Abstract
We describe recent progress towards defining neuronal cell types in the mouse retina, and attempt to extract lessons that may be generally useful in the mammalian brain. Achieving a comprehensive catalog of retinal cell types now appears within reach, because researchers have achieved consensus concerning two fundamental challenges. The first is accuracy—defining pure cell types rather than settling for neuronal classes that are mixtures of types. The second is completeness—developing methods guaranteed to eventually identify all cell types, as well as criteria for determining when all types have been found. Case studies illustrate how these two challenges are handled by combining state-of-the-art molecular, anatomical and physiological techniques. Progress is also being made in observing and modeling connectivity between cell types. Scaling up to larger brain regions, such as the cortex, will require not only technical advances but careful consideration of the challenges of accuracy and completeness.
When President Obama announced his BRAIN Initiative, the NIH enlisted a “dream team” of prominent neuroscientists to formulate a plan. In its final report, these advisors proclaimed that “It is within reach to characterize all cell types in the nervous system,” and named this as the number one goal out of seven for the BRAIN Initiative (BRAIN Initiative Working Group, 2014).
In this perspective piece, we describe methods currently being used to identify cell types, and discuss the prospects of extending them to catalog all cell types in the nervous system. We will restrict ourselves to neuronal cell types, though non-neuronal cell types are important too. The term “cell type” will refer to classification at the finest granularity, analogous to “species” in biological taxonomy (Masland, 2004), and accordingly the term “subtype” will be avoided. Ideally, one would also define higher ranks for neuronal taxonomy, analogous to “genus” and so on. In the absence of accepted terminology, we will use “class” to refer to any level of the hierarchy above cell type (Masland, 2004).
Our exposition focuses on the example of the retina, a region of the mammalian central nervous system in which cell types have been intensively investigated for well over a century (Cajal, 1893). We will discuss only the mouse retina, which has emerged as an important model system due to the power of mouse genetics. While mice have low visual acuity, they exhibit interesting visually guided behaviors, and visual regions of the mouse brain are being explored by many researchers (Huberman & Niell, 2011). The advances to be reviewed here build on previous work with rabbits, cats, monkeys, and nonmammalian species, but space does not permit inclusion of this previous literature.
Progress has been hindered not only by technical limitations, but also by two fundamental difficulties of methodology. The first is accuracy: how do we know when we have found a true cell type? The second is completeness: how can we identify all cell types, and how will we know when we are done? As will be explained below, provisional answers to these two questions have emerged for the retina, so finishing the catalog of retinal cell types does truly seem within reach. We will conclude this piece by speculating about whether and how impending success in the retina will generalize to the brain.
The retina is composed of three layers of cell bodies and two layers of neurites (Figure 1). You could imagine it as a club sandwich with somata as bread and neurites as meat. The three bread layers are the outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL). The two meat layers are the outer and inner plexiform layers (OPL and IPL). The retina contains five classes of neurons: photoreceptor, horizontal, bipolar, amacrine, and ganglion cells. Photoreceptor and horizontal cells are divided into just a handful of types, and will not be discussed here. The challenges of defining retinal cell types and determining their connectivity mainly involve the bipolar, amacrine, and ganglion cells, which synapse with each other in the IPL (Masland, 2012).
Figure 1. The retina is composed of three nuclear layers, containing cell bodies, and two plexiform layers, containing neurites.
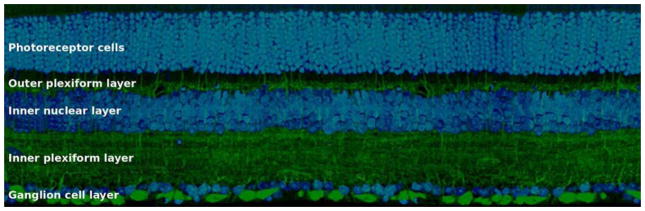
The inner and outer plexiform layers are sandwiched between the three layers of somata; the outer nuclear layer, the inner nuclear layer, and the ganglion cell layer. The nuclear layers contain the somata of the five classes of neurons of the retina: photoreceptor, horizontal, bipolar, amacrine, and ganglion cells. Figure adapted from Masland (2012).
The intricate structure of the IPL depends on depth, which is measured along the axis perpendicular to the retina. IPL depths 0 and 1 are conventionally placed at the IPL borders adjacent to the INL and GCL, respectively. The IPL depth is divided at roughly the halfway mark into Off and On zones that are adjacent to the INL and GCL respectively. The IPL depth is more finely divided into five (Cajal, 1893) or ten (Roska & Werblin, 2001) “strata.” A recent study has shown that the precision of IPL structure is even finer than these conventional divisions (Sümbül et al., 2014).
1 What is a cell type?
Roughly speaking, a cell type is defined as a population of cells with similar molecular, anatomical, and physiological properties. These three kinds of definition are illustrated by the starburst amacrine cell (SAC), a class of retinal neuron that comes in On and Off types (Figure 2):
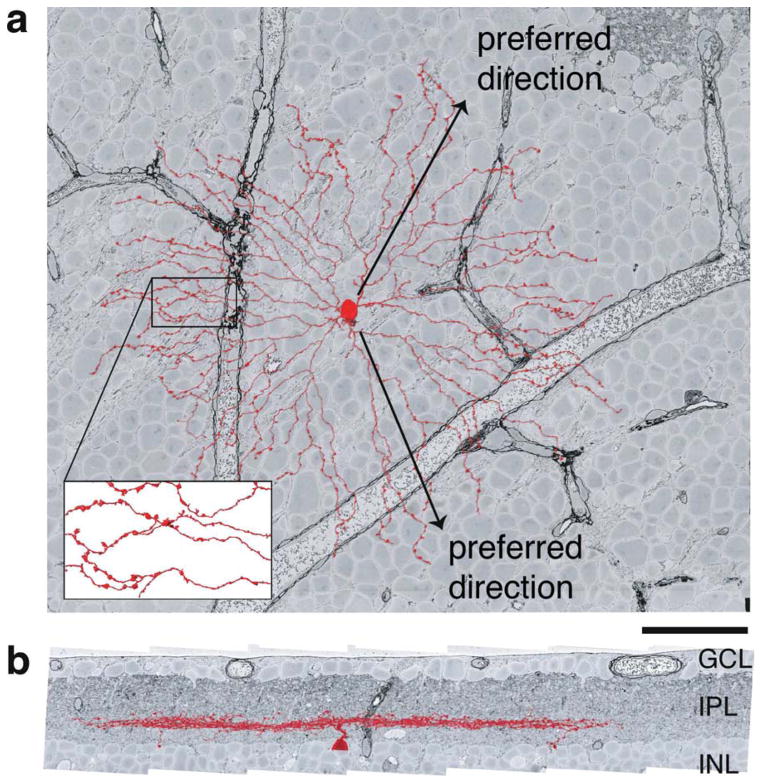
Figure 2. SAC dendrites extend roughly radially about the soma and are activated by outward motion.
a, Projection of an Off SAC onto the plane tangential to the retina shows the “starburst” arrangement of its dendrites. The preferred directions of individual dendrites are radially outward from the soma. Swellings of distal dendrites are presynaptic boutons (inset), so that SAC dendrites are output elements as well as input elements. b, Projection onto a plane perpendicular to the retina shows the thin stratification of the Off SAC at a characteristic IPL depth, between the INL and GCL. The neuron was reconstructed from a volume imaged by serial EM (Briggman et al., 2011). Scale bar, 50μm. Figure adapted from Kim et al. (2014).
The SAC is molecularly defined as the only cholinergic neuron in the mammalian retina (Masland & Tauchi, 1986). On SACs express semaphorin 6A (Sema6A), while Off SACs do not (Sun et al., 2013).
Anatomists define SACs as neurons that are almost planar, with dendrites extending roughly radially and symmetrically about the soma (Famiglietti, 1983). Off and On types of SACs arborize in strata located at roughly 1/3 and 2/3 of the IPL depth, respectively.
On SACs are activated when light turns on, while Off SACs are activated when light turns off. Physiologists have discovered that a SAC dendrite is more activated by outward motion than inward motion. Here “outward” refers to visual stimuli that move from the soma towards the tip of the dendrite (Euler et al., 2002; Hausselt et al., 2007).
