Abstract
Purpose
The aim of this study was to develop a pragmatic nomogram for prediction of progressionfree survival (PFS) for the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) in EGFR mutant non-small cell lung cancer (NSCLC).
Materials and Methods
A total of 306 recurred or metastatic NSCLC patients with EGFR mutation, who received EGFR TKIs, were enrolled in this study. We developed the nomogram, using a Cox proportional hazard regression model for PFS.
Results
The median PFS was 11.2 months. Response rate to EGFR TKI was 71.9%. Multivariate Cox model identified disease status, performance status, chemotherapy line, response to EGFR TKI, and bone metastasis as independent prognostic factors, and the nomogram for PFS was developed, based on these covariates. The concordance index for a nomogram was 0.708, and the calibration was also good.
Conclusion
We developed a nomogram, based on clinical characteristics, for prediction of the PFS to EGFR TKI in NSCLC patients with EGFR mutation.
Keywords: Nomograms, Lung neoplasms, Epidermal growth factor receptor, Tyrosine kinase inhibitor, Prognosis
Introduction
Somatic mutations in the epidermal growth factor receptor (EGFR) gene are present in a subset of non-small cell lung cancer (NSCLC). EGFR mutations occur in the tyrosine kinase domain, mostly involving in-frame deletions in exon 19 and a point mutation (L858R) in exon 21, leading to constitutive activation of EGFR [1,2]. EGFR mutations are associated with increased sensitivity to specific EGFR tyrosine kinase inhibitors (TKIs), and repeatedly confirmed as a predictive and prognostic factor for EGFR TKIs [3-5].
Mutation analysis for EGFR is essential for the guidance of treatment decisions, regarding the use of EGFR TKIs, and is becoming a standard recommendation in the pretreatment work-up of patients with lung adenocarcinoma [6,7]. Although EGFR mutation status is associated with a high response rate, all patients eventually develop acquired resistance, and progression-free survival (PFS), reaching approximately 10 months [8,9].
However, predictive or prognostic factors for EGFR TKI, other than EGFR mutation, have not been well elucidated. A pragmatic prognostic model, which integrates potential relevant factors, has not been developed in EGFR mutant NSCLC. Based on these findings, there is an urgent need for a robust prognostic model for prediction of PFS for EGFR TKI in patients with NSCLC. The aim of this study was to develop a pragmatic nomogram for prediction of PFS for EGFR TKI in EGFR mutant NSCLC.
Materials and Methods
1. Study population
We retrospectively analyzed a consecutive database of NSCLC patients treated with either gefitinib or erlotinib at Seoul National University Hospital, between January 2002 and December 2011. Inclusion criteria were as follows: 1) pathology-confirmed, 2) recurred or metastatic NSCLC, 3) underwent EGFR mutation test, and 4) receiving gefitinib or erlotinib as a palliative chemotherapy. Gefitinib was administered orally, at a dose of 250 mg daily, and erlotinib was administered at a dose of 150 mg daily, until tumor progression, death, significant uncontrolled toxicity, or patient refusal. Mutational analysis of EGFR exons 18, 19, 20, and 21 was performed, as previously described [10,11]. In brief, coding sequences from exon 18 to 21 were amplified by polymerase chain reaction (PCR) with both forward and reverse sequence-specific primers [10,11]. PCR fragments were sequenced and analyzed in both sense and antisense directions. All sequence variants were confirmed by sequencing the products of independent PCR amplifications in both directions. Chest computed tomography scans were performed every 8 to 12 weeks as a routine clinical procedure, and additionally as needed to confirm patient response and for assessment of disease progression. The treatment response was evaluated using Response Evaluation Criteria in Solid Tumors criteria [12]. PFS was measured from the first day of TKI treatment until the first objective sign of disease progression or death. This study was reviewed and approved by the Institutional Review Board of Seoul National University Hospital.
