Abstract
Autophagy is an intracellular degradation process that delivers cytosolic material to lysosomes and vacuoles. To investigate the mechanisms that regulate autophagy, we performed a genome-wide screen using a yeast deletion-mutant collection, and found that Npr2 and Npr3 mutants were defective in autophagy. Their mammalian homologs, NPRL2 and NPRL3, were also involved in regulation of autophagy. Npr2-Npr3 function upstream of Gtr1-Gtr2, homologs of the mammalian RRAG GTPase complex, which is crucial for TORC1 regulation. Both npr2∆ mutants and a GTP-bound Gtr1 mutant suppressed autophagy and increased Tor1 vacuole localization. Furthermore, Gtr2 binds to the TORC1 subunit Kog1. A GDP-bound Gtr1 mutant induced autophagy even under nutrient-rich conditions, and this effect was dependent on the direct binding of Gtr2 to Kog1. These results revealed that 2 molecular mechanisms, Npr2-Npr3-dependent GTP hydrolysis of Gtr1 and direct binding of Gtr2 to Kog1, are involved in TORC1 inactivation and autophagic induction.
Keywords: autophagy, GTPase-activating protein, Gtr1, Gtr2, RAG, TORC1
Introduction
Autophagy, an intracellular process that mediates the bulk degradation of cellular components, is conserved from yeast to mammals.1 This process is induced under stressful conditions, such as nutrient starvation. Autophagy is characterized by cytosolic membranous structures called autophagosomes, and the delivery of cytosolic materials to lysosomes and vacuoles for degradation. By supplying the products of degradation as resources for the cell, autophagy helps cells adapt to stressful conditions.
The TORC1 protein kinase complex, which in the yeast Saccharomyces cerevisiae consists of the Tor1/2 protein kinase, Kog1, Lst8, and Tco89, plays a central role in the regulation of autophagy, which is induced by TORC1 inactivation.2,3 TORC1 contributes to cell growth and homeostasis in response to environmental cues. Various factors, including nutrients, stress, and intracellular energy levels, affect the kinase activity of TORC1; TORC1 is activated under conditions favorable to cellular growth, whereas stressful conditions, including nutritional limitation, inactivate TORC1.4 In yeast autophagy, inactivation of TORC1 leads to dephosphorylation of Atg13, a substrate of TORC1, and induction of autophagy.5
The heterodimeric G proteins Gtr1 and Gtr2 play a critical role in the regulation of TORC1 in yeast.6 In mammals, RRAGA and RRAGB are orthologs of Gtr1, and RRAGC and RRAGD are orthologs of Gtr2.7,8 Together with Ego1 and Ego3, Gtr1 and Gtr2 are components of the EGO complex;9,10 Ego1 and Ego3 are the putative functional counterparts of the mammalian Ragulator complex.10-12
The nucleotide-binding state of Gtr1-Gtr2 is crucial for the regulation of TORC1. GTP-bound Gtr1 activates TORC1, whereas GDP-bound Gtr1 inactivates TORC1 even under conditions that promote wild-type cell growth.6 Gtr1 and mammalian RRAGB are thought to switch from the GTP-bound form to the GDP-bound form in response to amino-acid starvation.6,13 Thus, switching of the nucleotide-binding state of Gtr1, and the resultant effects on TORC1 activity, are key events in cellular adaptation to extracellular conditions. However, the mechanisms by which TORC1 is inactivated are poorly understood.
We performed a genome-wide screen that identified Npr2 and Npr3 as key regulators of autophagy in yeast; this function is also conserved in mammals. Genetic analyses revealed that Npr2-Npr3 regulates autophagy via Gtr1-Gtr2. An NPR2 deletion mutant mimicked the phenotypes produced by a GTP-bound Gtr1 mutant with respect to both autophagy and Tor1 localization, suggesting that Npr2-Npr3 regulates GTP hydrolysis in Gtr1. Furthermore, we found that Gtr2 binds to Kog1. GDP-bound Gtr1 mutant negatively regulates TORC1, and the Gtr2-Kog1 interaction is necessary for this regulation. These 2 molecular mechanisms are involved in inactivation of TORC1 and induction of autophagy.
Results
The roles of Npr2 and Npr3 in TORC1 regulation are functionally conserved from yeast to mammals
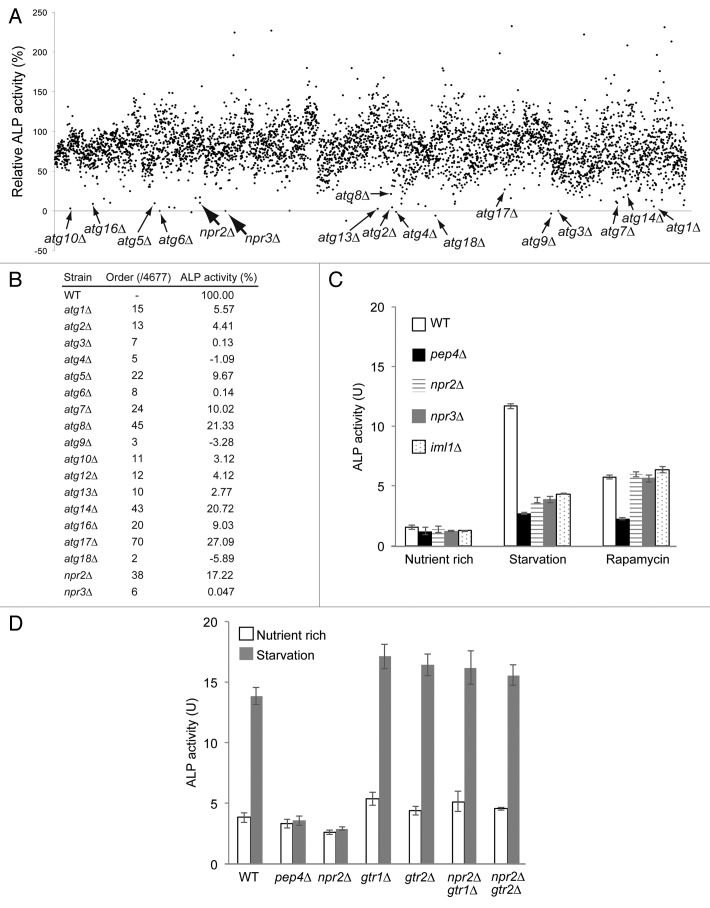
The alkaline phosphatase (ALP) assay is widely used to quantitatively assess the amount of cytosol delivered to vacuoles during autophagy in Saccharomyces cerevisiae.14,15 To examine the mechanisms regulating autophagy, we extended the ALP assay to a large-scale format, using 96-well microtiter plates to perform a genome-wide screen in a yeast mutant collection (see Materials and Methods). In this screen, approximately 4,600 mutant strains from a nonessential gene-deletion mutant collection were starved of nitrogen sources for 4 h, and then subjected to the ALP assay. All core atg mutants, which have been previously characterized as defective in autophagy,16 exhibited quite low levels of ALP activity, indicating that our large-scale assay system was capable of measuring the autophagic capacity of each strain in this collection (Fig. 1A and B). Deletions of NPR2 and NPR3 also resulted in significant defects in nitrogen-starvation-induced autophagy (Fig. 1B and C). Recent studies have shown that Npr2 and Npr3, along with Iml1, are components of the SEA complex.17,18 Consistent with this, the iml1∆ deletion mutant (which was not included in the original mutant collection) also exhibited defects in autophagy (Fig. 1C). Treatment with the TORC1-specific inhibitor rapamycin abolished the defect caused by deletion of Npr2 or Npr3, and induced autophagy at a level similar to that observed in wild-type cells (Fig. 1C). No additive defect in nitrogen-starvation-induced autophagy was observed in npr2∆ npr3∆ double mutants (Fig. S1A). These results are consistent with a previous report that Npr2 and Npr3 function as negative regulators upstream of TORC1.19 Thus, cells lacking Npr2, Npr3, and Iml1 exhibit defects in induction of autophagy through TORC1 inactivation.18
Figure 1. Gtr1 contributes to the regulation of autophagy by Npr2-Npr3. (A) ALP activities in the mutant strains bearing pho8∆60 after 4 h of starvation. Activity measured in the vacuolar proteinase A mutant strain pep4∆ (SKY001) was subtracted as the background signal. Activities are shown as the percentage values for each strain relative to that of the parental TNY509 strain. Deletion mutants lacking the core ATG genes, NPR2, or NPR3 are denoted with arrows. (B) Detailed ALP activities for representative atg mutants shown in (A). (C) ALP activities in the wild-type (SKY084), pep4∆ (SKY100), npr2∆ (SKY091), npr3∆ (SKY131), and iml1∆ (SKY264) strains. Before the cells were subjected to the ALP assay, the strains were incubated in YPD medium at log phase, under nitrogen starvation conditions for 3 h, or in the presence of rapamycin for 3 h. Data represent means ± standard deviation from 3 independent experiments. (D) Wild-type (BY4741, SKY084) and pep4∆ (SKY100), gtr1∆ (SKY244), gtr2∆ (SKY246), gtr1∆ npr2∆ (SKY245), gtr2∆ npr2∆ (SKY247) yeast cells were grown in YPD, starved for 3 h, and subjected to ALP assays. Data represent means ± standard deviation from 3 independent experiments.
