Abstract
Necroptosis has emerged as an important pathway of programmed cell death in embryonic development, tissue homeostasis, immunity and inflammation1–8. RIPK1 is implicated in inflammatory and cell death signalling9–13 and its kinase activity is believed to drive RIPK3-mediated necroptosis14,15. Here we show that kinase-independent scaffolding RIPK1 functions regulate homeostasis and prevent inflammation in barrier tissues by inhibiting epithelial cell apoptosis and necroptosis. Intestinal epithelial cell (IEC)-specific RIPK1 knockout caused IEC apoptosis, villus atrophy, loss of goblet and Paneth cells and premature death in mice. This pathology developed independently of the microbiota and of MyD88 signalling but was partly rescued by TNFR1 (also known as TNFRSF1A) deficiency. Epithelial FADD ablation inhibited IEC apoptosis and prevented the premature death of mice with IEC-specific RIPK1 knockout. However, mice lacking both RIPK1 and FADD in IECs displayed RIPK3-dependent IEC necroptosis, Paneth cell loss and focal erosive inflammatory lesions in the colon. Moreover, a RIPK1 kinase inactive knock-in delayed but did not prevent inflammation caused by FADD deficiency in IECs or keratinocytes, showing that RIPK3-dependent necroptosis of FADD-deficient epithelial cells only partly requires RIPK1 kinase activity. Epidermis-specific RIPK1 knockout triggered keratinocyte apoptosis and necroptosis and caused severe skin inflammation that was prevented by RIPK3 but not FADD deficiency. These findings revealed that RIPK1 inhibits RIPK3-mediated necroptosis in keratinocytes in vivo and identified necroptosis as a more potent trigger of inflammation compared with apoptosis. Therefore, RIPK1 is a master regulator of epithelial cell survival, homeostasis and inflammation in the intestine and the skin.
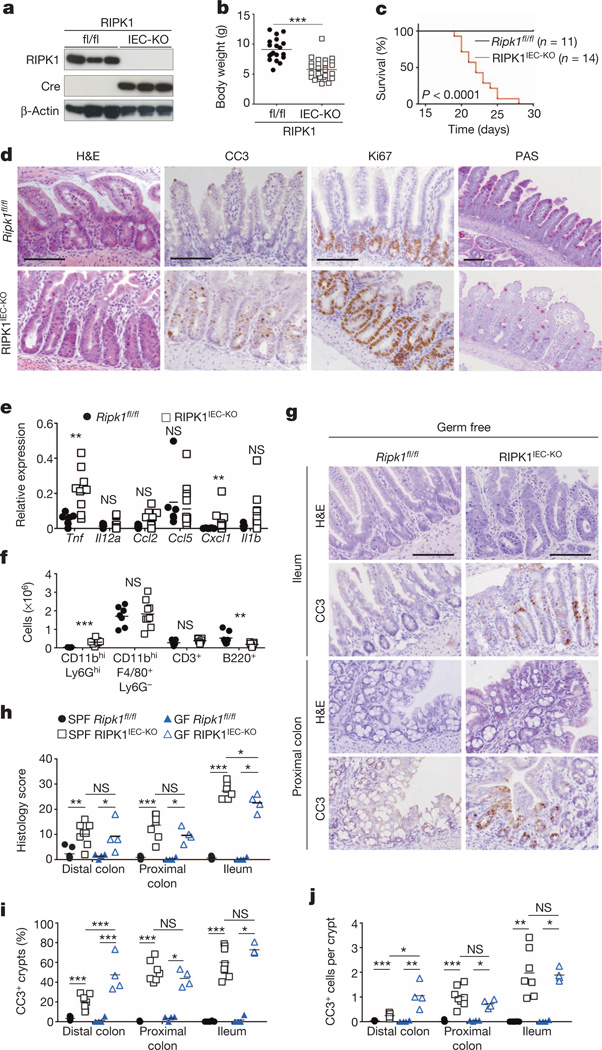
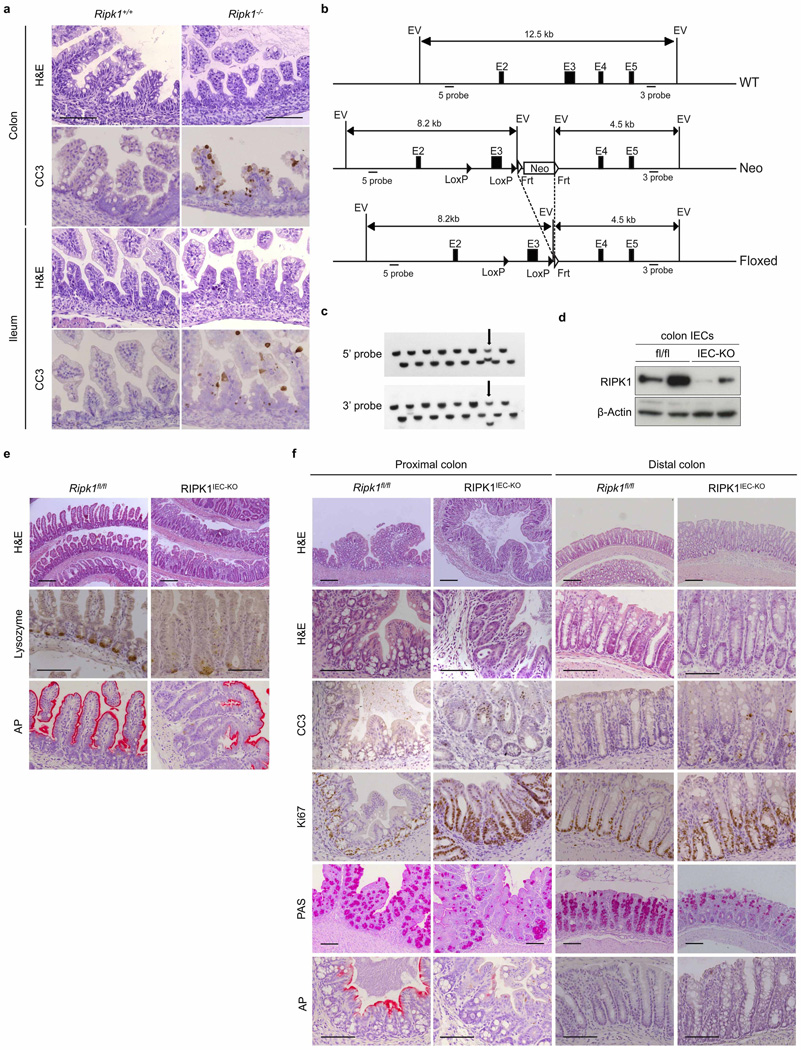
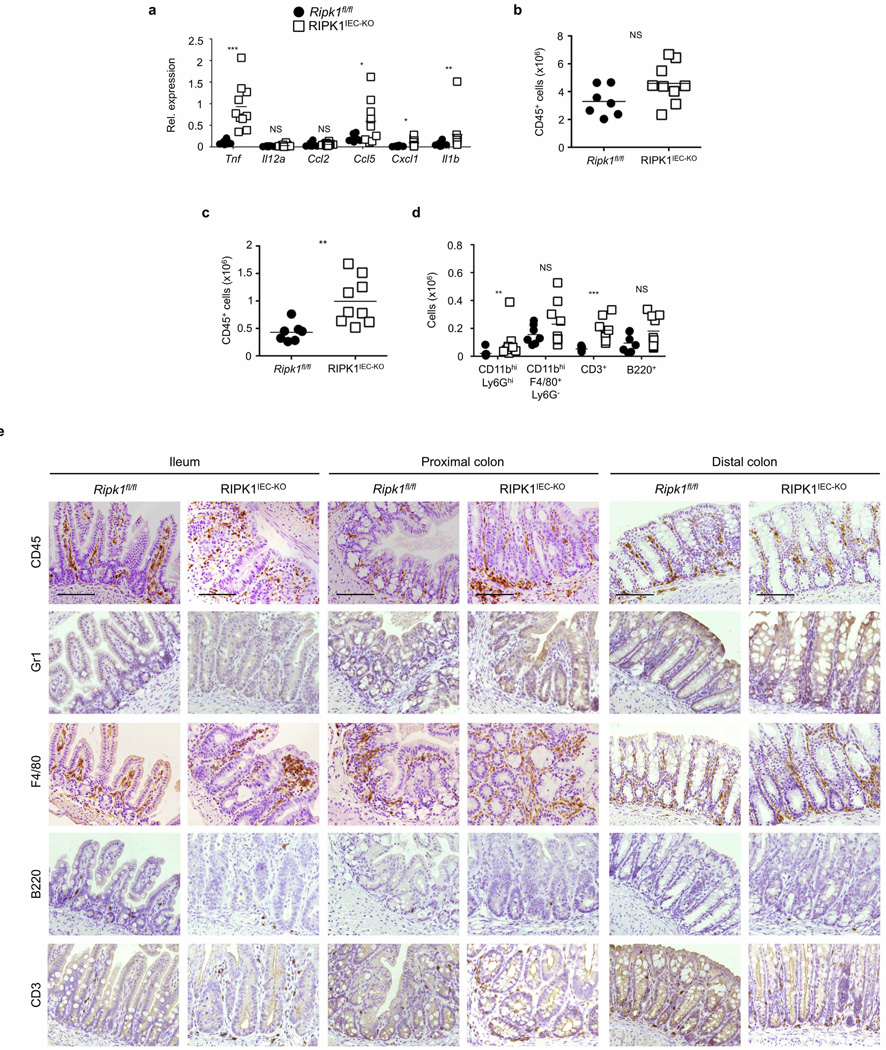
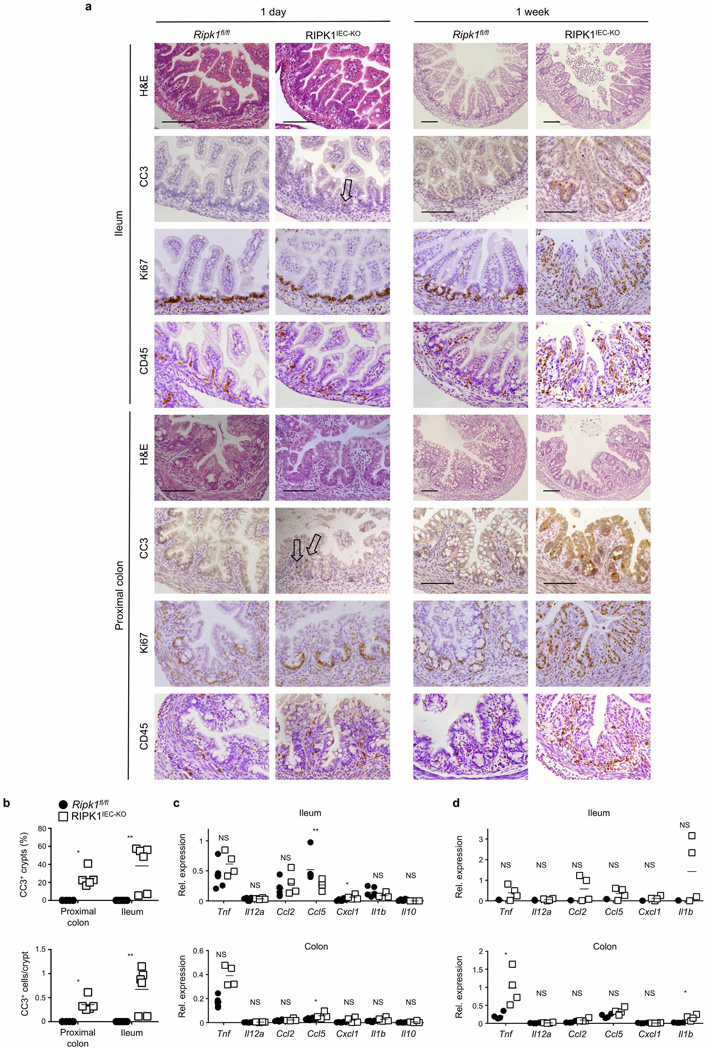
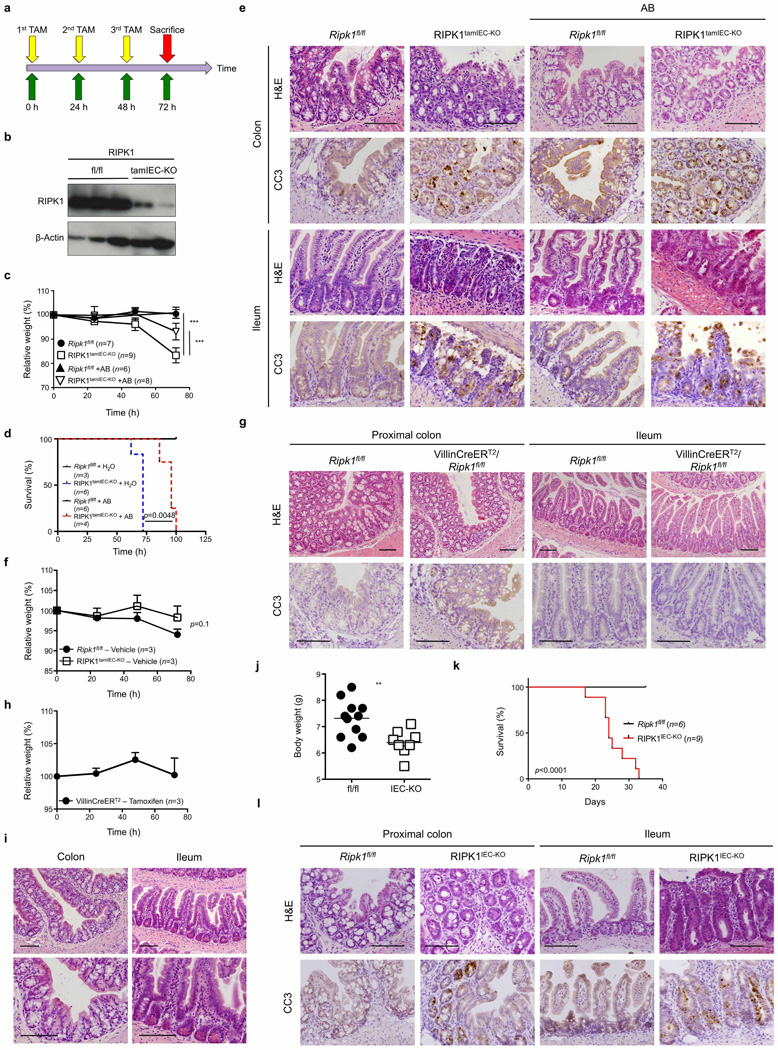
Mice lacking RIPK1 die perinatally, exhibiting apoptosis in multiple tissues including the intestine9 (Extended Data Fig. 1a). We generated mice with IEC-specific knockout of RIPK1 (RIPK1IEC-KO) (Fig. 1a and Extended Data Fig. 1b–d) and found that they showed reduced body weight and died within the first 4 weeks of life (Fig. 1b, c). Intestinal sections from 3-week-old RIPK1IEC-KO mice revealed pronounced villus atrophy and Paneth cell loss in the ileum, as well as reduced numbers of goblet cells and differentiated enterocytes, crypt elongation and IEC hyperproliferation in both the ileum and colon (Fig. 1d and Extended Data Fig. 1e, f). RIPK1IEC-KO intestines contained highly increased numbers of cleaved caspase-3-positive (CC3+) apoptotic IECs, but did not show signs of epithelial erosion, suggesting that the epithelial barrier remained largely intact (Fig. 1d and Extended Data Fig. 1e, f). Messenger RNA levels of Tnf, Il1b, Ccl5 and Cxcl1 and leukocyte numbers were moderately increased in the ileum and colon of RIPK1IEC-KO mice (Fig. 1e, f and Extended Data Fig. 2), indicating the presence of a mild inflammatory