2. Constructing a model
Details of model construction are described in our previous report [13]. We constructed the nomogram, using the Cox proportional hazard regression (PHR) model for the survival data [14]. Beta-coefficients from the model were used for allocation of points. Univariate Cox PHR analyses were performed to evaluate the prognostic values of each variable, followed by multivariate Cox PHR analysis. Multicollinearity between variables was also tested, and one of the variables, which showed multicollinearity, was removed in the model. The final multivariate model was chosen on the basis of the stepwise procedure, as well as consideration of the clinical or biologic importance of the variables in the model. Based on the prediction model with identified predictive and prognostic factors, a nomogram was built for prediction of PFS.
3. Evaluating model performance
The model performance was evaluated, in terms of the discrimination and calibration performance. Discrimination is the ability of the predictor to separate patients with different responses or events. Discrimination for survival data was evaluated, using the C statistic with concordance index (C-index) [15], which is similar in concept to the area under the receiver operating characteristic (ROC) curve in the logistic model [16], but appropriate for the censored data [17,18]. The concordance index provides the probability that given two randomly selected patients, the patient with the worse outcome will in fact have a worse outcome prediction. The C-index ranges from 0 to 1, with 1 indicating perfect concordance, 0.5 indicating no better concordance than chance, and 0 indicating perfect discordance. In general, the model is considered relatively good for discrimination with values above 0.70. ROC curve for the survival data was drawn, using the methods proposed by Heagerty et al. [19].
Calibration is the agreement between the observed probability and predicted probability produced by the model [15]. The nomogram was calibrated by plotting the nomogram’s predicted 12-month PFS rate against the actually observed PFS rate, as calculated using the Kaplan-Meier method. Patients were divided into eight groups, using the quartiles of the predicted risk as the cutoff points. We used the bootstrapping re-sampling method (400 repetitions) in order to obtain relatively unbiased estimates and to check the interval validation.
Statistical analyses were performed, using STATA statistical software ver. 11.0 (STATA, College Station, TX) and R software ver. 2.13.2 (http://www.r-project.org). R package with the Design, and survcomp libraries (available at URL: http://cran.r-project.org/web/packages/) were used.
Results
Clinical characteristics of the 306 patients are shown in Table 1. The median PFS was 11.2 months (95% confidence interval [CI], 9.9 to 12.5 months), and 181 progression events had occurred. The 6-, 12-, and 18-month PFS rates, for the entire patient cohort, were 73.5%, 46.0%, and 25.1%, respectively. Response rate to EGFR TKI was 71.9% (95% CI, 66.9% to 76.9%).
Table 1.
Characteristics of the 306 patients who received gefitinib or erlotinib
| No. (%) (n=306) | |
|---|---|
| Age (yr) | |
| Median (range) | 61 (31-85) |
| Gender | |
| Male | 108 (35.3) |
| Female | 198 (64.7) |
| Disease status | |
| Recurred | 67 (21.9) |
| Initial stage wet IIIB, IV | 239 (78.1) |
| Smoking | |
| Never-smoker | 222 (72.5) |
| Current or ex-smoker | 76 (24.8) |
| Unknown | 8 (2.6) |
| Pathology | |
| Adenocarcinoma | 275 (89.9) |
| Non-small cell carcinoma NOS | 23 (7.5) |
| Squamous cell carcinoma | 4 (1.3) |
| Others | 4 (1.3) |
| ECOG PS | |
| 0 | 16 (5.2) |
| 1 | 209 (68.3) |
| 2 | 27 (8.8) |
| 3 | 10 (3.3) |
| 4 | 2 (0.7) |
| Unknown | 42 (13.7) |
| Liver metastasis | |
| No | 260 (85.0) |
| Yes | 46 (15.0) |
| Bone metastasis | |
| No | 177 (57.8) |
| Yes | 129 (42.2) |
| Brain metastasis | |
| No | 196 (64.1) |
| Yes | 110 (35.9) |
| EGFR TKI | |
| Gefitinib | 274 (89.5) |
| Erlotinib | 32 (10.5) |
| Line | |
| 1st | 108 (35.3) |
| 2nd | 178 (58.2) |
| 3rd or more | 20 (6.5) |
| EGFR mutation | |
| Del-19 or L858R | 290 (67.6) |
| Rare mutationa) | 16 (3.7) |
| Treatment response | |
| Complete response | 12 (3.9) |
| Partial response | 208 (68.0) |
| Stable disease | 46 (15.0) |
| Progressive disease | 24 (7.8) |
| Not evaluable | 16 (5.2) |
NOS, not otherwise specified; ECOG PS, Eastern Cooperative Oncology Group performance status; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Rare mutations were defined as any mutation other than del-19 or L858R in exon 21 of the EGFR gene.