The physiological definition is interesting because it is closely related to the function of SACs, suggesting that SACs are involved in the visual detection of motion. Indeed, ablation of SACs results in loss of the optokinetic reflex (Yoshida et al., 2001). However, it is difficult to arrive at the physiological definition without means of visualizing, recognizing, and manipulating SACs. This is where the molecular and anatomical definitions come in.
The molecular definition of SACs is useful for visualization by antibody staining. SACs are selectively stained by antibodies against choline acetyltransferase (ChAT), the enzyme that synthesizes acetylcholine, or against vesicular acetylcholine transporter (VAChT). The molecular definition is also useful for genetic manipulations, for example, through transgenic mouse lines that express Cre in ChAT-positive cells (Yonehara et al., 2011; Duan et al., 2014). The ablation of SACs mentioned above was accomplished by genetic targeting of SACs based on their expression of the mGluR2 receptor, another molecule characteristic of SACs (Yoshida et al., 2001). In principle, a cell type is defined by its entire transcriptomic or proteomic state. In practice, just a few molecules are expected to be sufficient to distinguish any given cell type from other cell types in its vicinity. For example, as explained above, acetylcholine and sema6A appear to be sufficient to distinguish On and Off SACs from other retinal cell types.
The anatomical definition of SACs was used to recognize them after intracellular dye fills, so that they could be targeted for the physiological experiments that revealed direction selectivity of SAC dendrites (Euler et al., 2002; Hausselt et al., 2007).
In our opinion, it is important to cross-validate these three definitions. We can be most sure of the validity of a cell type if it has three independent definitions based on molecular properties only, anatomical properties only, and physiological properties only, and these three definitions agree with each other. This ideal situation has been partially achieved for SACs. A SAC can currently be identified by purely molecular criteria, or purely anatomical criteria. The physiological property of direction selectivity is not specific enough to stand on its own as an independent definition of SACs, but one can imagine that further research could yield a larger set of physiological properties that fully defines SACs.
Why do we expect that these definitions should agree for a genuine cell type? If a cell type serves a function, then we expect evolution to adapt the properties of the cell type to serve the function. It was already noted that the physiological property of direction selectivity appears to serve visual behaviors like the optokinetic reflex. Likewise, the anatomical properties of SACs support their role in visual function. To understand this point, it is helpful to consider the relation of SACs with another class of retinal neuron, On-Off direction selective ganglion cells (ooDSGCs). Each ooDSGC is bistratified, meaning that its dendrites stratify at two IPL depths. These depths correspond exactly to those of SACs, so that ooDSGCs co-stratify with On and Off SACs (Famiglietti, 1992). Co-stratification means that contact is possible, and contact is a necessary (though not sufficient) condition for synaptic coupling. Indeed, it turns out that SACs provide synaptic input to ooDSGCs (Fried et al., 2002), and the direction selectivity of ooDSGCs is thought to be inherited from their SAC inputs (Fried et al., 2002; Briggman et al., 2011). More generally, stratification depth is an important constraint on the connectivity of retinal circuits, which in turn is an important determinant of visual function (Masland, 2004). Therefore it makes sense for stratification depth to be crucial for the anatomical definition of not only SACs but virtually every type of retinal neuron.
The anatomical properties of a neuron not only influence its function by constraining its connectivity to inputs and outputs, but also by shaping the single-neuron biophysics underlying input-output relations. Additional distinctive anatomical properties not mentioned above—the existence of both input and output synapses on SAC dendrites, the lack of an axon, and the small diameter of SAC dendrites (suggesting weak electrical coupling)—were used as the basis for speculation that SAC dendrites function independently (Miller & Bloomfield, 1983), as was later confirmed by two-photon calcium imaging (Euler et al., 2002; Hausselt et al., 2007).
Molecules expressed in SACs support their anatomical properties by participating in developmental processes. MEGF10 is important for the regular spacing of SAC cell bodies across the retina (Kay et al., 2012). Protocadherins are important for SAC self-avoidance, the apparent repulsion between the dendrites of a single SAC that is important for its characteristic “starburst” shape (Lefebvre et al., 2012). Sema6A, mentioned above, is important for SAC stratification at 1/3 and 2/3 of the IPL depth (Sun et al., 2013)
SAC molecules also support the intrinsic and synaptic physiology of these cells. While this is the case, the link to function is often nonobvious. Cholinergic transmission in SACs is thought to exert excitatory influences on other cells, but its role in SAC function has not been explained. Most accounts of direction selectivity instead emphasize inhibitory GABAergic transmission by SACs. The mGluR2 receptor is expressed in SACs, a fact used for the genetically targeted ablation mentioned earlier (Yoshida et al., 2001). Pharmacological manipulation of mGluR2 receptors appears to have an effect on direction selectivity, but the mechanism and relevance for function are not altogether clear (Jensen, 2006).
The physiological definition of SACs given above is based on responses to visual stimuli. There is also considerable research on the intrinsic and synaptic physiology of retinal neurons, and such properties could potentially be distinctive enough to be used in cell type definitions. This approach is popular in the physiological definition of cortical cell types (Markram et al., 2004; Petilla Interneuron Nomenclature Group et al., 2008).
2 Why are cell types important?
Identifying cell types might be denigrated as a tedious exercise in “descriptive” neuroscience, akin to stamp collecting. Indeed, many publications on this subject have been relegated to “archival” journals with low impact factors. Why then have cell types become so prominent a topic, as evidenced by the priorities of BRAIN Initiative? One reason is the recent revolution in applying genetic methods to systems neuroscience. Identifying a cell type is now a prelude to manipulating it, enabling experiments that rely on visualizing neurons with fluorescent reporters or controlling their activity with optogenetics.
A second reason is conceptual rather than technological. Without knowledge of cell types, one cannot even frame many important questions of neuroscience. Ganglion cells are the outputs of the retina, the only class of neuron that extends axons from the eye to the brain. According to textbook accounts, a ganglion cell is the output of a computation that is well approximated by a linear filter with center-surround structure. If this simple picture were accurate, it would be hard to understand why ganglion cells come in at least 20 types. It seems more plausible that each ganglion cell type is the output of a distinct visual computation performed by the retina (Gollisch & Meister, 2010). Once a cell type is defined, then retinal physiologists can set to work characterizing the visual computation that it serves. Before the cell type is defined, it would be difficult to even formulate the question. Similarly, SACs must be anatomically defined before developmental neuroscientists can find molecules that are important for establishing the starburst shape (Lefebvre et al., 2012) and stratification depth (Sun et al., 2013).
Third, neurodegenerative disorders may not affect all cell types uniformly. For example, glaucoma researchers have long known that ganglion cells degenerate in the disease, while other classes of retinal neurons remain unaffected. A recent study on a mouse model of the disease suggests that individual ganglion cell types may be affected differentially (Della Santina et al., 2013), while a previous study of a different mouse model failed to detect differences across types (Jakobs et al., 2005).
Fourth, future therapies for diseases may depend on genetic targeting of specific cell types. For example, optogenetic control of On bipolar cells (Lagali et al., 2008) has been explored as a means of restoring visual function in a mouse model of retinal degeneration.
3 Bipolar cell types
Bipolar cell (BC) bodies are located in the inner nuclear layer (INL). Their dendrites extend into the outer plexiform layer (OPL), and axons into the inner plexiform layer (IPL). BCs are the only conduit from the OPL to the IPL, and therefore the only way for signals to travel from photoreceptors to ganglion and amacrine cells. BCs are functionally classified into On and Off classes, which are activated by light and dark stimuli, respectively. While BCs have been studied in many species, much recent work has focused on the mouse due to the availability of genetic methods. The classification of mouse BCs seems very close to complete, with about a dozen types (Euler et al., 2014). There is a single rod BC type, which receives its input from rod photoreceptors. Cone BC types are more numerous.
Ghosh et al. (2004) defined nine cone BC types in the mouse retina based on anatomical criteria. They filled BCs by microinjection, imaged them with light microscopy, and examined the IPL depth at which the axons stratified. Roughly speaking, Types 1 through 9 stratify progressively deeper in the IPL (Figure 1).
Wässle et al. (2009) defined 11 cone BC types by the presence and absence of certain molecules, as determined by a combination of antibody staining and transgenic mouse lines. For validation, cells were imaged using light microscopy to reveal their anatomical properties. This molecular classification mostly matched the anatomical classification of Ghosh et al. (2004). However, two of the anatomical types were further subdivided, as explained below.