response. Newborn RIPK1IEC-KO mice showed only a few apoptotic IECs, while 7-day-old mice showed increased numbers of apoptotic IECs, loss of goblet cells, elongated hyperproliferative crypts and increased immune cell infiltration concomitant with moderately increased expression of tumour necrosis factor (TNF) and interleukin (IL)-1β (Extended Data Fig. 3). Tamoxifen-inducible ablation of RIPK1 in IECs of adult mice (RIPK1tamIEC-KO) caused rapid weight loss and death of the animals (Extended Data Fig. 4a–d) due to extensive IEC apoptosis resulting in severe intestinal pathology (Extended Data Fig. 4e–i). Collectively, these results revealed that epithelial-cell-intrinsic RIPK1 is essential for IEC survival and the maintenance of intestinal tissue structure and homeostasis.
Figure 1. Epithelial RIPK1 ablation causes microbiota-independent intestinal pathology.
a, Immunoblot of small intestine IECs from Ripk1fl/fl (fl/fl) and RIPK1IEC-KO (IEC-KO) mice. b, c, Body weight (b) and Kaplan–Meier survival curve (c) of Ripk1fl/fl and RIPK1IEC-KO mice. d, Representative images of ileal sections from Ripk1fl/fl and RIPK1IEC-KO mice stained as indicated. H&E, haematoxylin and eosin. e, f, Quantitative polymerase chain reaction with reverse transcription (qRT–PCR) of cytokine and chemokine expression (e) and fluorescence-activated cell sorting (FACS) of lamina propria leukocytes (f) in the small intestine of Ripk1fl/fl and RIPK1IEC-KO mice. g, Representative images of H&E- or CC3-stained intestinal sections from germ-free Ripk1fl/fl and RIPK1IEC-KO mice. h–j, Quantification of histological pathology score (h), crypts containing CC3+ cells (i) and CC3+ cells per crypt (j) in intestinal sections of the indicated mice. GF, germ free; SPF, specific pathogen free. Scale bars, 100 µm. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
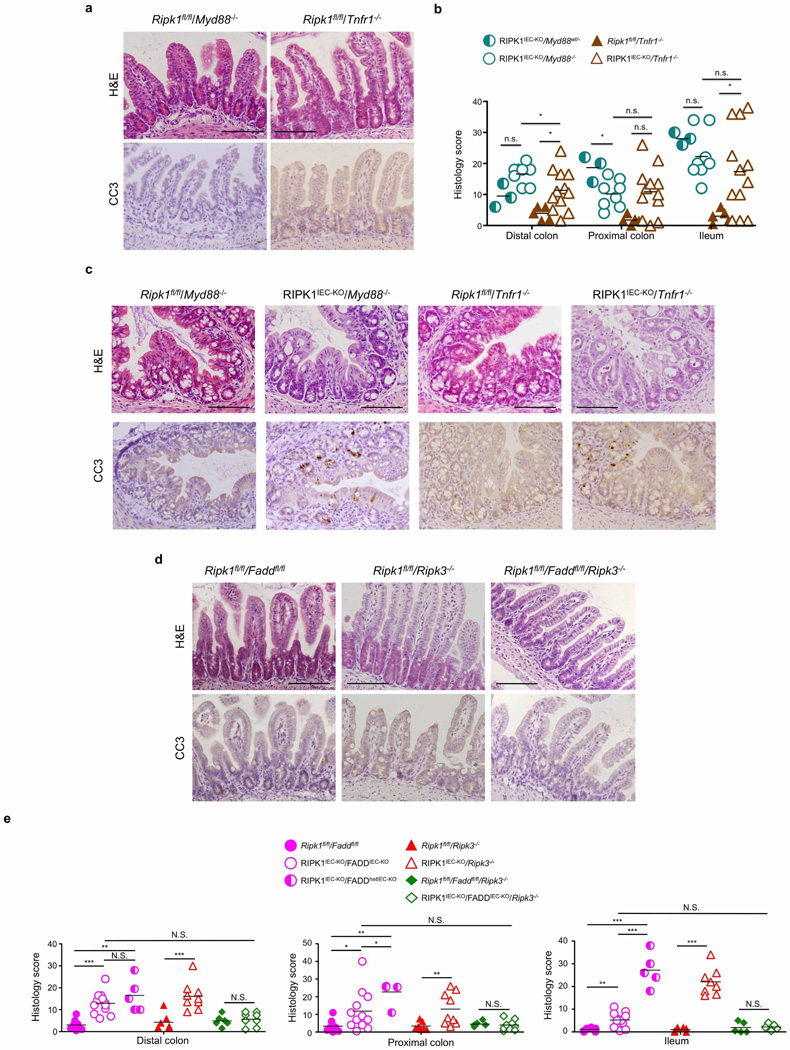
The luminal microbiota regulates intestinal homeostasis and contributes to inflammation both in human inflammatory bowel disease and in mouse models16,17. Antibiotic treatment briefly delayed but did not prevent severe intestinal pathology, weight loss and death of mice with inducible or constitutive IEC-specific RIPK1 knockout (Extended Data Fig. 4c–e, j–l). Moreover, RIPK1IEC-KO mice raised under germ-free conditions developed similar intestinal pathology, although some of the mice survived up to the age of 5 weeks, indicating that the microbiota contributes to disease severity (Fig. 1g–j). In addition, MyD88 deficiency did not prevent intestinal pathology in RIPK1IEC-KO mice (Fig. 2a, b, d and Extended Data Fig. 5a–c). Therefore, epithelial cell death and intestinal pathology in RIPK1IEC-KO mice develop independently of the microbiota and of MyD88-dependent TLR signalling. RIPK1IEC-KO/Tnfr1−/− mice were partially protected from severe wasting, with approximately 50% of the mice surviving to at least 3 months of age, and showed reduced IEC apoptosis and ameliorated intestinal pathology (Fig. 2a–e and Extended Data Fig. 5a–c). Thus, IEC death and intestinal pathology in RIPK1IEC-KO mice is induced mainly by TNFR1 signalling but TNFR1-independent pathways also contribute.
