of Ago1 is required for the deposition and spreading of H3K9me, particularly in reporter genes12. Catalytic activity is required for passenger strand release from Ago1 bound dsRNA, and thus is required to facilitate base pairing between loaded siRNA and their targets, explaining this observation27. This suggests that nuclear RNAi, specifically siRNA-target base pairing, is required for the spreading of heterochromatin, a phenomenon originally described as position effect variegation. These interactions place the RITSC in a central role, integrating transcription and chromatin modification. They also create a positive feedback loop between siRNA generation, RITSC localization and H3K9 methylation. A fascinating consequence of this is that H3K9 methylation itself is required for siRNA generation. The coupling of transcription, siRNA production, and silencing in S. pombe suggests that TGS occurs in cis, however examples from plants discussed in Box 1 show that it can also occur in trans. The coupling of transcription, siRNA production, and silencing in S. pombe suggests that TGS occurs in cis, however examples from plants discussed in Box 1 show that it can also occur in trans.
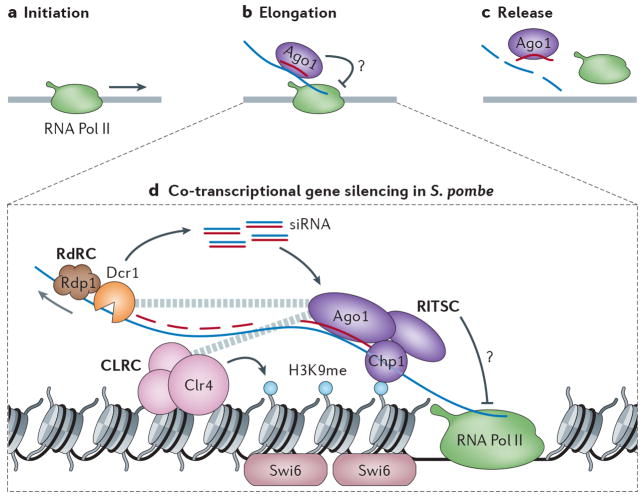
Figure 2. Co-Transcriptional Gene Silencing in S. pombe.
a | RNA Pol II initiates transcription at loci targeted by RNAi. b | During the elongation phase of transcription a Ago1 is guided to the nascent transcript and inhibits RNA Pol II transcription via an unknown mechanism. c | RNAi can lead to a release of RNA Pol II. d | A mechanistic model of RNAi acting during the elongation phase (2b) in S. pombe. The RITSC is localized through siRNA base-pairing with the nascent transcript, and chromatin interaction mediated by the chromodomain of Chp1. The RDRC couples dsRNA production by Rdp1 and siRNA cleavage by Dcr1 and is also associated with the nascent RNA Pol II transcript. The RITSC interacts with the CLRC that catalyzes H3K9 methylation at target loci. This histone modification serves as a binding site for Swi6, the S. pombe ortholog of the highly conserved heterochromatin protein 1 (HP1), which is a defining feature of heterochromatin. The RITSC promotes RNA Pol II release via an unknown mechanism. The dashed grey lines indicate interactions between complexes.
Box 1. Cis versus trans silencing: paramutation in maize.
While the concept of CTGS in S.pombe implies a role for small RNA silencing in cis, there is considerable evidence that RdDM in plants can act in trans. Perhaps the best known example is the phenomenon of paramutation, first discovered in maize. Paramutation refers to the silencing of one allele (the paramutated allele) in heterozygous combinations with a silent allele (the paramutagenic allele) in trans. The paramutagenic allele is converted by this process into a paramutagenic allele, and silencing is trans-generational, in some cases permanently so. Paramutation is allele-specific, and occurs classically at 3 loci in maize, namely Booster (B), Plant color (pl) and Red (R) (for review see Hollick, 2012)117. Paramutant alleles have rearrangements such as transposon insertions and tandem repeats, which in some cases had been identified as the sequences responsible for the effect. Importantly, extensive screens for mutants deficient in either the establishment or the maintenance of paramutation at the B locus have identified mutants in RdDM, including mediator of paramutation 1 (mop1) and mediator of paramutation 2 (mop2) which encode orthologs of RDR2 and the RNA Pol IV/V subunit Nrpd2, repsectively118. Similar screens for genes required to maintain repression (rmr) of paramutant alleles of Pl have recovered subunits of PolIV as well as homologs of the chromatin remodelers DRD1 and CLSY1119. DNA methylation changes at paramutant loci have been reported, and some examples at least involve transcriptional silencing120, but it is likely that both TGS and PTGS are involved.
Nuclear RNAi in S. pombe: CTGS
The dependency of RITSC localization on base pairing with ncRNA transcripts presents an interesting paradox in that loci targeted by RNAi for TGS must be transcribed in order to be silenced. Supporting this idea, genetic screens for loss of silencing in S. pombe have identified two point mutations in RNA Pol II subunits that decouple transcription and the RITSC at the pericentromeres9,28. A model linking transcription, RNAi and heterochromatin formation can be formed when these observations are taken in the context of the cell-cycle. Studies have shown that transcription of pericentromeric repeats targeted by RNAi occurs during S-phase, the same time at which DNA is replicating and chromatin modifications must be re-established29,30. DNA replication and transcription must also be coordinated to prevent collision of the two processes and subsequent replication fork stalling. We found that RNAi is required to facilitate the release of RNA Pol II and prevent read-through transcription into replicating DNA31. This suggests that RNAi once recruited to an actively transcribing Pol II may be able to inhibit transcription during the later elongation phase, resulting in the release of Pol II, as shown in figure 2c. These observations support a model of co-transcriptional gene silencing (CTGS) in S. pombe (Fig. 2d) that was first termed by Bühler et al. 32.