Ghosh et al. (2004) lumped Types 3a and 3b together in Type 3 because they stratify in the same IPL sublayer. Wässle et al. (2009) distinguished between Types 3a and 3b using antibodies against HCN4 and PKARIIb, following the lead of Mataruga et al. (2007). Wässle et al. (2009) also argued that Type 5 should be divided into Types 5a and 5b, but provided no molecular criteria by which the distinction could be made. Then they declared victory, saying that “our proposed catalog of 11 cone bipolar cells and one rod bipolar cell is complete, and all major bipolar cell types of the mouse retina appear to have been discovered.”
This brings up an important methodological question. When classifying cell types, how do we know when homogeneity is achieved? For example, how do we know that Types 3a and 3b are the final answer? Perhaps they are also mixtures of types, and should be further subdivided. It has become accepted that the axons of a BC type cover the area of the retina with little overlap, a property known as “tiling” (Figure 3). Wässle et al. (2009) showed that the axons of Type 3 BCs overlap heavily, violating the tiling property. They also showed that Type 3a axons tile the retina, and the same is true for Type 3b. Therefore they concluded that Types 3a and 3b are pure types rather than mixtures, and should not be subdivided further. Similarly, Wässle et al. (2009) showed that Type 5 cells overlap excessively, and on this basis proposed that they should be divided into Types 5a and 5b.
Figure 3. Bipolar cells of the same type tile the retina with little overlap, while ganglion cells of the same type form an overlapping mosaic.
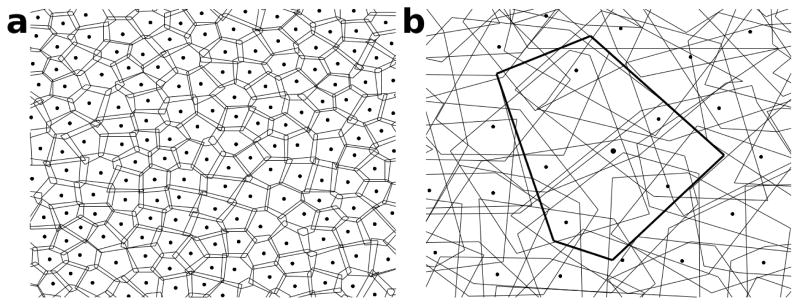
Dots represent somata, and the polygons around them represent the hulls of the arbors in an axial view. Somata of cells of the same type are arranged quasi-periodically. a, Cartoon of bipolar cells of a given type covering the area of the retina with little overlap. b, Ganglion cells of a given type cover the retina with substantial overlap, while their somata are arranged as if they repelled each other. The hull and the soma location of one ganglion cell are highlighted.
Ghosh et al. (2004) stained BCs by microinjection, while Wässle et al. (2009) used antibody staining and transgenic mouse lines. These techniques could yield a biased sample of BCs, causing some types to be missed. To test for this possibility, Wässle et al. (2009) computed the density of each BC type, i.e., cell bodies per square millimeter. When they added the densities together, they found good agreement with the total density of all BC types (Jeon et al., 1998), and concluded that they had identified all “major bipolar cell types.” In other words, any hypothetical missing type would have to be rare.
To summarize, declaring victory in cell type classification involves two claims, one of accuracy and the other of completeness. Accuracy means that cell types are pure rather than mixtures, and pure types are not further split into multiple clusters. Completeness means that no types are missing. For BCs, Wässle et al. (2009) verified accuracy using the tiling property, and completeness by counting cells.
More recently, Helmstaedter et al. (2013) used serial electron microscopy (EM) to reconstruct all BCs in a (0.1 mm)2 patch of mouse retina, and classified the cells into types based purely on anatomical criteria. They mostly reproduced the molecular classification of Wässle et al. (2009), with a few differences (Figure 4). A new BC type called XBC was defined, and Type 5 was subdivided into two groups, of which one, 5a, was a pure type. The other group most likely comprised 2 more types. So the number of cone BC types increased yet again, in spite of prior claims of accuracy and completeness. Kim et al. (2014) mostly reproduced the Off BC classification of Helmstaedter et al. (2013) using a different serial EM dataset, except that Types 1 and 2 were transposed.
Figure 4. Anatomical classification of bipolar cells reconstructed via serial EM.
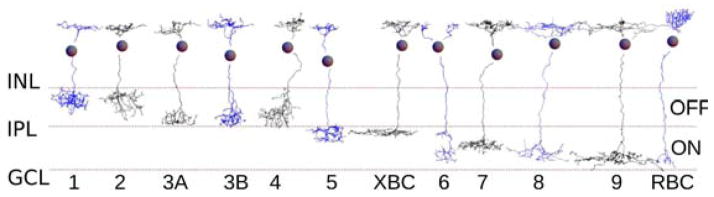
Bipolar cells were anatomically classified based mainly on stratification depth (Helmstaedter et al., 2013). The classification mostly agrees with a previous molecular classification (Wässle et al., 2009), except that a new type called XBC was defined, and Type 5 appears to be a mixture of more than two types (not shown). Figure adapted from Helmstaedter et al. (2013) by transposing types 1 and 2, following the classification in Kim et al. (2014).
Some general lessons about classification can be drawn. First, if serial EM can be used to reconstruct all cells, it has some advantage in achieving completeness compared to light microscopy (LM), which relies on sampling of cells. Second, if LM is used to combine molecular and anatomical information, it can be superior to EM for accuracy. Molecular reporters exist for EM, but are far more limited at the present time. Third, the molecular definition of a cell type should be validated by anatomical data. Fourth, the power of the anatomical approach is enhanced by the application of quantitative methods. For example, Helmstaedter et al. (2013) were able to distinguish between Types 3a and 3b using purely anatomical criteria, by utilizing arbor width as well as IPL depth in their classification scheme.
Efforts are also being made to define mouse BC types using physiological criteria. Baden et al. (2013) imaged calcium signals while illuminating with a full-field square-wave. They applied an automated clustering algorithm to separate the responses of 53 BCs into 8 clusters. The use of calcium imaging was important because it also enabled the visualization of axonal arbors, and hence the cross-validation of physiology with anatomy. The physiological clusters had some correspondence to the anatomical types, though there were some discrepancies. This approach seems promising, but it may be important to widen the ensemble of visual stimuli. For example, it may be helpful to add chromatic stimuli, which have already been used to make some distinctions between BC types (Breuninger et al., 2011).
4 Ganglion cell types
Ganglion cells are the only retinal neurons that project to the brain through the optic nerve. Each type of ganglion cell is believed to be the output of a distinct visual computation by the retina (Gollisch & Meister, 2010). Characterizing the visual responses of each type of ganglion cell would give us a complete specification of what the eye tells the brain. This goal is obviously important for visual neuroscience, but depends on our ability to classify ganglion cells into types.
Many efforts at anatomical classification have been made (Sun et al., 2002; Badea & Nathans, 2004; Kong et al., 2005; Coombs et al., 2006; Völgyi et al., 2009). These started out with LM imaging of neurons sparsely labeled by microinjection, transgenic mouse lines, or biolistics. The dendritic arbors of the cells were reconstructed by manually tracing their dendrites through the images. Finally, the reconstructions were classified into cell types. These studies yielded 12 to 22 ganglion cell types, showing that a purely anatomical approach had difficulty achieving consensus.
The genetic approach has shown promise for resolving these disagreements, as illustrated by recent progress in defining types of On-Off direction selective ganglion cells (ooDSGCs). Physiologists originally divided this neuron class into four types with anterior, posterior, superior, and inferior preferred directions (Oyster & Barlow, 1967). For a long time, it was not possible to distinguish between these types using molecular or anatomical properties. Huberman et al. (2009) showed that DRD4-expressing ganglion cells constitute an ooDSGC type that prefers posterior motion, and Rivlin-Etzion et al. (2011) showed that TRHR-expressing ganglion cells also prefer posterior motion. The BD transgenic line defines another ooDSGC type that prefers superior motion (Kim et al., 2010; Kay et al., 2011), and the same type was subsequently defined by expression of Hb9 (Trenholm et al., 2011, 2013a,b).