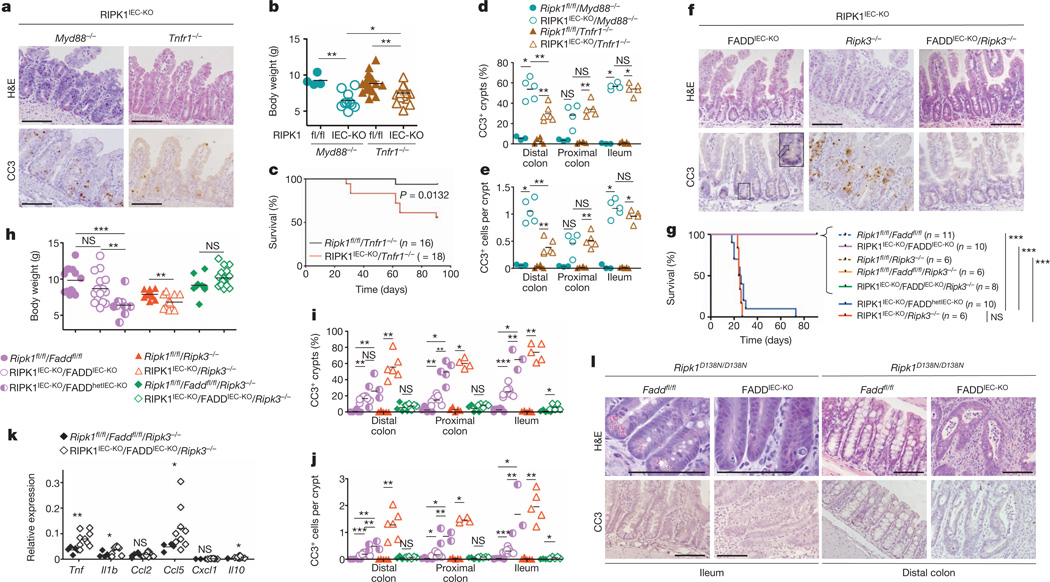
Figure 2. Death of RIPK1-deficient IECs depends on FADD and RIPK3.
a, Representative images of H&E- or CC3-stained ileal sections from the indicated mice. b, c, Body weight (b) and Kaplan–Meier survival curve (c) of the indicated mice. d, e, Quantification of crypts containing CC3+ cells (d) and CC3+ cell numbers per crypt (e) in intestinal sections of the indicated mice. f, Representative images of ileal sections from the indicated mice. Arrow depicts CC3− dying IEC. g, h, Kaplan–Meier survival curve (g) and body weight (h) of mice with the indicated genotypes. i, j, Quantification of crypts containing CC3+ cells (i) and CC3+ cell numbers per crypt (j) in intestinal sections of the indicated mice. k, qRT–PCR analysis of cytokine and chemokine expression in the small intestine of the indicated mice. l, Representative images of intestinal sections from 4-month-old mice with the indicated genotypes. Scale bars, 100 µm. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
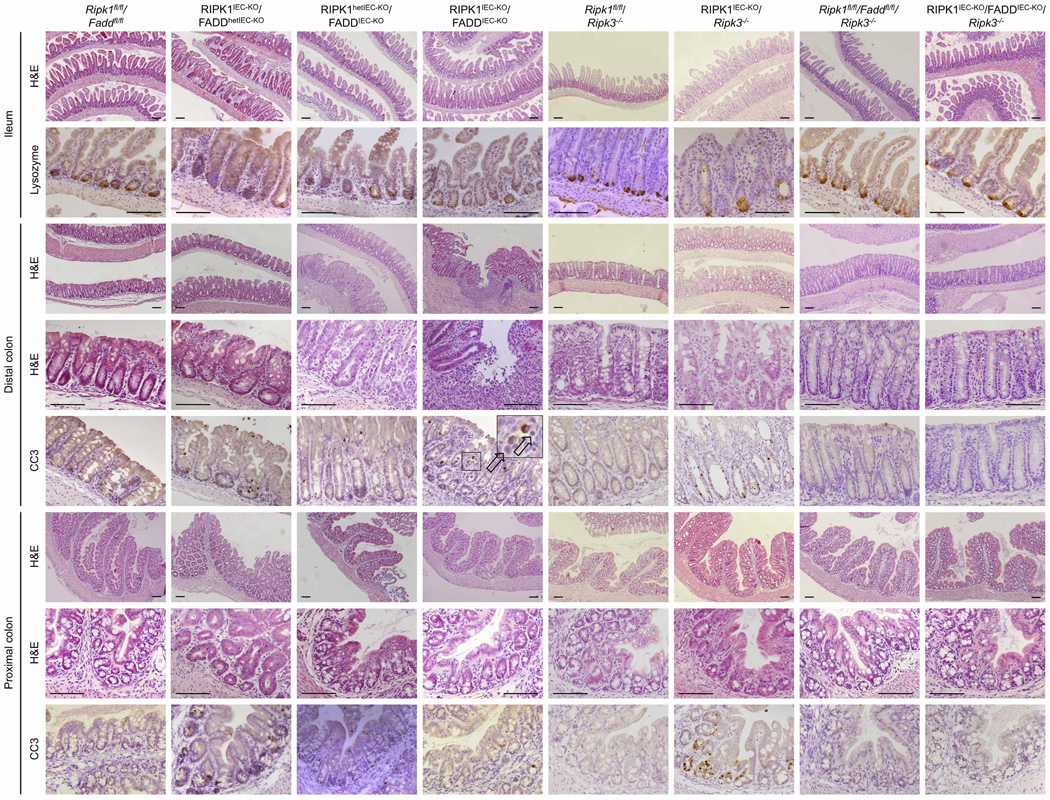
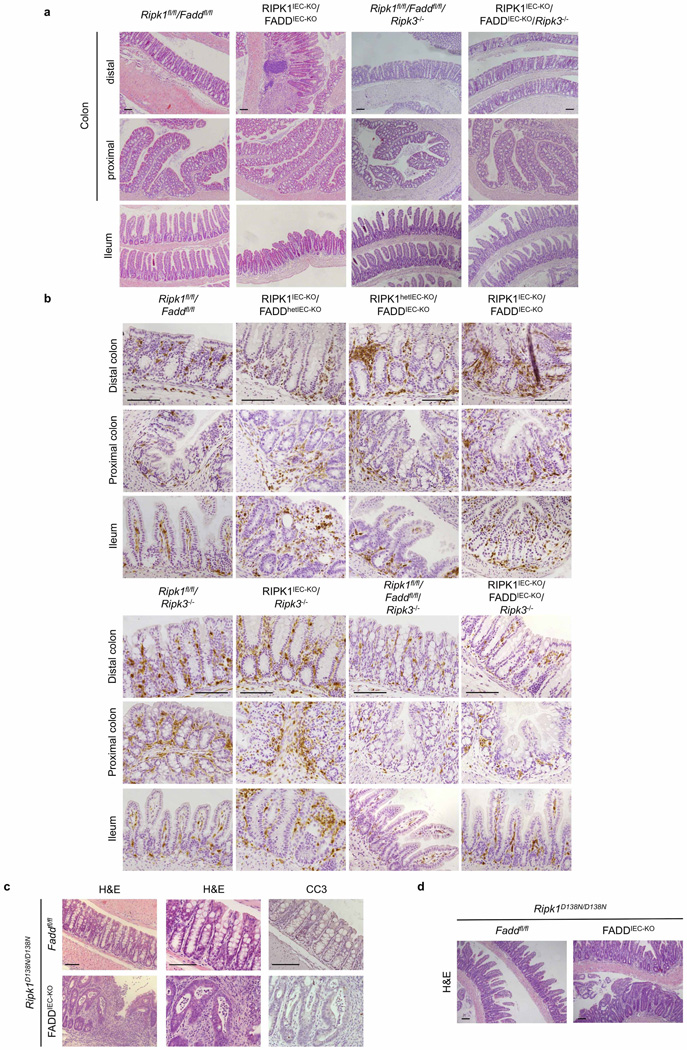
Epithelial FADD deficiency inhibited IEC apoptosis and villus atrophy and prevented the severe wasting and premature death of RIPK1IEC-KO mice (Fig. 2f–h), suggesting an important pathogenic role of FADD-dependent apoptosis. However, RIPK1IEC-KO/FADDIEC-KO intestines contained increased numbers of CC3− dying IECs and showed Paneth cell loss in the ileum and signs of colitis affecting primarily the distal colon (Fig. 2f, i, j and Extended Data Fig. 6). In contrast with RIPK1IEC-KO mice, in which the distal colon was only mildly affected and that did not show epithelial erosions (0 of 18 mice analysed), histological examination of Swiss roll sections revealed focal epithelial erosions in the distal colon of RIPK1IEC-KO/FADDIEC-KO mice (8 of 14 mice) and RIPK1IEC-KO mice with heterozygous epithelial FADD deficiency (FADDhetIEC-KO) (5 of 6 mice) at the age of 3 weeks (Extended Data Fig. 6). Adult RIPK1IEC-KO/FADDIEC-KO mice displayed villus shortening, loss of goblet and Paneth cells and crypt elongation in the small intestine, as well as focal inflammatory erosive lesions and crypt abscess formation in the distal colon in 5 of 7 mice examined (Extended Data Fig. 7a). Therefore, although epithelial FADD deficiency strongly reduced the number of apoptotic epithelial cells and prevented wasting and premature death of RIPK1IEC-KO mice, it did not normalize but rather altered their intestinal pathology, with RIPK1IEC-KO/FADDIEC-KO animals resembling the phenotype of FADDIEC-KO mice5.
We reasoned that RIPK3-dependent necroptosis could induce the death of IECs in RIPK1IEC-KO/FADDIEC-KO mice, as in FADDIEC-KO mice5. Indeed, triple-deficient RIPK1IEC-KO/FADDIEC-KO/Ripk3−/− mice did not show macroscopic or histological signs of intestinal pathology, although they displayed slightly increased mRNA levels of Tnf, Il1b, Ccl5 and Il10 in the colon (Fig. 2f–k and Extended Data Figs 5d, e, 6, 7a, b). In contrast, RIPK3 deficiency alone did not prevent wasting, early lethality, IEC apoptosis or intestinal pathology in RIPK1IEC-KO mice (Fig. 2f–j and Extended Data Figs 5d, e, 6, 7b). Therefore, RIPK3-dependent necroptosis does not constitute a primary pathway inducing IEC death and intestinal pathology in RIPK1IEC-KO mice, but becomes functionally important when FADD ablation inhibits apoptosis and sensitizes RIPK1-deficient IECs to necroptosis. These results suggested that RIPK3 induces RIPK1-independent necroptosis in FADD-deficient IECs, which is surprising as RIPK1 kinase activity is considered essential for RIPK3-mediated necroptosis, at least downstream of TNFR1 (refs 14, 15), which is the major driver of epithelial cell necroptosis in the colon of FADDIEC-KO mice5. To address specifically the role of RIPK1 kinase activity, we crossed FADDIEC-KO mice with knock-in mice expressing kinase-inactive RIPK1 (RIPK1 (D138N)) and found that lack of RIPK1 kinase activity did not prevent IEC necroptosis and intestinal inflammation in FADDIEC-KO mice (Fig. 2l and Extended Data Fig. 7c, d). Therefore, RIPK1 deficiency or loss of its kinase activity only partly inhibited RIPK3-dependent necroptosis of FADD-deficient IECs, providing genetic evidence that both RIPK1-dependent and RIPK1-independent pathways drive RIPK3-mediated necroptosis and intestinal inflammation in FADDIEC-KO mice.