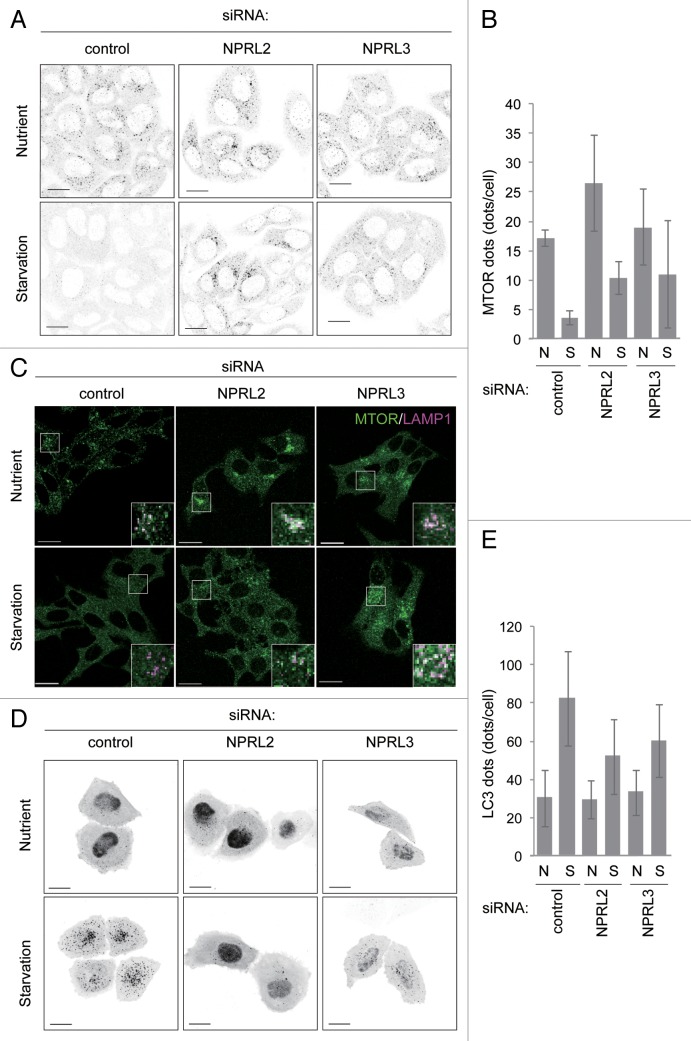
NPRL2 and NPRL3 are the mammalian homologs of Npr2 and Npr3.19 To investigate the roles of the mammalian proteins, we knocked down NPRL2 and NPRL3 expression in both HeLa and MCF7 cells. Specific small interfering RNA (siRNA) reduced NPRL2 and NPRL3 mRNA levels to less than 30% of the levels observed in control HeLa cells (Fig. S2A). siRNA treatment also reduced NPRL2 and NPRL3 protein levels in MCF7 cells (Fig. S2B). In control cells under nutrient-rich conditions, MTOR immunofluorescence signals were detected as multiple puncta in the perinuclear region (Fig. 2A and B; Fig. S3A and S3B). These MTOR puncta colocalized with a lysosomal protein, LAMP1 (Fig. 2C), whereas starvation conditions caused MTOR disperse throughout the cytosol, as reported previously.13 However, knockdown of NPRL2 or NPRL3 caused MTOR to remain on the lysosomes even under starvation conditions (Fig. 2A and B; Fig. S3A and S3B). Because lysosomal localization is important for MTORC1 activation,12 this observation suggested that knocking down NPRL2 or NPRL3 allowed MTORC1 to remain active even under starvation conditions, indicating that MTORC1 activity is regulated by NPRL2 and NPRL3. To further test this idea, we estimated the autophagic activity, as a read-out of MTORC1 activity, by counting the number of autophagosomes labeled with the specific marker GFP-LC3.20 In control cells under starvation conditions, numerous LC3-positive puncta were detected, indicating that autophagy was induced (Fig. 2D and E; Fig. S3C and S3D). On the other hand, markedly fewer LC3-positive puncta were observed following knockdown of NPRL2 or NPRL3, even under starvation conditions (Fig. 2D and E; Fig. S3C and S3D). These data indicate that deficiencies in NPRL2 and NPRL3 influenced both induction of autophagy and the starvation-dependent inactivation of MTORC1. Thus, Npr2 and Npr3 must function in some step of TORC1 regulation, which is conserved from yeast to mammals.
Figure 2. Effects of knockdown of NPRL2 and NPRL3 expression on MTOR localization and autophagy. (A) Two and a half d after siRNA transfection (clone #1), HeLa cells were incubated in nutrient-rich growth medium or EBSS (starvation medium) for 1 h. Cells were immunostained with anti-MTOR antibodies and subjected to fluorescence microscopy. Scale bar: 20 μm. (B) MTOR-positive puncta (A) were counted in NPRL2- or NPRL3-knockdown HeLa cells using the G-count software. Data represent means ± standard deviation of representative results (n > 40 cells). (C) Effect of NPRL2 or NPRL3 knockdown on MTOR localization. Knockdown MCF7 cells were incubated in growth medium (serum, amino acid: +) or EBSS (serum, amino acid: -) for 60 min, and then stained with MTOR (green) and LAMP1 (magenta) antibodies. Bars indicate 20 μm. Results shown are from siRNA clone #1 for NPRL2 knockdown and clone #2 for NPRL3 knockdown. (D) HeLa cells were cotransfected with siRNA (clone #1) and a vector harboring EGFP-LC3. Two and a half d after siRNA transfection, cells incubated in growth medium or starvation medium for 1 h were subjected to fluorescence microscopy. Scale bars: 20 μm. (E) GFP-LC3-positive puncta in each cell from (D) were counted. Data represent means ± standard deviation of representative results (n > 40 cells).