Other successes at defining ganglion cell types based on expression of a single gene include CB2 (Huberman et al., 2008) and JAM-B (Kim et al., 2008). The latter case was especially interesting because it revealed a cell type that had not been previously defined by anatomical or physiological means. Other ganglion cell types have been defined using transgenes containing Thy1 regulatory elements, and are not necessarily definable by the expression of an endogenous gene. For example, the W3 type was defined as the brightly labeled cells in a Thy1 mouse line (Kim et al., 2010; Zhang et al., 2012), and the BD type was already mentioned above.
Why are the above examples regarded as pure cell types, rather than mixtures? To rule out the latter possibility, it would be helpful to have a criterion like the tiling property, which was used for defining pure bipolar cell types. The dendritic arbors of a pure ganglion cell type typically do not tile, but rather overlap heavily. (See Borghuis et al., 2008 for an information theoretic explanation of how this overlap might contribute to visual function.) Fortunately, the tiling property can be generalized to the “mosaic property” (Figure 3). Namely, the somas of a pure cell type are assumed to be arranged quasiperiodically, something like the atoms of a crystal. In such an arrangement, somas should behave as if they repel each other. This is quantified by computing the autocorrelogram of the soma positions. For a pure cell type, the autocorrelogram should vanish as the separation approaches zero. On the other hand, the somas of different cell types appear not to repel each other, but rather are statistically independent (Rockhill et al., 2000). Therefore, for a mixture of cell types, the autocorrelogram should approach a non-zero value at zero separation. (It should be noted that “mosaic” is a misleading name, because it sounds like “tiling.” As noted above, many cell types satisfy the mosaic property but are non-tiling.) Five of the examples above (DRD4, TRHR, CB2, JAM-B, and bright W3) have been verified as pure cell types using the autocorrelogram.
In addition to these pure cell types, many other ganglion cell classes have been genetically defined. For example, PV-expressing ganglion cells come in at least eight types (Münch et al., 2009; Farrow et al., 2013). Although the PV class is a mixture of types, it has been useful, as specific types have been targeted for physiological studies using anatomical criteria.
All of these developments demonstrate that the genetic and anatomical approaches are complementary. If a pure cell type is genetically defined, it should be validated by anatomical criteria, and vice versa. Genetically defining a heterogeneous class of neurons makes it easier to distinguish the pure cell types in the class via anatomical criteria. Because genetics is facilitating fine distinctions between cell types that are anatomically similar, it has become important to modernize anatomy by making it more quantitative using modern computational techniques. This is illustrated by Sümbül et al. (2014), who undertook a more systematic attempt at cross-validation through anatomical classification of ganglion cells imaged from a number of transgenic lines. Two aspects of their computational approach are worth noting here: arbor density and alignment.
The arbor density is an answer to an old methodological question: on what anatomical features should distinctions between cell types be based? The arbor density is a spatially blurred version of the arbor (Figure 5). Blurring preserves the overall shape of the arbor, but effectively discards detailed information about individual dendrites, including many classical anatomical properties such as tortuosity and branch angle. The arbor density was previously used to classify invertebrate neurons (Jefferis et al., 2007). It is motivated by theoretical work on the idea that overlap between arbor densities is a good estimate of contact between neurons (Kalisman et al., 2003; Stepanyants & Chklovskii, 2005). Since contact means potential for synaptic coupling, overlap between arbor densities is said to be proportional to the number of “potential synapses.” In other words, the arbor density of a neuron is a predictor of its connectivity with other neurons.
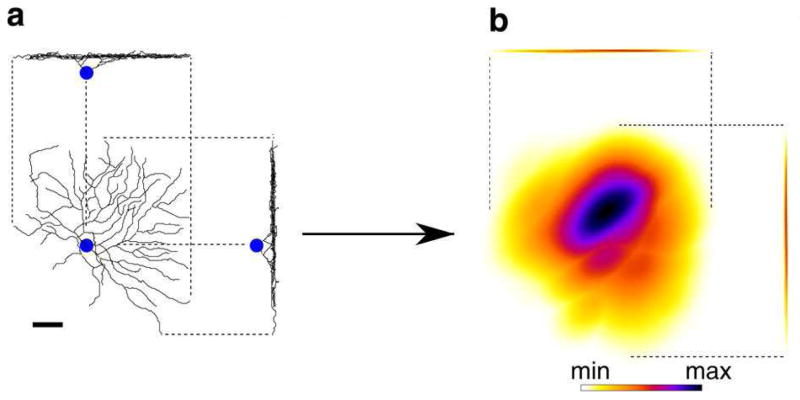
Figure 5. The arbor density is obtained by blurring the neuronal arbor.
RGC arbor reconstructed from LM image (a), and RGC arbor density produced by blurring (b), shown as projections onto the plane tangential to the retina (large), and projections onto two orthogonal planes perpendicular to the retina. The blurring is anisotropic, effective only in the tangential plane. No blurring is applied along the axis perpendicular to the retina, to preserve IPL depth information. The overall shape of the arbor and its IPL depth are preserved, but detailed information about individual dendrites is effectively discarded. Scale bar, 40μm. Figure adapted from Sümbül et al. (2014).
We prefer the term “anatomy” over the more commonly used “morphology,” because the latter is inappropriate for classifying cell types. Morphology refers to shape, which is mathematically defined as those geometric properties that are independent of the location, orientation, and size of an object. In other words, the shape of an object has to do with the relations of its parts with each other, rather than the relations between different objects.
Knowing the shape of a neuronal arbor is not enough for classifying it. The connectivity between neurons depends on the relations between their arbors, which are lost if information about location and orientation is discarded. To retain information about spatial relations, it is crucial to align all arbors to a common coordinate system.
Accordingly, Sümbül et al. (2014) paid careful attention to the problem of alignment. On and Off starburst amacrine cells were stained for ChAT and used as landmarks to define the depth coordinate, following previous work (Manookin et al., 2008; Siegert et al., 2009). Computational methods were used to flatten the two starburst amacrine cell layers in every image stack, which resulted in aligning every ganglion cell to a common depth coordinate (Figure 6). Because of this reliance on computational flattening, there was no effort made to physically flatten the retina by cover slip pressure before imaging.
Figure 6. Aligning ganglion cells to a common IPL depth coordinate reveals a remarkable reproducibility of stratification depth within a cell type.
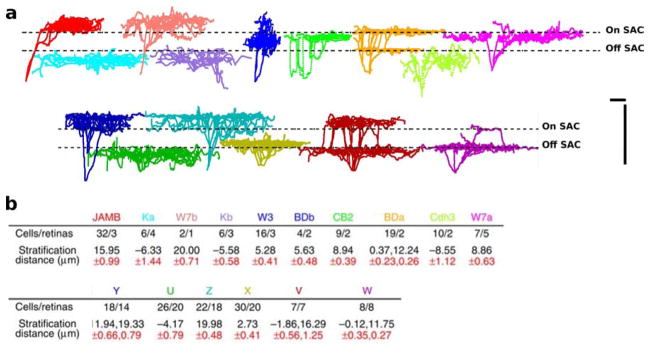
a, The arbor traces in the top row were obtained from transgenic lines in which a relatively homogeneous class of cells was labeled (Kim et al., 2008, 2010; Huberman et al., 2008; Osterhout et al., 2011). The ones in the bottom row were obtained by sampling from highly heterogeneous classes labeled in GFP-M, YFP-H and YFP-12 mice (Feng et al., 2000). The dashed lines indicate the positions of the On and Off SAC layers after alignment. b, Stratification properties of all 15 types. Colors of cell types match with the colors of the representative traces in a. Stratification distance refers to the distance of peak stratification plane to the On SAC layer. Scale bars, 40μm. Figure adapted from Sümbül et al. (2014).
The payoff of careful alignment became evident when examining the stratification depth of genetically defined types of neurons. The standard deviation of the depth across neurons of a given type from multiple retinas was typically less than a micron. This outstanding reproducibility enabled most ganglion cell types to be distinguished based only on the stratification profile, the projection of the arbor density onto the depth axis. However, in some cases cell types had very similar stratification profiles, so they could only be distinguished based on the full arbor density. In all, 15 types of ganglion cells were anatomically defined. This is encouraging, but is still not a complete classification.
Helmstaedter et al. (2013) identified 12 ganglion cell types using the same serial EM dataset that they used to classify bipolar cells. They demonstrated the potential of EM, but only the smallest types of ganglion cells could be fully reconstructed, due to the limited size of the retinal patch studied, about (0.1 mm)2. Therefore this classification is still provisional, but it at least placed a lower bound on the number of ganglion cell types.