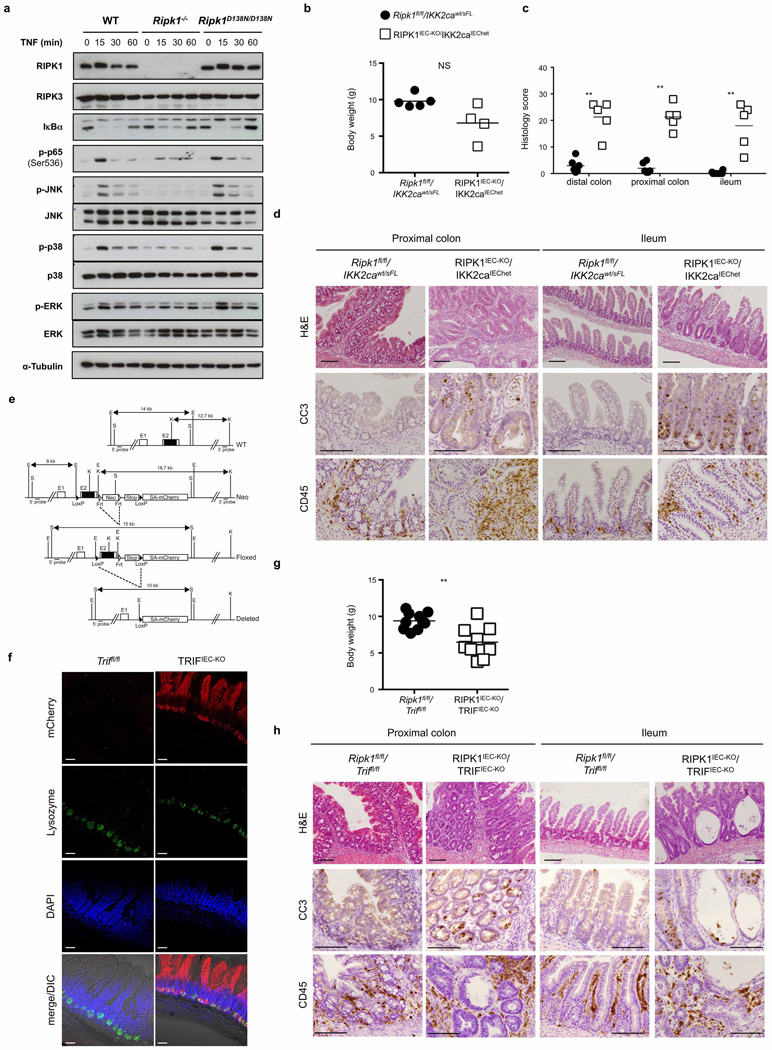
As shown previously9,18, RIPK1 deficiency but not loss of its kinase activity partially inhibited TNF-mediated NF-κB activation in mouse embryonic fibroblasts (MEFs) (Extended Data Fig. 8a), suggesting that loss of RIPK1 scaffolding function might trigger epithelial cell apoptosis by inhibiting NF-κB. However, NF-κB inhibition in the intestinal epithelium did not phenocopy the intestinal pathology of RIPK1IEC-KO mice19–21. Moreover, sustained NF-κB activation in RIPK1-deficient epithelial cells by expression of a constitutively active Ikk2 transgene22,23 did not prevent IEC death and rather aggravated the intestinal pathology of RIPK1IEC-KO mice, suggesting that RIPK1 deficiency does not sensitize IECs to apoptosis by inhibiting NF-κB (Extended Data Fig. 8b–d).
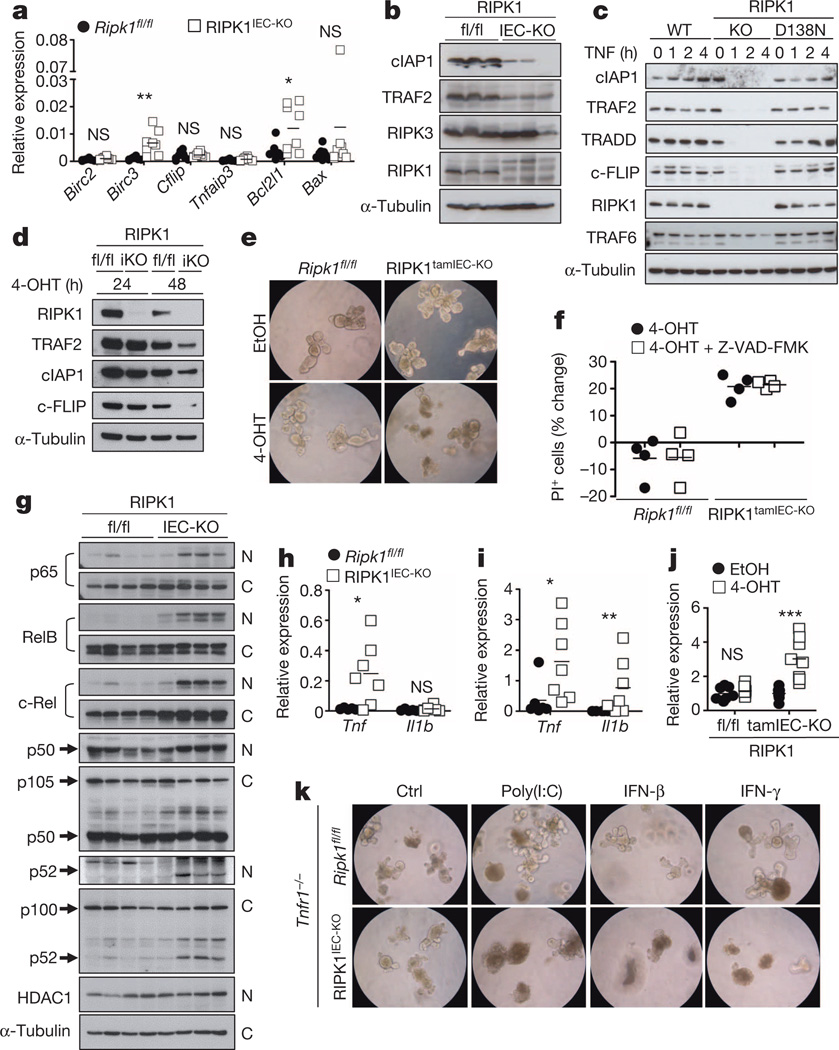
RIPK1-deficient IECs did not show reduced mRNA expression of pro-survival genes, but expressed reduced amounts of cIAP1 (also known as BIRC2) and TRAF2 proteins (Fig. 3a, b), suggesting that RIPK1 might control their stability. Indeed, consistent with previous reports24,25, we found that TNF stimulation induced rapid degradation of TRADD, cIAP1, TRAF2 and c-FLIP (also known as CFLAR), but not of TRAF6, in RIPK1-deficient but not in RIPK1(D138N) primary MEFs (Fig. 3c), suggesting that lack of the RIPK1 scaffolding function triggers degradation of proteins recruited to the TNFR1 signalling complex. We then sought to address whether RIPK1-deficient intestinal epithelial cells show similar responses. However, small intestinal crypts from RIPK1IEC-KO mice died shortly after isolation and failed to form organoids (data not shown). Nevertheless, tamoxifen-inducible RIPK1 deletion resulted in reduced expression of cIAP1, TRAF2 and c-FLIP proteins and caused rapid organoid death (Fig. 3d–f), suggesting that loss of pro-survival protein expression contributes to death of RIPK1-deficient IECs. Interestingly, TRAF2-deficient mice develop a similar intestinal phenotype and early lethality that was not prevented by antibiotic treatment and was only partially rescued by TNFR1 deficiency, suggesting that loss of TRAF2 could contribute to the intestinal pathology of RIPK1IEC-KO mice26.
Figure 3. RIPK1 prevents the degradation of pro-survival proteins.
a, b, qRT–PCR of pro-survival genes (a) and immunoblot of the indicated proteins (b) in small intestine IECs from Ripk1fl/fl (fl/fl) and RIPK1IEC-KO (IEC-KO) mice. c, Immunoblot of wild-type (WT), Ripk1−/− (KO) and Ripk1D138N/D138N (D138N) MEFs stimulated with 10 ng ml−1 recombinant murine TNF. d, Immunoblot of small intestine organoids from Ripk1fl/fl (fl/fl) and RIPK1tamIEC-KO (iKO) mice treated with 4-hydroxytamoxifen (4-OHT) for 24 or 48 h. e, Ripk1fl/fl and RIPK1tamIEC-KO organoids were treated with vehicle (ethanol; EtOH) or 4-OHT for 20 h and photographed 48 h later. Original magnification, ×400. f, FACS quantification of propidium iodide (PI)+ cells in organoids 43 h after treatment with 4-OHT in the presence or absence of z-VAD-FMK. g, Immunoblot of cytoplasmic (C) and nuclear (N) proteins from small intestine IECs of four Ripk1fl/fl and four RIPK1IEC-KO mice with the indicated antibodies. h, i, qRT–PCR of Tnf and Il1b expression in IECs from small intestine (h) or colon (i) of Ripk1fl/fl and RIPK1IEC-KO mice. j, qRT–PCR of Tnf expression in small intestine organoids from Ripk1fl/fl and RIPK1IEC-KO mice 30 h after treatment with EtOH or 4-OHT. k, Representative images of Ripk1fl/fl/Tnfr1−/− and RIPK1IEC-KO/Tnfr1−/− organoids 16 h after treatment with 100 µg ml−1 poly(I:C), 1,000 U ml−1 IFN-β and 1,000 U ml−1 IFN-γ. Original magnification, ×400. Ctrl, control. Representative data are shown of four (e, f, k), three (c) or two (d, j) independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
SMAC (also known as DIABLO) mimetic-compound-mediated cIAP1/2 degradation induced activation of both non-canonical and canonical NF-κB and the expression of TNF and other cytokines27,28, suggesting that loss of cIAP proteins could trigger the expression of TNF and other cytokines by RIPK1-deficient IECs that could act in an autocrine fashion to induce their death. Indeed, RIPK1-deficient primary IECs showed increased NF-κB activation and expressed elevated levels of TNF, while tamoxifen-inducible deletion of RIPK1 upregulated TNF expression in organoids (Fig. 3g–j). Moreover, intestinal crypts from RIPK1IEC-KO/Tnfr1−/− mice did not die rapidly, as did crypts from RIPK1IEC-KO mice, and although they did not grow as well as control organoids they could be maintained for a short period in culture (Fig. 3k), consistent with a role of autocrine TNF production in triggering the death of RIPK1-deficient IECs. RIPK1/TNFR1 double-deficient organoids were more sensitive to death induced by polyinosinic:polycytidylic acid (poly(I:C)), interferon (IFN)-β and IFN-γ (Fig. 3k), indicating that TIR-domain-containing adapter-inducing interferon-β (TRIF)- and IFN-dependent pathways also contribute to the death of RIPK1-deficient IECs. However, IEC-specific ablation of TRIF did not considerably ameliorate intestinal pathology in RIPK1IEC-KO mice (Extended Data Fig. 8e–h), suggesting that TRIF-mediated signalling is not a major driver of IEC apoptosis but could contribute in a redundant fashion together with TNFR1 and IFN signalling. Together, these findings suggest that RIPK1 deficiency results in degradation of cIAP proteins and TRAF2, triggering NF-κB-mediated expression of TNF and other cytokines that act in a partly redundant fashion to induce the death of RIPK1-deficient IECs. The mild upregulation of Tnf in the colon of RIPK1IEC-KO/FADDIEC-KO/Ripk3−/− mice (Fig. 2k) is compatible with this epithelial cell death independent mechanism inducing Tnf expression, which also provides a microbiota-independent trigger of IEC death in RIPK1IEC-KO mice.
