Abstract
Background
Biodefense vaccines against Category B bioterror agents Burkholderia pseudomallei (BPM) and Burkholderia mallei (BM) are needed, as they are both easily accessible to terrorists and have strong weaponization potential. Burkholderia cepaciae (BC), a related pathogen, causes chronic lung infections in cystic fibrosis patients. Since BPM, BM and BC are all intracellular bacteria, they are excellent targets for T cell-based vaccines. However, the sheer volume of available genomic data requires the aid of immunoinformatics for vaccine design. Using EpiMatrix, ClustiMer and EpiAssembler, a set of immunoinformatic vaccine design tools, we screened the 31 available Burkholderia genomes and performed initial tests of our selections that are candidates for an epitope-based multi-pathogen vaccine against Burkholderia species.
Results
Immunoinformatics analysis of 31 Burkholderia genomes yielded 350,004 9-mer candidate vaccine peptides of which 133,469 had perfect conservation across the 10 BM genomes, 175,722 had perfect conservation across the 11 BPM genomes and 40,813 had perfect conservation across the 10 BC genomes. Further screening with EpiMatrix yielded 54,010 high-scoring Class II epitopes; these were assembled into 2,880 longer highly conserved ‘immunogenic consensus sequence’ T helper epitopes. 100% of the peptides bound to at least one HLA class II allele in vitro, 92.7% bound to at least two alleles, 82.9% to three, and 75.6% of the binding results were consistent with the immunoinformatics analysis.
Conclusions
Our results show it is possible to rapidly identify promiscuous T helper epitopes conserved across multiple Burkholderia species and test their binding to HLA ligands in vitro. The next step in our process will be to test the epitopes ex vivo using peripheral leukocytes from BC, BPM infected humans and for immunogenicity in human HLA transgenic mice. We expect that this approach will lead to development of a licensable, pan-Burkholderia biodefense vaccine.
Background
Due to their exceptionally high virulence in animals and humans, and their potential for weaponization as aerosols, BP and BPM are both classified as category B bio-threat agents. In addition to use as a countermeasure, Burkholderia vaccines would also contribute to improving human health in certain patient populations (such as immunocompromised patients) and sectors of the globe (such as Thailand) most affected by exposure to these pathogens.
Attempts to develop both whole-cell killed and live attenuated vaccines against Burkholderia species, have failed to result in a complete protective immune response in mice. Ulrich et al. developed two differently attenuated strains of B. mallei (a capsule-negative mutant and a branched-chain amino acid auxotroph) to protect against aerosolized B. mallei challenge. No protection was observed to the capsule-negative mutant, but the auxotroph conferred a slight protective advantage although the mice did not clear the infection [1]. Other vaccine targets include the capsular polysaccharides and LPS, as there is significant genetic and structural conservation between the capsular polysaccharides of these species [2]. Recently, subunit vaccines against BC have shown promise. Mice nasally immunized with Burkholderia multivorans outer membrane proteins rapidly resolved pulmonary infections following B. multivorans challenge and also elicited cross-protection against B. cenocepacia [3, 4]. Although B. cepaciae proteins that appear to be protective have been identified, no vaccine against BC currently exists [5]. To date, no vaccine for any pathogenic Burkholderia species is approved for human use.
Although antibodies can protect against severe infection by BM, passive prophylaxis has not been shown to confer sterilizing protective immunity. This is likely due to Burkholderia’s capability of latent long-term intracellular infections. Cell-mediated immune response, in conjunction with a humoral response, may be required to successfully protect against infection with Burkholderia species, and to clear intracellular infections. In general, it is believed that robust cell-mediated immune responses to Burkholderia will be required for an effective protective or therapeutic vaccine [6].
Evidence for protective cellular immune response to BPM infection comes from several live attenuated vaccine studies in mice and suggests that cell-mediated immunity is critical. Immunization of C57BL/6 mice with a mutant of BPM (aroC) deficient in aromatic amino acid synthesis resulted in sterile immunity [7]. BALB/c mice inoculated with a BPM transposable 2D2 insertion mutant (ilvl) auxotrophic for branched chain amino acids induced a protective response and 85% survived a lethal wild type BPM challenge. However, BPM persisted in spleen, liver, kidney and lung tissues up to 30 days post challenge [8]. Splenic BPM-specific T cells, detected in immunized mice, proliferated and produced interferon-gamma in vitro in response to dead bacteria. Assessment of T cell antigen specificity indicated that subpopulations of BPM-specific T cells were responsive to secreted proteins. Adoptive immunization of severe combined immunodeficiency mice with T cells from 2D2 live-attenuated BPM mutant-immunized mice resulted in increased survival compared to naïve T cell recipients. This suggests that 2D2 immunization can generate T cell-mediated immunity [9]. CD4+ and CD8+ cell depletion studies argue that CD4+ cells, but not CD8+ cells, mediated this protection in vivo.
Cell mediated immune response to antigens produced by live organisms are important to protection from Burkholderia. In a separate study, immune responses and resistance following subcutaneous immunization with live BPM were compared with exposure to heat-killed culture filtrate and sonicated BPM antigens. Compared to heat-killed BPM, significant protection was generated in BALB/c mice following exposure to live bacteria. Thus, CD4+ T cells can mediate vaccine-induced immunity to experimental melioidosis [9]. These studies suggest CD4+ T cell recognition of processed and secreted proteins from live bacteria are crucial for disease protection. These results also suggest that the type of immune response generated in vivo is influenced by the nature of the BPM antigens, and that immune responses to those proteins that are actively secreted may be required to stimulate a protective immune response [10].
T cell epitopes are critical mediators of cellular immunity and are derived from a pathogen‘s proteins via two pathways. In one, a protein derived from an intracellular pathogen is processed and its constituent peptides bind to major histocompatability complex (MHC) Class I molecules. Alternatively, proteins derived from pathogens external to the antigen presenting cells (APCs) are processed in the proteolytic compartment; these constituent peptides bind to MHC Class II molecules. After processing and binding, MHC Class I and Class II peptide complexes are then transported to the surface of an APC, where they are exposed to interrogation by passing T cells (CD8+ and CD4+ T cells, respectively). From these different antigen processing and presentation pathways, two different T cell responses are generated: a CD4+ T helper immune response and a CD8+ cytotoxic lymphocyte immune response. After initial exposure to pathogen (or vaccine), memory T cells are established that respond more rapidly and efficiently upon subsequent exposure.