The autophagic defect of npr2 mutant cells is rescued by deletion of GTR1 or GTR2
The heterodimeric G-protein complex Gtr1-Gtr2, the yeast ortholog of mammalian RRAGA/B-RRAGC/D, is an upstream regulator of TORC1.7,8,21 Therefore, we assessed the potential relationships between Gtr1-Gtr2 and Npr2-Npr3 by examining double-deletion mutants under nitrogen-starvation conditions. Intriguingly, the autophagic defects observed in npr3∆ and npr2∆ cells were abrogated by further deletion of GTR1 or GTR2, which resulted in wild-type levels of autophagy (Fig. 1D; Fig. S1A). Next, we monitored the phosphorylation status of Atg13, which is a direct substrate of TORC1 kinase and essential for autophagy.5,22 In wild-type cells, Atg13 was dephosphorylated in response to starvation conditions (Fig. S1B and S1C).22 By contrast, Atg13 remained phosphorylated in npr2∆ or npr3∆ cells, even under starvation conditions (Fig. S1B and S1C). Meanwhile, deletion of GTR1 or GTR2 from npr2∆ or npr3∆ cells caused dephosphorylation of Atg13 under starvation conditions (Fig. S1B and S1C). These results indicate that Npr2 and Npr3 function upstream of the Gtr1-Gtr2 complex in the regulation of TORC1.
GTP-bound Gtr1 suppresses autophagy under nitrogen starvation
Our results suggested that Gtr1 is involved in the regulation of autophagy. Unexpectedly, however, deletion of neither GTR1 nor GTR2 had any effect on induction of autophagy (Fig. 1D and Fig. S1A, see ref. 9). Therefore, to ask whether the Gtr1-Gtr2 complex played a role in the regulation of autophagy, we tested the effects of Gtr1 and Gtr2 mutants that mimic various nucleotide-bound states.23 As a result, autophagy induction in response to nitrogen-starvation was severely deficient in gtr1∆ gtr2∆ cells exogenously expressing both the GTP-bound form Gtr1 mutant and wild-type Gtr2, driven from their native promoters (Fig. 3, column 8). These data indicate that the GTP-bound Gtr1 mutant exerts a dominant effect on autophagy in the gtr1∆ background. This effect was also observed in cells expressing both the GTP-bound Gtr1 mutant and the GDP-bound form of Gtr2 (Fig. 3, column 10). On the other hand, when cells completely lacked Gtr2, autophagy was induced normally in response to nitrogen-starvation (Fig. 3, column 12). Thus, Gtr2, at least in the GDP form, appears to be critical for the dominant effects of the GTP-bound Gtr1 mutant. We also found that vacuolar localization of Gtr16 was disrupted by deletion of Gtr2 (Fig. S4A). Likewise, deletion of GTR1 disrupted vacuolar localization of Gtr2 (Fig. S4A). Therefore, Gtr2 seems to be important for functional localization of Gtr1, and vice versa. We also noticed that the combination of the GTP-bound forms of Gtr1 and Gtr2 inhibited starvation-induced autophagy (Fig. S5A, column 18), but we interpret the role of Gtr2 in another way (see below and Discussion)
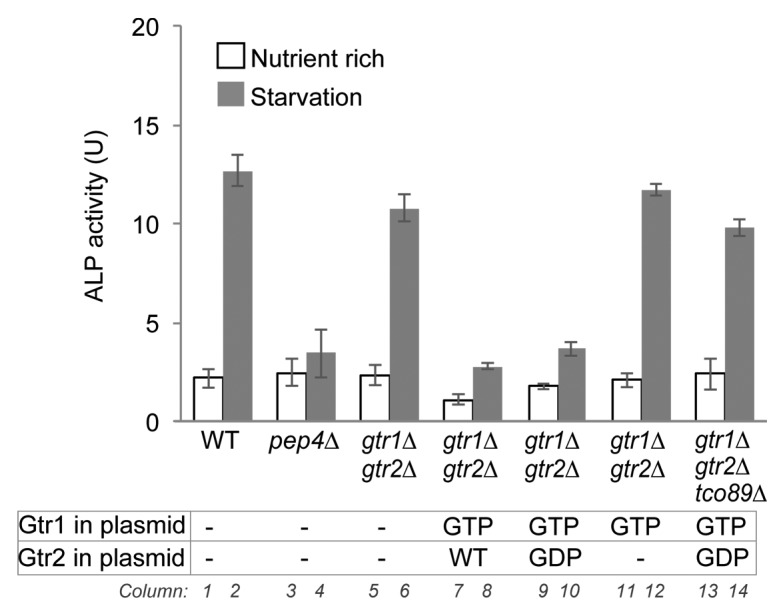
Figure 3. A GTP-bound Gtr1 mutant suppresses autophagy in a Gtr2-dependent manner. gtr1∆ gtr2∆ double-deletion cells (SKY167) harboring empty vector (-), plasmid encoding GTP-bound mutant Gtr1 (GTP), and/or plasmid encoding wild-type (WT) or GDP-bound mutant Gtr2 (GDP) and tco89∆ gtr1∆ gtr2∆ triple-mutant cells (SKY277) expressing GTP-bound Gtr1 mutant (GTP) and GDP-bound Gtr2 mutant (GDP) were grown in SD medium containing 0.5% casamino acids, incubated under nitrogen-starvation conditions for 3 h, and then subjected to ALP assays. Data represent means ± standard deviation from 3 independent experiments. The ALP activities of wild-type (SKY084) and pep4∆ (SKY100) cells harboring empty vector were also measured as control samples.
To determine whether TORC1 played a role in these effects, we monitored autophagy in cells lacking the nonessential TORC1 subunit Tco89.24 The tco89 mutant cells expressing GTP-bound Gtr1 and GDP-bound Gtr2 mutants exhibited starvation-induced autophagy at levels comparable to those in wild-type cells (Fig. 3, column 14). These data indicate that GTP-bound Gtr1 causes defects in autophagy by activating TORC1 even under nitrogen-starvation conditions.
Tor1 vacuolar localization is increased in both GTP-bound Gtr1 and npr2∆ mutants
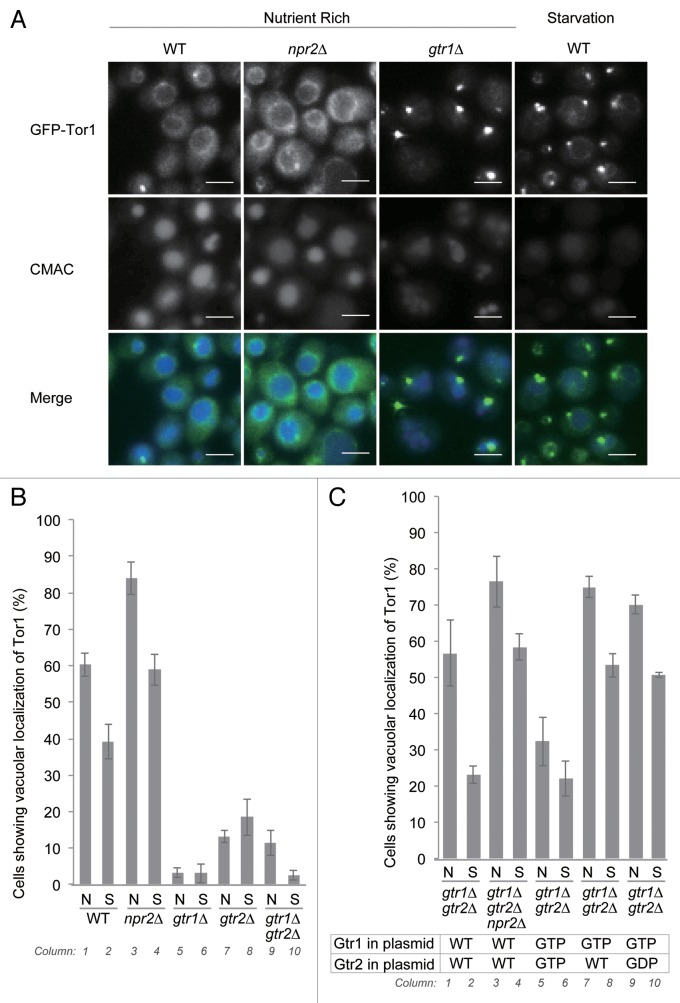
Next, we observed the localization of N-terminally GFP-tagged Tor1, expressed from the genome under the control of the native promoter. This GFP-Tor1 best replicated the native Tor1 function among several constructs including previously reported ones (Fig. S4B). As previously reported, GFP-Tor1 was detected on vacuolar membranes, and some puncta associated with vacuolar membranes under nutrient-rich conditions (Fig. 4A).6,25 The identity of these perivacuolar puncta remains obscure; they did not colocalize with markers of other compartments such as the phagophore assembly site (Ape1-mCherry) or the endosome (Snf7-mCherry) (data not shown). Deletion of GTR1 or GTR2 significantly reduced the vacuolar localization of Tor1 (Fig. 4A and B, columns 5 to 10), although perivacuolar localization of Tor1 persisted. The expression level of native Tor1 was not affected in these mutants (Fig. S4C). Thus, Gtr1 and Gtr2 appear to maintain Tor1 localization on the vacuoles.