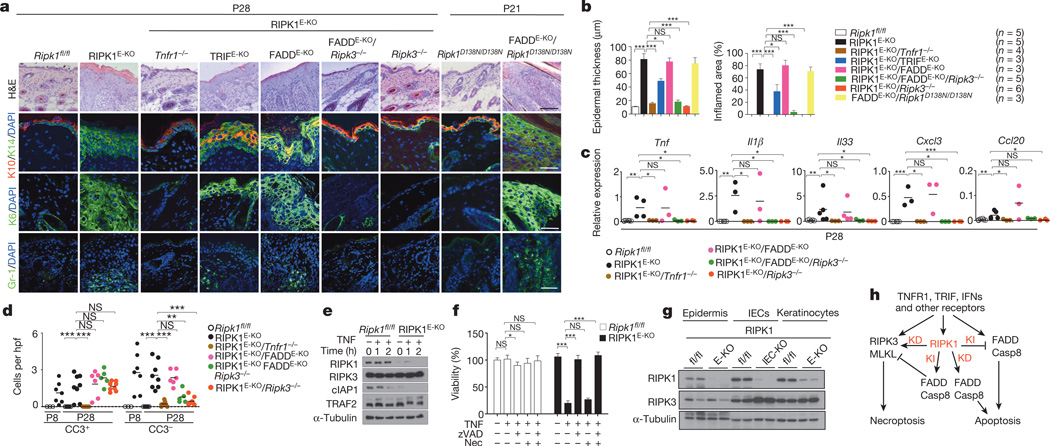
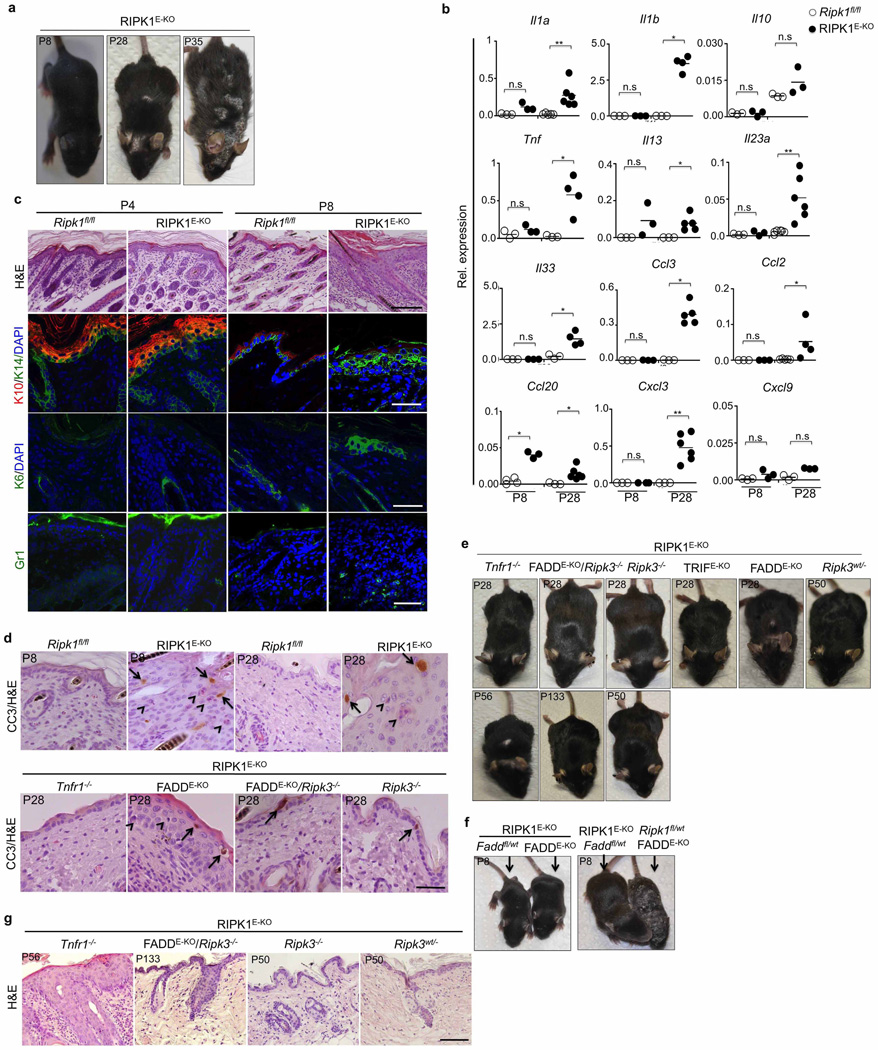
To investigate the function of RIPK1 in the epidermis, we generated mice with keratinocyte-specific RIPK1 deficiency (RIPK1E-KO mice) (Extended Data Fig. 9a). RIPK1E-KO mice progressively developed severe inflammatory skin lesions starting at around 1 week of age, characterized by epidermal thickening, increased keratin 14 (K14; also known as KRT14) and reduced K10 expression and upregulation of K6 in the interfollicular epidermis, as well as infiltration of Gr-1-positive cells and increased inflammatory cytokine and chemokine expression (Fig. 4a–c and Extended Data Fig. 9a, b). Histological analysis revealed the presence of CC3+ but also CC3− dying keratinocytes, identified by their pyknotic nuclei and eosinophilic cytoplasm in the epidermis of RIPK1E-KO mice (Fig. 4d and Extended Data Fig. 9d). To address the role of FADD-mediated apoptosis, we generated FADDE-KO/RIPK1E-KO mice and found that they did not show early postnatal skin inflammation and death, as did FADDE-KO mice4, but developed severe inflammatory skin lesions later, similarly to RIPK1E-KO mice (Fig. 4a–c and Extended Data Fig. 9c, d). Importantly, FADDE-KO/Ripk1D138N/D138N mice showed a similar skin phenotype to FADDE-KO/RIPK1E-KO mice (Fig. 4a, b). Therefore, lack of RIPK1 or its kinase activity delayed but could not prevent keratinocyte necroptosis and inflammation caused by FADD deficiency. Additional deficiency in RIPK3 fully prevented skin inflammation in RIPK1E-KO/FADDE-KO/Ripk3−/− mice (Fig. 4a–c and Extended Data Fig. 9e, g), showing that RIPK1-independent RIPK3-dependent keratinocyte necroptosis drives skin inflammation.
Figure 4. RIPK1 ablation causes keratinocyte necroptosis and skin inflammation.
a, Representative images of skin sections from the indicated mice stained with H&E or the indicated antibodies. Nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 100 µm (H&E); 50 µm (immunostainings). b, Microscopic quantification of epidermal thickness and inflamed skin area in the indicated mice. Error bars represent mean values ± standard error of the mean (s.e.m.). c, qRT–PCR of cytokine and chemokine expression in total skin from the indicated mice. d, Quantification of CC3+ and CC3− dying cells per high power field (hpf) in skin sections from the indicated mice. e, Immunoblot of primary keratinocytes stimulated with 20 ng ml−1 recombinant murine TNF for the indicated time points. f, Quantification of cell viability in primary keratinocytes treated with 50 ng ml−1 recombinant murine TNF in the presence or absence of z-VAD-FMK (zVAD; 20 µM) and necrostatin-1 (Nec; 30 µM). Mean values ± s.e.m. from biological triplicates (n = 3) are shown. g, Immunoblot of protein extracts from P4 epidermis, IECs or keratinocytes from mice of the indicated genotypes. h, Schematic model of the kinase-dependent (KD) and -independent (KI) functions of RIPK1 in apoptosis and necroptosis. Representative data of three independent experiments are shown in e, f. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
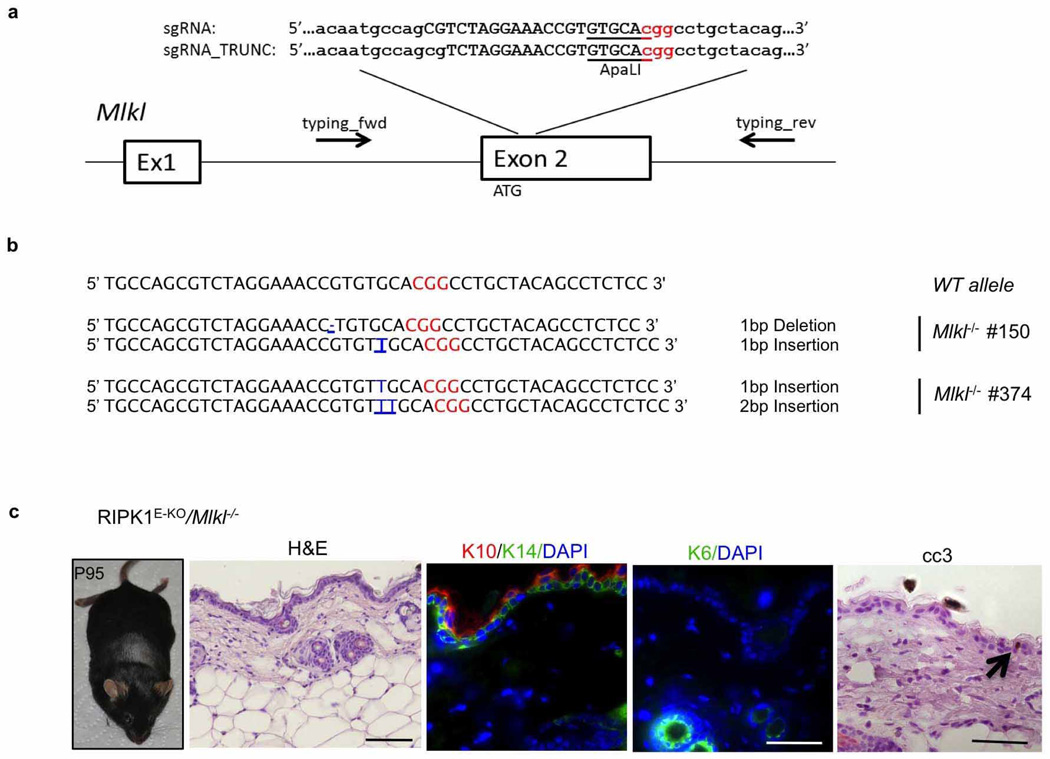
Surprisingly, RIPK1E-KO/Ripk3−/− mice did not develop skin inflammation and their epidermis contained strongly reduced numbers of CC3− but similar numbers of CC3+ keratinocytes compared with RIPK1E-KO mice (Fig. 4a–d and Extended Data Fig. 9d, e, g), showing that RIPK3-mediated necroptosis of RIPK1-deficient keratinocytes causes the inflammatory skin lesions. CRISPR/Cas9-mediated knockout of MLKL also prevented skin inflammation in RIPK1E-KO mice, further supporting an essential role of RIPK3/MLKL-dependent necroptosis in this model (Extended Data Fig. 10). Therefore, at least in the context of keratinocyte-specific RIPK1 deficiency, necroptosis of keratinocytes triggers skin inflammation but apoptosis does not, highlighting the different immune regulatory properties of these programmed cell death pathways in a disease-relevant in vivo experimental setting. Considering that Ripk1D138N/D138N mice did not develop skin lesions (Fig. 4a), these results show that RIPK1 acts through a kinase-independent scaffolding function to restrain RIPK3-mediated necroptosis in keratinocytes and prevent skin inflammation.