We have previously used this genome-derived epitope-based vaccine design approach to develop a prototype Francisella tularensis Type A (subsp. tularensis) vaccine that confers 60% protection against heterologous lethal respiratory challenge with the live vaccine strain (LVS), an attenuated subsp. holarctica derivative [11, 12]. To our knowledge no subunit vaccine for tularemia has achieved a comparable level of protection in this well-developed lethal respiratory challenge model in HLA transgenic mice. In parallel studies, we developed an epitope-based vaccine composed of T cell epitopes derived from sequences conserved between vaccinia and variola. This vaccine was 100% protective against intra-nasal small pox challenge in HLA transgenic mice and occurred in the absence of detectable antibody response [13]. Seven poxvirus genomes were previously the maximum number submitted for analysis by our vaccine design tools. Here we employed the same approach to a much larger set of genomic sequences, with the goal of selecting the optimal sequences for a vaccine that could protect against multiple Burkholderia species.
Methods
We utilized bioinformatics and immunological tools to identify candidate proteins from 31 Burkholderia genomes for inclusion in a multipathogen-specific prophylactic vaccine as previously published [11, 13, 14]. Details on the approach used for the multipath vaccine are provided below. We then used T cell epitope mapping tools (Conservatrix, EpiMatrix) to identify 9-mer amino acid sequences that were both highly conserved in Burkholderia genomes and potentially immunogenic. These putative epitopes were then assembled into immunogenic consensus sequence clusters (using EpiAssembler) and their in vitro binding properties tested against 5 human class II HLA alleles. We then hand-selected the best 70 clusters, of which 41 were synthesized for further testing in soluble HLA binding assays as previously described [11, 13].
Genome Collection
Genomes from 31 strains of Burkholderia were obtained from Pathema, a proprietary genome database from the J. Craig Venter Institute (http://pathema.jcvi.org/cgi-bin/Burkholderia). These included protein-coding genomes from 10 B. mallei strains (FMH: 5600 ORFS; NCTC10229: 5519 ORFS; 2002721280: 5519 ORFS; ATCC10399: 5746 ORFS; ATCC23344: 5229 ORFS; GB8: 5936 ORFS; JHU: 5559 ORFS; NCTC10247: 5869 ORFS; PRL-20: 5469 ORFS; and SAVP1: 5200 ORFS), 11 B. pseudomallei strains (1106a: 7180 ORFS; 1106b: 7223 ORFS; 1655: 6908 ORFS; 1710a: 7540 ORFS; 406e: 6866 ORFS; 576: 7400 ORFS; 668: 7135 ORFS; K96243: 6304 ORFS; MSHR346: 7588 ORFS; PAS-TEUR52237: 7140 ORFS; S13: 7253 ORFS), 2 B. ambifaria strains (AMMD: 6976 ORFS; MC40-6: 7163 ORFS), 2 B. cenocepacia strains (AU1054: 7109 ORFS; HI2424: 7227 ORFS), 5 B. multivorans strains (ATCC17616-JGI: 6779 ORFS; ATCC17616-Tohoku: 6699 ORFS; CGD1: 6572 ORFS; CGD2: 6653 ORFS; CGD2M: 6646 ORFS) and 1 B. vietnamiensis strain. B. ambifaria, B. cenocepacia, B. multivorans and B. vietnamiensis (G4: 8423 ORFS) comprise the Burkholderia cepaciae complex group [15].
Genome Alignment and Cross-walk
In order to identify proteins conserved within various Burkholderia species, the B. mallei, B. pseudomallei and B. cepaciae strains were aligned using GB8, MSHR346 and G4 as reference genomes, respectively. These intra species conserved proteins were then analyzed for inter-species conservation using a comparative genomics tool from Pathema (http://pathways.jcvi.org/comp-genomics). This identified proteins in each of the three reference genomes that have hits (defined as any two proteins with a sequence identity greater than or equal to 80%) among the selected comparison genomes.
Secretion Analysis and Conservatrix
The Phobius program was used to identify single peptides and transmembrane segments and the LipoP program was used to identify lipoprotein attachment sites in proteins from each of the 31 Burkholderia genomes (http://phobius.sbc.su.se/; http://www.cbs.dtu.dk/services/LipoP/) [16, 17]. Proteins with a signal sequence, no lipoprotein attachment sites and no more than 1 transmembrane segment were selected for further analysis (Figure 1). In order to target functionally or structurally important epitopes that are conserved between Burkholderia species, the Conservatrix algorithm parsed input sequences into component strings, typically comprised of overlapping 9-mer segments, then searched the input database for matching segments found in at least two of the three Burkholderia species and ultimately produced a sequence conservation frequency table for each 9-mer.
Figure 1. Selection of conserved and secreted Burkholderia proteins.
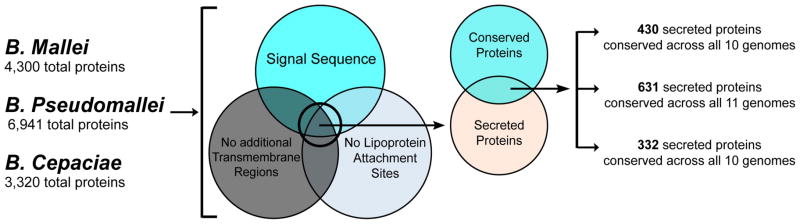
Proteins containing a signal sequence, lacking lipoprotein attachment sites and lacking predicted transmembrane domains were analyzed for conservation across 3 Burkholderia reference genomes (B. mallei GB8, B. pseudomallei MSHR346 and B. cepaciae G4).
EpiMatrix Analysis
EpiMatrix, a matrix-based epitope-mapping algorithm, was used to identify Class II HLA epitopes from the conserved 9-mer peptides identified. Potential binding of the 9-mer sequences was scored for 8 common HLA alleles that cover >90% of the human population (DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*0801, DRB1*1101, DRB1*1301 and DRB1*1501) [18, 19]. While assessment scores (Z-scores) range from approximately −3 to +3, Z-scores equal to or greater than 1.64 are generally comprise the top 5% of any given peptide set, are defined as “Hits” and considered potentially immunogenic. Z-scores above 2.32 are in the top 1% and are extremely likely to bind MHC molecule. A 9-mer frame predicted to react to at least 4 different HLA alleles is considered an EpiBar. EpiBars may be the signature feature of highly immunogenic, promiscuous class II epitopes (Figure 2); in previously published studies we have observed that these epitopes tend to be more immunogenic than epitopes that do not contain EpiBars [11–13].
Figure 2. EpiBar located on peptide MP-ICS-CLUSTERS-31-02A.
EpiMatrix analysis of the BPM amino ABC transporter, periplasmic amino acid-binding protein (GenBank ID# 237814370) identified residues 210–220 within the MP-ICS-CLUSTERS-31-02A peptide as an immunogenic EpiBar. High Z-scores (above 1.64) across 4 or more human class II MHC alleles are considered hits and constitute an EpiBar.