The CTGS model explains the paradox behind TGS. A nascent RNA transcript is required for the initial targeting of RNAi to a locus, once this occurs the nuclear RNAi complex can promote both transcriptional silencing at the chromatin level, and can co-transcriptionally silence by releasing RNA Pol via an unknown mechanism. It will be interesting to understand how transcription is initiated in what has previously been thought of as a restrictive heterochromatic environment and the mechanism by which the RITSC can promote Pol II release.
There is growing evidence that nuclear RNAi may co-transcriptionally regulate loci outside of constitutive heterochromatin in S. pombe. It has been shown to play a role in preventing read-through transcription at convergently transcribed genes, presumably though RNA Pol II release33–35. Additionally Dcr1 physically interacts with chromatin at euchromatic genes suggesting a role in gene regulation without histone modification36. Indeed, nuclear Dcr1 plays a role in regulating heat stress responsive genes through a “thermoswitch”37. In unstressed cells Dcr1 is nuclear localized and negatively regulates stress response genes, however under heat stress it is exported out of the nucleus and stress response genes are activated.
RNA Directed DNA Methylation in Arabidopsis
Transgene DNA methylation directed by viral RNA was discovered in plants long before a role for RNAi was known38, and later the involvement of small RNA and RNAi pathways in mediating TGS through cytosine methylation was first proposed in Arabidopsis39,40. There are many parallels between RNA Directed DNA Methylation (RdDM) in Arabidopsis and CTGS in S. pombe. For example, the requirement of transcription for silencing is common to both9,28,41, and both direct silencing at repetitive heterochromatic loci. RdDM differs from CTGS in S. pombe in that stepwise transcription by two RNA polymerases (Pol IV and Pol V) is required. Transcripts from Pol IV serve as substrates for siRNA generation, while nascent transcripts from Pol V are targeted by RNAi (figure 3, reviewed extensively in Haag et al. 2011)42. The initial template for Pol IV is not known, however it would presumably be loci that will be subject to RdDM. Pol IV physically interacts with the RNA-dependent polymerase 2 (RDR2) which produces dsRNA from transcripts43, that is subsequently processed into 24nt siRNA by Dicer Like 3 (DCL3) 44. These 24nt siRNA are exported into the cytoplasm where they are loaded into an Argonaute complex15.
Figure 3. The RNA-directed DNA Methylation pathway in Arabidopsis.
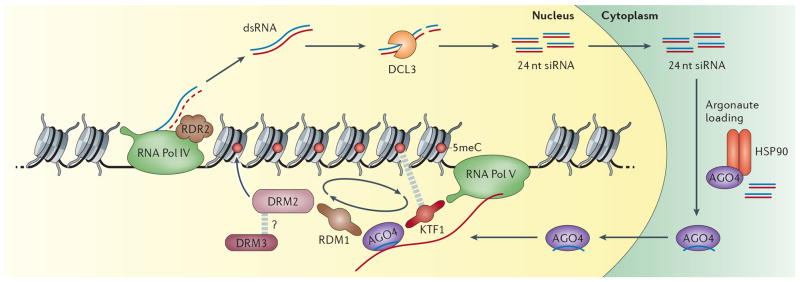
RNA Pol IV transcribes ssRNA from repetitive heterochromatic loci. RDR2 physically associates with RNA Pol IV to produce dsRNA. DCL3 cleaves dsRNA to produce siRNA that are transported to the cytoplasm for AGO4 loading, facilitated by HSP90, which is then imported back into the nucleus. In the nucleus AGO4 targets nascent RNA Pol V transcripts through complementarity to siRNA and forms the RdDM complex presumably containing the catalytically active de novo DNA methyltransferase DRM2. The Pol V associated GW/WG protein KTF1 may act as an organizer by interacting with AGO4 and 5meC. Similarly, the AGO4 associated protein RDM1 can bind single stranded methylated DNA and interacts with DRM2. Both could contribute to a positive feedback loop between AGO4 localization and DNA methylation (circular arrows). DRM3, a catalytically inactive paralog of DRM2 is required for RdDM however its role is unknown. Once localized, DRM2 catalyzes methylation of cytosine in all sequence contexts. The dashed grey lines indicate interactions.
At least 3 of the 10 Argonautes found in Arabidopsis are involved in RdDM, but AGO4 was the first to be identified 45. Once loaded with 24nt siRNA in the cytoplasm AGO4 is imported into the nucleus and guided to complementary Pol V intergenic non-coding transcripts through siRNA target base pairing46,47, and likely aided by direct protein-protein interaction with the Pol V subunit NRPE148 and the Pol V associated GW/WG protein KTF149,50.
This co-transcriptional silencing by RNAi ultimately leads to the deposition of repressive cytosine methylation at loci transcribed by Pol V. In Arabidopsis de novo cytosine methylation is catalyzed by the enzyme DRM2 at loci targeted by RdDM51. It might thus be expected to be a member of the RdDM effector complex alongside an Argonaute protein. Biochemical studies of a new complex member, RDM1 support this notion, as it interacts with both AGO4 and DRM2, and is required for RdDM, bridging RNAi and cytosine methylation52. The presence of a catalytically inactive DRM2 paralog DRM3 is also required for RdDM however its role is not known53. Once targeted DRM2 directs cytosine methylation in all cytosine contexts including at asymmetric CHH sites, to facilitate heterochromatin formation and TGS54. Perhaps analogous to the role of Chp1 in localizing the RITSC to heterochromatin in S. pombe, the AGO4 associated protein RDM1 binds single stranded methylated DNA52, and thus localizes AGO4 to methylated regions, creating a re-enforcing positive feedback loop.