1. Nomogram predicting PFS
We performed a univariate and multivariate Cox PHR analysis; Table 2 shows the results. In the univariate Cox PHR model, disease status, performance status, chemotherapy line, EGFR mutation status, response to TKI, liver metastasis, and bone metastasis showed association with PFS. Before constructing a multivariate Cox PHR model, colinearity was assessed, and the association between the variables was calculated, using the chi-square test for categorical variables. Significant association was observed between EGFR mutation status and response to TKI (chi-square p < 0.001). Liver metastasis and bone metastasis showed significant multicollinearity (chi-square p < 0.031). Therefore, liver metastasis and EGFR mutation status were not included in the multivariate Cox PHR model. Based on the results of multivariate analysis, the final nomogram model demonstrated the best fit and included all 5 variables (Fig. 1). The nomogram was developed for prediction of PFS, and can assign numeric predictions points for the risk of progression at 6, 12, and 18 months. Its C-index was 0.708 (95% CI, 0.659 to 0.758), and it appeared to be accurate. Fig. 2 shows the ROC curve and Fig. 3 shows the calibration plot.
Table 2.
Univariate and multivariate Cox proportional hazard regression analysis between clinicopathologic variables and progression-free survival
| Variable | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Disease status | ||||||
| Recurred | 1 | 1 | ||||
| Initial metastatic | 1.906 | 1.325-2.742 | 0.001 | 2.126 | 1.434-3.152 | < 0.001 |
| Pathology | ||||||
| ADC | 1 | - | ||||
| Non-ADC | 0.886 | 0.548-1.434 | 0.623 | - | - | - |
| Smoking | ||||||
| Never | 1 | - | ||||
| Current or ex-smoking | 1.237 | 0.880-1.739 | 0.220 | - | - | - |
| ECOG PS | ||||||
| 0-1 | 1 | 1 | ||||
| 2-4 | 1.740 | 1.088-2.783 | 0.021 | 1.978 | 1.210-3.235 | 0.007 |
| EGFR TKI | ||||||
| Gefitinib | 1 | - | ||||
| Erlotinib | 0.910 | 0.590-1.402 | 0.668 | - | - | - |
| Line | ||||||
| 1st | 1 | 1 | ||||
| 2nd | 1.270 | 0.908-1.777 | 0.163 | 1.233 | 0.860-1.767 | 0.254 |
| 3rd or more | 1.925 | 1.116-3.321 | 0.019 | 2.198 | 1.228-3.936 | 0.008 |
| EGFR mutation | ||||||
| Del-19 or L858R | 1 | - | ||||
| Rare | 4.158 | 2.358-7.331 | < 0.001 | - | - | - |
| Response to TKI | ||||||
| Responding | 1 | 1 | ||||
| Non-responding | 3.056 | 2.223-4.200 | < 0.001 | 3.141 | 2.228-4.426 | < 0.001 |
| Liver metastasis | ||||||
| No | 1 | - | ||||
| Yes | 1.546 | 1.067-2.240 | 0.021 | - | - | - |
| Bone metastasis | ||||||
| No | 1 | 1 | ||||
| Yes | 1.730 | 1.286-2.327 | < 0.001 | 1.384 | 1.001-1.913 | 0.049 |
| Brain metastasis | ||||||
| No | 1 | - | ||||
| Yes | 1.241 | 0.919-1.677 | 0.160 | - | - | - |
HR, hazard ratio; CI, confidence interval; ADC, adenocarcinoma; ECOG PS, Eastern Cooperative Oncology Group performance status; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Fig. 1.