Figure 4. Vacuolar localization of Tor1 is regulated by Gtr1, Gtr2, Npr2, and nutrient availability. (A) Strains expressing N-terminally GFP-tagged Tor1 under the control of the native promoter (WT, SKY222; npr2∆, SKY226; gtr1∆, SKY278; gtr2∆, SKY279; gtr1∆ gtr2∆, SKY299) were incubated in SD medium containing 0.5% casamino acids and 100 µM CMAC for 30 min, and either washed once with SD medium containing 0.5% casamino acids or starved for nitrogen (SD-N) for 1 to 2 h, and then subjected to fluorescence microscopy. Scale bar: 5 μm. (B) Percentage of cells in (A) exhibiting vacuolar localization of Tor1. Cells with GFP-Tor1 signals surrounding vacuoles stained by CMAC were counted. More than 100 cells were counted. Data represent means ± standard deviation from 3 independent experiments. (C) Strains expressing N-terminally GFP-tagged Tor1 (gtr1∆ gtr2∆, SKY299; gtr1∆ gtr2∆ npr2∆, SKY320) and expressing the indicated combinations of Gtr1 and Gtr2 mutants were subjected to fluorescence microscopy under the same conditions as in (A). Percentage of cells exhibiting vacuolar localization of Tor1 is presented in (B).
We noticed that the number of cells with vacuolar localization of Tor1 was reduced in response to starvation conditions, although a significant fraction of cells (39%) still exhibited vacuolar localization of Tor1 (Fig. 4A and B, columns 1 and 2). The GTP-bound Gtr1 mutants also increased vacuolar localization of GFP-Tor1 (Fig. 4C, columns 7 and 8). For this increase to occur, Gtr2 was needed to be in the GDP-bound form rather than the GTP-bound form (Fig. 4C, columns 5, 6, 9, and 10). Thus, dynamics of Tor1 localization are regulated by Gtr1 and Gtr2.
Furthermore, we found that deletion of NPR2 increased the cells with vacuolar localization of Tor1 (Fig. 4A and B, columns 3 and 4; Fig. 4C, columns 3 and 4). Thus, the npr2∆ mutant is similar to the GTP-bound Gtr1 mutant with respect to Tor1 localization and defects in induction of autophagy. These results support the model that the npr2∆ mutant is defective in hydrolysis of GTP bound to Gtr1.
The GDP-bound Gtr1 mutant induces autophagy even under nutrient-rich conditions
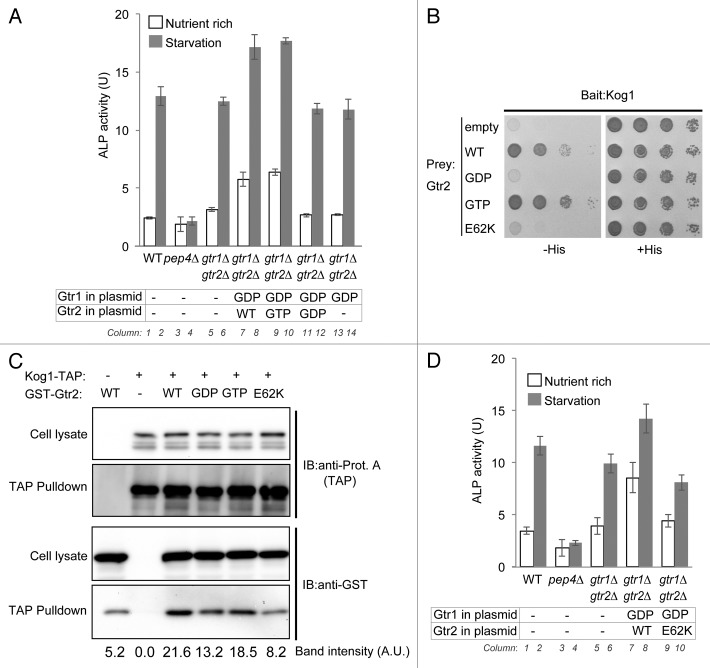
Next, we investigated the phenotype of a Gtr1 mutant mimicking a permanently GDP-bound state. Cells expressing the GDP-bound Gtr1 mutant induced autophagy even under nutrient-rich conditions (Fig. 5A, columns 7 and 9), but this phenotype was abrogated in cells lacking Gtr2 (Fig. 5A, column 13). Autophagic induction was still observed when wild-type Gtr2 was replaced with the GTP-bound mutant, but absent when Gtr2 was replaced by the GDP-bound form (Fig. 5A, columns 9 and 11). Thus, GTP-bound Gtr2 is required for the dominant effect of the GDP-bound Gtr1 mutant. Autophagic induction by this dominant effect was not affected by the deletion of NPR2 (Fig. S5A, columns 27 and 29), supporting the idea that Npr2 functions upstream of Gtr1-Gtr2.
Figure 5. Gtr2 interacts with Kog1 and inactivates TORC1. (A) Wild-type (SKY084), pep4∆ (SKY100), and gtr1∆ gtr2∆ double-mutant cells harboring empty vector (-), and gtr1∆ gtr2∆ cells harboring the GDP-bound mutant Gtr1 (top row: GDP) along with wild-type, GTP-bound mutant, or GDP-bound mutant Gtr2 (bottom row: WT, GTP, and GDP, respectively), were either grown in SD medium supplemented with 0.5% casamino acids or starved of nitrogen for 3 h, and then subjected to ALP assays. Data represent means ± standard deviation from 3 independent experiments. (B) Cells of the strain for yeast 2-hybrid analysis (PJ69-4A) harboring pGBD-C1 vector encoding wild-type, GTP-bound mutant, GDP-bound mutant, or E62K mutant Gtr2 (pSK150, pSK151, pSK152, or pSK222, respectively) and pGAD-C1/Kog1 (pSK156) were serially 10-fold diluted and spotted onto SC medium plates either lacking histidine and leucine or histidine, leucine, and uracil. Results after 2 d of culture at 30 °C are shown. (C) Cells endogenously expressing TAP-tagged Kog1 and harboring multicopy vector encoding wild-type, GTP-bound mutant, GDP-bound mutant, or E62K mutant Gtr2 were subjected to TAP affinity isolation assays. The lysates and the affinity isolates were subjected to western blot analysis with anti-protein A and anti-GST antibodies. (D) Strains (wild-type, SKY084; pep4∆, SKY100; and gtr1∆ gtr2∆, SKY167) harboring empty vector (-), and gtr1∆ gtr2∆ (SKY167) harboring GDP-bound Gtr1 mutant (GDP) along wild-type (WT) or E62K mutant Gtr2E62K, were either grown in SD medium supplemented with 0.5% casamino acids or starved of nitrogen for 3 h, and then subjected to ALP assays. Data represent means ± standard deviation from 3 independent experiments.