Variations on the canonical RdDM pathway have been observed. AGO6 plays a partially redundant role with AGO455, and AGO9 is loaded with 24nt siRNA in the female germline, where its activity is required for transposon silencing, but a direct role in DNA methylation has not yet been established56. There is also evidence that transcripts from RNA Pol II (which chiefly transcribes euchromatic genes) as opposed to Pol V, are targeted by RdDM however the significance of this remains unclear52,57.
The RdDM pathway may be involved in H3K9 methylation, although it is uncertain if nuclear RNAi plays a direct role as in S. pombe. There is significant cross-talk between the two pathways as DNA methylation is required for the recruitment of the H3K9 methyltransferase SUVH4 / KYP58. At least two SUVH homologs are required for RdDM59 and small RNA from inverted repeats has been shown to influence H3K9 methylation to a greater extent than cytosine methylation suggesting a direct role60.
RdDM may not be the only example of nuclear RNAi in Arabidopsis. There is evidence that another nuclear RNAi pathway involving DCL4 plays a co-transcriptional role in transcriptional termination. DCL4 was found to interact directly with chromatin in the 3′ region of a Pol II transcribed endogenous gene to promote cleavage of the nascent transcript and transcription termination61. Further study is needed to identify novel nuclear roles for other RNAi pathways.
A few examples outside of Arabidopsis indicate that siRNA may not be the only small RNA to direct DNA methylation in plants. In rice 24nt small RNA that arise from miRNA precursors termed long miRNA (lmiRNA) are RDR2-independent, processed by DCL3, and loaded into Ago4, which is normally associated with RdDM in Arabidopsis62. These lmiRNA are able to direct highly sequence specific cytosine methylation at their own locus (in cis) and at complementary loci (in trans). Some lmiRNA have been identified in Arabidopsis however they have not been shown to direct DNA methylation63. Similarly, in the moss Physcomitrella patens several 21nt miRNA have been shown to direct cytosine methylation at their targets64. While both examples show that other classes of small RNA can direct DNA methylation neither uncover a novel effector pathway outside of RdDM.
Metazoan Somatic Nuclear RNAi
While the germlines of metazoans have a clear role for nuclear RNAi (see Nuclear RNAi in the Germline), some evidence suggests that TGS also occurs in somatic cells, however the subject is controversial. Feeding C. elegans with dsRNA targeting an endogenous gene triggers H3K9 methylation at the target locus in somatic cells that is dependent on the nuclear RNAi pathway (NRDE)17,65–67 and on the RDRP RRF-168. There are many genes targetted by endogenous siRNA, and some but not all show a reduction of H3K9me in nrde mutants68. In Drosophila somatic cells, mutations in siRNA pathway members dcr2 or ago2 affect expression of a centromeric reporter and result in a marked reduction of centromeric H3K9 methylation 69–71.
As in the fission yeast, proteins required for nuclear RNAi interact with the transcriptional machinery in metazoan somatic cells, suggesting that CTGS may be conserved. In human and Drosophila cells, Ago1 interacts directly with RNA Pol II by co-immunoprecipitation72,73. In Drosophila S2 cells Ago2 and Dcr2 associate directly with both chromatin and RNA Pol II, and are required to inhibit the expression of heat-shock response genes under non-stress conditions by maintaining paused Pol II and preventing elongation13.
In C. elegans, loci targetted by RNAi show a downstream decrease in RNA Pol II occupency that is dependent on the nuclear RNAi factor NRDE-2 and Argonaute NRDE-3, suggesting that siRNA may facilitate transcription termination65. Overall current evidence suggests a conserved interaction of nuclear RNAi and the transcriptional machinery fitting a co-transcriptional model, however the role of these interactions needs further exploration.
Nuclear RNAi in the Germline
The germline is the battlefield on which evolutionary wars between selfish DNA elements and their hosts are played out because transposable element (TE) mobilization here would be inherited by future generations. Nuclear RNAi – the piRNA pathway in animals and various siRNA pathways in plants - is a front line defense.
Germline Nuclear RNAi in Arabidopsis
In plants, germline cells arise late in development from somatic stem cells (unlike in animals, in which the germline is specified early in development), and so transposons must be silenced extensively throughout development. Generally, chromatin marks that are present during somatic development must be reset in the germline. How this occurs selectively is a question that is actively being pursued. In somatic cells both the RdDM pathway and maintenance DNA methyltransferases keep transposons silent; however, this changes in the companion cells of the germline that will not contribute genetically to the next generation. The heterochromatin remodeler ddm1 is a master regulator of transposons74 and is down-regulated in the supportive vegetative nucleus (VN), leading to transposon mobilization and the production of 21nt sRNA antisense to transposons75 (figure 4a). These 21nt sRNA can silence reporters expressed in sperm cells so they appear to act non-cell-autonomously. With regards to DNA methylation, unlike mammals which undergo whole genome demethylation during spermatogenesis76, the Arabidopsis male germline retains symmetric methylation at levels similar to somatic cells77,78, but shows a reduction in the levels of asymmetric methylation specifically at a subset of retrotransposons that are later re-methylated in the developing embryo79.
In the female gametophyte the maintenance DNA methyltransferase met1 is repressed80 and the DNA glycosylase demeter, which removes cytosine methylation, is expressed81 in the diploid central cell (CC), that will later become the “extra-embryonic” endosperm (figure 4b). This leads to global cytosine demethylation in the endosperm, accompanied by increased production of 24nt siRNA leading to non-CG hypermethylation at target sites, which are primarily retroelements82. These 24nt siRNA are bound by AGO9 in the central cell, and act non-cell-autonomously to control specification of gametic egg cells56. Currently there is no direct experimental evidence showing the movement of either 24nt siRNA or AGO9 from the central cell to the egg cell, however in ago9 mutants transposable elements are activated in the egg cell where ago9 is not expressed supporting this hypothesis. These observations suggest a hypothetical model by which transposons are revealed in companion cells, and are then used to generate small RNA that enforces transposon silencing in the germ cells83, however it is not know if they can also direct TGS through nuclear RNAi. This movement of small RNA between germ cells has implications for epigenetic inheritance that are discussed in Box 2.