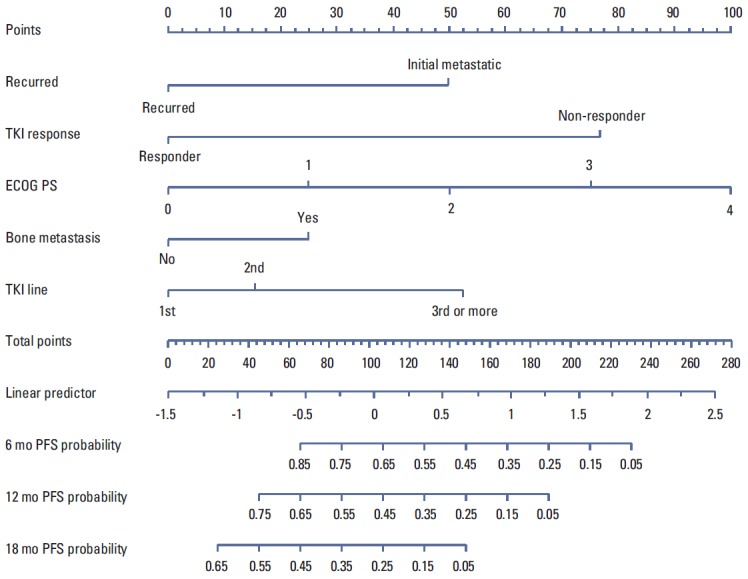
Nomogram for prediction of progression-free survival (PFS) to epidermal growth factor receptor tyrosine kinase inhibitor (TKI) in non-small cell lung cancer. The nomogram is used by totaling the points identified on the top scale for each independent covariate. The total points projected to the bottom scale indicate the % probability of 6-, 12-, and 18-month PFS. ECOG PS, Eastern Cooperative Oncology Group performance status.
Fig. 2.
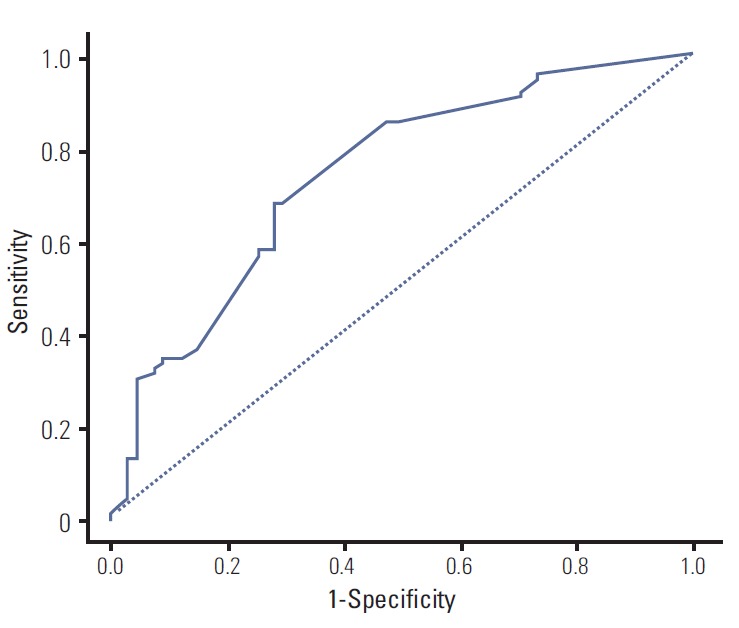
Receiver operating characteristic curve of the Cox proportional hazard regression model. Harrell’s C-index was 0.708 (95% confidence interval, 0.659 to 0.758).
Fig. 3.