The TORC1 subunit Kog1 is an effector of Gtr2
Based on these results, we investigated the role of Gtr2 in the autophagic induction dependent on GDP-bound Gtr1. In a 2-hybrid analysis, we found that Gtr2 binds to Kog1 (Fig. 5B), a subunit of TORC1 and a mammalian ortholog of RAPTOR.26 Kog1 also strongly bound to the GTP-bound Gtr2 mutant, but not to the GDP-bound mutant (Fig. 5B). To confirm this interaction biochemically, we performed affinity isolation assays. Lysates of Kog1-TAP-expressing cells were precipitated by IgG beads, and the affinity isolated fractions were subjected to western blotting. As shown in Figure 5C, wild-type GST-Gtr2 was precipitated in a Kog1-TAP-dependent manner (relative amount, 21.6); the amount of material pulled down in the specific precipitation was significantly larger than the amount pulled down by nonspecific binding, possibly to the resin (see control lane; relative amount 5.2), which could not be entirely prevented. The GTP-bound form of GST-Gtr2 was also efficiently precipitated, whereas the GDP-bound form of GST-Gtr2 was precipitated to a lesser extent (Fig. 5C). These data indicate that Kog1 is a direct effector of Gtr2.
TORC1 is inactivated by binding to Gtr2
A recent structural analysis of Gtr1 revealed the critical amino acids on its effector-binding surface.27 Based on its structural similarity to Gtr1, Gtr2 is predicted to bind to Kog1 via a similar surface. Accordingly, we made 9 point-mutants on the possible surface area of Gtr2; in one of these mutants, glutamine 62 of Gtr2, which corresponds to Gtr1 asparagine 61, was mutated to lysine. This E62K mutation of Gtr2 abolished the interaction to Kog1, as demonstrated by yeast 2-hybrid analysis (Fig. 5B). Furthermore, coprecipitation with Kog1-TAP was severely defective in the Gtr2E62K mutant, although the expression level of the E62K mutant was not affected (Fig. 5C). Using the E62K mutant, we asked whether the binding of Gtr2 to Kog1 is required for autophagic induction by expression of the GDP-bound form Gtr1. Indeed, the autophagic induction was abrogated in the Gtr2E62K mutant (Fig. 5D, columns 7 and 9). Collectively, these data indicate that direct binding of Gtr2 to Kog1 inactivates TORC1 and eventually induces autophagy, even under nutrient-rich conditions.
Discussion
In this study, we elucidated the mechanism by which Gtr1-Gtr2 regulates autophagy via TORC1. Npr2-Npr3 functions upstream of Gtr1-Gtr2, and the npr2∆ mutant replicates the phenotypes of the GTP-bound Gtr1 mutant with respect to both autophagy and Tor1 localization. Based on these findings, we reasoned that the function of Npr2-Npr3 is closely linked to the hydrolysis of GTP in Gtr1. Indeed, it has recently been reported that Iml1, a subunit of the SEA complex including Npr2 and Npr3,17 possesses GAP (GTPase activating protein) activity toward Gtr1.28 In mammals, a protein complex called GATOR1, which contains NPRL2 and NPRL3, exert GAP activity toward RRAGA/B.29 Therefore, our results are consistent with the idea that the Npr2-Npr3 complex is required for GAP activity toward Gtr1. In addition, our results demonstrate that Gtr2 directly binds the TORC1 subunit Kog1, resulting in inactivation of TORC1. These 2 molecular mechanisms are involved in TORC1 inactivation and autophagic induction.
In mammals, RRAGB activates MTORC1 through direct binding to RAPTOR.13 In yeast, Gtr1 is believed to bind to the Kog1 in the same manner, although direct binding has not been confirmed.6 Our results showed that GTP-bound Gtr2 is an inhibitor, whereas GTP-bound Gtr1 is an activator, of TORC1 (Fig. 5). What happens when both Gtr1 and Gtr2 are in the GTP-bound form? In that case, we found that autophagy was not induced under nutrient-rich conditions and faintly induced in starvation condition (Fig. S5A, columns 17 and 18). We reasoned that Gtr1-dependent activation and Gtr2-dependent inactivation occur simultaneously, but the effect of Gtr1 is epistatic to that of Gtr2, at least in regard to autophagy. The best evidence that Gtr2-dependent inactivation occurs in the cells harboring both GTP-Gtr1 and GTP-Gtr2 is that it exhibited rapamycin-sensitive growth, indicating weakened TORC1 function relative to wild-type and Gtr1-GTP Gtr2-GDP cells (Fig. S6). Further, Tor1 vacuolar localization was decreased in comparison to the Gtr1-GTP Gtr2-GDP cells (Fig. 4C, columns 5 to 8). Thus, there may be some mechanism that determines whether Gtr1 or Gtr2 will be dominant when both are in GTP-bound form.
In mammals, MTOR is localized on the lysosome when amino acids are available, and this localization depends on RRAGA/B, and RRAGC/D. We showed that Gtr1 and Gtr2 are also important for Tor1 localization on vacuole in yeast, just as in mammals (Fig. 4A and B). Furthermore, the nucleotide-bound state of Gtr1-Gtr2 was critical in determining the dynamics of Tor1 vacuolar localization: the Gtr1-GTP form increased vacuolar localization, whereas the Gtr2-GTP form decreased it (Fig. 4C, columns 5 to 10; Fig. S4C, columns 5, 6, 11, and 12). Cells in which Tor1 was localized to the vacuole became less abundant in response to starvation (Fig. 4A and B). Via regulation of Gtr1, Npr2 seems to play a critical role in Tor1 dynamics. This Npr2-dependent regulation must be important, because even in mammals, NPRL2 and NPRL3 regulate MTOR localization (Fig. 2A–C; Fig. S3A and S3B). Where does this inactivation occur? Despite the dynamic changes in Tor1 localization in response to starvation, a significant fraction of Tor1 remained on the vacuole under nitrogen-starvation (Fig. 4A and B), as in the case of Gtr2 (our unpublished observation). Based on our findings that Gtr2 binds to Kog1 (Fig. 5B and C), it is possible that Gtr2 interacts with TORC1 on the vacuole and/or the perivacuolar dot.
A similar dominant effect of the GDP-bound mutant of Gtr1 is also observed in mammalian RRAGB: overexpression of RRAGB-GDP inactivates MTORC1 irrespective of the availability of amino acids. This RRAGB-GDP-dependent dominant effect has also been observed in cells overexpressing GTP-bound RRAGD, but not GDP-bound RRAGD.30,31 Therefore it is possible that RRAGD negatively regulates MTORC1, as we have observed in yeast. However, the target is unlikely to be a component of MTORC1: the GDP-bound form of RRAGA/B in complex with the wild-type or GTP-bound form of RRAGC fails to bind RPTOR.32,33 Moreover, under starvation conditions, MTORC1 is not located on lysosomes, but is instead dispersed throughout the cytosol, whereas wild-type and GTP-bound mutant RRAGD were detected on lysosomes irrespective of the availability of amino acids.12 SH3BP4, which binds to RRAG proteins when MTOR is not localized on lysosomes,34 is one of the possible targets of RRAGD, and there may exist some mechanism by which such a target could inactivate TORC1.
Regardless of the findings described above, it is noteworthy that deletion of neither GTR1 nor GTR2 markedly affected autophagy induced by nitrogen-starvation conditions (Fig. 1D; Fig. S1A).9 Thus, autophagy can be induced under nitrogen-starvation by an as-yet-unidentified signaling pathway independent of the Npr2-Npr3 and Gtr1-Gtr2 axis. Expression of the GTP- or GDP-bound mutants of Gtr1-Gtr2 supersedes this unknown Gtr1-Gtr2-independent pathway in the regulation of TORC1 and autophagy by locking TORC1 in an activated or inactivated state irrespective of upstream signals. We reasoned that deletion of NPR2 or NPR3 could provide these effects. One candidate for such an Npr2-Npr3 and Gtr1-Gtr2-independent pathway is the cAMP-dependent protein kinase pathway, which plays a role in autophagic regulation.2,35,36 Alternatively, Gcn4-dependent amino-acid metabosim may be involved, although that pathway is also distinct from the effects of nitrogen starvation.37 In addition, our results do not necessarily support a model in which Npr2-Npr3 complexes are regulated by nitrogen availability. Rather, our data are consistent with the possibility that the Npr2-Npr3 and Gtr1-Gtr2-dependent pathway is regulated by other factors.38 Indeed, autophagy and TORC1 activity are regulated by variety of environmental factors, including a lack of carbon sources or sulfates.38,39 A recent study shows that the Npr2-Npr3-Iml1 complex is regulated by sulfur-containing amino acids.40 Uncovering the relationship between Npr2-Npr3 and various environmental stimuli will help to elucidate the functional and regulatory mechanisms of TORC1.