EpiAssembler and Blastimer
EpiAssembler was then used to identify sets of overlapping and conserved epitopes from selected 9-mer peptides, as well as assemble them into extended immunogenic consensus sequences (ICS) [20]. This algorithm iteratively identifies core highly conserved sequences that contain multiple putative T cell epitopes (clusters) and extends the core sequence right and left culling from a database of similarly highly conserved, putatively epitope rich sequences (Figure 3). The EpiMatrix scores within these ICS clusters are then aggregated to create an EpiMatrix Cluster Immunogenicity Score. As cross-reactivity with self may lead to deleterious immune responses, we evaluated the ICS clusters for homology to the human genome by BLAST analysis [21]. Peptides sharing greater than 70% identity with sequences in the human genome were eliminated from consideration. None of the 2,880 Burkholderia ICS clusters were found to have significant homology (>90%) to the human genome.
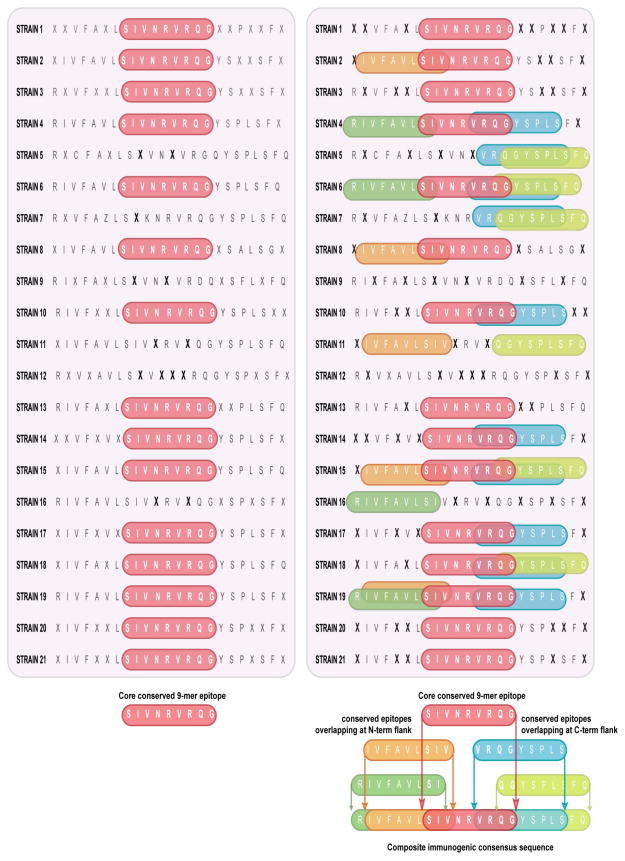
Figure 3. Constructing an Immunogenic Consensus Sequence.
(A) EpiAssembler identified a core conserved 9-mer epitope (red) and identified naturally overlapping N- and C-terminal flanking regions from other 9-mer epitopes (orange, green and blue) in a serial fashion to generate a composite immunogenic consensus sequence.
ICS Selection and Peptide Synthesis
In order to minimize technical difficulties with peptide synthesis and low water solubility stemming from hydrophobic peptides, the amino acid hydropathy score was assessed for each ICS cluster by GRAVY [22]. Each ICS sequence was constructed to contain a minimal set of T cell epitopes, as well as cover a maximum number of observed Burkholderia strains. This was accomplished by comparing the remaining ICS clusters for cross-species conservation. Selected epitopes were synthesized, purified by HPLC and verified by mass spectrometry (21st Century Biochemicals, Marlboro, MA).
Class II HLA binding assay
Purified, soluble HLA Class II DR competition binding assays were performed as previously described [23]. Briefly, non-biotinylated ICS peptides over a wide range of concentrations competed with biotinylated influenza hemagglutinin 306–318 standard peptide (0.1 M) for binding to purified DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, and DRB1*1501 (50 nM) in 96-well plates for 24 hours at 37°C. ELISA plates coated with pan anti-Class II antibodies (L243, anti-HLA-DR; BioXCell, West Lebanon, NH) were blocked with 5% FBS in PBS-0.05% Tween-20 and then bound to the DR/ peptide complexes for 1 hour at 37°C. Following extensive washing in PBS-0.05% Tween-20, the ELISA plates were developed by addition of streptavidin-europium and analyzed on a Victor3V Microtiter Plate Reader. Percent inhibition and IC50 values of the biotinylated peptide binding were calculated using SigmaPlot 11.1 software.
Results
In silico epitope mapping
Conservatrix, EpiMatrix
Comparative genomic alignment analysis of the B. mallei GB8, B. pseudomallei MSHR346 and B. cepaciae G4 genomes as references yielded a total of 3,288 proteins conserved across all 10 genomes of B. mallei, 4,682 proteins conserved across all 11 genomes of B. pseudomallei and 2,823 proteins conserved across all 10 genomes of B. cepaciae. LipoP and Phobius analyses identified 10,793 secreted core ORFs. Conservatrix analysis of these ORFs yielded 350,004 9-mer peptides; of which 133,469 had perfect conservation across the 10 BM genomes, 175,722 had perfect conservation across the 11 BPM genomes and 40,813 had perfect conservation across the 10 BC genomes. EpiMatrix analyses of these conserved Burkholderia 9-mer peptides yielded 54,010 putative Class II HLA epitopes.
EpiAssembler
Using the 54,010 unique 9-mer peptides as a starting point, EpiAssembler produced 2,880 candidate ICS clusters. Figure 3 shows a conceptual example of ICS assembly from conserved and overlapping HLA peptide epitopes using EpiAssembler.
Blastimer
Cross-conservation with the human genome may lead to deleterious anti-self immune responses to vaccines. Therefore, we used BLAST analysis to confirm that none of these ICS clusters had any significant homology to the human genome. GRAVY analysis removed 19 ICS clusters with extremely hydrophobic properties. Cross-species conservation analysis yielded 90 ICS epitopes >70% conserved between B. mallei and B. pseudomallei, 42 ICS clusters >70% conserved between B. mallei and B. cepaciae, 32 ICS clusters >70% conserved between B. pseudomallei and B. cepaciae and 20 ICS clusters >70% conserved among all 3 Burkholderia species.
Protein ontology
The top-scoring 70 Class II ICS clusters were selected for further analysis (Table 1). These ICS cluster peptides indeed correspond to Burkholderia proteins predicted to have a variety of cellular functions (Figure 4). Many of these proteins, such as transmembrane transporters, transmembrane and extracellular receptors, cell wall and membrane biogenesis proteins and flagellar proteins, are predicted to function at the bacterial cell surface, even though they passed the initial screen for putative secreted proteins. In at least four cases, transcription factors have been identified as secreted proteins [24–28], thus while rare, this is not an unprecedented observation. Based on the previous examples, secretion of these proteins may be indicative of a highly immunogenic protein, therefore we have elected to retain these epitopes in our vaccine development program.
Table 1. Class II Epitopes selected for HLA-binding analyses.