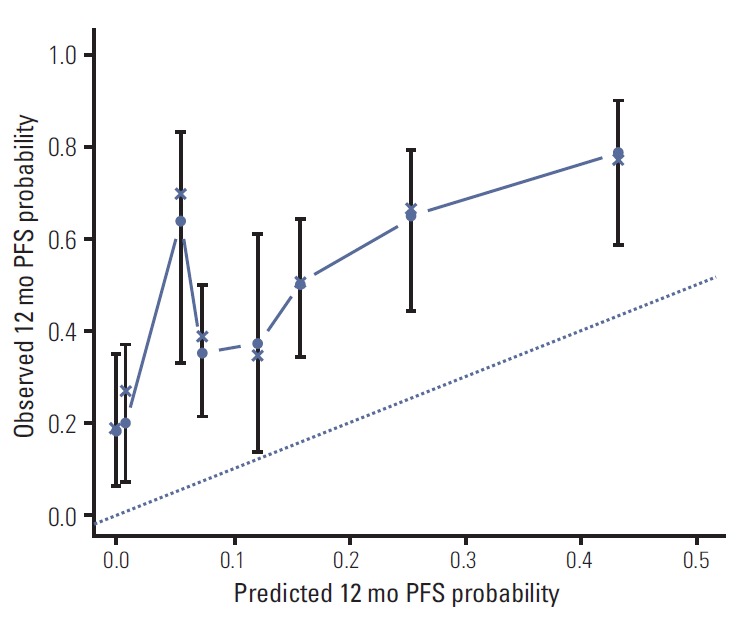
Calibration plot for 12-month progression-free survival (PFS) from the nomogram. On the calibration plot, the x-axis is nomogram predicted probability of PFS. The y-axis is observed PFS. Vertical bars indicate 95% confidence interval calculated using Kaplan-Meier analysis.
Discussion
In the current study, we developed the nomogram for prediction of PFS for EGFR TKI in NSCLC patients with EGFR mutation, based on the clinical and molecular characteristics. These practical models were internally validated and showed a good model performance in terms of calibration and discrimination. This nomogram may be useful in predicting when primary or acquired resistance to EGFR TKI will develop.
To date, several prognostic models [20,21] have been reported in NSCLC patients receiving EGFR TKI. Florescu et al. [20], who analyzed patients treated with erlotinib in the National Cancer Institute of Canada Clinical Trial Group Study BR21, identified the ten following clinical factors associated with overall survival: Eastern Cooperative Oncology Group performance status, smoking, weight loss, anemia, lactate dehydrogenase, time from diagnosis, response to prior treatment, ethnicity, and number of prior regimens. Florescu et al. [20] developed a clinical prognostic index, which allocates the scores to ten clinical factors, and categorized patients according to four subgroups. Among these 10 factors, Eastern Cooperative Oncology Group (ECOG) performance status and smoking were also valid in our model.
In addition to the clinical factors, several molecular markers are also known to be associated with PFS or resistance for EGFR TKI. Expression of thyroid transcription factor-1 [22] and estrogen receptor beta [23] showed an association with longer PFS for EGFR TKI, while K-ras mutation [24,25] and phosphoinositide-3-kinase catalytic alpha mutation [24] were associated with shorter PFS. Incorporation of these molecular markers into the nomogram can result in construction of a better prediction model.
The current study has some limitations. First, our nomogram was not externally validated using an independent data set. We performed only the internal validation, using the bootstrap method. It will be necessary to validate our nomogram by application in independent patients in other patient populations particularly Western patients. Second, response evaluation was repeatedly performed every 8-12 weeks, based on routine practice pattern. PFS might not be accurate due to inconsistent evaluation interval. Despite these limitations, the current study first developed the practical nomogram, regarding EGFR TKIs efficacy.
Conclusion
We developed a simple and precise nomogram that can be used for prediction of the PFS to EGFR TKIs in NSCLC patients with EGFR mutant. This prognostic nomogram can enable physicians to provide patients with an approximation of their prognosis before the initiation of EGFR TKIs treatment. In the clinic, this nomogram may be useful in prediction of when resistance to EGFR TKIs will occur, and provide guidance for identification of patients who will have long or short duration benefit from EGFR TKIs.