From these observations, Gtr1-Gtr2-dependent inactivation of TORC1 and induction of autophagy occurs when the Gtr2 is able to adopt its GTP form. In that case, which molecule is responsible for exchanging GDP for GTP in Gtr2? It is unlikely that Npr2-Npr3 is directly involved in this exchange. Expression of Gtr1-GDP caused induction of autophagy when Gtr2 is in the WT or GTP form, even under nutrient-rich conditions, suggesting that Gtr2 can adopt its GTP form in a cell expressing Gtr1-GDP (Fig. 5A, columns 7 and 9; Fig. S5A, columns 19 and 23). If Npr2 is involved in the exchange of GDP for GTP in Gtr2, Gtr2 is not able to adopt its GTP form, and autophagy will not be induced in a npr2∆ cell expressing Gtr1-GDP. However, in npr2∆ cells expressing Gtr1-GDP and Gtr2-WT, autophagy was induced even under nutrient-rich conditions (Fig. S5A, column 29). The same results were obtained in npr3∆ and npr2∆ npr3∆ cells (data not shown). Some unknown mechanism seems to be involved in these phenomena, and this issue should be addressed in future work.
Hyperactivation of TORC1 has been observed in cancer cells.41,42 Intriguingly, NPRL2 has been reported to be a tumor suppressor.43,44 Furthermore, mice lacking NPRL3 die between 15 d after conception and birth, and these animals exhibit developmental defects in the heart.45 Mice bearing only the permanently GTP-bound form of RRAGA die during a neonatal period of starvation,46 underscoring the physiologic requirement for proper GTP and GDP cycling by RRAGA. Our findings will help to provide a foundation for research into clinical approaches that target TORC1 and autophagy.
Materials and Methods
Yeast growth media
Yeast cells were grown at 30 °C in YPD (1% yeast extract, 2% peptone, and 2% glucose) or SD (0.67% yeast nitrogen base and 2% glucose) medium supplemented with amino acids and 0.5% casamino acid. For nitrogen starvation, cells were washed twice with SD-N (0.17% yeast nitrogen base without amino acids and ammonium sulfate, and 2% glucose) and resuspended in SD-N. Rapamycin (Sigma, R0395) in stock solution (1 mg/ml ethanol and Triton X-100 at a ratio of 9:1 [v/v]) was added to YPD to achieve a final concentration of 0.2 μg/ml.
Plasmid and yeast strains
The PHO8 locus from the knockout collection was replaced with pho8∆60 using a synthetic genetic-array method.47 Briefly, the TNY509 parental strain harboring pho8∆60 was crossed with each mutant strain on YPD in a rectangular plate (OmniTray, Nunc) using a 96-pin replicator (V&P Scientific, VP408). Haploid cells containing the pho8∆60 allele and one of the mutant alleles were selected using auxotrophic and drug-resistance markers and sequential replica plating.15,48
Yeast strains and plasmids used in this study, except for strains made using the synthetic genetic-array method, are listed in Table 1 and Table 2. All gene disruptions, tagged constructs, and other modifications were confirmed by PCR.53-56 N-terminal GFP-tagged Tor1 strains were generated by integrating SpeI-digested pSK108, generated by cloning a fragment containing PTOR1 (1000 bases)-GFP-TOR1 (426 N-terminal bases), created in a 2-step PCR, into pRS305 using the SacI and ApaI sites. An internally 3 × GFP-tagged Tor1 strain was generated based on a previous report.25 GTR1 or GTR2 fragments containing approximately 1,000 bases of the promoter regions, the open reading frame, and 500 bases from the terminal noncoding region were digested with SacI and XbaI or BamHI and XhoI, respectively, and cloned into the single-copy vector pRS316. GST-GTR2 fragments, in which the GST coding region was integrated into the N terminus of GTR2, were amplified by PCR from the yeast genome, digested with NotI and EcoRI, and cloned into the multicopy vector pRS426. GTP- or GDP-bound GTR1 and GTR2 mutants, and other point mutants of GTR2, were generated using primer-based 2-step PCRs, based on previous reports.23 ATG13 fragments, containing approximately 1000 bases of the promoter regions and the open reading frame, were digested with SalI and HindIII and cloned into pRS426.
Table 1. Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| TNY509 | MATα, pho8Δ60::NatMX4Δcan1:: MFA1pr-HIS3-MF 1pr-LEU2 ura3Δleu2Δhis3Δmet15Δlys2Δ | 49 |
| BY4741 | MATa, his3D1leu2D0met15D0ura3D0 | 50 |
| BY4742 | MATα, his3D1leu2D0lys2D0ura3D0 | 50 |
| SKY084 | [BY4741] pho8D60::NatMX4 | This study |
| SKY100 | [SKY084] pep4Δ::kanMX6 | This study |
| SKY091 | [SKY084] npr2Δ::kanMX6 | This study |
| SKY131 | [SKY084] npr3Δ::hphMX4 | This study |
| SKY264 | [SKY084] iml1Δ::kanMX6 | This study |
| SKY244 | [SKY084] gtr1Δ::zeoNT3 | This study |
| SKY246 | [SKY084] gtr2Δ::hphMX4 | This study |
| SKY127 | [SKY084] npr2Δ::kanMX6npr3Δ::hphMX4 | This study |
| SKY167 | [SKY084] gtr1Δ::kanMX6 gtr2Δ::hphMX4 | This study |
| SKY245 | [SKY084] gtr1Δ::zeoNT3 npr2Δ::kanMX6 | This study |
| SKY247 | [SKY084] gtr2Δ::hphMX4 npr2Δ::kanMX6 | This study |
| SKY186 | [SKY084] npr2Δ::zeoNT3 gtr1Δ::kanMX6 gtr2Δ::hphMX4 | This study |
| SKY270 | [SKY084] npr3Δ::hphMX4 gtr1Δ::zeoNT3 | This study |
| SKY298 | [SKY084] gtr2Δ::zeoNT3 npr3Δ::hphMX4 | This study |
| SKY308 | [SKY084] gtr2Δ::kanMX6 npr3Δ::hphMX4 gtr1Δ::zeoNT3 | This study |
| SKY271 | [SKY084] npr2Δ::kanMX6 npr3Δ::hphMX4 gtr1Δ::zeoNT3 | This study |
| SKY291 | [SKY084] npr2Δ::kanMX6 npr3Δ::hphMX4 gtr2Δ::zeoNT3 | This study |
| SKY309 | [SKY084] gtr2Δ::LEU2 gtr2Δ::kanMX6 npr3Δ::hphMX4 gtr1Δ::zeoNT3 | This study |
| SKY277 | [SKY084] gtr1Δ::kanMX6 gtr2Δ::hphMX4 tco89Δ::zeo | This study |
| SKY222 | [BY4741] LEU2::GFP-TOR1 | This study |
| SKY226 | [SKY222] npr2Δ::natMX4 | This study |
| SKY278 | [SKY222] gtr1Δ::hphMX6 | This study |
| SKY279 | [SKY222] gtr2Δ::hphMX4 | This study |
| SKY299 | [SKY222] gtr2Δ::zeoNT3 gtr1Δ::hphMX4 | This study |
| SKY135 | [BY4741] npr2Δ::natMX4 | This study |
| NKR002 | [BY4741] ego1Δ::kanMX6 | EUROSCARF |
| NKR003 | [BY4741] tor1Δ::kanMX6 | EUROSCARF |
| SKY173 | [BY4742] Tor1-D330-3xGFP | This study |
| SKY112 | [BY4741] GTR1-GFP::hphMX4 | This study |
| SKY249 | [BY4741] GTR1-GFP::hphMX4, gtr2Δ::natMX4 | This study |
| SKY259 | [BY4741] GTR2-GFP::hphMX4 | This study |
| SKY260 | [BY4741] GTR2-GFP::hphMX4 gtr1Δ::zeoNT3 | This study |
| SKY287 | [BY4741] KOG1-TAP::kanMX6 | This study |