Column 1: Immunogenic consensus sequence peptide cluster ID; column 2: amino acid sequence for each peptide; column 3: protein description for the parent protein from GenBank; columns 4–6: GenBank ID reference numbers for the reference genomes B. mallei (BM) GB8, B. pseudomallei (BPM) MSHR346 and B. cepaciae (BC) G4.
| CLUSTER LABEL | SEQUENCE | SOURCE PROTEIN | GenBank ID | ||
|---|---|---|---|---|---|
| BM(GB8) | BPM (MSHR346) | BC(G4) | |||
| BM-BC-02 | GNRALLLLVLAAISLMASRGE | polyhydroxyalkanoate depolymerase | 67643796 | 237811476 | 134296512 |
| BM-BPM-07 | LKSVLKMVKVYLAVKRRLQPGD | DNA-directed RNA polymerase, beta subunit | 67640298 | 237814156 | 134294435 |
| BM-BPM-BC-03 | YVERVNVLKLNLALDAAQAALA | K+-transporting ATPase, C subunit | 67639435 | 237811537 | 134296470 |
| BM-BPM-01 | AFNFVRQFSLNLRADLIGA | oxidoreductase, short-chain dehydrogenase/reductase family protein | 67643921 | 237813600 | 134294849 |
| BM-BC-03 | FAHLKSVLKVLSAFNPSVKGK | BC nitrate/sulfonate/bicarbonate transporter, periplasmic ligand binding protein | 67641562 | 237814431 | 134294167 |
| BM-BPM-03 | KGEWKLYLHSIRRLLRIDRPGF | Beta-N-acetylhexosaminidase, chitobiase | 238561344 | 237810743 | 134297015 |
| BM-BPM-10 | PMGVYSKRVNSLAALRARPF | D-methionine ABC transporter, periplasmic D-methionine-binding protein | 67642900 | 237813281 | 134295093 |
| BM-BPM-05 | SKLIFSVNSVLAYQAAQQ | cystine-binding periplasmic protein FliY, putative amino acid ABC transporter | 238563032 | 237508413 | 134293667 |
| BM-BPM-06 | ALVLNVLNAALANNVTLD | glycosy hydrolase family protein | 67639146 | 237812240 | 134295413 |
| BM-BPM-BC-05 | ALMSILQAMKAAGALRAAAL | flagellar P-ring protein | 238562528 | 237810465 | 134297194 |
| BM-BC-04 | SGIFRGIKRYFSANPPLAKLS | N-acetylmuramoyl-L-alanine amidase AmiC | 238561654 | 237811107 | 134296748 |
| BM-BPM-22 | MRQFQSIVAQLRAKPLHLM | intracellular PHB depolymerase | 67640207 | 237812617 | 134288123 |
| BM-BPM-12 | VIPWQMGIVAKLLHVMNSGGFKMK | oxidoreductase, short chain dehydrogenase/reductase family | 67641011 | 237810610 | 134297108 |
| BM-BPM-18 | LEQWLRGVKANLARKATGG | ferric iron uptake ABC transporter (FeT) family, periplasmic iron-binding protein | 67639460 | 237811665 | 134292266 |
| BPM-BC-03 | RTALQRSRNLVSIRILSLAL | penicillin-binding protein, 1A family | 67642044 | 237812726 | 134294482 |
| BM-BPM-19 | GSMVASFVLMRLSSVTHTT | ricin-type beta-trefoil lectin domain/galactose oxidase domain protein | 238562996 | 237508926 | 134296706 |
| BM-BC-05 | LPMYTGLHGANRLASNSLLEMA | L-aspartate oxidase | 238562659 | 237811178 | 134296677 |
| BM-BPM-09b | ARGLVYADGILLSNLLGSSYSMT | TonB-dependent receptor | 238562879 | 237813367 | 134295041 |
| BM-BPM-15 | SKASKQALNLLVNAKVPTRRA | Phospholipase D (PLD) family protein | 67642027 | 237812695 | 134295648 |
| BM-BPM-11 | TEPFSAFINVLAHPSAVMI | Glutathione-binding protein GsiB | 67642278 | 237509488 | 134293406 |
| BM-BPM-BC-01 | KYNDRLSLRRAQAVKSYLV | ompA family protein | 238561689 | 237813303 | 134295071 |
| BM-BPM-02 | ANSMNAFRTNRSLSAAHL | hypothetical protein BURPS1655_A2260 | 67641082 | 237811791 | 134295372 |
| BM-BPM-20 | LTRYNRSFFYALAVYQLG | lytic murein transglycosylase B | 67642905 | 237813286 | 134295088 |
| BM-BPM-BC-04 | VCDKVLSQAKALLAKAG | conserved hypothetical protein | 238563606 | 237811018 | 134293794 |
| BPM-BC-05a | QRHLQRMFKVYLNVSPTHYYR | transcriptional regulator, AraC family protein | 238563940 | 237510183 | 134292187 |
| BPM-BC-01 | KIYEMLLAYRIERALTKDQI | penicillin-binding protein, 1A family | 238562280 | 237814104 | 134294482 |
| BM-BPM-09a | QAGFRFQHVSNAGILLS | lipid A 3-O-deacylase (PagL) superfamily | 238561336 | 237810753 | 134297010 |
| BM-BC-06a | NEFFVPLNVRVQKARRVGAAL | N-acetylmuramoyl-L-alanine amidase | 238561654 | 237811107 | 134296748 |
| BM-BC-07 | RNMWQSHRIKLIALSAFTLAE | conserved hypothetical protein | 67643124 | 237814304 | 134294271 |
| BM-BPM-08 | LFDVSALLNQMRRNPSAYGLST | GDSL-like lipase/acylhydrolase domain/outer membrane autotransporter barrel domain protein | 67642947 | 237812898 | 134293351 |
| BPM-BC-08 | NPFMMQQALNKLEGKPV | carboxyl-terminal protease | 238563023 | 237810682 | 134297049 |
| BM-BPM-13 | RDQVLIRFANRSCSVLVM | ATP-dependent dead/deah box RNA helicase DbpA | 67639524 | 237510128 | 134292614 |
| BPM-BC-02 | LRTLEELARLVRLSQRHLQRLQ | transcriptional regulator, AraC family | 238563940 | 237510183 | 134292187 |
| BPM-BC-07 | QEQFKLLLIRTYSGALAQ | ABC transporter, periplasmic ligand binding protein, toluene tolerance TtgeD-like protein | 67641601 | 237814073 | 134294509 |
| BM-BPM-BC-06 | AAAIKYLEALRVELRPA | oxidoreductase, short chain dehydrogenase/reductase family | 67641011 | 237810610 | 134297108 |