Box 2. Systemic TGS and Epigenetic Inheritance.
The hypothesis that siRNA can move into Arabidopsis germ cells (see Germline Nuclear RNAi in Arabidopsis) has implications for epigenetic inheritance. Outside of the gametophytes, grafting experiments have shown that nuclear silencing signals can be transmitted from the root to shoot121 and vice versa122. Mobile 21 to 24nt siRNA are the effectors of this systemic silencing and can guide epigenetic modification through RdDM in recipient cells123,124. These 24nt siRNA have been demonstrated to direct DNA methylation in meristematic root stem cells122, and it is therefore tempting to speculate that they may act similarly in the shoot meristems (where germ cells are produced) to direct heritable epigenetic modification.
Systemic RNAi is well known in C. elegans, and there is recent evidence for small RNA mediated epigenetic inheritance. The progeny of animals exposed to dsRNA show H3K9 methylation of target loci and generate complementary small RNA for multiple generations66. The appearance of siRNA precedes H3K9me in progeny so it’s likely that this inheritance is indirect and is instead re-established by inherited siRNA in each generation. This process is dependent on the nuclear RNAi pathway, including the Argonaute NRDE-3. Furthermore small RNA produced against viral RNA can be transgenerationally inherited, and continue to persist even in the absence of the viral template itself125. These studies both point to small RNA as an epigenetic vector, which can be inherited and through nuclear RNAi direct chromatin modifications in progeny. Once established, these chromatin modifications can be maintained and transmitted across generations even in the absence of the dsRNA trigger67.
In Drosophila the makeup of cellular piRNA is epigenetically inherited. Reciprocal crosses have shown that progeny inherit the maternal piRNA composition, and this composition persists into adulthood126. The maternally deposited piRNA may prime the ping-pong cycle and determine its specificity, or could potentially direct epigenetic modification to enforce a specific piRNA transcription program. A similar situation is seen in the Arabidopsis endosperm where maternally deposited 24nt siRNA silence TEs and TE-associated genes during its development 127,128.
The Drosophila piRNA Pathway
In animals, the role of the piRNA pathway in TE silencing has been best described in Drosophila ovaries. Within the ovaries piRNA silence transposons in somatic follicle cells surrounding the oocyte, germline nurse cells and the oocyte itself20,21 (figure 4c). The somatic follicle cells produce only antisense piRNA from the flamenco locus that do not participate in the ping-pong cycle and are instead processed and loaded solely into Piwi. These piRNA predominantly target elements from the gypsy family of long terminal repeat (LTR) retroviruses. Gypsy family elements are able to propagate by producing viral packages in follicle cells that can infect germline cells, thus the flamenco derived piRNA pathway is thought to be an evolutionary counter to this class of transposons20. In nurse cells and ovaries the ping-pong cycle defends against a wide variety of TEs using input from all piRNA clusters and mRNA of active transposons18,21. Here the piRNA pathway degrades transposon transcripts, and directs H3K9 methylation to transcriptionally silence transposons and prevent their mobilization84. Piwi has been shown to specifically interact with heterochromatin protein 1 (HP1a), a defining component of heterochromatin, and is chromatin associated itself85. Furthermore, the nuclear localization of Piwi is required for chromatin-mediated repression of a subset of transposons suggesting a direct role86. Silencing of piRNA clusters themselves would be detrimental, as this would prevent primary piRNA from entering the cycle. This is solved by the HP1 variant Rhino that is restricted to germline nuclei and specifically localizes to piRNA clusters and promotes transcription of the heterochromatic clusters87. How Rhino is localized to piRNA clusters and not active transposons remains unexplained.
The Mouse piRNA Pathway
The Piwi pathway is highly conserved in animals and plays a similar role in the mouse germline. In mouse two Piwi homologs, MILI and MIWI2 are required for transposon silencing in the male germline. Loss of either causes transposon mobilization and sterility88,89. The piRNA pathway however operates differently from Drosophila. In the mouse male germline transposons are globally derepressed by cytosine demethylation during early development. The piRNA pathway is then primed with individual transposons and re-establishes methylation patterns during development90,91. As MIWI2 is found in the nucleus it is likely to be the effector Argonaute of RNA directed DNA methylation in mouse92. The role of MIWI2 in establishing DNA methylation in the germline may not be direct as with AGO4 in Arabidopsis (or Ago1 for H3K9me in S. pombe) since co-immunoprecipitation experiments have failed to show interaction between MIWI2 and the de novo methyltransferases Dnmt3a and Dnmt3b. The role of nuclear RNAi in directing DNA methylation in mammals is nicely demonstrated at the imprinted rasgfr1 locus where the piRNA pathway is required for de novo methylation in the male germline91. Upstream of the differentially methylated region is an LTR that matches piRNAs with a typical pong-pong signature; probably these piRNAs can be generated due to the presence of another copy of the LTR in a piRNA cluster. The LTR is contained within a non-coding RNA that is transcribed specifically during spermatogenesis when de novo methylation occurs. This nascent ncRNA is targeted by piRNA and co-transcriptionally silenced by the deposition of DNA methylation. This may facilitate the spread of targeted silencing into the nearby rasgfr1 locus, leading to imprinting, similarly to CTGS in S. pombe. Again the authors do not rule out the possibility that silencing by piRNA may be indirect, and this is a single locus example. The rasgfr1 locus is however unlikely to be the only example of RNAi directing imprinting or silencing of an endogenous gene, and hints that nuclear RNAi and transposon acquisition play a role in imprinting across organisms. Further genetic and biochemical dissection is needed to discern if the piRNA pathway plays a direct role in DNA methylation, and if so what the mechanistic details are. Specifically interactions between piRNA effectors and cytosine methyltransferases, and the use of exogenous reporters containing sequences complementary to known piRNA would provide convincing evidence.