Farrow & Masland (2011) reported a physiological clustering of ganglion cells based on their responses to four classes of visual stimuli. This study rejected the null hypothesis of a few physiological ganglion cell types by providing a 12-cluster partitioning, and set another lower bound on the number of ganglion cell types. The study used a multielectrode array to record visual responses, which did not allow cross-validation of physiology with anatomy. Future work would ideally use calcium imaging and rich stimulus ensembles to arrive at physiological definitions of ganglion cell types that correspond to the molecular and anatomical definitions.
5 Amacrine cell types
The project of dividing amacrine cells into types is farthest from completion, compared to bipolar and ganglion cells. Yet amacrine cells are crucial for understanding the complexity of IPL circuitry. Using the CreER technique for sparse labeling, Badea & Nathans (2004) distinguished between 23 types of amacrine cells in the mouse retina. Zhu et al. (2014) identified additional amacrine cell types by combining viral sparse labeling with Cre driver lines originally created for classes of cortical GABAergic neurons. In their serial EM study of the mouse retina, Helmstaedter et al. (2013) were able to distinguish between at least 40 amacrine cell types. Extension of this approach to a larger patch should yield a complete classification of narrow-field and medium-field cells in the near future. However, the arbors of wide-field amacrine cells extend for millimeters, and sometimes over virtually the entire retina (Lin & Masland, 2006). Identification of such cell types will have to depend on LM rather than EM in the foreseeable future. Larger retinal cells are generally less numerous, so it will be difficult to know when the catalog is complete.
6 Rules of connectivity
Cell types simplify the study of neural connectivity, because there are fewer cell types than cells. To understand this point, consider two neurons i and j of cell types ti and tj, and let Wij be some measure of the strength of connectivity from neuron j to neuron i. We expect Wij to fluctuate randomly about some mean value given by
| (1) |
The randomness is due to biological variation across the nervous systems of different individuals, and E denotes an average across individuals. The variability around the mean is expected to be small in some sense, if cell types are indeed a useful concept. If there are N cells and n cell types, Eq. (1) implies that the N × N matrix E[Wij] reduces to an n × n matrix W. If n ≪ N, this is a drastic reduction in the number of parameters, meaning that fewer observations are required to characterize connectivity.
The model of Eq. (1) is applicable to a small nervous system like that of C. elegans, in which each neuron is identified, and every normal individual has the same number of neurons. Since the neurons of any two worms can be placed in one-to-one correspondence, and averaging Wij over individuals is well-defined. White et al. (1986) divided the 302 neurons of C. elegans into 118 cell types. The introduction of cell types does help to reduce complexity, but not by much because cell types are almost as numerous as neurons.
Cell types are much more essential for grappling with the complexity of the large nervous systems of mammals. However, Eq. (1) should not be interpreted as an average over individuals, because neurons of different individuals cannot be placed in one-to-one correspondence. Instead, one could regard this as an average over neuron pairs drawn from the same cell types, and this average could be within a single individual.
Furthermore, to model the connectivity of certain sheet-like structures, like the retina and neocortex, the model should be modified so that each neuron has not only a cell type but also a 2D location in the sheet (Seung, 2009). The “location” of a neuron most commonly refers to its cell body, but might also refer to its dendritic or axonal arbor. If two neurons i and j are of cell types ti and tj and are located at ri and rj, Wij is expected to fluctuate randomly about some mean value given by
| (2) |
This depends on the locations of the cells only through their separation ri − rj. This dependence is equivalent to the assumption of translation invariance, which is a reasonable first approximation for a nonfoveal retina like that of the mouse.
Most commonly, the dependence on separation is treated in a qualitative, binary fashion. Nearby neurons obey rules like Eq. (1), while distant neurons are not connected at all because their arbors do not overlap. This sort of model is left implicit in much experimental work on connectivity, but can be explicitly formalized by adding a distance-based cutoff to Eq. (1),
| (3) |
where H is the Heaviside step function. Note that the threshold θtitj depends on the cell types, as the arbor of each cell type has a characteristic size. However, the more general dependence on separation given in Eq. (2) is likely to be important for function. For example, Kim et al. (2014) proposed that the connectivity between an Off SAC and a bipolar cell depends on the distance of the latter from the SAC soma. More specifically, one bipolar cell type prefers to connect close to the SAC soma, and the other prefers to connect further from the soma. This wiring specificity can be modeled by Eq. (2) but not Eq (3), and is potentially important for function as it could support the direction selectivity of SAC dendrites (Kim et al., 2014).
Fried et al. (2002) found physiological evidence that an ooDSGC preferentially receives input from SACs on its null side. This rule can be expressed in the form of Eq. (2), provided that the function Φ depends on the direction of the separation, not only its magnitude. Note that Φ depends on the preferred direction of the ooDSGC through its dependence on the type of the ooDSGC.
Briggman et al. (2011) found anatomical evidence that the connectivity of a SAC dendrite and an On-Off direction selective ganglion cell depends more on the direction of the SAC dendrite than on the location of the SAC soma. This connectivity rule cannot be expressed in the form of Eq. (2), demonstrating the need for models of connectivity that are more sophisticated. In particular, since the dendrites of a single SAC can function independently, connectivity between subcellular units rather than between neurons may have to be modeled.
7 Methods for observing connectivity
Many experimental methods have been used to extract rules of connectivity governing retinal neurons. The electrophysiological approach is to observe synaptic coupling with two intracellular electrodes, simultaneously stimulating one cell and recording responses from another, and anatomically identify the cell types with LM after dye-filling. This approach was used, for example, to investigate connectivity between SACs (Zheng et al., 2004). As cell types come under genetic control, it is becoming possible to extend this approach by combining optogenetic stimulation of one cell with intracellular recording from another (Yonehara et al., 2011; Duan et al., 2014).
Anatomical evidence for connectivity can be gained through LM and various techniques for marking neurons. Transsynaptic tracing with viruses has become popular recently. Beier et al. (2013) used a pseudotyped vesicular stomatitis virus to examine connectivity between SACs and ganglion cell types. Cruz-Martín et al. (2014) used a modified rabies virus to demonstrate the existence of a disynaptic pathway from direction selective retinal ganglion cells to the superficial layers of mouse V1. Alternatively, one can look for appositions of neurites and co-localization with synaptic markers (Schwartz et al., 2012). A putative synapse can be further verified by targeted EM (Bleckert et al., 2013), in a correlative LM-EM approach.
Serial EM has long been used to discover rules of retinal connectivity. In recent years, this technique was used to examine synaptic partners of the AII amacrine cell (Anderson et al., 2011), and of the SAC (Briggman et al., 2011; Kim et al., 2014).
It should be noted that synaptic coupling can be electrical rather than chemical. Electrical synapses have been investigated by methods similar to those mentioned above, as illustrated by the case of the AII amacrine cell (see review by Demb & Singer, 2012). Transsynaptic tracing has been accomplished by dye injections rather than viruses, and electrical synapses have also been visualized via high resolution images from serial EM.
All techniques above have generally been applied piecemeal, probing only a few cell types at a time. Serial EM offers the exciting prospect of reconstructing the connectivity between all neurons in a retinal sample, i.e., finding the retinal connectome. The feasibility of this approach was demonstrated by Helmstaedter et al. (2013), who reconstructed all neurons with cell bodies contained in a small patch of mouse retina. As mentioned earlier, this study lacked full reconstructions of large cells, because the patch was only about (0.1 mm)2 in area. Another deficiency was the use of an unconventional staining technique that marked extracellular space, but left intracellular organelles like vesicles and postsynaptic densities invisible. This made it impossible to positively identify synaptic connections. Instead, the researchers quantified contact between the roughly 1000 neurons in the study, resulting in a “contactome.” Although this work fell short of delivering the retinal connectome, it demonstrated that the goal is within reach.
