Abstract
Helices are important structural/recognition elements in proteins and peptides. Stability and conformational differences between helices composed of α- and β-amino acids as scaffolds for mimicry of helix recognition has become a theme in medicinal chemistry. Furthermore, helices formed by β-amino acids are experimentally more stable than those formed by α-amino acids. This is paradoxical because the larger sizes of the hydrogen-bonding rings required by the extra methylene groups should lead to entropic destabilization. In this study, molecular dynamics simulations using the second-generation force field, AMOEBA [1], explored the stability and hydrogen-bonding patterns of capped oligo-β-alanine, oligoalanine and oligoglycine dodecamers in water. The MD simulations showed that oligo-β-alanine has strong acceptor+2 hydrogen bonds, but surprisingly did not contain a large content of 312-helical structures, possibly due to the sparse distribution of the 312-helical structure and other structures with acceptor+2 hydrogen bonds. On the other hand, despite its backbone flexibility, the β-alanine dodecamer had more stable and persistent <3.0 Å hydrogen bonds. Its structure was dominated more by multicentered hydrogen bonds than either oligoglycine and oligoalanine helices. The 31 (PII) helical structure, prevalent in oligoglycine and oligoalanine, does not appear to be stable in oligo-β-alanine indicating its competition with other structures (stacking structure as indicated by MD analyses). These differences are among the factors that shape helical structural preferences and the relative stabilities of these three oligopeptides.
Keywords: AMOEBA force field, hydrogen bond, helical conformation, beta amino acid, replica-exchange MD, polyglycine, polyalanine, H-bond persistence, H-bond interchange
Introduction
Chemists have sought to mimic the helical recognition motifs of biologically active peptides with semi-rigid structures, often non-peptidic, to optimize affinity by preorganization. Useful peptide-like helices can be formed from the amino acid homologs, β-amino and γ-amino acids [2–5]. Non-peptidic helix mimetics have been designed and prepared [6–10]. Since one seldom knows the exact conformational requirements (both static and dynamic) required for helix recognition by its cognative receptor, determination of the conformational flexibility available to helices and their mimetics is a first step to the design of a more rigid non-peptide mimetics; how much conformational flexibility should be retained in the mimetic? It has become increasingly apparent that binding entropy dominates many associations between ligands and proteins, and remains a limiting factor in molecular design [11].
The dynamic stability of helical conformations composed of α– and β-amino acid have been characterized experimentally and summarized in a review [4].
“Together with the beautiful regularity of right-to-left-to-right-handed helix and from N+→ C− to C+→N− to N+→C− oriented dipoles in every homologation step, the great surprise encountered on comparison of the helices of the proteinogenic α-amino acids, and those of their β- and γ-amino acid homologues is the fact that the stabilities of these helices increase with homologation of their component moieties! In an α-hexapeptide in MeOH or H2O solution, no 3.613-helix (α-helix) was detectable; in protein/enzyme structures a helix length of 15 to 20 amino acids is typical (ca. five turns and ten H-bridges). The 314-helix has been detected by NMR spectroscopy in solution with a β-peptide consisting of only six acyclic amino acids, and in γ-peptides a chain length of a mere four amino acids appears to suffice for a solution helix structure to be detectable. Helix stability thus increases with homologation of the amino acids, even though the number of possible conformations becomes larger with each homologation step, with the introduction of an additional C-C bond, while, at the same time, the number of H-bonds per atom in the chain decreases.” Modified from Seebach et al. review [12].
The enhanced stability of helices formed by β-and γ-amino acids seems problematic as Prof. Dieter Seebach pointed out, because of the larger sizes of the hydrogen-bonding rings required by the extra methylene groups that should lead to entropic destabilization. In order to examine possible explanations of Seebach’s paradox, MD simulations of model peptides in water were done. The difference in stabilities might simply be due to the proximity of alternative hydrogen-bonding partners in the helical structures, and/or the ease with which transitions of hydrogen bonds could be made between hydrogen-bonding partners. Also, the structural distribution of oligo-β-alanine in water, which was unclear experimentally according to Seebach et al. [12], was also a subject worth testing. In order to probe these issues, replica-exchange MD simulations of capped oligoglycine, oligoalanine and oligo-β-alanine dodecamers (12-residues) were performed in explicit water with the second-generation force field, AMOEBA, which includes multipole electrostatics and polarizability [1]. Obviously, interpretation of dynamics from simulations requires the most accurate force field available if the results are to be creditable.
One measure of stability is the persistence of individual hydrogen bonds throughout the MD simulations. No previous experiments to our knowledge have estimated hydrogen-bond persistence in glycine, alanine, or β-alanine helices. Another metric is the persistence of structural motifs common in proteins, α-helix, 310-helix, polyglycine (31 or PGII) helix, etc. Previous experiments have shown that polyglycine helices adapt multiple conformations. In the crystal, two primary structures of polyglycine (PG) have been found [13]: an extended β-sheet form known as PGI with backbone torsional angles nominally φ = − 150° and ψ =+147°, and a 31-helix form (PGII) with approximate torsion angles φ = − 77° and ψ = +145 as also shown by solid-state NMR [14]. A simulation study in vacuo by Itoh et al. [15] showed that polyglycine prefers an α-helical structure in order to maximize intermolecular hydrogen bonding in the absence of solvent. The NMR study by Ohnishi et al. and Raman study by Bykov et al. [16] indicated that the polyglycine chain prefers an extended structure 31 helix (PGII) in water (Fig. 1). Bykov et al. used high concentrations of Li(+) to enhance solubility and stabilize the PGII conformation in solution [16]. In this study, the structure and H-bonding properties of oligoglycine and oligoalanine was characterized and used to compare with oligo-β-alanine.
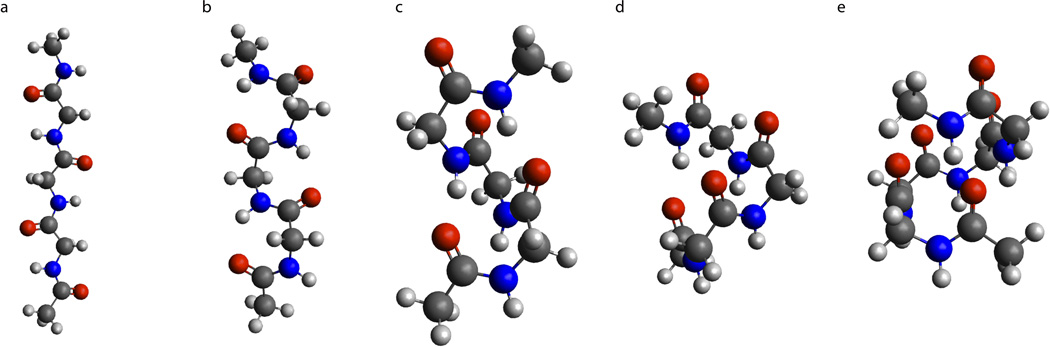
Figure 1.
Schematic drawing of possible polyglycine helical conformers; (a) C2-5 conformer (PGI); (b) C2-7 (PGII); (c) C310 helix; (d) α-helix (Cn-13); (e) π-helix (Cn-16)[17].
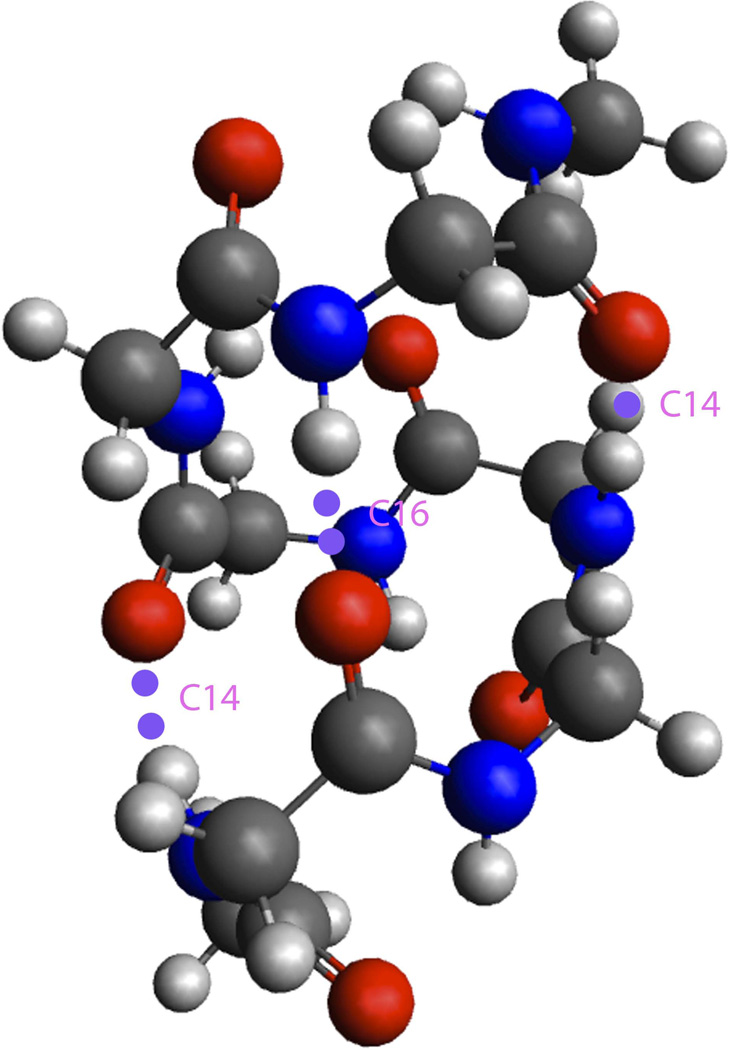
As for the β-peptide helix, previous experiments have suggested that the 314 helix is the most preferred structure in solvent [18, 19]. A set of Ψ, θ, Φ angles distribution have been defined for oligo-β-peptides in a computational study by Glättli et al. [20]. In this experiment, we will focus on a simplified β-peptide without side chains, the oligo-β-alanine helix, and study its structure and hydrogen-bonding properties. In vacuo spectroscopic studies of Dean et al. [21], however, were most consistent with the work of Baldauf et al. [22] as they identified and characterized a nonstandard helix that could be formed by polyglycine chains, referred to as a “mixed” H14/16 helix, composed of alternating C14 and C16 H-bonded rings with a helical pitch of about 4 residues per turn. As an illustration, Figure 2 shows a 14/16 mixed helix structure for Ac-(Gly)5-NHMe.