Germline Nuclear RNAi in C. elegans
A class of small RNA termed 21U has been proposed to be the piRNA of C. elegans93–95. They associate with the Piwi-family protein PRG-1 which is required to silence Tc3 mariner transposons in the germline and for fertility93,96.
C. elegans 21U RNA originate from over 5700 loci dispersed over two broad clusters on chromosome IV94, however no evidence of a ping-pong cycle has been observed. The 21U pathway has been suggested to function by determining the specificity of the 22G siRNA and nrde pathways (see Metazoan Somatic Nuclear RNAi and figure 1b) that direct TGS in the form of H3K9me at piRNA targets (figure 5). Two avenues of study have validated this model. In C. elegans single copy transgenes with long exogenous DNA sequences, such as GFP, are stably silenced at a high frequency. This silencing correlates with H3K9me3 accumulation and is dependent on PRG-1 and 21U RNA accumulation for its establishment, and the germline specific nuclear Argonautes WAGO-9 and WAGO-10 that bind 22G RNA for its maintenance97. Studies with reporter transgenes that contain sequences complementary to known 21U small RNAs (piRNA sensors) have revealed identical requirements for silencing and additionally implicated the HP1 ortholog HPL-2 and putative methyltransferases SET-25 and SET-32 in establishing H3K9me3 at loci targeted by piRNA98. Outside of transgenes, silencing at endogenous loci mediated by piRNA likely functions by the same mechanism. Indeed many endogenous loci that are targeted by 21U small RNA and silenced exhibit increased mRNA expression and a loss of corresponding 22G RNA in a prg1 mutant background99. RNAi also acts to establish repressive heterochromatin during meiosis at unpaired chromosomal regions in C. elegans. Specifically the RdRP EGO-1 and the Piwi family Argonaute protein CSR-1 are required for this process100,101.
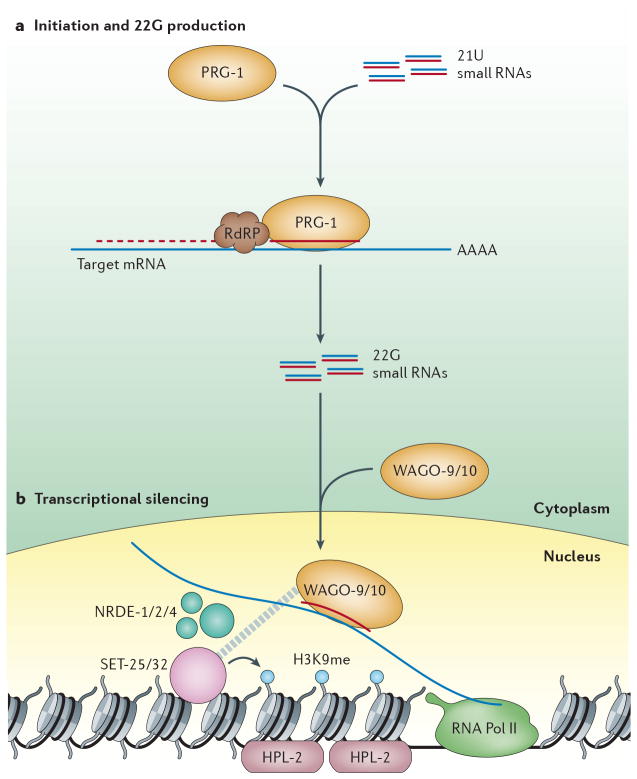
Figure 5. piRNA (21U) Pathway in the C. elegans germline.
a | The “21U” piRNA of C. elegans originate from two broad clusters on chromosome IV, however little is known about their biogenesis. They act with the Piwi family Argonaut PRG-1 to target mRNA in the cytoplasm. Targeting of PRG-1 to mRNA recruits a RdRP to produce abundant 22G siRNA. b | 22G siRNA is loaded into the germline specific nuclear Argonautes WAGO-9/10, which are closely related to NRDE-3, the nuclear Argonaute involved in somatic TGS. Loaded WAGO-9/10 is transported into the nucleus where it targets nascent transcripts of RNA Pol II and directs H3K9me that is dependent on the nuclear RNAi components NRDE-1/2/4. H3K9 methylation is catalyzed by two putative histone methyltransferases SET-25/32. The HP1 ortholog HPL-2 binds H3K9me and is required for multi-generational silencing.
Nuclear RNAi in Genome Maintenance and Repair
Nuclear RNAi plays a critical role in maintaining genome integrity by preventing transposon mobilization, however more direct roles in genome maintenance and DNA repair are emerging.
Chromosome structure and function
Proper chromosome condensation is required for segregation during mitosis. In S. pombe the loss of RNAi causes a high incidence of lagging chromosomes and sensitivity to a microtubule inhibiting drug102. Also, in the Drosophila germline the piRNA DEAD-box RNA helicase Vasa facilitates condensin I localization, which promotes chromosome condensation and is dependent on the piRNA components aub and spindle-E103. A vasa paralog belle acts analogously in somatic cells and requires the siRNA components ago2 and dcr2104. Interestingly, a role for RNAi in cohesin localization has also been proposed in S. pombe35, suggesting a conserved role for RNAi in facilitating cohesin/condensin localization ensuring proper chromosome condensation.