| BM-BPM-BC-09 | HHRIYYRLHKLAKQWHAQV | Phosphate regulon sensor protein PhoR | 67641079 | 237811788 | 134295369 |
| BM-BPM-BC-08 | VTTYLKAVQAAGSTDSLLV | putative amino acid uptake ABC transporter, periplasmic amino acid-binding protein | 238561553 | 237814370 | 134294218 |
| BM-BPM-16b | RRRFDVSIALSSLAGGI | conserved hypothetical protein | 238561938 | 237810872 | 134295646 |
| BPM-BC-10 | YGVIKLNWVILEENVLRQR | mannitol ABC transporter, periplasmic mannitol-binding protein | 67641206 | 237811073 | 134296800 |
| BM-BPM-BC-11 | NGNVSSLGNAKSLRGGTL | flagellar basal body P-ring protein (Flgl) | 238562528 | 237810465 | 134297194 |
| BM-BC-08 | HPMLYYRALTQGLNQTLA | tRNA delta(2)-isopentenylpyrophosphate transferase | 67642140 | 237813667 | 134294835 |
| BM-BPM-BC-13 | NKILRAFLKSKGVKDS | putative amino acid ABC transporter, periplasmic amino acid-binding protein | 67644003 | 237813471 | 134294953 |
| BM-BPM-BC-14 | LQNFYVLTRYNRSADVKRFI | lytic murein transglycosylase B | 67642905 | 237813286 | 134295088 |
| BM-BC-09 | DYIYQKLLGVRGVFA | Tol-Pal system beta propeller repeat protein Tol | 238561025 | 237813605 | 134294844 |
| BM-BPM-24b | ARGFILGLANAGGD | branched-chain amino acid ABC transporter | 238561553 | 237814370 | 134294218 |
| BM-BPM-BC-15 | TLRIRGLHNAANALAAVS | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase | 238561283 | 237813921 | 134294645 |
| BM-BPM-04a | NAAFNFNNAIRFTSVFP | outer membrane porin protein | 238561395 | 237509809 | 134295282 |
| BPM-BC-05b | GTAIYYRLHKLAKQWHAQV | phosphate regulon sensor kinase PhoR | 67641079 | 237811788 | 134295369 |
| BPM-BC-09b | AEVALQRSLPAPAAH | tetratricopeptide repeat protein | 238561327 | 237810775 | 134296995 |
| BM-BPM-21 | NVCQLRVSRNALASIAVL | putative lipoprotein, parallel beta-helix repeat protein | 67642171 | 237811603 | 134296638 |
| MP-ICS-CLUSTERS-31-02A | ASDFSSFLLQAQSSKAQILGLA | ABC branched-chain amino acid family transporter | 238561553 | 237814370 | 134294218 |
| MP-ICS-CLUSTERS-31-03 | GEPRVNVLKLNLALDAAQAAH | K+-transporting ATPase, C subunit (kdpC) | 67639435 | 237811537 | 134296470 |
| MP-ICS-CLUSTERS-31-01 | KKNFTAMLRLDHNRALSQLAA | malate dehydrogenase | 67642757 | 237508279 | 134293103 |
| MP-ICS-CLUSTERS-31-06 | TQTLANMLANLGISINNGSANG | flagellar P-ring protein Flgl (flgl) | 238562528 | 237810465 | 134297194 |
| MP-ICS-CLUSTERS-31-02B | RAAAKVILNARTGSIVMNQ | flagellar P-ring protein (flgl) | 238562528 | 237810465 | 134297194 |
| MP-ICS-CLUSTERS-31-04B | SNPYKLDVAKAKALLAKAG | ABC transporter, periplasmic substrate-binding protein | 238563343 | 237508894 | 134293794 |
| MP-ICS-CLUSTERS-31-05 | PRSYFRGLNFLLNEQP | tetratricopeptide repeat protein | 67642911 | 237813292 | 134295082 |
| MP-ICS-CLUSTERS-31-12 | LERFGRYHATLSPGLNIV | SPFH domain protein/band 7 family protein | 67641689 | 237812784 | 134296009 |
| MP-ICS-CLUSTERS-31-11B | ALNDRLMLAYLTKNAQLPLRPG | amino acid ABC transporter, periplasmic amino acid-binding protein | 67640271 | 237812103 | 134295624 |
| MP-ICS-CLUSTERS-31-08 | TAAIKGMVSSLDPHSSYLDK | carboxyl-terminal protease [3.4.21.-] | 238563023 | 237810682 | 134297049 |
| MP-ICS-CLUSTERS-31-11A | TVELHFALGNLFRRR | tetratricopeptide repeat protein | 67642911 | 237813292 | 134295082 |
| MP-ICS-CLUSTERS-31-09 | VPTLQKNAVPKLVDGA | conserved hypothetical protein | 238561685 | 237813300 | 134295073 |
| MP-ICS-CLUSTERS-31-16 | ARGVRGIINFSGGLRQDLCEG | conserved hypothetical protein | 67641022 | 237810595 | 134297116 |
| MP-ICS-CLUSTERS-31-20 | GETYTASVRMQLDTALMPKPF | conserved hypothetical protein | 67642458 | 237810345 | 134297371 |
| MP-ICS-CLUSTERS-31-04A | ARQFKSLEDLKGKKL | amino acid ABC transporter, periplasmic amino acid-binding protein | 67640271 | 237812103 | 134295624 |
| MP-ICS-CLUSTERS-31-13 | DDGIKGMLGHFNSGTTIPASRI | putative periplasmic substrate-binding protein | 67643100 | 237814333 | 134294246 |
| MP-ICS-CLUSTERS-31-18 | REKMKKLYAPFNRNHERTIYMDV | UDP-glucose 6-dehydrogenase (udg) | 67642910 | 237813291 | 134295083 |
| MP-ICS-CLUSTERS-31-15 | DNNFRYSRAVLDACL | ADP-L-glycero-D-manno-heptose-6-epimerase (rfaD) | 67642908 | 237813289 | 134295085 |
| MP-ICS-CLUSTERS-31-14 | VEGVRVLAGVNLPMLVRA | PTS system, mannose/fructose/sorbose (Man | 67640741 | 237810677 | 134297053 |
| MP-ICS-CLUSTERS-31-19 | VPSYVYQAGLKSFDDI | ABC transporter, quaternary amine uptake transporter (QAT) family | 238563533 | 237509058 | 134292111 |
Figure 4. Functional classification of identified immunogenic consensus sequence cluster source proteins.
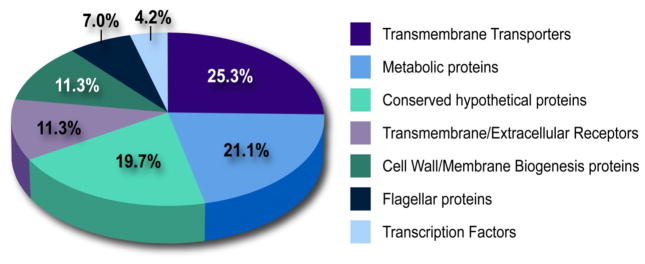
Functional categories are based on cellular biological processes ascertained from manual gene ontology analysis using the UniProt protein database.