Figure 2.
Acetyl-(Gly)5-NHMe shows the H14/16 conformation in vacuo [21, 22].
Ohnishi et al. combined NMR and SAXS to study glycine oligomers of different lengths capped with tripeptides to improve solubility [23]. They found elongated solution conformations of the Ac-YES-Gn-ATD (where n = 0, 1, 2, 6 and 9) peptides that were distinct from β-strand, α-helix, and polyglycine II conformations. In particular, the calculated peptide lengths from the SAXS data were significantly shorter that those expected for a PGII conformation. Thus, the diverse experimental and theoretical studies of polyglycine have yet to converge on a self-consistent view.
Oligo-β-alanine (Nylon-3 oligomers [24–26]) - experimental and theoretical studies
As the simplest member of the β-alanine class of foldamers [8, 27], much information regarding the conformational preferences of derivatives of this β-amino acid is available [2, 3, 12, 28–31], but little on oligo-β-alanine itself except as polymers designated Nylon-3. Seebach et al. have shown that β-substituted, β-amino acids form a “314-helix”, which is defined by 14-membered H-bonded ring between backbone amide groups [28]. The Gellman group have shown that inclusion of 6-membered ring constraints in the β-amino acid stabilizes this conformer [2]. Hybrid oligomers of α- and β-amino acids have also been extensively explored [5]. Seebach has also described a 2.710,12 helix for peptides with alternating β2 and β3 amino acids (Fig. 3). This is analogous to the mixed helix (Fig. 2) described above for oligoglycine again emphasizing the conformational adaptation of these flexible oligomers to environmental effects, including substitution patterns.
Figure 3.
Model of a 2.712/10-helix. This helix is characterized by alternating wide twelve-membered and narrow ten-membered rings with the C=O groups pointing alternatively up and down the helix axis [32].
Force-field dependence
Because oligoglycine and oligo-β-alanine are so inherently flexible and multiple helical forms have been discussed depending on the experimental/theoretical approach, the choice of force field for our MD simulations was crucial. A previous study by Zheng et al. [33] compared three monopole force fields in common use with AMOEBA, a second-generation force field using multipole electrostatics and polarizability; a significant improvement in the ability of the AMOEBA force field to predict experimental observations on peptide dynamics was observed and justified the choice of AMOEBA for these studies.
Methods
Construction of oligopeptides dodecamers
AMEOBAbio09 parameters within the Tinker 6.1 package were used in this study. Most of the vdW, angle and torsional parameters for the amino acids were taken directly from the existing AMOEBA force field. The torsional parameters of the C(O)-N-C-C and N-C-C-C(O) bonds in β-alanine were not available, and were generated by fitting the torsional-energy curves generated by quantum mechanics. The wave functions were obtained from Gaussian09 calculations at the MP2/6–311G(d1,p1) level of theory. Electrostatic parameters were derived with Stone’s distributed multipole analysis program GDMA and TINKER POLEDIT program. GDMA was configured to calculating multipoles with the fully analytical integrals using only the original DMA protocol (the parameter SWITCH in GDMA was set to 0) [1]. Charges on individual atoms were adjusted such that the total charge on the β-alanine dodecamer was zero. The AMOEBA electrostatic potentials were validated with the TINKER POTENTIAL program via a potential grid-fitting procedure as described [1].
Generation of starting conformations
Right-handed α-helix for oligo-α-amino-acids
The starting glycine helix of 12 residues was directly built using the TINKER PROTEIN program with Φ = −60° and Ψ = −43° torsion angles to generate a right-handed α-helix with ACE and NME capping groups at each end. Then it was minimized using TINKER MINIMIZE program with a 0.01 cutoff. The final angles were approximately (−63°, −39°). The dodecamer of oligoalanine was constructed the same way, and the angles were approximately the same.
Right-handed 416-helix for oligo-β-alanine
The β-alanine starting helix structure was built by adding CH2 groups to each residue of the glycine helix. Each +4 O-H-N angle was constrained to 180 degrees and minimized using TINKER MINIMIZE program with a 0.01 cutoff. After this, another TINKER MINIMIZE with a 0.01 cutoff was performed on the structure with the angle constraints released to produce the starting structure for simulation. The Ψ, θ, Φ (e.g., C-C-C(O)-N, N-C-C-C(O), C(O)-N-C-C angles) torsional angles were approximately (−20°, −50°, −20°).
Generation of solvent box
The TINKER XYZEDIT program was used to tailor a water box with 40.7487 Å on each side and to solvate the starting helices were solvated within the solvent. Since the TIREX replica exchange program offered only NVT options, the box boundaries were manually adjusted to achieve an average pressure close to 1 atm at 300° K. The final water-box sizes were set to 40.7617 Å, 40.7777 Å and 40.7800 Å for oligo-β-alanine, oligoalanine and oligoglycine, respectively.
Isothermal-isobaric MD simulations
All solvent-solute systems were optimized via the TINKER MINIMIZE program with an 0.01 kcal/mol/Ang RMS gradient cutoff prior to dynamics simulation. All MD simulations were performed with elf clusters (Prof. Jay Ponder) and the Washington University Medical School Center for High Performance Computing nodes. Since the replica-exchange MD simulation only offered structure samplings, rather than true dynamic trajectories, a total of 24 isothermal-isobaric simulations, 8 each for the oligoglycine, oligoalanine and oligo-β-alanine helices, at 300° K and under 1.0 atmospheres were run for 17 ns to obtain trajectory data. In the key file, the boundary size was set up as described in the above section. The vdw cutoff was set to be 12.0 Å and the particle-mesh Ewald real-space cutoff to 7.0 Å. Induced dipoles were converged to an RMS change of less than 0.01 Debye per iteration. The simple bonded/non-bonded RESPA integration scheme was used to enable a larger simulation step size of 2.0 fs. Trajectory frames were recorded every 1 ps. The resulted hydrogen-bond matrix was compared to the matrix from the replica-exchange, and found to be reasonably similar. This indicated that the trajectory sampling was adequate.
Replica-exchange MD
Replica-exchange MD simulation was performed with the protocol of Sugita and Okamoto [34] to get extensive structural sampling for both oligoglycine, oligoalanine and oligo-β-alanine helices. Twenty-four replicas exponentially distributed from 300° K to 400° K were set up with the help of an automated temperature generator for REMD-simulations [35] to obtain an exchange rate near 20% for each structure. The simulation was carried out with the TIREX replica-exchange program developed by Penev et al. [36]. All the setups were the same as the isobaric-isothermal simulations as described above, except the NVT ensemble was used and the trajectories were recorded every 0.5 ps. Each replica was simulated for 10 ns with exchanges attempted every 1000 steps.
Analyses of simulations
Clustering
The 300° K pseudo-trajectory data isolated from the replica-exchange data was analyzed. To make analysis simpler, sampled structures rather than the complete data were used in the analysis. For oligo-β-alanine, structures were taken every 4 frames from the whole data of 20,000 frames to get 5,000 samples. The sampled structures for oligoglycine were taken every 2 frames with a total of 10,000 structures due to its higher structural variance than oligo-β-alanine. The same number of structures were used from the oligoalanine trajectory. In order to classify the structures and simplify the analysis, a clustering based on pairwise RMSD was performed. The RMSD value was calculated between each pair of structure using the TINKER SUPERPOSE program. Then a PCA (Principle Component Analysis) with the princomp package in R was done on the RMSD matrix to extract principle components for clustering. Gaussian-mixture clustering was performed with the Mclust package in R on the principle components extracted above. The center structure in each cluster was determined by computing the RSMD between each pair of structures within the cluster and taking the structure with the minimum sum of squared RMSD. For hydrogen-bond clustering, distances were computed between each pair of amide hydrogens and oxygens on amide carbonyls. All the distances smaller than 2.5 Å were set to 1 in a hydrogen-bond matrix. Then the Jaccard distance as described by the formula below [37] was calculated between the hydrogen-bond matrices of each structure. The PCA and clustering were performed the same way as in the RMSD case.
Determination of H-bond persistence
For the persistence of single hydrogen bonds, the average time each donor-acceptor bonding persisted in the isothermal-isobaric simulations was used as the measure. The existence of donor-acceptor bonding (e.g., the corresponding cell in the hydrogen-bonding matrix was 1) were determined by:
The average of all the existence period of each donor-acceptor bond was used as the hydrogen-bond persistence of that bond. The hydrogen-bonding state persistence was computed in the same way, except the bonding state was considered lost only if the donor or acceptor had no hydrogen bond at all.