The telomeres of Drosophila are unique in that they rely on a transposon based elongation mechanism105. The piRNA pathway has been found to regulate these telomeric transposons in the germline, and thus can regulate telomere length106. Specifically ago3 mutant embryos show an increase in telomeric transposition, and a subsequent increase in telomere length107. Additionally, aub and the RNA helicase armitage are involved in the production of telomere specific piRNA and their loss results in increased telomere fusion, suggesting another role for the piRNA pathway in telomere cap assembly107. Nuclear RNAi is also required for proper telomere function in S. pombe. Subtelomeric regions contain a region that is homologous to the pericentromeric repeats and this region facilitates RNAi-dependent heterochromatin formation108. It’s possible that nuclear RNAi may have a conserved role in telomere maintenance across organisms.
Eukaryotic genomes contain extensive regions of repetitive DNA which if engaged in recombination can cause detrimental changes to chromosome structure. There is evidence that RNAi pathways may act to repress recombination in repetitive regions. The loss of RNAi in S. pombe cells leads to both an increase in meiotic recombination109, and a dependence on mitotic recombination in repetitive pericentromeric regions, as double mutants between RNAi components and the master regulator of homologous recombination rhp51 are synthetic lethal31. This observation has also been made in Drosophila where RNAi mediated suppression of recombination is required to maintain stability of repetitive DNA71.
DNA damage response
A role for small RNA in the DNA damage response was first observed in Neurospora, where small RNA is generated from rDNA repeats when cells are treated with DNA damaging agents110. More recently, RNAi has been shown to directly mediate DNA repair in Arabidopsis. Double strand breaks (DSBs) were found to induce a population of 21nt small RNA111. These small RNA originate from the vicinity of the double strand breaks and their biogenesis requires the siRNA biogenesis factors RNA Pol IV and dicer-like proteins. They are recruited to DSBs by AGO2 and mediate repair, as mutants in ago2 or biogenesis factors cause a reduction in DSB repair efficiency. The authors suggest that AGO2 recruits the DSB repair complex to damaged loci, analogously to the localization of DNA methylation complexes in RdDM. Importantly the results were validated in human cell lines pointing to a conserved role for RNAi in DSB repair. A similar finding has been reported in Drosophila cells where double strand breaks induce a localized production of siRNA that is dependent on Ago2 and Dcr2, members of the endo-siRNA pathway112. Upon DSB formation the DNA-damage response (DDR) pathway is activated and can arrest cell proliferation. Focus on this pathway has revealed that DICER and DROSHA-dependent small RNA are required for DDR activation in human, mouse, and zebrafish113. It is therefore likely that the DDR pathway may link RNAi and DNA repair, although the specific function of the small RNAs themselves remains a mystery.
Targeted genome elimination
Perhaps the most extreme role for nuclear RNAi in genome stability is in targeted genome elimination in Tetrahymena. Tetrahymena species retain two nuclei, a germline micro-nucleus (Mic) and a somatic macro-nucleus (Mac). After zygote formation a new Mac develops by the deletion of ~6000 internal eliminated sequences (IES). These IES are enriched for H3K9 methylation before deletion114 and produce a population of 28nt scan RNA (scnRNA) that associate with the Argonaute Twi1p115. A RNA helicase Ema1p facilitates the interaction between loaded Twi1p and chromatin by promoting base-pairing with nascent transcripts, fitting a co-transcriptional model116. It is hypothesized that this leads to the deposition of H3K9 methylation by a mechanism similar to S. pombe that then serves as a mark for DNA elimination in the Mac.
These examples show that in addition to silencing transposons nuclear RNAi has a conserved role in maintaining genome stability by participating in a variety of pathways across different organisms. In particular the link to DSB repair shows that Argonaute effector complexes can be directly involved in DNA repair. In other examples it is not clear if RNAi plays a direct role or if it simply maintains genome integrity through H3K9 methylation. Higher eukaryotes have numerous Argonaute proteins many of which are uncharacterized. Further investigation of these Argonautes may reveal novel roles in genome maintenance outside of classical RNAi.
Conclusions
Although a role for RNAi in the nucleus was first described in Arabidopsis and S. pombe, observations in key model organisms suggest that it is evolutionarily conserved. RNAi mediated transcriptional gene silencing has now been observed in plants, fungi, and metazoans, and evidence is mounting that it operates co-transcriptionally as in S. pombe. Across organisms nuclear RNAi operates predominantly at heterochromatic loci where it facilitates sequence specific silencing through the direction of histone H3K9 methylation and/or cytosine methylation. Differences are however seen in small RNA biogenesis particularly in the subcellular localization of small RNA processing, and loading of Argonaute proteins, and could represent alternative approaches to regulating nuclear RNAi. Mechanistically it is still unclear in the context of the co-transcriptional model how nuclear RNAi complexes regulate transcriptional machinery. Outside of constitutive heterochromatin RNAi co-transcriptionally regulates some genes, and experiments are underway to determine if this is a widespread phenomenon across organisms.
The role played by nuclear RNAi in the germline to prevent the propagation of selfish DNA elements in future generations is significant and highly conserved. There is often a link between imprinted genes and nearby transposons, in mammals as well as in plants, which may be important in the evolution of some aspects of imprinting from germline transposon control. This field of study will be particularly fruitful in parallel with work on co-transcriptional models that could explain the spreading of silencing at transposon targets into nearby genes associated with non-coding RNA and RNAi. Outside of imprinting it is likely that small RNA themselves play a conserved role in epigenetic inheritance. As the ability to profile germline cells improves these question will be addressed.
Finally, the participation of nuclear RNAi in genome maintenance and DNA repair shows that there are other roles that nuclear small RNA and their effectors play outside of those involved in classical transcriptional silencing. Biochemical purification of novel Argonaute effectors in the context of DNA repair will help to identify the players. The more we learn about nuclear RNAi, the more apparent it becomes that RNAi plays a fundamental role in gene regulation and genome maintenance from one generation to the next.