RIPK1E-KO/Tnfr1−/− mice showed largely normal skin until postnatal day (P)28, but developed patchy inflammatory skin lesions by the age of 7–8 weeks (Fig. 4a–d and Extended Data Fig. 9d, e, g). Therefore, TNFR1 critically contributes but is not the only trigger of keratinocyte necroptosis and inflammation in RIPK1E-KO mice. Keratinocyte-specific TRIF ablation also mildly ameliorated skin lesion development in RIPK1E-KO mice (Fig. 4a, b and Extended Data Fig. 9e). Collectively, these results showed that TNFR1- and TRIF-mediated signalling contribute to RIPK3-dependent keratinocyte necroptosis and skin inflammation in RIPK1E-KO mice.
cIAP1 was expressed at very low levels at steady state and was undetectable after TNF stimulation in RIPK1-deficient primary keratinocytes (Fig. 4e), similar to our findings in IECs. Moreover, RIPK1-deficient primary keratinocytes were highly sensitive to TNF-induced death that was inhibited by the caspase inhibitor z-VAD-FMK, demonstrating that in vitro these cells died primarily by apoptosis and were resistant to necroptosis even though they expressed RIPK3 (Fig. 4f, g). This result is surprising considering that RIPK1 deficiency in vivo triggers skin inflammation by sensitizing keratinocytes to RIPK3-dependent necroptosis, and suggests that mechanisms related to keratinocyte differentiation and the in vivo tissue context could determine the sensitivity of keratinocytes to apoptosis and necroptosis.
Collectively, our results revealed a novel kinase-independent scaffolding function of RIPK1 as an inhibitor of epithelial cell apoptosis and necroptosis in vivo that is essential for the maintenance of physiological tissue homeostasis in the intestine and the skin. The mechanisms determining whether RIPK1 deficiency sensitizes epithelial cells to apoptosis or necroptosis remain unclear and do not seem to depend on differential expression of RIPK3 (Fig. 4g). Our experiments also showed that necroptosis of FADD-deficient IECs and keratinocytes only partly depends on RIPK1 kinase activity, demonstrating that both RIPK1-dependent and -independent pathways drive RIPK3-mediated necroptosis. Therefore, RIPK1 is a central regulator of cell death that has the capacity to either inhibit or induce apoptosis and necroptosis (Fig. 4h). Considering its key role as a molecular hub integrating and decoding multiple ubiquitination (K63, K48, linear) signals to determine cellular responses to stimuli that are derived (for example, lipopolysaccharide, double-stranded RNA) or induced (for example, TNF, IFNs) bymicrobes and are capable of triggering both inflammation and cell death, RIPK1 emerges as a key regulator of immunity and homeostasis in barrier tissues.
Note added in proof: An accompanying paper (Takahashi, N. et al., Nature http://dx.doi.org/10.1038/nature13706) reports similar results on the role of RIPK1 in preventing IEC apoptosis, with some differences on the effect of antibiotic treatment and MYD88 deficiency on the intestinal pathology of RIPK1IEC-KO mice.
METHODS
Mice
Ripk1fl/fl, Ripk1D138N/D138N (ref. 29), Triffl/fl, Faddfl/fl (ref. 30), FADD-IRES-eGFPfl/fl (ref. 4) and R26-StopflIkk2ca22 mice expressing a constitutively active Ikk2 (Ikk2ca) transgene were generated by gene targeting in C57BL/6 embryonic stem (ES) cells. FLPe-Deleter31 mice were used to delete the FRT-flanked neo cassette and Cre-Deleter32 mice were employed to delete the RIPK1 floxed sequences in the germ line and generate Ripk1−/− mice and MEFs. Villin-Cre33, VillinCreERT2 (ref. 34), K14-Cre35, Tnfr1−/− (ref. 36), Myd88−/− (ref. 37) and Ripk3−/− (ref. 38) mice were backcrossed for at least ten generations into the C57BL/6 genetic background. Mice were maintained at the SPF animal facilities of the Institute for Genetics, University of Cologne, and of the University of Massachusetts Medical School and kept under a 12 h light cycle, and given a regular chow diet (Harlan diet no. 2918 or Prolab Isopro RMH3000 5P76) ad libitum. Germ-free mice were produced at the gnotobiotic facilities of the University of Ulm and of the Hannover Medical School. All animal procedures were conducted in accordance with European, national and institutional guidelines and protocols were approved by local government authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, Germany). Animals requiring medical attention were provided with appropriate care and excluded from the experiments described. No other exclusion criteria existed. For antibiotic treatment of RIPK1IEC-KO mice one of two different broad spectrum antibiotic mixtures was added to the drinking water starting from embryonic day (E)17.5: 1 g l−1 ampicillin (ICN Biomedicals), 1 g l−1 neomycin (Sigma), 0.5 g l−1 Meronem (AstraZeneca) and 0.5 g l−1 ciprofloxacin (Fluka) or 1 g l−1 ampicillin (ICN Biomedicals), 1 g l−1 metronidazole (Sigma), 0.5 g l−1 vancomycin (Eberth) and 0.2 g l−1 ciprofloxacin (Fluka). In VillinCreERT2/Ripk1fl/fl mice a modified version of the first protocol was employed, where Meronem was replaced by 0.5 g l−1 vancomycin (Eberth). As neither the overall physiological response nor tissue pathology differed between the two regimens, mouse groups receiving either antibiotic mixture were analysed together and data from individual experiments were pooled. VillinCreERT2 recombinase activity was induced by three daily intraperitoneal administrations of 1 mg tamoxifen. Littermates not carrying the Villin-Cre, Villin-CreERT2 and K14-Cre transgenes were used as controls in all experiments. Mice of the indicated genotype were assigned at random to groups. Mouse studies were performed in a blinded fashion. Unless otherwise indicated, mice were analysed at 3 weeks of age. Groups included male and female animals.
Targeting of the Mlkl gene in zygotes from RIPK1E-KO mice
Fertilized oocytes obtained from breedings of K14-Cre; Ripk1fl/wt males with Ripk1fl/fl females were microinjected with Mlkl short guiding (sg)RNA and Cas9 mRNA. sgRNA and truncated sgRNA were prepared by in vitro transcription of T7-promoter and sgRNA targeting sequence containing PCR products. These sgRNA templates were generated using a long sgRNA forward primer containing the T7 promoter (5′-TTAATACGACTCACTATAGG-3′) used for in vitro transcription, 20 bp sgRNA(5′-CGTCTAGGAAACCGTGTGCA-3′) or 18 bp sgRNA_TRUNC39 (5′-TCTAGGAAACCGTGTGCA-3′) targeting sequence and 20 bp homology to the px330 template (5′-GTTTTAGAGCTAGAAATAGC-3′; Zhang laboratory, Addgene plasmid 42230). The forward primer for MLKL_sgRNA is 5′-TTAATACGACTCACTATAGGCGTCTAGGAAACCGTGTGCAGTTTTAGAGCTAGAAATAGC-3′ and the forward primer for MLKL_sgRNA_TRUNC is 5′-TTAATACGACTCACTATAGGTCTAGGAAACCGTGTGCAGTTTTAGAGCTAGAAATAGC-3′. The long forward primers were used together with sgRNA_rev (5′-AAAAAGCACCGACTCGGTGCC-3′) to amplify the chimaeric sgRNA template from px330. The resulting 121 bp or 119 bp PCR product (MLKL_sgRNA or MLKL_sgRNA_TRUNC)was used for in vitro transcription using the T7 High Yield in vitro transcription kit (NEB) and sgRNA was purified using RNA isolation columns (Macherey Nagel). Fertilized oocytes were injected at 0.5 days post-coitum (dpc) with 100 ng µl−1 Cas9 mRNA (TriLink) together with 50 ng µl−1 sgRNA and the surviving two-cell-stage embryos were transferred the next day to foster mothers. Progeny was genotyped using the following primers: typing_fwd, 5′-GTCTTGCACGGTGGAGGTAT-3′; typing_rev, 5′-CCCAGACGTCTCTCAGCTTC-3′, and the T7 endonuclease I assay. Briefly, PCR product was boiled for 10 min at 98 °C and annealed by slowly cooling down to room temperature. The reaction was subsequently incubated with T7 endonuclease I (NEB) and analysed on a 2% agarose gel. Appearance of additional bands indicated mutations at the Mlkl locus. Additionally the PCR product was subjected to ApaLI restriction (NEB) with a lack of cleavage indicating the presence of a mutation. Mutations in the Mlkl locus were identified by sequencing the PCR product (GATC-Biotech).