Overlapping Zone
Since most of the structures were interconvertible, the RMSD cutoff boundary was not a hard cutoff. Some structures are ambiguous and can fit into both of the two classes as defined, and their number increases as the similarity between the structural classes increased. To estimate this, the size of the overlapping zone was calculated with the formula:
Determination of conformational motifs
At first, helical-structure detection on oligo-β-alanine was performed. The Ψ, θ, Φ angles (e.g. C-C-C(O)-N, N-C-C-C(O), C(O)-N-C-C angles) were computed in each segments from the center structures obtained by clustering. Then, the Ψ = −120°, θ = 60°, Φ = −120° for the 314 helix structure [11] were used as the average of Ψ, θ, Φ angles. This approach did not yield as many valid helical structures within the ±40° range as observed manually. When two helical structures with the angles detected by the method above were separated by a non-helical structure, they were difficult to detect. Thus, an alternate structural approach, computing the RMSD between a 3-residue standard with each segment of helices was used. The standard segment was obtained by restrained the Ψ, θ, Φ angles to the angles mentioned above and minimized. 1 Å is used as the cutoff for oligo-β-alanine. The persistence of the segments was determined by the length of time the segment maintained its structure, the same method used to measure hydrogen-bond persistence.
Results
β-Alanine Parameterization
In Fig. 4, the AMOEBA-parameterized torsional energies for the Ψ, θ, Φ torsion angles are of acetyl-β-alanine-methylamide compared to those derived from quantum calculations.
Figure 4.
The relative potential energy of the 3-backbone torsional bonds in a capped-β-alanine residue, the color codes are blue (Ψ, RMSD=0.36 kcal/mol), green (θ, RMSD=0.30 kcal/mol), cyan (Φ, RMSD=0.25 kcal/mol). The energies from the quantum calculation (points) overlap the energies (curves) from the AMOEBA MINIMIZE program.
The relative torsional energy from AMOEBA agrees very well with the torsional energy from quantum mechanics (Fig. 4). The parameters were verified by superposing the minimized structure from Gaussian MP2/aug-cc-pVTZ optimization with the structure minimized with TINKER MINIMIZE program with 0.01 cutoff. The small RMSD of 0.0175 Å between the two structures that resulted provided further confidence in the derived parameter set.
PCA and Clustering
To make data easier to manipulate, the structures in the 300° K replica were extracted into a pseudotrajectory. The original RMSD matrix for oligo-β-alanine was a 5,000 by 5,000 matrix; for oligoglycine, it was a 10,000 by 10,000 matrix; for oligoalanine, it was a 10,000 by 10,000 matrix. This amount of data complicated the computing, but also made the clustering inefficient due to the noise introduced by large amounts of factors. To solve this problem, principal component analyses were used. For oligo-β-alanine RMSD matrix, the PCA yielded the following result:
Since the principal components after Component 7 explained less than 1% variance each and Components 1–7 together explained ~ 94.2% of the total variance, the first 7 PCs were used as factors in clustering. The further a conformation deviated from its closest local optimal conformation, the less stable it tended to be. As for structures, we assumed that the structures belong to the same cluster were normally distributed around its local optimal structure, the center. For this reason, a Gaussian-mixture clustering method [38] was used. The G term in Mclust program, the possible number of clusters was set to 130, and 89 clusters were found with the best BIC (Bayesian information criterion). Clusters were checked to insure that the structures within each cluster were similar. Then, another pair-wise RMSD was computed within the clusters. The structure with the smallest sum of RMSD squared in each cluster was considered the center of the cluster (the central and most stable conformation) and represented all the structures in that cluster. Then structure fitting was performed as follow: a 3-residue standard segment was fit (by computing the backbone RMSD) to each segment in the center-structure helices. If the backbone RMSD was smaller than a certain cutoff value, for instance, 1 Å for β-alanine, it was considered a helical segment of the same type as the standard segment. The total number of helical segments divided by total number of segments (10 here) was the helical component, the helicity of the structure. For instance, the standard 314 helical segment of oligo-β-alanine was constructed by optimizing the structure of a 12-residue β-alanine chain with its Ψ, θ, Φ angles (e.g., C-C-C(O)-N, N-C-C-C(O), C(O)-N-C-C angles) restrained to Ψ = −120°, θ = 60°, and Φ = −120° [20]. The other standard segments (26, 28, 210, 10/12, 31, Stacking, 312, 314, 416) were constructed by first constraining hydrogen-bond distances, and then restraining the torsional angles with the angles from the hydrogen-bond restrained structures.
Hydrogen Bonding and Structure
To study the structure and hydrogen-bonding pattern of these three oligopeptides, their hydrogen-bonding matrices and structural contents were examined. The hydrogen-bonding matrices were constructed by calculating intersegmental O-H distance and filtering with a 2.5 Å or 3.0 Å cutoff. The structural content was determined as described in the last section. For the RMSD cutoff, the RMSD for oligo-β-alanine was chosen to be 1 Å, a value enough to distinguish between major structures, e.g. 31 and α helices. For oligoalanine and oligoglycine, since they have 3 backbone atoms instead of 4, was used as RMSD cutoff. The results are presented below:
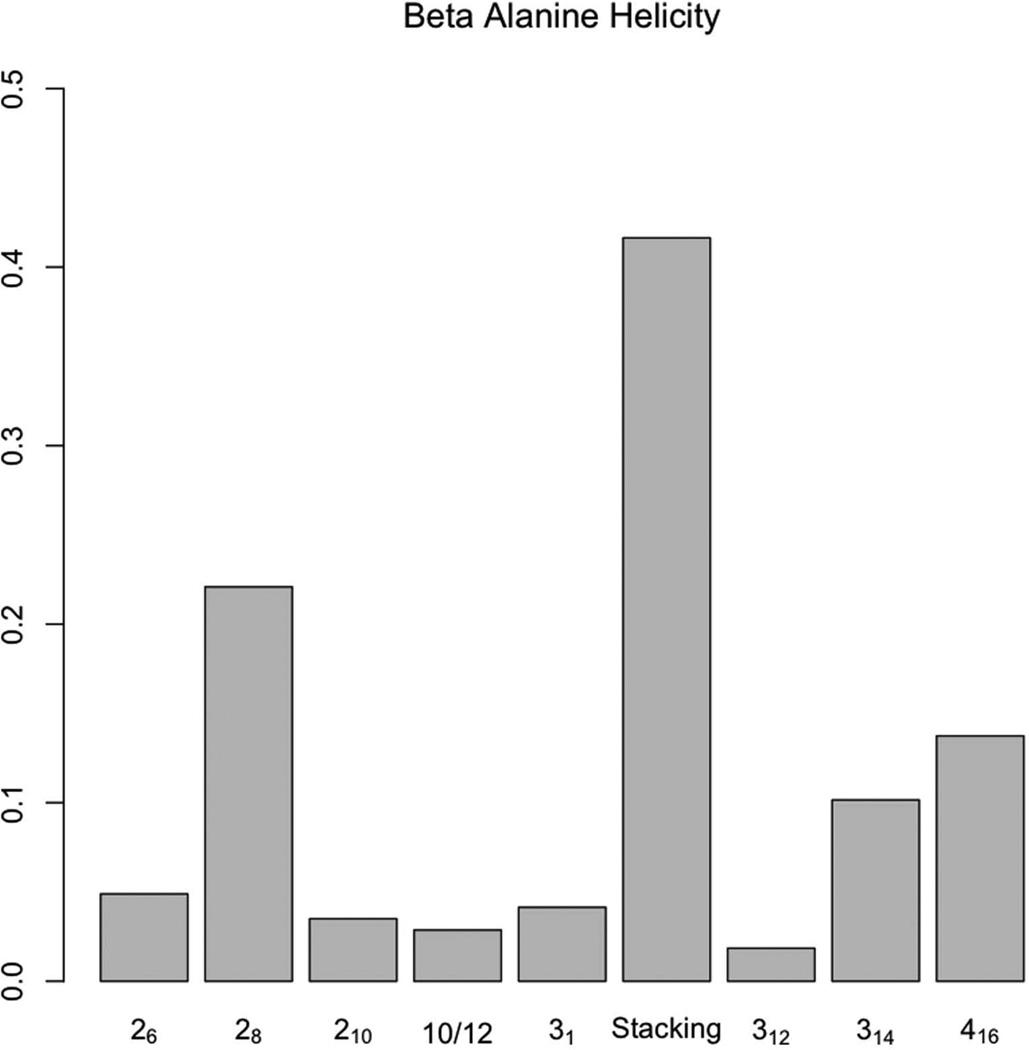
When the cutoff increased from 2.5 Å to 3.0 Å, oligo-β-alanine had some non-significant increases in hydrogen bonding in the acceptor+1 and donor+1 diagonal while the acceptor+2 diagonal remained almost the same. The 2.5 Å cutoff matrix suggested some strong (<2.5 Å) acceptor+2 bonding. The donor+1, donor+2 and acceptor+1 diagonals also showed non-trivial numbers of hydrogen bonds, but mostly in the weak (2.5 Å-3.0 Å) region. To examine this effect, the center structures from the clustering were scanned for helical content as shown in Figure 6.
Figure 6.
Average helical content in the center structures from clustering.
The sum of the total structural content was 104.8%, due to the fact that some structures in the overlapping zone were double counted (especially between 28 and stacking, which have very similar backbone structures). After an overlapping zone computation and removal of ambiguous structures, about 60% of the total structures remained. About 40% of the structures were hard to classify uniquely, which suggested that structures are interconverting and highly interleaved. About 40% percent of the segments cannot be identified as any of the above 3-segment backbones within a 1 Å cutoff.