BLAST against other bacterial proteins
A BLAST search was performed for the epitopes identified in this manner against non-human, bactetrial protein. No hits were identified that had greater than 70% conservation (six out of nine amino acid residues conserved). Therefore, these epitopes are relatively unique and unlikely to be cross reactive with other commensals and other pathogens. Vaccination with these epitopes would be expected to drive a pan-Burkholderia immune response; that will be the focus of the next stage of our gene-to-vaccine program.
Class II HLA-binding analyses
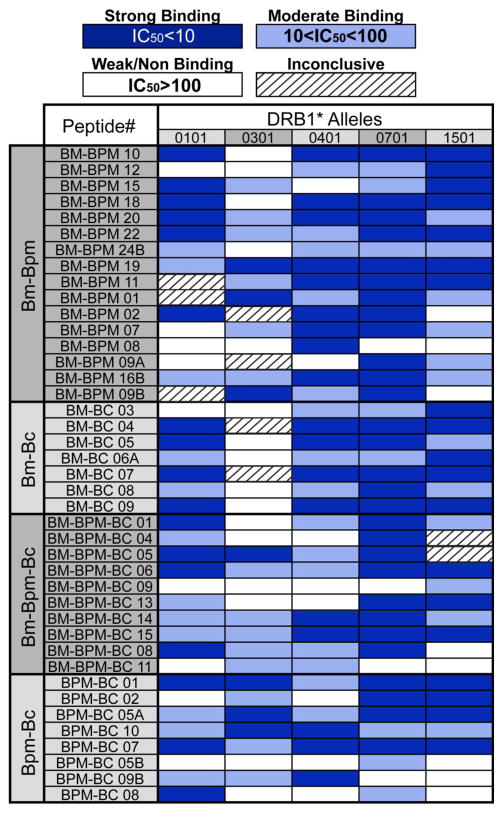
Peptide binding affinities for HLA DRB*0101, DRB1*0301, DRB1*0401, DRB1*0701, and DRB1*1501 were determined in competitive binding assays. Of the 205 ICS peptide-HLA binding interactions assayed, 44% displayed strong binding (IC50<10), 30% showed moderate binding (10<IC50<100) and 22% displayed weak or non-binding (IC50>100). In only 9 cases, the HLA binding results were inconclusive (Figure 5).
Figure 5. MHC Class II HLA-binding analysis for promiscuity.
Column 1: Inter-Burkholderia species conservation of immunogenic consensus sequence (ICS) peptides (B. mallei = BM, B. pseudomallei = BPM, B. cepaciae = BC); column 2: ICS peptide ID; columns 3–7: ICS peptide binding affinities to the human HLA class II alleles DRB1*0101, DRB1*0301 DRB1*0401 DRB1*0701 DRB1*1501. Weak or no affinity (IC50>100 M = white); moderate affinity (100 M >IC50>10 M = light blue); strong affinity (IC50<10 M = dark blue); inconclusive binding (hash).
All peptides bound to at least one of the HLA alleles for which they were predicted, 92.7% bound to two alleles for which they were predicted, 82.9% bound to three alleles for which they were predicted. These data support the use of this approach for the high-volume genomic screening for vaccine candidates. Therefore, we proceeded to the next step in our development process with this highly conserved, highly promiscuous candidate epitope cohort.
Comparison between computational predictions and actual in vitro HLA binding results show 75.6% overall predictive success rate when excluding inconclusive results (Figure 6). Epitope prediction success was also compared for each class II MHC allele. Successful binding prediction was 76.3% for DRB1*0101, 59.5% for DRB1*0301, 82.9% for DRB1*0401, 78.6% for DRB1*0701 and 79.5% for DRB1*1501. A lack of accord between positive binding predictions and actual binding data was observed at 23.7% for DRB1*0101, 40.5% for DRB1*0301, 17.1% for DRB1*0401, 9.5% for DRB1*0701 and 20.5% for DRB1*1501 (Figure 6). This could be due to the affinity of the competitor peptide (bound too tightly to compete off), peptide synthesis, problems with peptide aggregation in the in vitro assay, or lack of predictive accuracy by the EpiMatrix tool. In a large, retrospective comparison of the EpiMatrix with other online tools, EpiMatrix was as accurate or more accurate than other available epitope prediction tools [29]. Therefore, it is likely that much of the discrepancy between predictions and HLA binding is due to physical interference in the in vitro assay.
Figure 6. EpiMatrix binding prediction success.
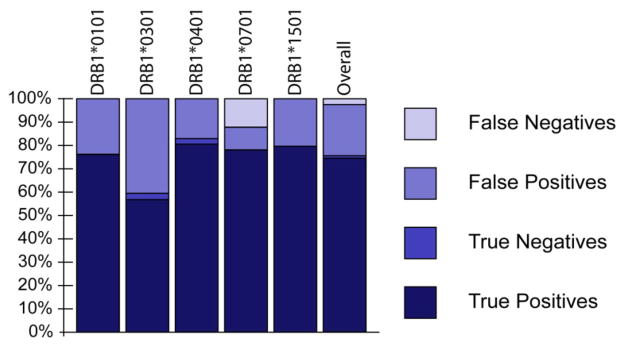
The HLA class II binding result for each ICS peptide was compared to its EpiMatrix predictive binding scores for each human HLA class II allele. True positives (dark blue) reflect correctly predicted HLA-binding peptide results. False positives (medium blue) reflect incorrectly predicted HLA-binding peptide results. True negatives (light blue) reflect correctly predicted non-HLA-binding peptide results. False negatives (grey) indicate incorrectly predicted non-HLA-binding peptide results.
Discussion
Using publically accessible bioinformatics tools we identified secreted proteins conserved between 31 different Burkholderia genomes and used our validated vaccine design toolkit to select highly conserved class II epitope clusters as potential T cell epitopes for a T cell directed vaccine. These peptides were then evaluated for their in vitro binding properties to 5 different human class II HLA alleles.
Burkholderia mallei (BM) and Burkholderia pseudomallei (BPM) are responsible for the severe diseases glanders and melioidosis, respectively. Burkholderia mallei is a Gram-negative, non-motile bacillus that requires a mammalian host environment for survival (Whitlock et al., 2007). BM is the etiological agent of glanders in donkeys, mules, horses and occasionally humans. Horses are the predominant natural reservoir for BM and transmission to humans occurs through direct contact with infected animals [30]. While BM is generally confined to animal species, it can cause severe respiratory infection when aerosolized and for that reason is considered, along with BPM, a Category B pathogen by the NIAID Biodefense Research Agenda [31].