One might question whether a connectome is a worthwhile goal. If retinal connectivity is governed by a relatively small number of rules, as in Eqs. (1)–(3), then piecemeal observations should be enough for discovering the rules. One answer is that the number of observations might still have to be large to average out biological variability, in which case high throughput techniques are desirable. Some criticize serial EM as slow, because reconstruction of neural circuits currently requires significant human labor for image analysis. Reconstruction speed is expected to increase owing to ongoing improvements in image quality and artificial intelligence. But it should be noted that even with the current throughput, serial EM is competitive with other techniques. For example, Kim et al. (2014) investigated contact between 195 BCs and 97 SACs, for a total of 15,405 BC-SAC pairs. For comparison, one could point to a physiological study like that of Lee & Zhou (2006), who performed dual intracellular recordings from 26 SAC-SAC pairs to study the dependence of SAC-SAC connectivity on distance.
More data about connectivity will be useful not only for averaging out biological variability, but also for enabling more sophisticated techniques for modeling connectivity, as will be explained in the following.
8 Cell types from connectivity
In the traditional approach, neuronal cell types are defined using information other than connectivity, such as molecular markers or the anatomy of single cells. Consequently, cell types and their rules of connectivity are extracted from distinct sources of information.
Alternatively, cell types can be defined using connectivity information, using the principle that cells of the same type have similar connectivity with other cells, as implied by the models of Eq. (1)–(3). This approach was previously used to define cell types using the C. elegans connectome (White et al., 1986). It may seem dangerously circular to both define cell types and discover their rules of connectivity using the same source of information. Inferring cell type from connectivity amounts to regarding cell type as a latent variable (Seung, 2009), rather than a directly observable variable. The field of latent variable modeling is well-established, and it should be possible to learn models like Eqs. (1)–(3), given sufficient amounts of connectivity data. The only danger of circularity lies in model selection, i.e., in deciding on the number of cell types.
In the traditional approach, the anatomy of single cells can be regarded as a proxy for information about connectivity (Masland, 2004). In line with this idea, Sümbül et al. (2014) used arbor densities to classify ganglion cells, because overlap between arbor densities predicts contact, or the potential for connectivity (Kalisman et al., 2003; Stepanyants & Chklovskii, 2005). If connectomic information becomes more plentiful than information about the anatomy of single neurons, then it will become advantageous to base the definitions of cell types mainly on the former (Seung, 2012).
For example, Helmstaedter et al. (2013) were unable to subdivide Type 5 BCs based on single cell anatomy, and Wässle et al. (2009) were unable to find molecular markers that could do this. Instead, Helmstaedter et al. (2013) distinguished Type 5a BCs from other Type 5 BCs based on the ratio of contact with two types of ganglion cells. They validated this anatomical criterion by showing that it was consistent with tiling of Type 5a dendritic arbors, and also predicted contact with a type of amacrine cell. Ideally, this approach would be extended to define all neuronal cell types in the retina. However, the approach is still data-limited, because Helmstaedter et al. (2013) used an unconventional stain that does not allow positive identification of connectivity, and because the reconstructed volume is still relatively small. Once these barriers are overcome, computational methods for learning latent variable models like Eqs. (1)–(3) will become useful (Jonas & Kording, 2014).
9 Discussion
What are the prospects for finally completing the catalog of neuronal cell types in the retina? As mentioned earlier, it has not yet been possible to define all types of On-Off direction selective ganglion cells based on expression of single genes. However, Kay et al. (2011) used a combination of transgenic and endogenous molecular markers to define four ooDSGC types corresponding to four preferred directions. Similarly, identifying all ganglion cell types may require definitions involving combinatorial expression of two or more genes, rather than a single gene. This may also be the case for amacrine cell types.
On the anatomical front, over the next few years serial EM should be able to deliver the retinal connectome, at least on the scale of a (0.5 mm)2 patch. This will require improvements in the quality of the serial EM images (Briggman & Bock, 2012), as well as in the accuracy of the artificial intelligence used (Jain et al., 2010). The retinal connectome will make it possible to define many cell types based on connectivity. However, LM will remain essential for reconstructing large cells, such as wide-field amacrine cells, in sufficiently large numbers. LM will also be important for comparing anatomical and molecular definitions of cell types, and the adoption of computational methods for image analysis will make LM-based anatomy more quantitative.
The physiological classification of ganglion cells and cross-validation with anatomical classification will be achieved by combining two-photon calcium imaging with structural imaging by LM and/or serial EM. This goal is technically more difficult for bipolar and amacrine cells, because the somata are often non-spiking and may not exhibit strong calcium signals.
Can lessons from classifying retinal cell types be applied to other regions of the mammalian brain? Many GABAergic neuron classes have been genetically defined in the cortex (Taniguchi et al., 2011), but these are generally mixtures of cell types. For example, the widely studied PV-expressing class of cortical neurons includes at least two anatomically distinct cell types, large basket cells and chandelier cells. These are known to make inhibitory synapses on the somata and axon initial segments of pyramidal neurons, respectively, and thus may have very different physiological effects (Kubota, 2014). Such heterogeneity may not matter for some experiments, because chandelier cells are less numerous than basket cells, but could be problematic under other circumstances. It seems safe to say that researchers of cortical cell types should focus on identifying accurate cell types, as retinal researchers have been doing for some time.
Skeptics say that lessons from the retina will not generalize to the cortex, because the retina is highly ordered, whereas the cortex looks more random (Braitenberg & Schüz, 1998). It is commonly believed that cortical neurons of the same type display more anatomical variability than retinal neurons. Actually, retinal neurons display a surprising degree of variability when projected onto the plane tangential to the retina. This is demonstrated for the mouse by Figure 5 and the Supplementary Information of Sümbül et al. (2014), though the variability is said to be much less for the rabbit.
The anatomic stereotypy of mouse retinal neurons only becomes obvious after precise registration and projection of arbors onto the axis perpendicular to the retina. The resulting stratification profile is outstandingly reproducible for neurons of the same type, to within submicron resolution (Sümbül et al., 2014). This is not only the case for thinly stratified cell types, but also for thickly stratified ones. It is possible that cortical neurons, which are thickly stratified, may also possess a comparable degree of precision. To reveal such precision, it will be important to register cortical neurons to a common coordinate system based on their arbors, as Sümbül et al. (2014) did for retinal neurons. Previous efforts have generally used the cell bodies for registration (Oberlaender et al., 2012), consistent with the traditional identification of the cell type of a pyramidal neuron based on the layer in which its cell body is situated.
In an 1857 letter, Charles Darwin wrote “Those who make many species are the ‘splitters,’ and those who make few are the ‘lumpers.”’ The same can be said for those who study cortical cell types. In the lumper camp, Braitenberg & Schüz (1998) recognize only pyramidal neurons and two types of non-pyramidal neurons in the cortex, for a total of just three cell types. Markram et al. (2004) divided non-pyramidal neurons in the cortex into over 50 types. Stevens (1998) speculated that eventually more than 500 cortical cell types would be identified.
The divergence between lumpers and splitters shows that one cannot succeed in cataloging cortical cell types without also solving the more fundamental problem of defining a cortical cell type. In the retina, the mosaic property plays a pivotal role in verifying the accuracy of a cell type. There is also evidence that cell types in the avian tectum and the cerebellum are organized in mosaics (Cook & Chalupa, 2000). An intriguing possibility is that the mosaic property will hold for cortical cell types. It is critical to test this idea, or to identify some other defining characteristic of an accurate cell type in the cortex.
Discreteness is central to the notion of cell types. However, there is increasing awareness of the interplay between discrete cell types and continuous variation across the retina. Mouse ganglion cells of the On alpha type exhibit a nasal-to-temporal gradient in cell density, and arbor size (Bleckert et al., 2014). Most JAM-B ganglion cells exhibit a remarkable asymmetry, with the dendritic arbor extending downward from the cell body. However, those cells at the dorsal and ventral margins of the retina have more symmetric arbors than the majority of JAM-B cells (Kim et al., 2008).
Are cell types more like the seven colors (ROYGBIV) of the rainbow, discrete categories used by convention to describe a physical variable (wavelength) that is actually continuous? Or are cell types more like the entries of the periodic table, which seem objectively discrete? These questions have to do with the concept of “natural kinds” introduced by analytic philosophers, and should be addressed by them. Our practical viewpoint is that cell types are variables in models like Eqs. (1)–(3), which are used to predict some properties of neurons given other properties. Ultimately, cell types are useful only insofar as they enable us to make accurate predictions.