IEC isolation and immunoblotting
IECs were isolated by sequential incubation of intestinal tissue in 1 mM dithiothreitol (DTT) and 1.5 mM EDTA solutions as described previously40. Cell lysates from IECs and keratinocytes were prepared as described41,42. Lysis buffer was supplemented with protease and phosphatase inhibitor tablets (Roche). Cell lysates were separated on SDS–PAGE and transferred to PVDF membranes (IPVH00010, Millipore). Membranes were probed with primary antibodies against the following proteins: RIPK1 (610459, BD Biosciences), RIPK3 (ADI-905-242-100), cIAP1 (ALX-803-335-C100, Enzo), IκBα (sc-371), TRAF2 (sc-876), TRAF3 (sc-949), TRADD (sc-7868), p65 (sc-372), c-Rel (sc-71),HDAC1 (sc-7872), β-actin, (sc-1616, Santacruz), phospho-p65 (3033), JNK (9252), RelB (4922), phospho-p38 (9211), p38 (8690), phospho-ERK (9101), ERK (9102, Cell Signaling), phospho-JNK (44-682G, Invitrogen), p100/52 (NR1495), p105/50 (NR1157, NCI BRB), α-tubulin (T6074, Sigma), TRAF6 (597, MBL), c-FLIP (AG-20B-0005-C100, Adipogen), followed by secondary HRP-coupled antibodies (GE Healthcare and Jackson ImmuneResearch) and developed with chemiluminescent detection substrate (GE Healthcare and Thermo Scientific).
Isolation of lamina propria cells and flow cytometry
Colon and small intestine lamina propria cells were isolated using a modified version of a previously described protocol43. In brief, after removal of Peyer’s patches, small intestine and colon tissues were incubated at 37 °C in HBSS containing 4% FCS, 10 mM HEPES and 5 mM EDTA. The remaining tissue was cut into small pieces and digested in HBSS containing 4% FCS, 10 mM HEPES, Dispase II (0.15 mg ml−1; Roche), Collagenase D (0.4 mg ml−1; Roche) and DNase I (0.5 mg ml−1; Sigma-Aldrich). The isolated cells were purified by Percoll gradient centrifugation (GE Healthcare Life Sciences). Single-cell suspensions were preincubated with CD16/CD32 antibody (BD Biosciences) for Fc-receptor blocking and subsequently stained with the following fluorescently labelled antibodies: CD3e (145-2C11), CD11b (M1/70), CD45.2 (104), Ly6G (1A8), B220 (RA3-6B2), F4/80 (BM8) (BD Biosciences or eBioscience). Experiments were acquired on a BD FACS Canto I (BD Biosciences) interfaced to FACSDiva software (BD), and analysed with FlowJo software (Tree Star).
Histology
Intestinal tissues were fixed in 4% paraformaldehyde, skin samples in 10% formalin and embedded in paraffin and cut in 3–5 µm sections. Paraffin sections were rehydrated and heat-induced antigen retrieval was performed in citrate buffer, TRIS buffer or by proteinase K treatment. Primary antibodies for immunohistochemistry were anti-CD45 (550566, eBioscience), anti-Gr-1 (551459, BD Biosciences and MCATTIGA, Serotec), anti-F4/80 (clone A3-1, homemade), anti-B220 (clone RA36B2, homemade), anti-CD3(ab5690, Abcam), anti-active caspase 3 (9661, Cell signalling and AF835, R&D Systems), anti-Ki67 (M724901, Dako), anti-human lysozyme (A0099, Dako). Biotinylated secondary antibodies were purchased from Perkin Elmer and Dako. Stainings were visualized with ABC Kit Vectastain Elite (Vector Laboratories) or Streptavidin-HRP (Millipore) and DAB substrate (DAKO and Vector Laboratories). Incubation times with DAB substrate were equal for all samples. Periodic acid–Schiff (PAS) reaction was performed according to standard protocols. Endogenous alkaline phosphatase activity was visualized using Vector Red Alkaline Phosphatase Substrate Kit according to the manufacturer’s instructions (SK-5100, Vector Laboratories). Immunostainings on skin sections were performed as previously described42, Gr-1 staining was performed on cryo-sections. For assessment of intestinal pathology, H&E-stained intestinal sections were subjected blindly to a semiquantitative composite histological pathology scoring system as described previously44. Microscopic quantification of epidermal thickness was performed by measurement of epidermal thickness in four different back skin areas for each mouse. In each field, four measurements were performed. The percentage of the inflamed area was determined by measuring the cumulative number of the inflamed areas in relation to the total area of one section from back skin of each mouse. The number of CC3+ and CC3− dying cells was determined on the same skin section by staining with anti-CC3 antibodies and counterstaining with H&E. Dying CC3− keratinocytes were identified by their pyknotic nuclei and eosinophilic cytoplasm.
Near native imaging
Sections for imaging of mCherry at near native conditions were prepared as described45. Briefly, the intestine was dissected, thoroughly rinsed and fixed in 4% PFA for 20 min at room temperature. Tissue was embedded in 4% low melting agarose and sectioned with a vibrating blade microtome (Leica) at 150 µm. Permeabilization and staining were performed in PBS supplemented with 1% (wt/vol) BSA, 1% (vol/vol) dimethylsulphoxide (DMSO) and 2% (vol/vol) Triton X-100. Paneth cells were visualized with anti-lysozyme antibody and goat anti-rabbit IgG Alexa-488 (Molecular Probes, A11008). Images of fluorescent stainings were acquired with a Zeiss Meta 510 confocal laser-scanning microscope.
Isolation and culture of primary murine keratinocytes
Isolation and culture of primary murine keratinocytes was performed as described previously46.
Isolation and culture of intestinal organoids
Small intestinal crypts were isolated and grown to organoids as described previously47. Deletion of RIPK1 in organoid cultures from RIPK1tamIEC-KO mice was induced by addition of 100 nM 4-OH-tamoxifen (4-OHT) to the culture medium for 20–24 h. Cell death of organoid cultures was determined by quantification of propidium iodine (PI)-positive cells by flow cytometry acquired on a FACSCalibur, BD. For qRT–PCR analysis organoids were harvested 36 h after 4-OHT removal. For each genotype, organoids from two different mice were enrolled, seeded out in quadruplicates and processed independently.
qRT–PCR
Total RNA was extracted with Trizol Reagent (Life Technologies) and RNeasy Columns (Qiagen) and cDNA was prepared with Superscript III cDNA-synthesis Kit (Life Technologies). qRT–PCR was performed with TaqMan probes (Life Technologies) with TATA-box-binding protein as reference gene for intestinal samples and HRPT for skin tissue. Data were analysed according to the ΔCT method. Primer sequences are available upon request.
Statistics
Data shown in column graphs represent the mean ± s.d. or ± s.e.m., as indicated in the figure legends. To determine the group size necessary for adequate statistical power, power analysis was performed using preliminary data sets. When data fulfilled the criteria for Gaussian distribution, unpaired Student’s t-tests were performed; otherwise the nonparametric Mann–Whitney test was chosen. For the keratinocyte viability assay, a paired Student’s t-test was performed. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005. Statistical analysis was performed with Prism6, GraphPad or Excel, Microscoft software.
Extended Data
Extended Data Figure 1. Intestinal pathology in Ripk1−/− and RIPK1IEC-KO mice.
a, Representative images of colonic and ileal sections from newborn Ripk1+/+ and Ripk1−/− mice stained with H&E or immunostained for CC3. b, Targeting strategy for the generation of mice with loxP-flanked (floxed (fl)) Ripk1 alleles. Exon 3 of the Ripk1 gene was flanked with loxP sites and an FRT-flanked neomycin (Neo) selectable cassette was introduced after the 3′ loxP site. The Neo cassette was excised by crossing the Ripk1neoFloxed mice with Flp-deleter mice. c, Representative Southern blots depicting the identification of correctly targeted embryonic stem (ES) cell clones by using 5′ and 3′ external probes. Arrows indicate a correctly targeted ES cell clone. Screening for the loxP site in intron 2 was performed by PCR (data not shown). Double combs allowing loading samples at two levels were used to maximize loading capacity of the gels for screening ES clones in 96-well plates. d, Immunoblot of colon IEC protein extracts from Ripk1fl/fl and RIPK1IEC-KO mice. e, f, Representative images of ileal (e) or colon (f) sections from Ripk1fl/fl and RIPK1IEC-KO mice stained with H&E, PAS or alkaline phosphatase (AP), or immunostained against lysozyme, CC3 or Ki67. Scale bars, 100 µm.
Extended Data Figure 2. Mild intestinal inflammation in RIPK1IEC-KO mice.
a, qRT–PCR analysis of cytokine and chemokine expression in colon tissue from Ripk1fl/fl and RIPK1IEC-KO mice. b–d, FACS analysis of lamina propria leukocytes in the small intestine (b) and colon (c, d) of Ripk1fl/fl and RIPK1IEC-KO mice. e, Representative images from intestinal sections from Ripk1fl/fl and RIPK1IEC-KO mice immunostained with the indicated antibodies. Scale bars, 100 µm. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
Extended Data Figure 3. Assessment of intestinal pathology in newborn and 1-week-old RIPK1IEC-KO mice.
a, Representative images of intestinal sections from Ripk1fl/fl and RIPK1IEC-KO mice stained with H&E or immunostained with the indicated antibodies. Arrows indicate sparse CC3+ IECs in sections from 1-day-old animals. Scale bars, 100 µm. b, Quantification of crypts containing CC3+ cells and the number of CC3+ cells per crypt in intestinal sections of 1-week-old Ripk1fl/fl and RIPK1IEC-KO mice. c, d, qRT–PCR analysis of cytokine and chemokine expression in small intestinal and colon tissue from 1-day-old (c) and 1-week-old (d) Ripk1fl/fl and RIPK1IEC-KO mice. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
Extended Data Figure 4. Antibiotic treatment does not prevent intestinal pathology in RIPK1IEC-KO and RIPK1tamIEC-KO mice.
a, Experimental outline of tamoxifen injections (TAM; 1 mg, intraperitoneally). b, Immunoblot of small intestine IEC protein extracts from tamoxifen-treated Ripk1fl/fl and RIPK1tamIEC-KO mice. c–e, Body weight change (c), Kaplan–Meier survival curve (d) and representative images of H&E- and CC3-stained intestinal sections (e) of tamoxifen-treated Ripk1fl/fl and RIPK1tamIEC-KO mice receiving antibiotics (+AB) or normal drinking water starting 4 weeks before tamoxifen administration. f, g, Body weight change (f) and representative images of H&E- and CC3-stained intestinal sections (g) in vehicle-injected Ripk1fl/fl and RIPK1tamIEC-KO mice. h, i, Body weight changes (h) and representative images of H&E-stained intestinal sections (i) of tamoxifen-injected Villin-CreERT2 mice (n = 3). j–l, Body weight (j), Kaplan–Meier survival curve (k) and representative images of H&E- and CC3-stained intestinal sections (l) of Ripk1fl/fl and RIPK1IEC-KO mice treated with antibiotics from E17.5 to 3 weeks of age. Scale bars, 100 µm. Error bars represent mean values ± s.d. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
Extended Data Figure 5. Role of MyD88, TNFR1, FADD and RIPK3 in the intestinal pathology of RIPK1IEC-KO mice.
a, Representative images of ileal sections from the indicated mice as controls for the mice shown in Fig. 2a. b, Quantification of histological pathology score of the indicated mice. c, Representative images of H&E- or CC3-stained colon sections from the indicated mice. d, Representative images of H&E- or CC3-stained ileal sections from the indicated mice. Scale bars, 100 µm. e, Quantification of histological pathology score of the indicated mice. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
Extended Data Figure 6. Epithelial-specific FADD deficiency reduces IEC apoptosis, ameliorates crypt atrophy but triggers erosive lesions in the colon of RIPK1IEC-KO mice.