Burkholderia pseudomallei, the etiological agent of melioidosis, is a Gram-negative, facultatively anaerobic, motile bacillus that is responsible for a broad spectrum of illnesses in both humans and animals. The incidence of disease is particularly high in Southeast Asia. In Thailand, an estimated 20% of community-acquired septicemias and approximately 40% of deaths due to complications associated with bacterial sepsis can be attributed to this organism [32, 33]. Antibiotic therapy is the first line of defense post-exposure but faces significant challenges. Despite extensive antibiotic regimens, recurrence of infection ranges from 13% to 26% and therapy choice is limited by antibiotic resistance [32, 34, 35]. As a result, mortality rates as high as 50% in northeast Thailand and ~20% in Northern Australia have been observed [32, 34, 36]. However, infection with this pathogen in tropical regions of the world may be underreported. Recrudescence may occur: reactivation of latent BPM in Vietnam veterans up to 18 years after their last exposure has been reported [37].
Burkholderia cepaciae (BC) is a Gram-negative, non-sporulating motile bacillus found in a variety of both aquatic and terrestrial environments [38]. BC is an opportunistic human pathogen associated with life-threatening pulmonary infections in immunocompromised individuals and individuals with cystic fibrosis [39].
Based on their highly infectious properties in aerosol form and extremely high virulence, BPM and BM are both classified as category B bioterrorist agents. There is currently no vaccine available for any Burkholderia species. Due to the potential bioterrorism threat, the development of a safe and effective Burkholderia vaccine is a national and worldwide goal. Addition of BC sequences may contribute to the development of a vaccine that could prevent disease in select cystic fibrosis patient populations in the United States.
Conventional vaccines using whole killed, whole protein, or live attenuated have offered success for over a century. However, development of a Burkholderia vaccine through this approach has proven elusive. Inactivated whole cell vaccines provided some Burkholderia protection in mouse models, but protection in intravenously challenged mice and sterile immunity was unsuccessful [10, 40–42]. Furthermore, killed, whole-cell BM vaccines did not protect the vaccinated mice from a live challenge (>300 50% lethal doses), suggesting that proteins or polysaccharides that are produced by live bacteria are critically important to protection from BPM and BM disease [43]. BPM vaccine studies using live attenuated virus, killed virus and adoptive immunization provide evidence for CD4+ T cell-mediated vaccine-induced immunity to melioidosis [9]. Despite this progress in vaccine development, Burkholderia’s propensity for latent infections along with the undefined mechanistic nature behind several attenuated Burkholderia strains pose significant challenges towards developing vaccines approved for human use. Contemporary immunome-derived vaccines have a significant advantage over conventional vaccines; the careful selection of the vaccine components through the use of computer-driven analysis should diminish undesired side effects as those observed with whole pathogen and protein subunit vaccines.
This study couples the current boon of genomic resources with our sophisticated bioinformatics and immunoinformatic tools to design candidate peptide epitopes for a multi-species Burkholderia vaccine. This approach moves away from whole protein, killed whole cell and attenuated pathogen-based Burkholderia vaccines for several reasons. Potentially dangerous cross-reactive or inert space-consuming epitopes present in canonical vaccines are not included in the vaccine. By eliminating superfluous components, epitope-based vaccines maximize their immunogenic payload as well as maximize the protective efficacy to direct a broad based immune response against multiple antigenic proteins associated with the pathogen(s) and also reduce formulation challenges and cost. Safety concerns stemming from the use of intact recombinant proteins that may have undesired biological activities, such as enzymes, immunomodulators, cross-reactivity or toxins, may also be mitigated through targeted epitope approach. These bioinformatics sequence analysis tools, epitope-mapping tools, microarrays and high-throughput immunology assays successfully identified the minimal essential vaccine components for smallpox, tularemia, Helicobacter pylori and tuberculosis vaccines [11–13]. As described here, we are also using this approach for the development of a vaccine for biodefense against multiple Burkholderia species. The tools enabling these vaccine development successes are described here, and the anticipated clinical development of immunome-derived and epitope-driven vaccines will be the subject of future reports.
Our results show it is possible to identify and in vitro validate T cell epitopes that are conserved across multiple Burkholderia species. These epitopes will be further tested in human PBMC and transgenic mice. We aim to use these epitopes for inclusion and further testing in a multi-pathogen-specific Burkholderia vaccine. We anticipate that a multi-epitope construct could be administered with an anti-LPS vaccine, already in clinical trial [44], resulting in an effective vaccine directed at providing both humoral and cellular immune response. The resulting multi-pathogen Burkholderia vaccine will benefit both the developing world and biodefense.
Acknowledgments
The authors wish to acknowledge the efforts of Ryan Tassone and Joe Desrosiers, who helped conduct the HLA-binding experiments. This study was supported by National Institutes of Health Research Grant: NIH 1U19AI082642 (PI: A.S. De Groot).
List of Abbreviations
- BPM
Burkholderia pseudomallei
- BM
Burkholderia mallei
- BC
Bukholderia cepaciae
- MHC
major histocompatability complex
- APC
antigen presenting cell
- ORF
open reading frame
- HLA
human leukocyte antigen
- ICS
immunogenic consensus sequence
- NIAID
National Institute of Allergy and Infectious Diseases
Footnotes
Disclosures
Anne S. De Groot and William Martin are senior officers and majority shareholders at EpiVax, Inc., a privately owned vaccine design company located in Providence, RI. Leonard Moise has options in EpiVax, Inc.