Acknowledgments
We are grateful to Tom Baden, Bart Borghuis, Winfried Denk, Thomas Euler, Moritz Helmstaedter, Andy Huberman, Alex Kolodkin, Wei-Chung Lee, Dick Masland, Josh Sanes, and Yongling Zhu for comments and suggestions. H.S.S. is grateful for research support from the Army Research Office, DARPA, the Human Frontiers Science Program, and NIH/NINDS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JR, Jones BW, Watt CB, Shaw MV, Yang JH, Demill D, Lauritzen JS, Lin Y, Rapp KD, Mastronarde D, Koshevoy P, Grimm B, Tasdizen T, Whitaker R, Marc RE. Exploring the retinal connectome. Mol Vis. 2011;17:355–79. [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–51. doi: 10.1002/cne.20304. [DOI] [PubMed] [Google Scholar]
- Baden T, Berens P, Bethge M, Euler T. Spikes in mammalian bipolar cells support temporal layering of the inner retina. Curr Biol. 2013;23:48–52. doi: 10.1016/j.cub.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Beier KT, Borghuis BG, El-Danaf RN, Huberman AD, Demb JB, Cepko CL. Transsynaptic tracing with vesicular stomatitis virus reveals novel retinal circuitry. J Neurosci. 2013;33:35–51. doi: 10.1523/JNEUROSCI.0245-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckert A, Parker ED, Kang Y, Pancaroglu R, Soto F, Lewis R, Craig AM, Wong ROL. Spatial relationships between gabaergic and glutamatergic synapses on the dendrites of distinct types of mouse retinal ganglion cells across development. PLoS One. 2013;8:e69612. doi: 10.1371/journal.pone.0069612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckert A, Schwartz GW, Turner MH, Rieke F, Wong ROL. Visual space is represented by nonmatching topographies of distinct mouse retinal ganglion cell types. Curr Biol. 2014;24:310–5. doi: 10.1016/j.cub.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghuis BG, Ratliff CP, Smith RG, Sterling P, Balasubramanian V. Design of a neuronal array. J Neurosci. 2008;28:3178–89. doi: 10.1523/JNEUROSCI.5259-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAIN Initiative Working Group. Brain 2025: A scientific vision. Tech rep 2014 [Google Scholar]
- Braitenberg V, Schüz A. Cortex: statistics and geometry of neuronal connectivity. 2 Springer; Berlin: 1998. [Google Scholar]
- Breuninger T, Puller C, Haverkamp S, Euler T. Chromatic bipolar cell pathways in the mouse retina. J Neurosci. 2011;31:6504–17. doi: 10.1523/JNEUROSCI.0616-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggman KL, Bock DD. Volume electron microscopy for neuronal circuit reconstruction. Curr Opin Neurobiol. 2012;22:154–61. doi: 10.1016/j.conb.2011.10.022. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–8. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- Cajal SRy. La rétine des vertébrés. Cellule. 1893;9:17–257. [Google Scholar]
- Cook JE, Chalupa LM. Retinal mosaics: new insights into an old concept. Trends Neurosci. 2000;23:26–34. doi: 10.1016/s0166-2236(99)01487-3. [DOI] [PubMed] [Google Scholar]
- Coombs J, van der List D, Wang GY, Chalupa LM. Morphological properties of mouse retinal ganglion cells. Neuroscience. 2006;140:123–36. doi: 10.1016/j.neuroscience.2006.02.079. [DOI] [PubMed] [Google Scholar]
- Cruz-Martín A, El-Danaf RN, Osakada F, Sriram B, Dhande OS, Nguyen PL, Callaway EM, Ghosh A, Huberman AD. A dedicated circuit links direction-selective retinal ganglion cells to the primary visual cortex. Nature. 2014;507:358–61. doi: 10.1038/nature12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Santina L, Inman DM, Lupien CB, Horner PJ, Wong ROL. Differential progression of structural and functional alterations in distinct retinal ganglion cell types in a mouse model of glaucoma. J Neurosci. 2013;33:17444–57. doi: 10.1523/JNEUROSCI.5461-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb JB, Singer JH. Intrinsic properties and functional circuitry of the aii amacrine cell. Vis Neurosci. 2012;29:51–60. doi: 10.1017/S0952523811000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. Type ii cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014;158:793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- Euler T, Detwiler P, Denk W. Directionally selective calcium signals in dendrites of starburst amacrine cells. Nature. 2002;418:845–852. doi: 10.1038/nature00931. [DOI] [PubMed] [Google Scholar]
- Euler T, Haverkamp S, Schubert T, Baden T. Retinal bipolar cells: elementary building blocks of vision. Nat Rev Neurosci. 2014;15:507–19. doi: 10.1038/nrn3783. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV. Dendritic co-stratification of on and on-off directionally selective ganglion cells with starburst amacrine cells in rabbit retina. J Comp Neurol. 1992;324:322–35. doi: 10.1002/cne.903240303. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV., Jr ’starburst’ amacrine cells and cholinergic neurons: mirror-symmetric on and off amacrine cells of rabbit retina. Brain Res. 1983;261:138–44. doi: 10.1016/0006-8993(83)91293-3. [DOI] [PubMed] [Google Scholar]
- Farrow K, Masland RH. Physiological clustering of visual channels in the mouse retina. J Neurophysiol. 2011;105:1516–30. doi: 10.1152/jn.00331.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow K, Teixeira M, Szikra T, Viney TJ, Balint K, Yonehara K, Roska B. Ambient illumination toggles a neuronal circuit switch in the retina and visual perception at cone threshold. Neuron. 2013;78:325–38. doi: 10.1016/j.neuron.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor R, Bernstein M, Keller-Peck C, Nguyen Q, Wallace M, Nerbonne J, Lichtman J, Sanes J. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of gfp. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fried SI, Münch TA, Werblin FS. Mechanisms and circuitry underlying directional selectivity in the retina. Nature. 2002;420:411–4. doi: 10.1038/nature01179. [DOI] [PubMed] [Google Scholar]
- Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wässle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004;469:70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- Gollisch T, Meister M. Eye smarter than scientists believed: neural computations in circuits of the retina. Neuron. 2010;65:150–64. doi: 10.1016/j.neuron.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausselt SE, Euler T, Detwiler PB, Denk W. A dendrite-autonomous mechanism for direction selectivity in retinal starburst amacrine cells. PLoS Biol. 2007;5:e185. doi: 10.1371/journal.pbio.0050185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013;500:168–174. doi: 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- Huberman A, Manu M, Koch S, Susman M, Lutz A, Ullian E, Baccus S, Barres B. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59:425–438. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman A, Wei W, Elstrott J, Stafford B, Feller M, Barres B. Genetic identification of an on-off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Niell CM. What can mice tell us about how vision works? Trends Neurosci. 2011;34:464–73. doi: 10.1016/j.tins.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Seung H, Turaga S. Machines that learn to segment images: a crucial technology for connectomics. Current Opinion in Neurobiology. 2010;20:1–14. doi: 10.1016/j.conb.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs TC, Libby RT, Ben Y, John SWM, Masland RH. Retinal ganglion cell degeneration is topological but not cell type specific in dba/2j mice. J Cell Biol. 2005;171:313–25. doi: 10.1083/jcb.200506099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GSXE, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RJ. Activation of group ii metabotropic glutamate receptors reduces directional selectivity in retinal ganglion cells. Brain Res. 2006;1122:86–92. doi: 10.1016/j.brainres.2006.08.119. [DOI] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–46. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas E, Kording K. Automatic discovery of cell types and microcircuitry from neural connectomics. 2014 doi: 10.7554/eLife.04250. arXiv preprint arXiv:1407.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisman N, Silberberg G, Markram H. Deriving physical connectivity from neuronal morphology. Biol Cybern. 2003;88:210–8. doi: 10.1007/s00422-002-0377-3. [DOI] [PubMed] [Google Scholar]
- Kay JN, Chu MW, Sanes JR. Megf10 and megf11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483:465–9. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, Sanes JR. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci. 2011;31:7753–62. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Zhang Y, Meister M, Sanes J. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. The Journal of Neuroscience. 2010;30:1452. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Zhang Y, Yamagata M, Meister M, Sanes J. Molecular identification of a retinal cell type that responds to upward motion. NATURE-LONDON- 2008;452:478. doi: 10.1038/nature06739. [DOI] [PubMed] [Google Scholar]