In Figure 7, the stacking structure (Ψ = −50°, θ = −55°, and Φ = 100°) was the most prevalent structure. It is stabilized by the partial charge interaction between carbonyl carbon and amide nitrogen described by Wu et al. [39] as well as internal hydrogen bonding. Evaluating other structures, 416 and 28 helices were found at 15–20% each. The rest of the helices were all below 10%. Surprisingly, the 312-helix, predicted by an acceptor+2 hydrogen-bonding pattern, showed very low content. This difference suggested that the acceptor+2 hydrogen bonds are sparsely distributed and do not form long segments of 312 helices. The same reasoning can also be applied to the three other hydrogen-bonding patterns.
Figure 7.
The stacking structure [39] was most prevalent in the oligo-β-alanine MD simulation.
On the other hand, the 416-helical backbone was prevalent, but its characteristic internal hydrogen bonding was not observed in the hydrogen-bonding matrix. This may be explained by the fact that only backbone RMSDs were used in our analysis: the 416-helical backbone structure found was maintained by water, where the amide and carbonyl groups utilized a bridging water molecule for interaction.
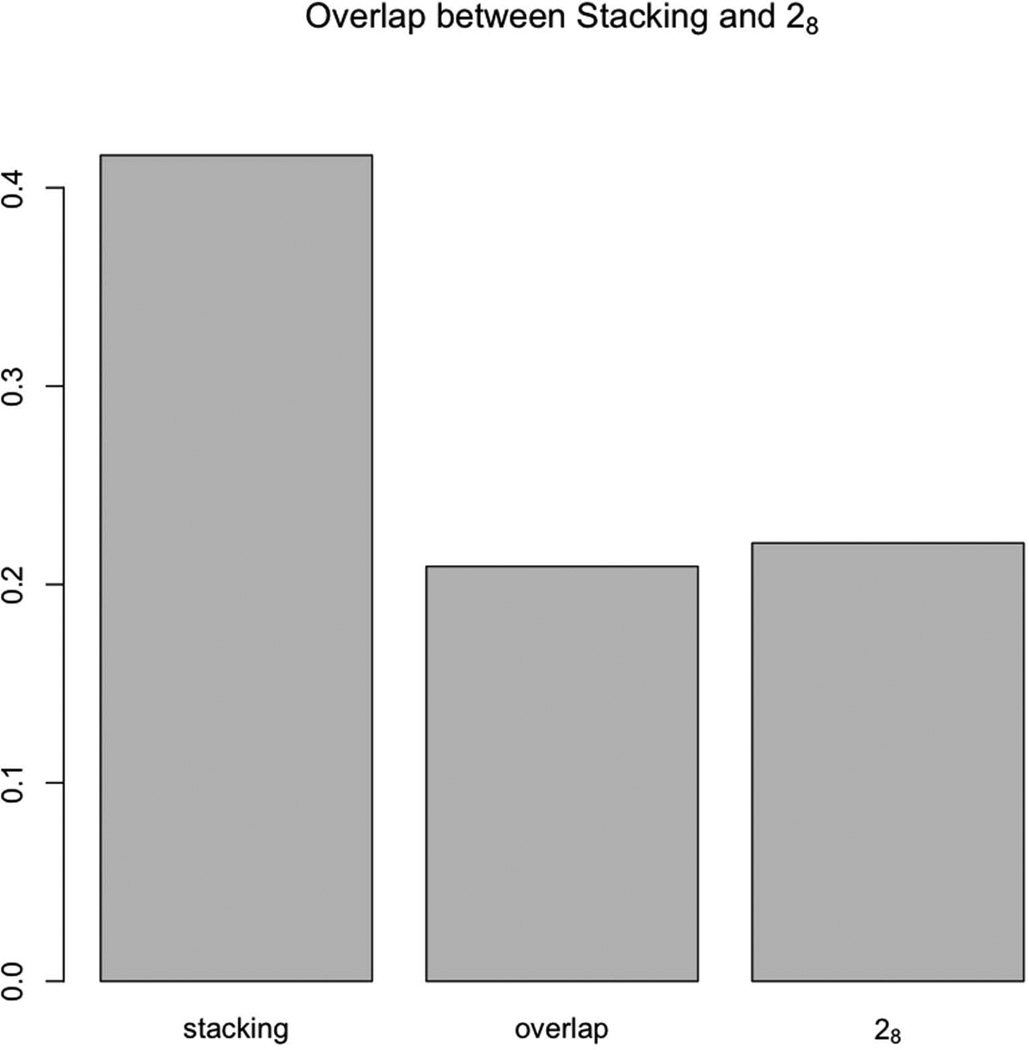
Due to the high similarity of the 28-helix and the stacking structure, an overlapping zone plot was used to check whether their structures are overrepresented as shown in Figure 8.
Figure 8.
The overlapping zone size between stacking and 28, was almost as large as the 28 structure.
The overlapping zone between stacking and 28 structures has almost the same size as the 28-helix. This indicates that the stacking structure and the 28 helix are almost the same structure at the resolution used in the analysis.
In the analysis of structure and hydrogen bonding of oligo-β-alanine, the result indicates an insignificant correlation between hydrogen bonds and superposition based on structure classification. This could primarily be due to the segmentation of structures, and the relatively ease of structure transformation to be further discussed in the dynamic section.
In the case of oligoglycine, the situation was much more straightforward:
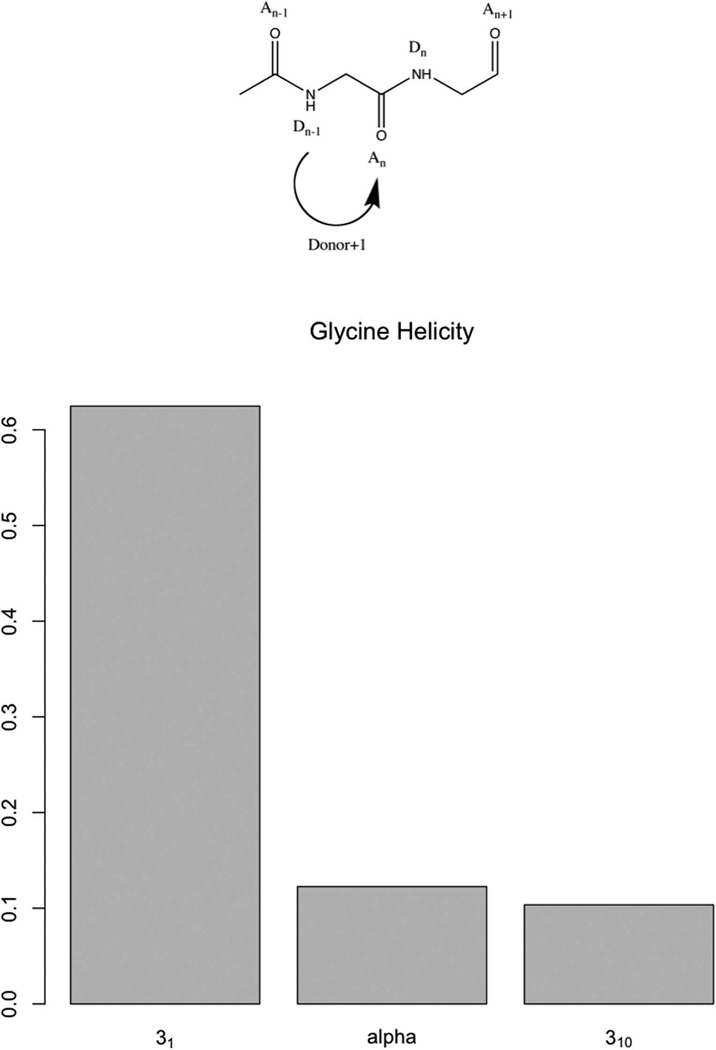
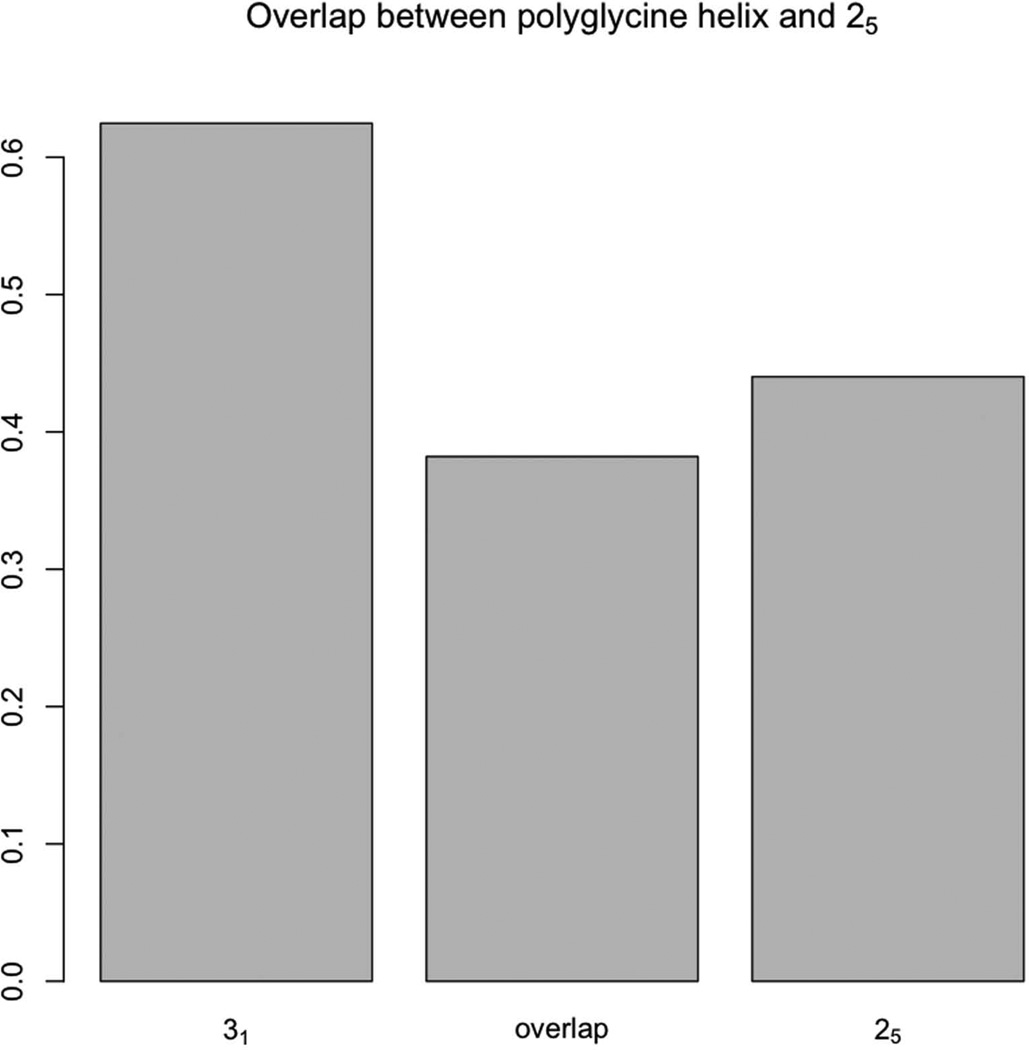
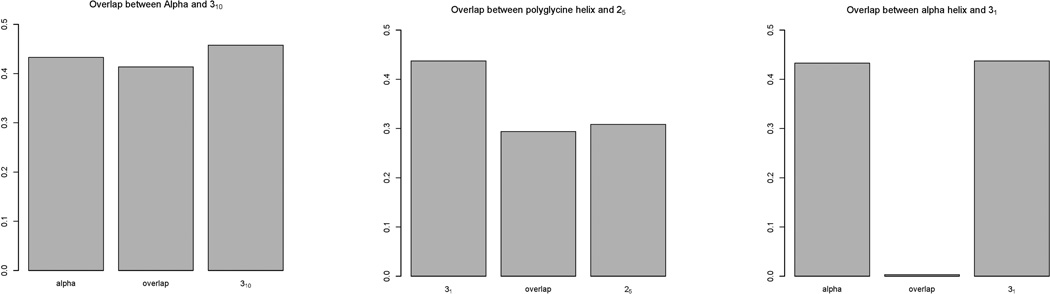
The structure content plot (Fig. 9 and Table III) shows the 31-helix as the dominant structure of oligoglycine as described by Bykov et al. [16]. But some discrepancy showed up in the hydrogen-bonding matrix that had a strong donor+1 diagonal in the weak (<3.0 Å), but not in the strong (<2.5 Å) part of the matrix. To find out whether that indicated 25 components, the 25-helical content was calculated in the sample. A 44% 25-helical content versus 62% 31-helical content was found. The zone overlap was also calculated and plotted below:
Figure 9.
Hydrogen-bonding pattern (31) of oligoglycine and percentage of types of helical hydrogen binding observed in MD simulations.