Representative images of ileal and colonic sections from the indicated mice stained with H&E or immunostained against lysozyme or CC3. Scale bars, 100 µm.
Extended Data Figure 7. FADD and RIPK3 deficiency restores intestinal homeostasis in RIPK1IEC-KO mice and IEC necroptosis in FADDIEC-KO mice occurs in the absence of RIPK1 kinase activity.
a, Representative images of H&E-stained colonic and ileal sections of adult mice with the indicated genotypes. b, Representative images of colonic and ileal sections of mice with the indicated genotypes immunostained for CD45. c, d, Representative images of colonic (c) and ileal (d) sections of adult Faddfl/fl/Ripk1D138N/D138N and FADDIEC-KO/Ripk1D138N/D138N mice stained with H&E or immunostained against CC3. Scale bars, 100 µm.
Extended Data Figure 8. Assessment of the role of NF-κB and TRIF signalling in RIPK1IEC-KO mice.
a, Immunoblot analysis of protein extracts from wild-type (WT), Ripk1−/− and Ripk1D138N/D138N MEFs after stimulation with 10 ng ml−1 recombinant murine TNF for the indicated time points. Data are representative of five independent experiments. b–d, Body weight (b), quantification of histological pathology score (c) and representative H&E-, CC3- or CD45-stained intestinal sections (d) of mice with the indicated genotypes. e, Targeting strategy used for the generation of Triffl/fl mice. Exon 2 of the Trif gene was flanked with loxP sites. FRT-flanked neo and a stop cassette were placed upstream of the 3′ loxP site. A splice acceptor (SA)-mCherry cassette was introduced downstream of the 3′ loxP site. Cre-mediated recombination removes the Trif coding sequences and the stop cassette inducing the expression of mCherry by the Trif locus. The FRT-flanked neo was excised by crossing TrifneoFloxed mice with Flp-Deleter mice. f, Confocal microscopy images of near native small intestinal sections of Triffl/fl and TRIFIEC-KO mice stained with anti-lysozyme and DAPI. g, h, Body weight (g) and representative images of H&E-, CC3- or CD45-stained intestinal sections (h) of the indicated mice. Scale bars, 100 µm. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
Extended Data Figure 9. Skin inflammation in RIPK1E-KO mice is dependent on RIPK3-mediated necroptosis.
a, Representative macroscopic images of RIPK1E-KO mice. b, qRT–PCR analysis of pro-inflammatory cytokines and chemokines on total skin mRNA from Ripk1fl/fl and RIPK1E-KO mice. c, d, Representative images of skin sections from the indicated mice stained as indicated. In d, arrows point to CC3+ cells and arrowheads depict CC3− dying cells identified by their pyknotic nuclei and eosinophilic cytoplasm. Scale bars, 100 µm in H&E stained; 50 µm in immunostained sections. e–g, Representative macroscopic pictures (e, f) and H&E-stained skin sections (g) of the indicated mice. Scale bars, 100 µm. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005; NS, not significant.
Extended Data Figure 10. CRISPR/Cas9-mediated knockout of MLKL prevents skin inflammation in RIPK1E-KO mice.
a, Schematic depiction of the Mlkl locus. The sequence targeted by the small guide RNAs (sgRNA and truncated TRUNC_sgRNA) is indicated by capital letters. The position of the ATG and the binding sites of the primers used for genotyping and sequencing are indicated. The sgRNAs were designed to target a sequence containing an ApaLI restriction site that is used for RFLP analysis (underlined). The protospacer-adjacent motif (PAM) sequence is depicted in red. b, Sequences of the wild-type (WT) Mlkl locus and of the targeted Mlkl alleles of the two obtained RIPK1E-KO/Mlkl−/− mice. Mouse #150 carries one allele with one base pair (bp) deletion and one allele with one bp insertion. Mouse #374 carries one allele with one bp insertion and one allele with two bp insertions. All mutations cause frame shift and ablate MLKL protein expression. Mouse #150 was obtained using the full-length MLKL-sgRNA and mouse #374 was obtained using the truncated MLKL_sgRNA_TRUNC. c, Macroscopic appearance and histological images of skin sections from RIPK1E-KO/Mlkl−/− mouse #150 at the age of P95. Scale bars, 100 µm (H&E); 50µm (keratins and CC3).
Acknowledgements
We are grateful to V. Dixit for Ripk3−/−, D. Gumucio for Villin-Cre and S. Robine for Villin-CreERT2 mice. We thank C. Uthoff-Hachenberg, J. Buchholz, E. Mahlberg, B. Kühnel, B. Hülser, P. Jankowski, S. Assenmacher and P. Scholl for technical assistance. M.P. acknowledges funding from the European Research Council (2012-ADG_20120314), the German Research Council (DFG; SFB670, SFB829, SPP1656), the European Commission (grants 223404 (Masterswitch) and 223151 (InflaCare)), the Deutsche Krebshilfe, the Else Krüner-Fresenius-Stiftung and the Helmholtz Alliance (PCCC). Research reported in this publication was also supported by the National Institute of Allergy and Infectious Diseases division of the National Institutes of Health under award RO1AI075118 to M.K.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Author Contributions M.D. together with K.V. performed and analysed the experiments related to the intestine and S.K. performed and analysed the experiments related to the skin. N.H. and M.K. designed and generated the targeting constructs for the Ripk1fl/fl and Ripk1D138N/D138N mice. A.P. performed the gene targeting in embryonic stem cells and generated the Ripk1fl/fl, Ripk1D138N/D138N and Triffl/fl mice. M.Z. contributed to the analysis of intestines from Ripk1−/− neonates. C.K. and J.L. performed biochemical analysis of RIPK1-deficient MEFs, IECs and keratinocytes. C.E. performed FACS analysis of intestinal immune cells. T.C. performed qRT–PCR analysis. L.W. designed and tested the short guiding RNAs for CRISPR/Cas9-mediated targeting of Mlkl. P.K., M.B. and A.B. generated germ-free RIPK1IEC-KO mice. M.P. coordinated the project and together with K.V., M.D. and S.K. wrote the paper.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Oberst A, et al. Catalytic activity of the caspase-8–FLIPL complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiser WJ, et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, et al. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet MC, et al. The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity. 2011;35:572–582. doi: 10.1016/j.immuni.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Welz PS, et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 6.Dillon CP, et al. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep. 2012;1:401–407. doi: 10.1016/j.celrep.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duprez L, et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35:908–918. doi: 10.1016/j.immuni.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Upton JW, Kaiser WJ, Mocarski ES. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe. 2010;7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelliher MA, et al. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 10.Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J. Biol. Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 11.Meylan E, et al. RIP1 is an essential mediator of Toll-like receptor 3-inducedNF-κB activation. Nature Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 13.Rajput A, et al. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity. 2011;34:340–351. doi: 10.1016/j.immuni.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu. Rev. Physiol. 2014;76:129–150. doi: 10.1146/annurev-physiol-021113-170259. [DOI] [PubMed] [Google Scholar]
- 15.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nature Rev. Mol. Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 16.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu. Rev. Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee TH, Shank J, Cusson N, Kelliher MA. The kinase activity of Rip1 is not required for tumor necrosis factor-α-induced IκB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J. Biol. Chem. 2004;279:33185–33191. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]
- 19.Greten FR, et al. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Nenci A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 21.Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J. Immunol. 2008;180:2588–2599. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki Y, et al. Canonical NF-κB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Vlantis K, et al. Constitutive IKK2 activation in intestinal epithelial cells induces intestinal tumors in mice. J. Clin. Invest. 2011;121:2781–2793. doi: 10.1172/JCI45349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentle IE, et al. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-κB and activation of caspase-8. J. Biol. Chem. 2011;286:13282–13291. doi: 10.1074/jbc.M110.216226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JY, et al. TNFα induced noncanonical NF-κB activation is attenuated by RIP1 through stabilization of TRAF2. J. Cell Sci. 2011;124:647–656. doi: 10.1242/jcs.075770. [DOI] [PubMed] [Google Scholar]
- 26.Piao JH, et al. Tumor necrosis factor receptor-associated factor (TRAF) 2 controls homeostasis of the colon to prevent spontaneous development of murine inflammatory bowel disease. J. Biol. Chem. 2011;286:17879–17888. doi: 10.1074/jbc.M111.221853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 28.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Polykratis A, et al. Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 2014 doi: 10.4049/jimmunol.1400590. http://dx.doi.org/10.4049/jimmunol.1400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mc Guire C, et al. Oligodendrocyte-specific FADD deletion protects mice from autoimmune-mediated demyelination. J. Immunol. 2010;185:7646–7653. doi: 10.4049/jimmunol.1000930. [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 32.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 34.El Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 35.Hafner M, et al. Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis. 2004;38:176–181. doi: 10.1002/gene.20016. [DOI] [PubMed] [Google Scholar]
- 36.Pfeffer K, et al. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 37.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 38.Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-κBs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell. Biol. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ukena SN, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE. 2007;2:e1308. doi: 10.1371/journal.pone.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt-Supprian M, et al. NEMO/IKKγ-deficient mice model incontinentia pigmenti. Mol. Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- 42.Kumari S, et al. Tumor necrosis factor receptor signaling in keratinocytes triggers interleukin-24-dependent psoriasis-like skin inflammation in mice. Immunity. 2013;39:899–911. doi: 10.1016/j.immuni.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Klose CS, et al. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 44.Adolph TE, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snippert HJ, Schepers AG, Delconte G, Siersema PD, Clevers H. Slide preparation for single-cell-resolution imaging of fluorescent proteins in their three-dimensional near-native environment. Nature Protocols. 2011;6:1221–1228. doi: 10.1038/nprot.2011.365. [DOI] [PubMed] [Google Scholar]
- 46.Tscharntke M, et al. Impaired epidermal wound healing in vivo upon inhibition or deletion of Rac1. J. Cell Sci. 2007;120:1480–1490. doi: 10.1242/jcs.03426. [DOI] [PubMed] [Google Scholar]
- 47.Sato T, et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]