- Kim JS, Greene MJ, Zlateski A, Lee K, Richardson M, Turaga SC, Purcaro M, Balkam M, Robinson A, Behabadi BF, Campos M, Denk W, Seung HS EyeWirers . Space-time wiring specificity supports direction selectivity in the retina. Nature. 2014;509:331–6. doi: 10.1038/nature13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Fish D, Rockhill R, Masland R. Diversity of ganglion cells in the mouse retina: unsupervised morphological classification and its limits. The Journal of Comparative Neurology. 2005;489:293–310. doi: 10.1002/cne.20631. [DOI] [PubMed] [Google Scholar]
- Kubota Y. Untangling gabaergic wiring in the cortical microcircuit. Curr Opin Neurobiol. 2014;26:7–14. doi: 10.1016/j.conb.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Lagali PS, Balya D, Awatramani GB, Münch TA, Kim DS, Busskamp V, Cepko CL, Roska B. Light-activated channels targeted to on bipolar cells restore visual function in retinal degeneration. Nat Neurosci. 2008;11:667–75. doi: 10.1038/nn.2117. [DOI] [PubMed] [Google Scholar]
- Lee S, Zhou ZJ. The synaptic mechanism of direction selectivity in distal processes of starburst amacrine cells. Neuron. 2006;51:787–99. doi: 10.1016/j.neuron.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–21. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Masland RH. Populations of wide-field amacrine cells in the mouse retina. J Comp Neurol. 2006;499:797–809. doi: 10.1002/cne.21126. [DOI] [PubMed] [Google Scholar]
- Manookin M, Beaudoin D, Ernst Z, Flagel L, Demb J. Disinhibition combines with excitation to extend the operating range of the off visual pathway in daylight. The Journal of Neuroscience. 2008;28:4136. doi: 10.1523/JNEUROSCI.4274-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Masland RH. Neuronal cell types. Curr Biol. 2004;14:R497–500. doi: 10.1016/j.cub.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Masland RH. The neuronal organization of the retina. Neuron. 2012;76:266–80. doi: 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH, Tauchi M. The cholinergic amacrine cell. Trends in neurosciences. 1986;9:218–223. [Google Scholar]
- Mataruga A, Kremmer E, Müller F. Type 3a and type 3b off cone bipolar cells provide for the alternative rod pathway in the mouse retina. J Comp Neurol. 2007;502:1123–37. doi: 10.1002/cne.21367. [DOI] [PubMed] [Google Scholar]
- Miller RF, Bloomfield SA. Electroanatomy of a unique amacrine cell in the rabbit retina. Proc Natl Acad Sci U S A. 1983;80:3069–73. doi: 10.1073/pnas.80.10.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch TA, da Silveira RA, Siegert S, Viney TJ, Awatramani GB, Roska B. Approach sensitivity in the retina processed by a multifunctional neural circuit. Nat Neurosci. 2009;12:1308–16. doi: 10.1038/nn.2389. [DOI] [PubMed] [Google Scholar]
- Oberlaender M, de Kock CPJ, Bruno RM, Ramirez A, Meyer HS, Dercksen VJ, Helmstaedter M, Sakmann B. Cell type-specific three-dimensional structure of thalamocortical circuits in a column of rat vibrissal cortex. Cereb Cortex. 2012;22:2375–91. doi: 10.1093/cercor/bhr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout JA, Josten N, Yamada J, Pan F, Wu S-w, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, Inoue T, Bloomfield SA, Barres BA, Berson DM, Feldheim DA, Huberman AD. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron. 2011;71:632–9. doi: 10.1016/j.neuron.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyster CW, Barlow HB. Direction-selective units in rabbit retina: distribution of preferred directions. Science. 1967;155:841–2. doi: 10.1126/science.155.3764.841. [DOI] [PubMed] [Google Scholar]
- Petilla Interneuron Nomenclature Group. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–68. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivlin-Etzion M, Zhou K, Wei W, Elstrott J, Nguyen PL, Barres BA, Huberman AD, Feller MB. Transgenic mice reveal unexpected diversity of on-off direction-selective retinal ganglion cell subtypes and brain structures involved in motion processing. J Neurosci. 2011;31:8760–9. doi: 10.1523/JNEUROSCI.0564-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockhill R, Euler T, Masland R. Spatial order within but not between types of retinal neurons. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2303. doi: 10.1073/pnas.030413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roska B, Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature. 2001;410:583–7. doi: 10.1038/35069068. [DOI] [PubMed] [Google Scholar]
- Schwartz GW, Okawa H, Dunn FA, Morgan JL, Kerschensteiner D, Wong RO, Rieke F. The spatial structure of a nonlinear receptive field. Nat Neurosci. 2012;15:1572–80. doi: 10.1038/nn.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS. Reading the book of memory: sparse sampling versus dense mapping of connectomes. Neuron. 2009;62:17–29. doi: 10.1016/j.neuron.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Seung S. Connectome: How the brain’s wiring makes us who we are. Houghton Mifflin Harcourt 2012 [Google Scholar]
- Siegert S, Scherf B, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nature neuroscience. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Chklovskii DB. Neurogeometry and potential synaptic connectivity. Trends Neurosci. 2005;28:387–94. doi: 10.1016/j.tins.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Stevens CF. Neuronal diversity: too many cell types for comfort? Curr Biol. 1998;8:R708–10. doi: 10.1016/s0960-9822(98)70454-3. [DOI] [PubMed] [Google Scholar]
- Sümbül U, Song S, McCulloch K, Becker M, Lin B, Sanes JR, Masland RH, Seung HS. A genetic and computational approach to structurally classify neuronal types. Nat Commun. 2014;5:3512. doi: 10.1038/ncomms4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LO, Jiang Z, Rivlin-Etzion M, Hand R, Brady CM, Matsuoka RL, Yau KW, Feller MB, Kolodkin AL. On and off retinal circuit assembly by divergent molecular mechanisms. Science. 2013;342:1241974. doi: 10.1126/science.1241974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li N, He S. Large-scale morphological survey of rat retinal ganglion cells. Visual neuroscience. 2002;19:483–493. doi: 10.1017/s0952523802194107. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ. A resource of cre driver lines for genetic targeting of gabaergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholm S, Johnson K, Li X, Smith RG, Awatramani GB. Parallel mechanisms encode direction in the retina. Neuron. 2011;71:683–94. doi: 10.1016/j.neuron.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholm S, Johnson K, Li X, Smith RG, Awatramani GB. Erratum: Parallel mechanisms encode direction in the retina. Neuron. 2013a;77:204–8. doi: 10.1016/j.neuron.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholm S, Schwab DJ, Balasubramanian V, Awatramani GB. Lag normalization in an electrically coupled neural network. Nat Neurosci. 2013b;16:154–6. doi: 10.1038/nn.3308. [DOI] [PubMed] [Google Scholar]
- Völgyi B, Chheda S, Bloomfield S. Tracer coupling patterns of the ganglion cell subtypes in the mouse retina. The Journal of Comparative Neurology. 2009;512:664–687. doi: 10.1002/cne.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wässle H, Puller C, Müller F, Haverkamp S. Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. The Journal of Neuroscience. 2009;29:106–117. doi: 10.1523/JNEUROSCI.4442-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yonehara K, Balint K, Noda M, Nagel G, Bamberg E, Roska B. Spatially asymmetric reorganization of inhibition establishes a motion-sensitive circuit. Nature. 2011;469:407–10. doi: 10.1038/nature09711. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Watanabe D, Ishikane H, Tachibana M, Pastan I, Nakanishi S. A key role of starburst amacrine cells in originating retinal directional selectivity and optokinetic eye movement. Neuron. 2001;30:771–80. doi: 10.1016/s0896-6273(01)00316-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim IJ, Sanes JR, Meister M. The most numerous ganglion cell type of the mouse retina is a selective feature detector. Proc Natl Acad Sci U S A. 2012;109:E2391–8. doi: 10.1073/pnas.1211547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J-j, Lee S, Zhou ZJ. A developmental switch in the excitability and function of the starburst network in the mammalian retina. Neuron. 2004;44:851–864. doi: 10.1016/j.neuron.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Xu J, Hauswirth WW, DeVries SH. Genetically targeted binary labeling of retinal neurons. J Neurosci. 2014;34:7845–61. doi: 10.1523/JNEUROSCI.2960-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]