Acknowledgments
This study was supported by grants from the Innovative Research Institute for Cell Therapy, Republic of Korea (A062260), and this work was also supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (2010-0009563).
Footnotes
Conflict of interest relevant to this article was not reported.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Han SW, Kim TY, Hwang PG, Jeong S, Kim J, Choi IS, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 4.Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–37. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–20. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 6.Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29:2121–7. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 7.Azzoli CG, Baker S, Jr, Temin S, Pao W, Aliff T, Brahmer J, et al. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27:6251–66. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 9.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 10.Keam B, Kim DW, Park JH, Lee JO, Kim TM, Lee SH, et al. Rare and complex mutations of epidermal growth factor receptor, and efficacy of tyrosine kinase inhibitor in patients with non-small cell lung cancer. Int J Clin Oncol. doi: 10.1007/s10147-013-0602-1. 2013 Aug 6 [Epub]. http://dx.doi.org/10.1007/s10147-013-0602-1. [DOI] [PubMed] [Google Scholar]
- 11.Kim YT, Kim TY, Lee DS, Park SJ, Park JY, Seo SJ, et al. Molecular changes of epidermal growth factor receptor (EGFR) and KRAS and their impact on the clinical outcomes in surgically resected adenocarcinoma of the lung. Lung Cancer. 2008;59:111–8. doi: 10.1016/j.lungcan.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 13.Keam B, Im SA, Park S, Nam BH, Han SW, Oh DY, et al. Nomogram predicting clinical outcomes in breast cancer patients treated with neoadjuvant chemotherapy. J Cancer Res Clin Oncol. 2011;137:1301–8. doi: 10.1007/s00432-011-0991-3. [DOI] [PubMed] [Google Scholar]
- 14.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26:1364–70. doi: 10.1200/JCO.2007.12.9791. [DOI] [PubMed] [Google Scholar]
- 15.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 17.Nam BH. Discrimination and calibration in survival analysis [Dissertation] Boston: Boston University; 2000. [Google Scholar]
- 18.Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–23. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 19.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–44. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 20.Florescu M, Hasan B, Seymour L, Ding K, Shepherd FA, National Cancer Institute of Canada Clinical Trials Group A clinical prognostic index for patients treated with erlotinib in National Cancer Institute of Canada Clinical Trials Group study BR.21. J Thorac Oncol. 2008;3:590–8. doi: 10.1097/JTO.0b013e3181729299. [DOI] [PubMed] [Google Scholar]
- 21.Kim ST, Lee J, Sun JM, Park YH, Ahn JS, Park K, et al. Prognostic model to predict outcomes in non-small cell lung cancer patients with erlotinib as salvage treatment. Oncology. 2010;79:78–84. doi: 10.1159/000320190. [DOI] [PubMed] [Google Scholar]
- 22.Chung KP, Huang YT, Chang YL, Yu CJ, Yang CH, Chang YC, et al. Clinical significance of thyroid transcription factor-1 in advanced lung adenocarcinoma under epidermal growth factor receptor tyrosine kinase inhibitor treatment. Chest. 2012;141:420–8. doi: 10.1378/chest.10-3149. [DOI] [PubMed] [Google Scholar]
- 23.Nose N, Uramoto H, Iwata T, Hanagiri T, Yasumoto K. Expression of estrogen receptor beta predicts a clinical response and longer progression-free survival after treatment with EGFR-TKI for adenocarcinoma of the lung. Lung Cancer. 2011;71:350–5. doi: 10.1016/j.lungcan.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Ludovini V, Bianconi F, Pistola L, Chiari R, Minotti V, Colella R, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2011;6:707–15. doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 25.Han SW, Kim TY, Jeon YK, Hwang PG, Im SA, Lee KH, et al. Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res. 2006;12:2538–44. doi: 10.1158/1078-0432.CCR-05-2845. [DOI] [PubMed] [Google Scholar]