| PJ69-4A | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δga180Δ LYS2::GALl-HIS3GAL2-ADE2 met2::GAL7-lacZ | 52 |
Table 2. Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pRS316 | CEN, URA3 | 51 |
| pRS426 | 2 micron, URA3 | 51 |
| pGBD-C1 | 2 micron, TRP1 | 52 |
| pGAD-C1 | 2 micron, LEU2 | 52 |
| pSK122 | [pRS316] Gtr1 Gtr2 | This study |
| pSK123 | [pRS316] Gtr1 Gtr2S23L | This study |
| pSK124 | [pRS316] Gtr1 Gtr2Q66L | This study |
| pSK125 | [pRS316] Gtr1S20L Gtr2 | This study |
| pSK126 | [pRS316] Gtr1S20L Gtr2S23L | This study |
| pSK127 | [pRS316] Gtr1S20L Gtr2Q66L | This study |
| pSK128 | [pRS316] Gtr1Q65L Gtr2 | This study |
| pSK129 | [pRS316] Gtr1Q65L Gtr2S23L | This study |
| pSK130 | [pRS316] Gtr1Q65L Gtr2Q66L | This study |
| pSK115 | [pRS316] Gtr1S20L | This study |
| pSK116 | [pRS316] Gtr1Q65L | This study |
| pSK256 | [pRS316] Gtr1S20L Gtr2E62K | This study |
| pSK150 | [pGBD-C1] Gtr2 | This study |
| pSK151 | [pGBD-C1] Gtr2Q66L | This study |
| pSK152 | [pGBD-C1] Gtr2S23L | This study |
| pSK222 | [pGBD-C1] Gtr2E62K | This study |
| pSK156 | [pGAD-C1] Kog1 | This study |
| pSK242 | [pRS426] PGTR2-GST-Gtr2 | This study |
| pSK243 | [pRS426] PGTR2-GST-Gtr2Q66L | This study |
| pSK244 | [pRS426] PGTR2-GST-Gtr2S23L | This study |
| pSK252 | [pRS426] PGTR2-GST-Gtr2E62K | This study |
| pSK048 | [pRS426] PATG13-ATG13 | This study |
ALP assays
For large-scale assays, YPD plates were inoculated with cells from each pool using a 96-pin replicator, and then incubated for 16 to 24 h at 30 °C. For conventional assays, cells were cultured in liquid medium as described above. Subsequently, the cells were collected by centrifugation, suspended in 200 μl of SD-N medium in 96-well plates, and incubated for 4 h at 30 °C. The plates were centrifuged, and the supernatant fraction was discarded. We then added 50 μl of ice-cold lysis buffer (10 mM Tris-Cl, pH 9.0, 10 mM MgSO4, 10 μM ZnSO4) and approximately 10 μl of 0.6-mm zirconia silica beads (Biomedical Science, ZS06-0001) to each well. The plates were sealed with Parafilm and mixed vigorously (2,500/min) on a microplate mixer (Taitec, MBR-022) for 10 min at 4 °C. After a brief centrifugation, the Parafilm was removed, 150 μl of ice-cold lysis buffer was added, and the plates were centrifuged for 15 min at 490 × g at 4 °C. Protein levels were quantified in 50-μl aliquots of the supernatant fractions using a bicinchoninic acid kit (Nacalai, 06385-00), and the enzymatic activity in the aliquots was measured as described previously with slight modifications.15 One unit of ALP activity was defined as one emission/μg protein/min.
Microscopy
For visualizing vacuolar lumen, 1 OD600 unit of cells were incubated for 30 min at 30 °C in SD medium containing 0.5% casamino acids and 100 mM CMAC fluorescent dye (Molecular Probes, C2110). Yeast cells were observed Leica AF6500 fluorescent imaging system (Leica Microsystems, Wetzlar, Germany) mounted on a DIM6000 B microscope (HCX PL APO 63.6×/1.40–0.60 oil-immersion objective lens, xenon lamp, Leica Microsystems, Wetzlar, Germany) under the control of LAS-AF software (Leica Microsystems). In Figure S4A, cells were attached on 35-mm glass bottom dishes (Matsunami, D110300) coated with concanavalin A (Sigma, C7275), and examined using a Leica TCS SP8 confocal system (Leica Microsystems, Wetzlar, Germany) mounted on a DMI 6000 CS microscope (HCX PL APO 100.6×/1.4 Oil STED objective lens, Leica Microsystems, Wetzlar, Germany). Mammalian cells were cultured on coverslips and fixed by incubation in 3% paraformaldehyde in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.2) for 20 min. For immunofluorescence microscopy, the cells were permeabilized with 50 μg/ml digitonin (Sigma, D141) in PBS for 10 min at room temperature. The permeabilized cells were incubated in blocking solution (0.1% gelatin in PBS), and then stained with anti-MTOR (Cell Signaling Technology, 7C10; 2983) and LAMP1 (Santa Cruz Biotechnology, Inc., H4A3; sc-20011). The coverslips were mounted in SlowFade Gold Reagent (Invitrogen, S36936), and images were obtained using an Olympus FV1000 laser-scanning confocal microscope (Olympus, Tokyo, Japan).
Yeast 2-hybrid analysis
Yeast 2-hybrid analysis was performed as described in a previous report.52 The open reading frames of Kog1 and Gtr2 mutants were cloned into the pGAD and pGBD vectors respectively.52 Two OD600 units of PJ69-4A cells harboring these vectors were dissolved in 200 µl of dH20, and then serially 10-fold diluted in a 96-well plate. The cells were inoculated, using a 48-pin replicator (V&P Scientific, VP407AH), onto SC medium lacking leucine and uracil or lacking leucine, uracil, and histidine.
TAP affinity isolation assay
From a culture of logarithmically growing cells in SD medium containing 0.5% casamino acids without uracil, 60 OD600 units of cells were collected. These cells were resuspended in 10 ml of SD medium containing 0.5% casamino acids, 1 M sorbitol, and 300 units of Zymolyase 100T (Nacalai, 07665-55), and then incubated for 1 h at 30 °C to convert cells into spheroplasts. The cells were washed twice with 10 ml of ice-cold 50 mM Tris-Cl, pH 7.5, containing 1 M sorbitol, and then resuspended in 1,200 µl of lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 20 mM MgCl2, 0.5% NP-40 [Wako, 160-21691]) supplemented with protease inhibitors (10 μg/ml pepstatin A [Peptide Instituite, 4397], 20 μg/ml leupeptin [Sigma, L2884], 20 μg/ml benzamidine [Wako, 028-09481], 40 μg/ml aprotinin [Sigma, A1153], 1 mM phenylmethylsulfonyl fluoride [Wako, 164-12181]) in order to burst the cells. Cell lysates were cleared by centrifugation at 20,000 g at 4 °C for 10 min. Fifteen microliters of 50% slurry of IgG–Sepharose 6 Fast Flow (GE Healthcare, 17-0969) were washed once with 1 ml of lysis buffer, mixed with 1 ml of the lysate, and incubated for 45 min at 4 °C. The beads were washed 10 times with 1 ml ice-cold lysis buffer, and boiled with 2 × SDS-PAGE sample buffer to elute the proteins. For immunoblot analysis, rabbit polyclonal anti-protein A (Sigma, P3775) and mouse monoclonal anti-GST (Cell Signaling Technology, 26H1; 2624S) were used. Intensities of the immunoreactive bands were measured using the ImageJ software.