Table III.
Hydrogen bonding shorter than 2.5 Å (before ‘/’) and 3.0 Å (after ‘/’) count matrix for each position for oligoglycine. The larger numbers in the matrix indicating frequent hydrogen binding between residues are highlighted by boxes
| acptr donor |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0/ 2 |
2283/ 9136 |
0/ 4 |
0/ 1 |
40/ 61 |
3/ 14 |
31/ 61 |
17/ 44 |
0/ 0 |
0/ 3 |
3/ 17 |
0/ 12 |
0/ 4 |
| 2 | 28/ 139 |
0/ 2 |
1384/ 6983 |
0/ 1 |
24/ 31 |
38/ 56 |
27/ 56 |
1/ 9 |
36/ 64 |
0/ 1 |
28/ 69 |
127/ 202 |
25/ 34 |
| 3 | 70/ 155 |
16/ 160 |
0/ 3 |
1400/ 7252 |
7/ 14 |
0/ 0 |
108/ 156 |
28/ 74 |
19/ 37 |
5/ 39 |
114/ 135 |
251/ 332 |
36/ 90 |
| 4 | 112/ 176 |
213/ 433 |
76/ 286 |
0/ 0 |
1326/ 6922 |
0/ 7 |
3/ 14 |
56/ 123 |
4/ 8 |
3/ 8 |
13/ 23 |
30/ 51 |
14/ 112 |
| 5 | 0/ 3 |
50/ 105 |
76/ 146 |
30/ 145 |
0/ 5 |
1447/ 7325 |
0/ 2 |
5/ 23 |
0/ 0 |
168/ 237 |
59/ 98 |
0/ 2 |
400/ 445 |
| 6 | 21/ 37 |
0/ 3 |
88/ 109 |
24/ 86 |
45/ 205 |
0/ 3 |
1083/ 6034 |
0/ 9 |
0/ 0 |
0/ 5 |
0/ 0 |
0/ 0 |
5/ 13 |
| 7 | 2/ 11 |
2/ 16 |
0/ 0 |
114/ 140 |
113/ 249 |
47/ 222 |
0/ 4 |
1705/ 8201 |
0/ 2 |
1/ 9 |
0/ 0 |
88/ 102 |
0/ 0 |
| 8 | 2/ 3 |
0/ 0 |
0/ 0 |
0/ 0 |
95/ 128 |
74/ 206 |
12/ 82 |
0/ 4 |
1063/ 5831 |
10/ 23 |
0/ 4 |
0/ 8 |
0/ 0 |
| 9 | 0/ 2 |
0/ 2 |
8/ 20 |
0/ 0 |
0/ 2 |
182/ 268 |
68/ 167 |
68/ 240 |
0/ 2 |
1550/ 7392 |
1/ 23 |
0/ 0 |
9/ 13 |
| 10 | 34/ 68 |
24/ 47 |
0/ 0 |
0/ 0 |
0/ 8 |
0/ 1 |
184/ 261 |
48/ 132 |
27/ 188 |
0/ 5 |
1449/ 7743 |
1/ 5 |
0/ 0 |
| 11 | 80/ 112 |
3/ 5 |
5/ 44 |
7/ 44 |
0/ 0 |
0/ 2 |
4/ 7 |
82/ 108 |
79/ 183 |
13/ 72 |
0/ 2 |
1400/ 7084 |
0/ 2 |
| 12 | 118/ 178 |
5/ 14 |
2/ 13 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 6 |
0/ 0 |
65/ 105 |
80/ 158 |
16/ 100 |
0/ 2 |
1410/ 7158 |
| 13 | 7/ 14 |
1/ 4 |
1/ 12 |
0/ 0 |
0/ 0 |
1/ 5 |
43/ 75 |
0/ 0 |
88/ 103 |
36/ 76 |
70/ 134 |
19/ 144 |
0/ 5 |
The same situation as between the 28 and the stacking structures of oligo-β-alanine was found. This evidence argues that the strong donor+1 diagonal was due to the structure proximity between 31 and 25 structures. This will be further discussed in the dynamic section. Without counting 25-helices, the 31- and α-helices represented ~75% of the total structures (Fig. 10).
Figure 10.
The overlapping zone size between 31 and 25 was almost as high as the 25 structure.
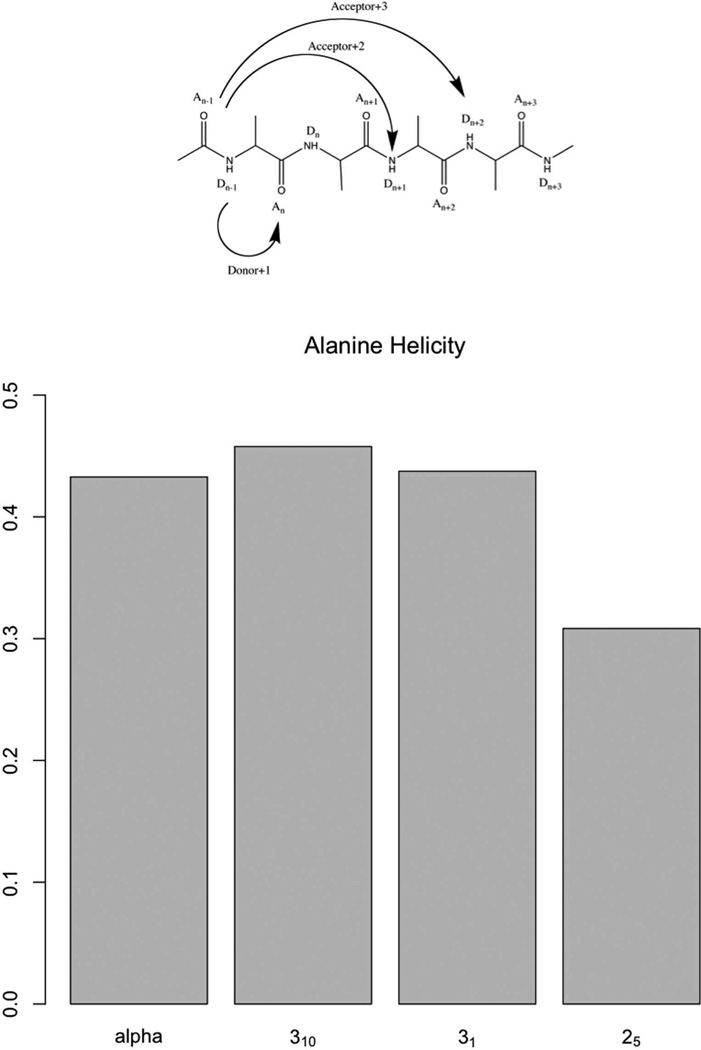
Alanine is known for its α-helix stability. Consistent with experimental studies, a large content of α-helix was also found by simulation:
In the hydrogen-bonding matrix, the acceptor+2 and acceptor+3 diagonal confirmed the fact that α-helical structures were a large component. The high percentage of 25-helix also satisfies the high frequency of hydrogen bonds on the donor+1 diagonal. The four structures in Figure 11 sum up to higher than 100%. This is due to the structural overlap between the α- and 310-helix and the 31- and 25-helix. To confirm this suspicion, the overlap was plotted in Figure 12.