Acknowledgments
We thank the members of the Martienssen Lab for discussion. S.E.C. is a Cashin Scholar of the Watson School of Biological Sciences, and is supported by a Natural Sciences and Engineering Research Council of Canada Post Graduate Scholarship.
Glossary
- RNAi
Silencing both at the post-transcriptional and transcriptional level that is directed by small RNA molecules
- Argonaute
The effectors proteins of RNAi that are composed of three characteristic domains, a PAZ domain and MID domain which bind the 3′ and 5′ ends of siRNA respectively, and a Piwi domain which may possess RNAse H like slicer activity if the protein is catalytically active
- Post-Transcriptional Gene Silencing
Silencing achieved by the degradation and/or prevention of translation of a target transcript targeted by small RNA
- Transcriptional Gene Silencing
Silencing achieved by the formation of a repressive chromatin environment at a locus targeted by small RNA making it inaccessible to transcriptional machinery
- Co-Transcriptional Gene Silencing
The coupling of repressive epigenetic modification with transcription by an RNA polymerase that produces a nascent RNA molecule targeted by small RNA
- RITSC
RNA Induced Transcriptional Silencing Complex, the effector of nuclear RNAi in S. pombe. Composed of an Argonaute protein and other co-factors that may aid in localization to chromatin
- Pericentromeres
Sites of constitutive heterochromatin that flank the central kinetochore-binding region of the centromere and are necessary for proper centromere function
- H3K9 Methylation
Mono, di, or tri methylation of histone H3 catalyzed by a histone methyltransferase and highly enriched in repressive heterochromatin. Acts as a binding site for Heterochromatin Protein 1 (HP1, Swi6 in S. pombe) the presence of which is the defining feature of heterochromatic loci
- Cytosine Methylation
Covalent modification of a cytosine base catalyzed by a DNA methyltransferase that is often associated with heterochromatic loci. Can occur in various “contexts” including CG, CHG, CHH which have implications for their establishment and inheritance
- Transposable Element
Genetic elements that can move their positions within the genome. The mechanism of transposition varies and defines transposon families
- Companion Cells
Cells in the germline of plants that will not contribute genetically to progeny but are produced by meiosis. These are the vegetative nucleus (VN) in the male germline and the central cell (CC) in the female germline. The CC is fertilized to produce the endosperm that acts as a supportive tissue to the developing embryo
- Cohesin
Large protein rings which predominantly localize to heterochromatic regions of the genome. They function to keep sister chromatids connected during mitosis, facilitate spindle attachment to chromosomes, and are involved in DNA repair through recombination
- Recombination
The joining of similar or identical DNA sequences to produce a novel molecule. Homologous recombination is used as a mechanism to repair damaged DNA in cells, however at repetitive regions it can be detrimental by leading to copy number changes of repetitive elements
- Double Strand Breaks
A deleterious form of DNA damage that occurs when the covalent bonds of both strands of a double helix are broken at a locus. Can be repaired by homologous recombination or error-prone nonhomologous end joining
Biography
Rob Martienssen leads the plant biology group at Cold Spring Harbor Laboratory, where he focuses on epigenetic mechanisms that shape and regulate the genome, and their impact on development and inheritance. His work on transposons or ‘jumping genes’ in plants and in fission yeast revealed a link between heterochromatin and RNA interference. He has also developed reverse genetics strategies using transposons in maize and Arabidopsis that have become powerful and widely used tools in plant genetics research.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests
References
- 1.Cox DN, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Colmenares SU, Buker SM, Bühler M, Dlakić M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 6.Lee HC, et al. Diverse Pathways Generate MicroRNA-like RNAs and Dicer-Independent Small Interfering RNAs in Fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 10.Emmerth S, et al. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell. 2010;18:102–113. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Barraud P, et al. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. EMBO J. 2011;30:4223–4235. doi: 10.1038/emboj.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irvine DV, et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 13.Cernilogar FM, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalzell JJ, et al. RNAi effector diversity in nematodes. PLoS Negl Trop Dis. 2011;5:e1176. doi: 10.1371/journal.pntd.0001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye R, et al. Cytoplasmic Assembly and Selective Nuclear Import of Arabidopsis ARGONAUTE4/siRNA Complexes. Mol Cell. 2012:1–12. doi: 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17:1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- 17.Guang S, et al. An Argonaute Transports siRNAs from the Cytoplasm to the Nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennecke J, et al. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 19.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 20.Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell research. 2012 doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 24.Motamedi MR, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 25.Verdel A. RNAi-Mediated Targeting of Heterochromatin by the RITS Complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 27.Buker SM, et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- 28.Djupedal I, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 31.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bühler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 33.Zofall M, et al. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gullerova M, Moazed D, Proudfoot NJ. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 2011;25:556–568. doi: 10.1101/gad.618611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 36.Woolcock KJ, Gaidatzis D, Punga T, Bühler M. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol. 2011;18:94–99. doi: 10.1038/nsmb.1935. [DOI] [PubMed] [Google Scholar]
- 37.Woolcock KJ, et al. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev. 2012 doi: 10.1101/gad.186866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wassenegger M, Heimes S, Riedel L, Sänger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 39.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aufsatz W, Mette MF, van der Winden J, Matzke AJM, Matzke M. RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA. 2002;99 (Suppl 4):16499–16506. doi: 10.1073/pnas.162371499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 43.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasschau KD, et al. Genome-Wide Profiling and Analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 46.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing. PLoS Genet. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He XJ, et al. An Effector of RNA-Directed DNA Methylation in Arabidopsis Is an ARGONAUTE 4- and RNA-Binding Protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao X, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 52.Gao Z, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henderson IR, et al. The De Novo Cytosine Methyltransferase DRM2 Requires Intact UBA Domains and a Catalytically Mutated Paralog DRM3 during RNA–Directed DNA Methylation in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001182. doi: 10.1371/journal.pgen.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pélissier T, Thalmeir S, Kempe D, Sänger HL, Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olmedo-Monfil V, et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng B, et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-Domain Proteins Required for DRM2-Mediated De Novo DNA Methylation. PLoS Genet. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enke RA, Dong Z, Bender J. Small RNAs Prevent Transcription-Coupled Loss of Histone H3 Lysine 9 Methylation in Arabidopsis thaliana. PLoS Genet. 2011;7:e1002350. doi: 10.1371/journal.pgen.1002350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu F, Bakht S, Dean C. Cotranscriptional role for Arabidopsis DICER-LIKE 4 in transcription termination. Science. 2012;335:1621–1623. doi: 10.1126/science.1214402. [DOI] [PubMed] [Google Scholar]