Cultured mammalian cells
HeLa and MCF7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma, D6546) containing 10% fetal bovine serum supplemented with 4 mM L-glutamine in an atmosphere containing 5% CO2 at 37 °C. Transient transfections were performed using Lipofectamine 2000 or RNAiMAX Reagent (Invitrogen, 12566014, 13778150).
siRNA-mediated knockdown in cultured mammalian cells
Cells were transfected with siRNA twice. In each transfection, the final RNA concentration was 10 nM. Cells were used in experiments 2.5 d after transfection. The siRNA sequences in this study were as follows: NPRL2, AGCCAGAGCU GCAGAACAAd TdT and GCAAUGCUCU CCUCUUCAAd TdT; NPRL3, GCGUAGUUCG GCUUCACAUd TdT and CCACUGAACC AGAGGAUGA- dTdT; and control, UUCUCCGAAC GUGUCACGUd TdT.
Immunoblot analysis
Yeast cells were harvested by centrifugation, resuspended in 0.3 M NaOH containing 1% β-mercaptoethanol and incubated for 5 min. Then, 15% (v/v) ice-cold trichloroacetic acid was added and the samples were incubated for 10 min on ice. Cells were washed once with 600 µl ice-cold acetone, and 2× SDS-PAGE sample buffer was added; the pellet fractions were dissolved by sonication. Proteins were extracted by boiling at 100 °C for 5 min. The samples were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 1% skim milk in 0.1% Tween-20/TBS (25 mM Tris, 150 mM NaCl, 2 mM KCl, pH 7.4), and then incubated with the following antibodies: rabbit polyclonal anti-Atg13 (gift from Dr Yoshinori Ohsumi, Tokyo Instituite of Technology), mouse monoclonal anti-Pgk1 (Life Technologies, 459250), anti-Tor1 antibody (Santa Cruz Biotechnology Inc, sc-11900). Immunoreactive bands were detected using horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, 111-035-003) and luminol solution (ECL plus; GE Healthcare, RPN2132).
Real-time quantitative PCR
Total RNA was isolated from cells using the TRIzol reagent (Invitrogen, 10296010) and converted to cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, 04379012001). The cDNA was amplified and quantified by real-time PCR on a PRISM 7900HT (ABI) using Power SYBR Green PCR master mix (ABI, 4367659). The primer sequences used in this study were as follows: NPRL2, TGATGCCCAG GCCAAGAC and GCCAGCCAGC TTTTTAACAA TG; NPRL3, TGCCCTAGTG CGGGTGAT and GGGCTAGCTG CTGCAGGTT; and ACTB (encoding β-actin), CCAGCTCACC ATGGATGATG and ATGCCGGAGC CGTTGTC.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr Ted Powers (University of California), Dr Kuninori Suzuki (University of Tokyo), Dr Koji Okamoto (Osaka University), Dr Scott D Emr (Cornell University), Dr Charles Boone (University of Toronto), Dr Hayashi Yamamoto and Dr Yoshinori Ohsumi (Tokyo Institute of Technology) for various yeast strains, antibodies, and plasmids. We would also like to express our gratitude to the members of our laboratories for helpful discussions about these experiments. This work was supported in part by the Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.
Glossary
Abbreviations:
- ALP
alkaline phosphatase
- ATG
autophagy-related
- CMAC
7-amino-4-chloromethyl coumarin
- GAP
GTPase-activating protein
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factor
- GTP
guanosine triphosphate
- GTR
GTp-binding protein Resemblance
- Kog1
Kontroller Of Growth 1
- NPR
nitrogen permease regulator
- PBS
phosphate-buffered saline
- siRNA
small interfering RNA
- TAP
tandem affinity purification TBS, Tris-buffered saline
- Tco89
Tor Complex One 89
- TORC1
target of rapamycin complex 1
- WT
wild-type
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 3.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–65. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 4.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–58. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binda M, Péli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–73. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111:11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 8.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–57. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 9.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, Péli-Gulli M-P, Yang H, De Virgilio C, Ding J. Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1. Structure. 2012;20:2151–60. doi: 10.1016/j.str.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Kogan K, Spear ED, Kaiser CA, Fass D. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol. 2010;402:388–98. doi: 10.1016/j.jmb.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 12.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–32. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 15.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 16.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 17.Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics. 2011;10:006478. doi: 10.1074/mcp.M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell. 2011;22:4124–33. doi: 10.1091/mbc.E11-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 2009;5:e1000515. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–67. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–67. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 24.Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–62. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 25.Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7:1819–30. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–68. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 27.Jeong J-H, Lee K-H, Kim Y-M, Kim D-H, Oh B-H, Kim Y-G. Crystal structure of the Gtr1p(GTP)-Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J Biol Chem. 2012;287:29648–53. doi: 10.1074/jbc.C112.384420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panchaud N, Péli-Gulli M-P, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal. 2013;6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- 29.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–6. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan K-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–24. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 32.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202:1107–22. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsun Z-Y, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y-M, Stone M, Hwang TH, Kim Y-G, Dunlevy JR, Griffin TJ, Kim DH. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol Cell. 2012;46:833–46. doi: 10.1016/j.molcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–71. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–54. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ecker N, Mor A, Journo D, Abeliovich H. Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy. 2010;6:879–90. doi: 10.4161/auto.6.7.12753. [DOI] [PubMed] [Google Scholar]
- 38.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–74. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 39.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–11. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutter BM, Wu X, Laxman S, Tu BP. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–15. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene. 2006;25:6436–46. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 42.Menon S, Yecies JL, Zhang HH, Howell JJ, Nicholatos J, Harputlugil E, Bronson RT, Kwiatkowski DJ, Manning BD. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci Signal. 2012;5:ra24. doi: 10.1126/scisignal.2002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji L, Nishizaki M, Gao B, Burbee D, Kondo M, Kamibayashi C, Xu K, Yen N, Atkinson EN, Fang B, et al. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2002;62:2715–20. [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Wang F, Haraldson K, Protopopov A, Duh FM, Geil L, Kuzmin I, Minna JD, Stanbridge E, Braga E, et al. Functional characterization of the candidate tumor suppressor gene NPRL2/G21 located in 3p21.3C. Cancer Res. 2004;64:6438–43. doi: 10.1158/0008-5472.CAN-03-3869. [DOI] [PubMed] [Google Scholar]
- 45.Kowalczyk MS, Hughes JR, Babbs C, Sanchez-Pulido L, Szumska D, Sharpe JA, Sloane-Stanley JA, Morriss-Kay GM, Smoot LB, Roberts AE, et al. Nprl3 is required for normal development of the cardiovascular system. Mamm Genome. 2012;23:404–15. doi: 10.1007/s00335-012-9398-y. [DOI] [PubMed] [Google Scholar]
- 46.Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–83. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–8. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 48.Tong AHY, Boone C. 16 High-Throughput Strain Construction and Systematic Synthetic Lethal Screening in. Methods in Microbiology. 2007;36:369–707. doi: 10.1016/S0580-9517(06)36016-3. [DOI] [Google Scholar]
- 49.Shirahama-Noda K, Kira S, Yoshimori T, Noda T. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J Cell Sci. 2013;126:4963–73. doi: 10.1242/jcs.131318. [DOI] [PubMed] [Google Scholar]
- 50.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 54.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–53. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 55.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–62. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 56.Nakatogawa H, Ishii J, Asai E, Ohsumi Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 2012;8:177–86. doi: 10.4161/auto.8.2.18373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.