Figure 11.
Hydrogen-bonding pattern and structural content of oligoalanine MD
simulations.
Figure 12.
The overlapping zone sizes between α and 310, 31 and 25, α and 31 helices.
The large overlap showed up as expected. Indeed, the α- and 310-helices were very similar in structure at this resolution, and i->i+3, i+4 bifurcation of hydrogen bonds are common in nature, e.g. membrane proteins[38]. As for 31- and 25-helices, a similar large overlap zone appeared as with oligoglycine above. The overlapping region between α- and 31-helices was very small, illustrating the significant difference between these two structures, which together add up to ~ 87% of the total structures in water. As explained in the analysis above, the oligo-β-alanine showed more structural flexibility: its structures were harder to be identified within cutoffs. This is explained by the flexibility introduced by the extra methylene in the backbone that favors kinks and structural transition intermediates. Zheng et al. had estimated the energy cost of α-amino-acid helical bending to be only 0.6 kcal/mol [33] consistent with our results.
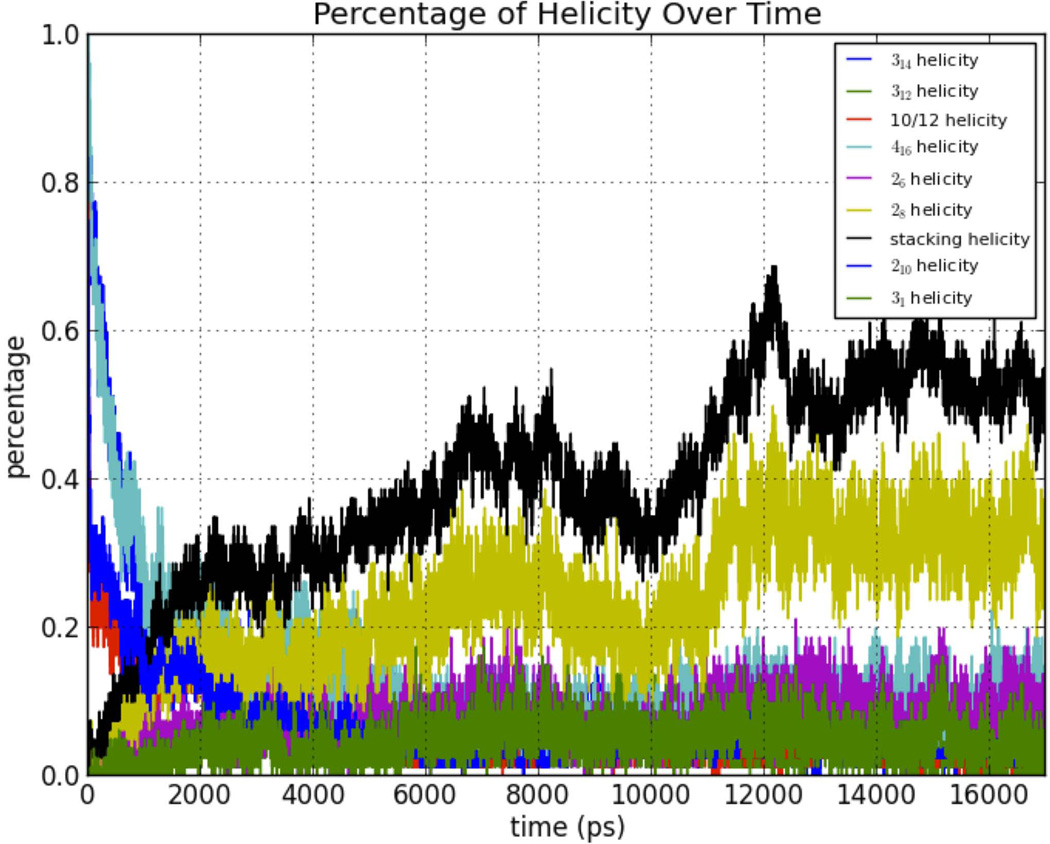
Another key observation was that both oligoglycine and oligoalanine have a large component of 31-helix while oligo-β-alanine does not. Since the stacking structure is preferable and consistently appears in the analysis above, a hypothesis can be made that the stacking structure is a competing structure in water with the 31-helix (both structures point their carbonyl oxygens and amide nitrogens toward water) that became dominant in the case of oligo-β-alanine. This structure competed, and became a destructive force for the continuity of other helices.
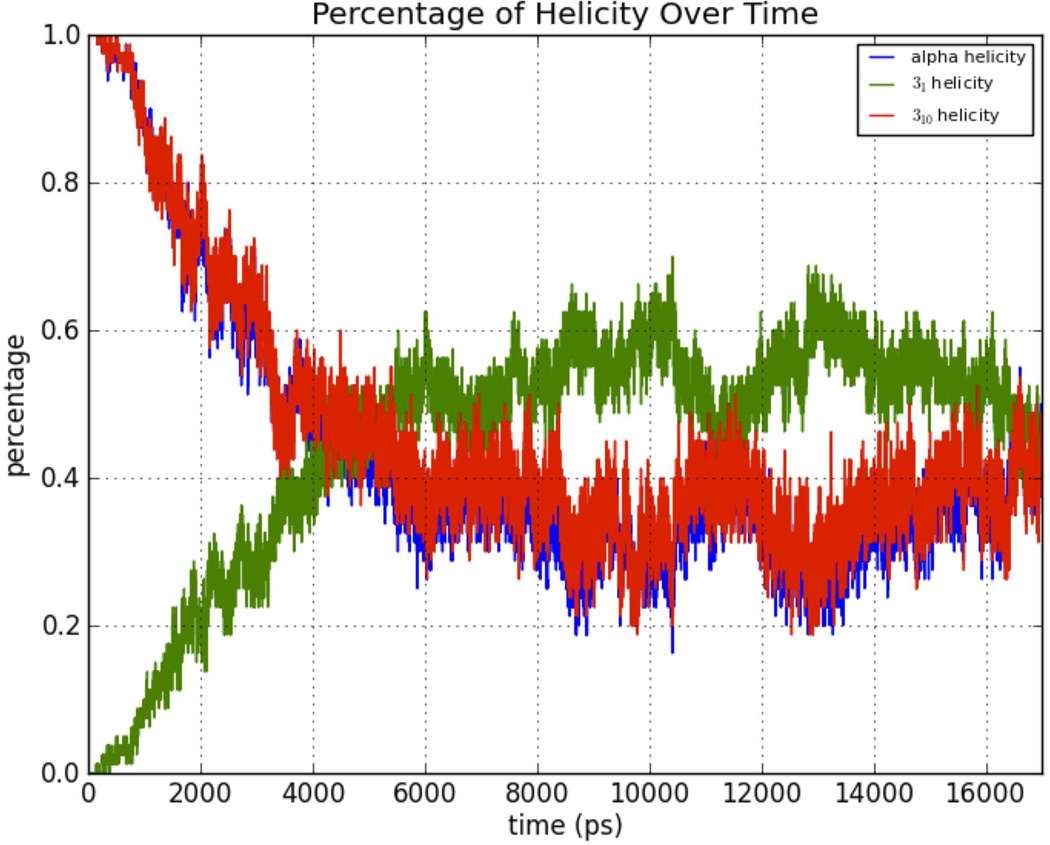
Conformational Transition
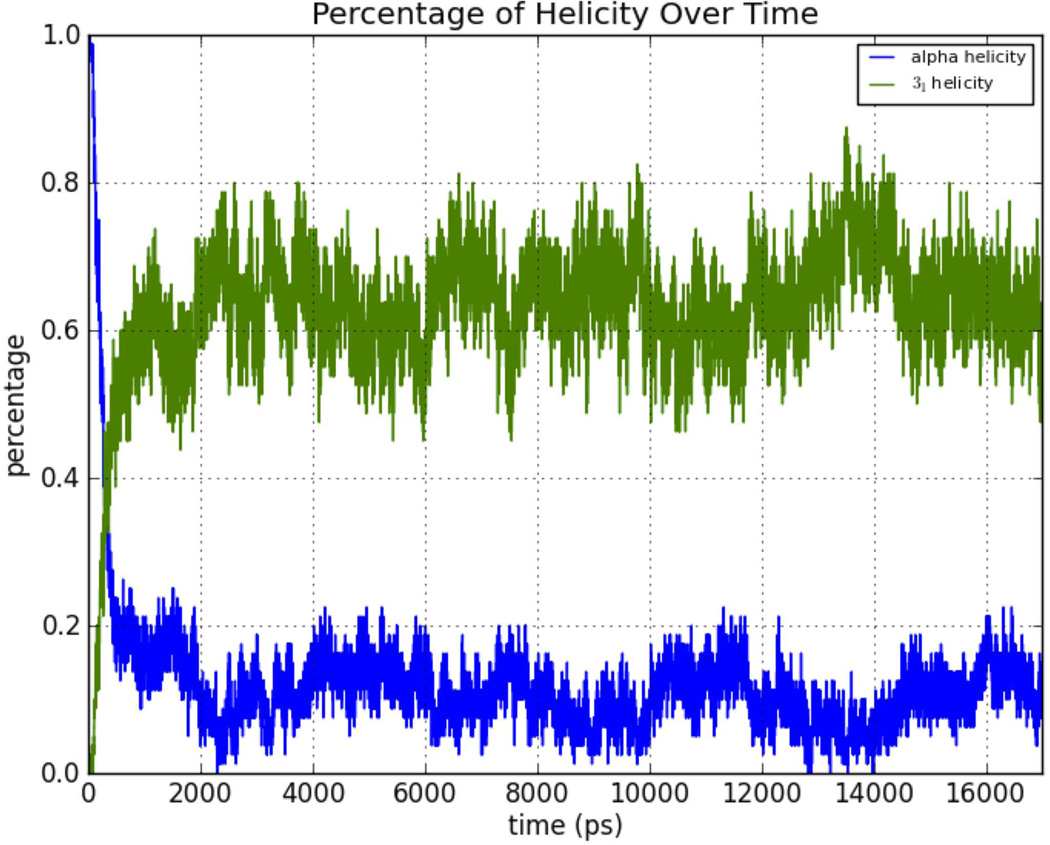
The oligoglycine helix was started in the α-helical conformation, but quickly made a transition to the PGII (31) helix as shown in Figure 13. For the oligo-β-alanine dodecamer, starting the MD simulation with a 416-helix led to a quick equilibration with other conformations (Fig. 14). The oligoalanine plot also showed equilibration with balancing of 31 and α-helix conformations (Fig. 15). These graphs are consistent with adequate sampling of conformational space by the AMOEBA MD simulations with structural equilibration.