- 62.Wu L, et al. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Vazquez F, Blevins T, Ailhas J, Boller T, Meins F. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 2008;36:6429–6438. doi: 10.1093/nar/gkn670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khraiwesh B, et al. Transcriptional Control of Gene Expression by MicroRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 65.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gu SG, et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burkhart KB, et al. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011;7:e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fagegaltier D, et al. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci USA. 2009;106:21258–21263. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deshpande G. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005;19:1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kavi HH, Birchler JA. Interaction of RNA polymerase II and the small RNA machinery affects heterochromatic silencing in Drosophila. Epigenetics Chromatin. 2009;2:15. doi: 10.1186/1756-8935-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 74.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 75.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calarco JP, et al. Reprogramming of DNA Methylation in Pollen Guides Epigenetic Inheritance via Small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibarra CA, et al. Active DNA Demethylation in Plant Companion Cells Reinforces Transposon Methylation in Gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. DNA Methylation Dynamics during Sexual Reproduction in Arabidopsis thaliana. Current Biology. 2012;22:1825–1830. doi: 10.1016/j.cub.2012.07.061. [DOI] [PubMed] [Google Scholar]
- 80.Jullien PE, et al. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008;6:e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 82.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martienssen RA. Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytol. 2010;186:46–53. doi: 10.1111/j.1469-8137.2010.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang SH, Elgin SCR. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brower-Toland B, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klenov MS, et al. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klattenhoff C, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally Regulated piRNA Clusters Implicate MILI in Transposon Control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 89.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 90.Aravin AA, et al. A piRNA Pathway Primed by Individual Transposons Is Linked to De Novo DNA Methylation in Mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watanabe T, et al. Role for piRNAs and Noncoding RNA in de Novo DNA Methylation of the Imprinted Mouse Rasgrf1 Locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aravin AA, et al. Cytoplasmic Compartmentalization of the Fetal piRNA Pathway in Mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Das PP, et al. Piwi and piRNAs Act Upstream of an Endogenous siRNA Pathway to Suppress Tc3 Transposon Mobility in the Caenorhabditis elegans Germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 95.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shirayama M, et al. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell. 2012 doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ashe A, et al. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee H-C, et al. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell. 2012 doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maine EM, et al. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of Heterochromatin Assembly on Unpaired Chromosomes during Caenorhabditis elegans Meiosis by Components of a Small RNA-Mediated Pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Volpe T, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 103.Pek JW, Kai T. A Role for Vasa in Regulating Mitotic Chromosome Condensation in Drosophila. Curr Biol. 2011;21:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- 104.Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci USA. 2011;108:12007–12012. doi: 10.1073/pnas.1106245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shpiz S, Kalmykova A. Role of piRNAs in the Drosophila telomere homeostasis. mge. 2011;1:274–278. doi: 10.4161/mge.18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Savitsky M. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct Functions for the Drosophila piRNA Pathway in Genome Maintenance and Telomere Protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere Binding Protein Taz1 Establishes Swi6 Heterochromatin Independently of RNAi at Telomeres. Current Biology. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 109.Ellermeier C, et al. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci USA. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee HC, et al. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei W, et al. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 112.Michalik KM, Böttcher R, Förstemann K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Francia S, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 115.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 116.Aronica L, et al. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hollick JB. Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr Opin Plant Biol. 2012 doi: 10.1016/j.pbi.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 118.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 119.Hale CJ, Stonaker JL, Gross SM, Hollick JB. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 2007;5:e275. doi: 10.1371/journal.pbio.0050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barbour JER, et al. required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize. THE PLANT CELL ONLINE. 2012;24:1761–1775. doi: 10.1105/tpc.112.097618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brosnan CA, et al. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Melnyk CW, Molnar A, Bassett A, Baulcombe DC. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol. 2011;21:1678–1683. doi: 10.1016/j.cub.2011.08.065. [DOI] [PubMed] [Google Scholar]
- 123.Molnar A, et al. Small Silencing RNAs in Plants Are Mobile and Direct Epigenetic Modification in Recipient Cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 124.Dunoyer P, et al. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Rechavi O, Minevich G, Hobert O. Transgenerational Inheritance of an Acquired Small RNA-Based Antiviral Response in C. elegans. Cell. 2011 doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brennecke J, et al. An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu J, Zhang C, Baulcombe DC, Chen ZJ. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1203094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mosher RA, et al. An Atypical Epigenetic Mechanism Affects Uniparental Expression of Pol IV-Dependent siRNAs. PLoS ONE. 2011;6:e25756. doi: 10.1371/journal.pone.0025756. [DOI] [PMC free article] [PubMed] [Google Scholar]