References
- 1.Ulrich RL, Amemiya K, Waag DM, Roy CJ, DeShazer D. Aerogenic vaccination with a Burkholderia mallei auxotroph protects against aerosol-initiated glanders in mice. Vaccine. 2005;23:1986–1992. doi: 10.1016/j.vaccine.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Perry MB, MacLean LL, Schollaardt T, Bryan LE, Ho M. Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect Immun. 1995;63:3348–3352. doi: 10.1128/iai.63.9.3348-3352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertot GM, Restelli MA, Galanternik L, Aranibar Urey RC, Valvano MA, Grinstein S. Nasal immunization with Burkholderia multivorans outer membrane proteins and the mucosal adjuvant adamantylamide dipeptide confers efficient protection against experimental lung infections with B. multivorans and B. cenocepacia. Infect Immun. 2007;75:2740–2752. doi: 10.1128/IAI.01668-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makidon PE, Knowlton J, Groom JV, 2nd, Blanco LP, LiPuma JJ, Bielinska AU, Baker JR., Jr Induction of immune response to the 17 kDa OMPA Burkholderia cenocepacia polypeptide and protection against pulmonary infection in mice after nasal vaccination with an OMP nanoemulsion-based vaccine. Med Microbiol Immunol. 2010;199:81–92. doi: 10.1007/s00430-009-0137-2. [DOI] [PubMed] [Google Scholar]
- 5.Mariappan V, Vellasamy KM, Thimma JS, Hashim OH, Vadivelu J. Identification of immunogenic proteins from Burkholderia cepacia secretome using proteomic analysis. Vaccine. 2010;28:1318–1324. doi: 10.1016/j.vaccine.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Bondi SK, Goldberg JB. Strategies toward vaccines against Burkholderia mallei and Burkholderia pseudomallei. Expert Rev Vaccines. 2008;7:1357–1365. doi: 10.1586/14760584.7.9.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srilunchang T, Proungvitaya T, Wongratanacheewin S, Strugnell R, Homchampa P. Construction and characterization of an unmarked aroC deletion mutant of Burkholderia pseudomallei strain A2. Southeast Asian J Trop Med Public Health. 2009;40:123–130. [PubMed] [Google Scholar]
- 8.Atkins T, Prior RG, Mack K, Russell P, Nelson M, Oyston PC, Dougan G, Titball RW. A mutant of Burkholderia pseudomallei, auxotrophic in the branched chain amino acid biosynthetic pathway, is attenuated and protective in a murine model of melioidosis. Infect Immun. 2002;70:5290–5294. doi: 10.1128/IAI.70.9.5290-5294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haque A, Chu K, Easton A, Stevens MP, Galyov EE, Atkins T, Titball R, Bancroft GJ. A live experimental vaccine against Burkholderia pseudomallei elicits CD4+ T cell-mediated immunity, priming T cells specific for 2 type III secretion system proteins. J Infect Dis. 2006;194:1241–1248. doi: 10.1086/508217. [DOI] [PubMed] [Google Scholar]
- 10.Barnes JL, Ketheesan N. Development of protective immunity in a murine model of melioidosis is influenced by the source of Burkholderia pseudomallei antigens. Immunol Cell Biol. 2007;85:551–557. doi: 10.1038/sj.icb.7100084. [DOI] [PubMed] [Google Scholar]
- 11.McMurry JA, Gregory SH, Moise L, Rivera D, Buus S, De Groot AS. Diversity of Francisella tularensis Schu4 antigens recognized by T lymphocytes after natural infections in humans: identification of candidate epitopes for inclusion in a rationally designed tularemia vaccine. Vaccine. 2007;25:3179–3191. doi: 10.1016/j.vaccine.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 12.Gregory SH, Mott S, Phung J, Lee J, Moise L, McMurry JA, Martin W, De Groot AS. Epitope-based vaccination against pneumonic tularemia. Vaccine. 2009;27:5299–5306. doi: 10.1016/j.vaccine.2009.06.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27:6471–6479. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Groot AS, McMurry J, Marcon L, Franco J, Rivera D, Kutzler M, Weiner D, Martin B. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine. 2005;23:2121–2131. doi: 10.1016/j.vaccine.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 15.Lipuma JJ. Update on the Burkholderia cepacia complex. Curr Opin Pulm Med. 2005;11:528–533. doi: 10.1097/01.mcp.0000181475.85187.ed. [DOI] [PubMed] [Google Scholar]
- 16.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Groot AS, Jesdale BM, Szu E, Schafer JR, Chicz RM, Deocampo G. An interactive Web site providing major histocompatibility ligand predictions: application to HIV research. AIDS Res Hum Retroviruses. 1997;13:529–531. doi: 10.1089/aid.1997.13.529. [DOI] [PubMed] [Google Scholar]
- 19.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 20.De Groot AS, Bishop EA, Khan B, Lally M, Marcon L, Franco J, Mayer KH, Carpenter CC, Martin W. Engineering immunogenic consensus T helper epitopes for a cross-clade HIV vaccine. Methods. 2004;34:476–487. doi: 10.1016/j.ymeth.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Reijonen H, Kwok WW. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods. 2003;29:282–288. doi: 10.1016/s1046-2023(02)00350-x. [DOI] [PubMed] [Google Scholar]
- 24.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 27.Urbanowski ML, Lykken GL, Yahr TL. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc Natl Acad Sci USA. 2005;102:9930–9935. doi: 10.1073/pnas.0504405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsot C, Ageron E, Penno C, Mavris M, Jamoussi K, d’Hauteville H, Sansonetti P, Demers B. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol Microbiol. 2005;56:1627–1635. doi: 10.1111/j.1365-2958.2005.04645.x. [DOI] [PubMed] [Google Scholar]
- 29.De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol. 2009;131:189–201. doi: 10.1016/j.clim.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Neubauer H, Sprague LD, Zacharia R, Tomaso H, Al Dahouk S, Wernery R, Wernery U, Scholz HC. Serodiagnosis of Burkholderia mallei infections in horses: state-of-the-art and perspectives. J Vet Med B Infect Dis Vet Public Health. 2005;52:201–205. doi: 10.1111/j.1439-0450.2005.00855.x. [DOI] [PubMed] [Google Scholar]
- 31.NIAID Biodefense Research Agenda for Catagory B and C Priority Pathogens.
- 32.White NJ. Melioidosis. Lancet. 2003;361:1715–1722. doi: 10.1016/s0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 33.Dance DA. Melioidosis: the tip of the iceberg? Clin Microbiol Rev. 1991;4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wuthiekanun V, Peacock SJ. Management of melioidosis. Expert Rev Anti Infect Ther. 2006;4:445–455. doi: 10.1586/14787210.4.3.445. [DOI] [PubMed] [Google Scholar]
- 36.White NJ, Dance DA, Chaowagul W, Wattanagoon Y, Wuthiekanun V, Pitakwatchara N. Halving of mortality of severe melioidosis by ceftazidime. Lancet. 1989;2:697–701. doi: 10.1016/s0140-6736(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 37.Koponen MA, Zlock D, Palmer DL, Merlin TL. Melioidosis. Forgotten, but not gone! Arch Intern Med. 1991;151:605–608. doi: 10.1001/archinte.151.3.605. [DOI] [PubMed] [Google Scholar]
- 38.Govan JR, Hughes JE, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 39.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar-Tyson M, Titball RW. Progress toward development of vaccines against melioidosis: A review. Clin Ther. 2010;32:1437–1445. doi: 10.1016/j.clinthera.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 41.Harding SV, Sarkar-Tyson M, Smither SJ, Atkins TP, Oyston PC, Brown KA, Liu Y, Wait R, Titball RW. The identification of surface proteins of Burkholderia pseudomallei. Vaccine. 2007;25:2664–2672. doi: 10.1016/j.vaccine.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Razak CE, Ismail G, Embim N, Omar O. Protection studies using whole cells and partially purified toxic material (PPTM) of Psedomonas pseudomallei. Malays Appl Biol. 1986;15:105–111. [Google Scholar]
- 43.Amemiya K, Bush GV, DeShazer D, Waag DM. Nonviable Burkholderia mallei induces a mixed Th1- and Th2-like cytokine response in BALB/c mice. Infect Immun. 2002;70:2319–2325. doi: 10.1128/IAI.70.5.2319-2325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cross AS. Development of an anti-endotoxin vaccine for sepsis. Subcell Biochem. 2010;53:285–302. doi: 10.1007/978-90-481-9078-2_13. [DOI] [PubMed] [Google Scholar]