Figure 13.
Transition of oligoglycine from a starting α-helical conformation to predominately 31-helix (PGII) during the MD simulation.
Figure 14.
Transition of oligo-β-alanine from a starting near 416-helical conformation to mixed conformations with less than 10% 416-helical conformation.
Figure 15.
Transition of oligoalanine from starting near α-helical conformation to mixed conformations of α-, 310-, and 31 helix.
Hydrogen Bonding and Structural Persistence
The previous analyses focused on the average static presence of hydrogen bonds by analyzing MD simulation frames. The dynamics of hydrogen bonding is another aspect that relates more directly to the activation energy required for intramolecular hydrogen-bond interchange. One metric to examine is the persistence of given hydrogen bonds from frame to frame. For internal hydrogen-bond persistence, the 2.5 Å and 3.0 Å persistence matrices are shown in Supplemental Information Tables SI, SII and SIII.
Analysis of the data in the Supplemental Material leads to the following conclusions. The persistence for the 3.0 Å-cutoff hydrogen bonds could be either higher or lower than 2.5 Å depending on how long the hydrogen bonds remain within the 2.5–3.0 Å length range. If the bonds in this range appear transiently, these bonds will have very low persistence and lower the overall persistence of that position. But if these hydrogen bonds persist and stabilize the structures, they will increase the average persistence. Comparison between the persistence matrices for 2.5 Å cutoff and 3.0 Å cutoff shows that the 3.0 Å cutoff matrices have an overall higher persistence, indicating that hydrogen bonds in the 2.5–3.0 Å range are very important in persistence: rather than appearing transiently, most of the interactions in this range are sufficient to maintain the overall structure. Comparison between the oligo-β-alanine, oligoalanine and oligoglycine persistence matrices shows that the oligo-β-alanine H-bonds have longer persistence than the oligoalanine H-bonds, and oligoalanine H-bonds have longer persistence than oligoglycine H-bonds. The higher persistence of oligoalanine compared to oligoglycine can be explained by the fact that the high content of α- and 310-helices in oligoalanine depends more on intramolecular hydrogen bonds than the 31 helix in oligoglycine which has fewer internal hydrogen bonds. Noticeably, only the acceptor+3 diagonal for oligoalanine, which is in the +4 hydrogen bond region has high persistence. This suggests that the α(3.613)-helices, have stronger +4 than +3 hydrogen bonds for oligoalanine consistent with experiment. Also, the low persistence of donor+1 diagonal in oligoalanine and oligoglycine indicates the transience of 25 helical structure which suggests the structures in the overlapping zone of Figure 7 have more stable 31 helix components.
The high hydrogen-bond persistence in oligo-β-alanine is more complicated. Only the donor+2 and acceptor+2 diagonal in the persistence matrix coincide with the hydrogen-bond matrix. The strong donor+2 region can be explained by the persistence of the 28 helix and stacking structures. The strong acceptor+2 is likely also due to the segmentation of 312 mentioned above: although 312 helix-like bonding is prevalent and stable, they were not present adjacent to each other to be defined as a 312 helix segment. There are also several random high-persistence regions that may represent hairpin structures, but since they did not coincide with high-value diagonals, which represent helices, they were not considered in this analysis.
The structural persistence of helices was also examined in these three oligopeptides and analyzed in the Supplemental Material. The persistence was calculated using conformation adjacent to each frame of the trajectory as mentioned above. If a structure was within the RMSD boundary in one frame, but not in the next, it was considered lost. The average period from the appearance frame to the disappearance frame of the structure were defined as the average persistence time.
In this analysis, a total of 8 trajectories of 17 ns each were used. Each frame was 1ps:
Most of the persistent values ranged from 10–20 frames, i.e. 10–20 ps. Noticeably, both 31 and α-helical structures were persistent in oligoalanine. While the average structural persistence of α-helical structures in oligoalanine varied between 7.5 and 14.2 psec with obvious fraying at the C-terminus, the persistence for stacking structures in oligo-β-alanine varied between 12.2 and 28.4 psec with no obvious differences between N- and C-terminal segments. Besides the stacking structure, all other structures of oligo-β-alanine have persistence lower than 10 ps. Thus, one potential rationalization for Seebach’s paradox is the enhanced persistence of the stacked structures in in oligo-β-alanine versus the stability of α-helical structures in oligoalanine.
Cooperative Hydrogen-Bond Interactions
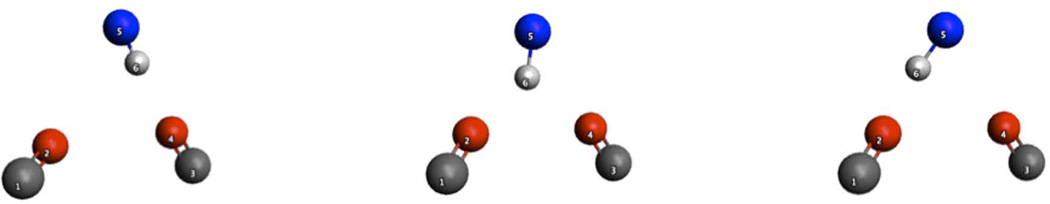
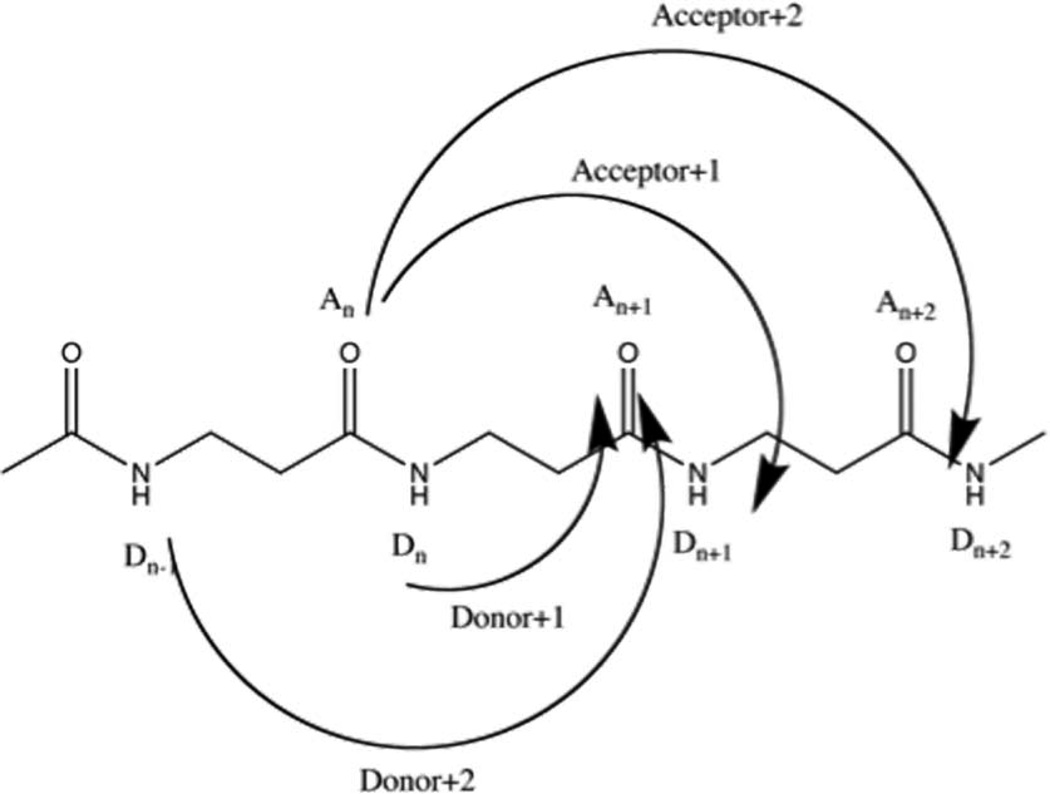
the persistence facilitated by multicenter hydrogen bonding was also explored. If the hydrogen bonds interact with each other by holding the structure in a state that is easier for other hydrogen bonds to form, donors or acceptors “switching” between their hydrogen-bonding partners (Figure 16 shows donor “switching”) will be observed. Thus, the position-stabilization effect can be quantified by measuring how long a donor, or acceptor, maintains its hydrogen-bonding state regardless of specific partner.
Figure 16.
Schematic of hydrogen bond “switching”: the hydrogen atom 6 interact with oxygen 4 (left), then it will be easy for it to move to the middle and interact with both oxygen 2 and 4 (middle), when it’s in the middle state, it can also swing to the left and interact with oxygen 2.
The average time each donor or acceptor stays in its hydrogen-bonding state is shown in Table V. An obvious observation is that the overall H-bonding persistence for oligo-β-alanine is much higher than both for oligoglycine and oligoalanine. This indicates that when multicenter bondings was considered, the oligo-β-alanine multicenter H-bonds were more persistent than oligoalanine H-bonds and oligoglycine H-bonds. But this metric does not provide a good comparison between 2.5 Å and 3.0 Å hydrogen bonds, because if more than 1 hydrogen bond includes a donor or acceptor simultaneously, it was only counted as one. Thus, another metric, the loss of H-bonding state, was used to measure the multicenter bond effect: If the H-bond formations did not interact with each other, the last H-bond break will not provide a good position for the next hydrogen bond to form. Thus, the total number of time a certain donor or acceptor lost its H-bonding state will be close to its total number of bond breaks, e.g. close to the situation that the hydrogen bonds are not continuous. Here an H-bond state was considered lost and discontinuous if the donor or acceptor was bonded, but was not present in the next ps of the simulation. This metric counted the change of bonding state of all the H-bonding partners. The results are shown in Figure 15–20.
Table V.
Donor-acceptor hydrogen-bonding state persistence (in unit of ps) for donor (upper values in each table) and acceptor (lower in each table) for oligo-β-alanine (A), oligoglycine (B), oligoalanine (C). The 2.5 Å are shown before the ‘/’ and 3.0 Å after the ‘/’.
| A | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| type | ||||||||||||||
| donor | 2.92/ 4.87 |
3.91/ 2.84 |
4.07/ 3.29 |
8.67/ 6.25 |
7.48/ 5.1 |
18.83/ 13.15 |
11.6/ 7.52 |
7.56/ 5.18 |
10.97/ 9.1 |
10.7/ 7.43 |
9.02/ 6.17 |
12.09/ 6.96 |
3.29/ 2.42 |
|
| acptr | 2.54/ 2.19 |
7.23/ 3.93 |
3.39/ 3.95 |
13.55/ 11.05 |
15.13/ 9.88 |
7.93/ 4.68 |
12.08/ 7.48 |
9.61/ 5.79 |
9.62/ 5.41 |
8.69/ 5.91 |
5.55/ 4.91 |
2.71 1.92 |
5.92/ 6.21 |
|
| B | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| type | ||||||||||||||
| donor | 1.81/ 4.72 |
1.84/ 3.92 |
1.68/ 3.92 |
1.85/ 3.92 |
1.82/ 4.03 |
2.18/ 4.18 |
1.86/ 4 |
1.81/ 3.84 |
1.78/ 3.83 |
1.7/ 3.87 |
1.71/ 3.69 |
1.67/ 3.61 |
2.67/ 2.68 |
|
| acptr | 3.39/ 2.85 |
2/ 4.91 |
1.99/ 4.1 |
1.71/ 4 |
1.71/ 3.92 |
1.76/ 4.04 |
1.79/ 4.06 |
1.78/ 3.99 |
1.78/ 3.84 |
1.73/ 3.79 |
1.79/ 3.92 |
1.66/ 3.63 |
1.58/ 3.55 |
|
| C | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| type | ||||||||||||||
| donor | 2.13/ 7.11 |
2.27/ 5.41 |
2.31/ 4.73 |
3.5/ 6.79 |
3.44/ 6.96 |
4.03/ 7.72 |
3.64/ 7.99 |
3.46/ 7.62 |
3.62/ 7.79 |
3.08/ 7.39 |
2.77/ 6.54 |
2.34/ 5.53 |
2.37/ 2.3 |
|
| acptr | 7.26/ 9.02 |
3.21/ 8.11 |
3.87/ 7.62 |
3.67/ 7.8 |
3.41/ 7.72 |
3.13/ 6.66 |
3/ 6.65 |
2.64/ 6.29 |
2.46/ 6.01 |
2.35/ 5.14 |
2.38/ 5.56 |
2.28/ 5.44 |
2.15/ 5.47 |
|
In Table VI, the <2.5 Å statistics, there is a significant decrease of H-bond breaking, mostly between 10–30%. In Table VII, the <3.0 Å statistics, most of the hydrogen bond-break decrease goes further to the 20–60% range. These observations indicated that multiple bond interactions for bond length between 2.5–3.0 Å also have significant stabilization effect on the helix structure, which did not necessarily appear in the bonding-state persistence in Table V. The same investigation was carried out for the H-bond break effect in the oligoglycine helix structure in Tables VIII and IX.
Table VI.
Oligo-β-alanine bond-break table for all the hydrogen bonds shorter than 2.5 Å. The upper table is the actual number of bond breaks (before the ‘/’) compare to the number of bond breaks if the hydrogen bonds do not interact (after the ‘/’). The down table is the percentage of bond break avoid due to hydrogen-bond interaction (before the ‘/’) and the sample standard deviations weighted by the number of bond breaks if the hydrogen bonds do not interact (after the ‘/’).
| A | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| type | ||||||||||||||
| donor | 4083/ 4538 |
1822/ 1947 |
2228/ 2526 |
2228/ 2526 |
1872/ 2216 |
2253/ 3462 |
2491/ 3148 |
2385/ 2693 |
2984/ 3837 |
2172/ 2836 |
2759/ 3109 |
1745/ 2058 |
1810/ 1904 |
|
| acptr | 1565/ 1619 |
2270/ 2718 |
4482/ 4541 |
4482/ 4541 |
1880/ 2820 |
2322/ 2741 |
2170/ 2925 |
2223/ 2686 |
2096/ 2763 |
3180/ 3579 |
2489/ 2792 |
1485/ 1564 |
2068/ 2198 |
|
| B | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| type | ||||||||||||||
| donor | 0.1/ 0.04 |
0.06/ 0.05 |
0.12/ 0.07 |
0.06/ 0.03 |
0.16/ 0.16 |
0.35/ 0.23 |
0.21/ 0.17 |
0.11/ 0.07 |
0.22/ 0.11 |
0.23/ 0.12 |
0.11/ 0.11 |
0.15/ 0.14 |
0.05/ 0.03 |
|
| acptr | 0.03/ 0.02 |
0.16/ 0.16 |
0.01/ 0.01 |
0.18/ 0.2 |
0.33/ 0.32 |
0.15/ 0.24 |
0.26/ 0.15 |
0.17/ 0.1 |
0.24/ 0.18 |
0.11/ 0.1 |
0.11/ 0.1 |
0.05/ 0.07 |
0.06/ 0.04 |
|
Table VII.
Oligo-β-alanine bond-break table for all the hydrogen bonds shorter than 3.0 Å.
| A | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 4081/ 6354 |
5253/ 6058 |
5570/ 7012 |
4438/ 5850 |
4078/ 5130 |
3719/ 8112 |
5065/ 9335 |
5138/ 7300 |
4515/ 8295 |
4140/ 6659 |
5441/ 7491 |
3941/ 5650 |
5506/ 6107 |
| acptr | 4837/ 5116 |
6847/ 8336 |
6823/ 7616 |
4544/ 6030 |
3517/ 7492 |
6070/ 7793 |
4472/ 6360 |
5368/ 8358 |
5355/ 7193 |
6582/ 8636 |
4536/ 5824 |
6660/ 6995 |
2739/ 3604 |
| B | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 0.36/ 0.1 |
0.13/ 0.08 |
0.21/ 0.17 |
0.24/ 0.17 |
0.21/ 0.19 |
0.54/ 0.37 |
0.46/ 0.35 |
0.3/ 0.21 |
0.46/ 0.23 |
0.38/ 0.22 |
0.27/ 0.12 |
0.3/ 0.27 |
0.1/ 0.11 |
| acptr | 0.05/ 0.04 |
0.18/ 0.12 |
0.1/ 0.03 |
0.25/ 0.13 |
0.53/ 0.43 |
0.22/ 0.18 |
0.3/ 0.24 |
0.36/ 0.25 |
0.26/ 0.18 |
0.24/ 0.18 |
0.22/ 0.19 |
0.05/ 0.01 |
0.24/ 0.24 |
Table VIII.
Oligoglycine bond-break table for all the hydrogen bonds shorter than 2.5 Å.
| A | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 8138/ 8598 |
6420/ 6703 |
5992/ 6076 |
5907/ 6096 |
6689/ 6953 |
6415/ 7177 |
6734/ 7079 |
6110/ 6365 |
5865/ 6060 |
6246/ 6412 |
6469/ 6646 |
7347/ 7559 |
607/ 704 |
| acptr | 503/ 589 |
8338/ 9199 |
6763/ 7415 |
5998/ 6143 |
5697/ 5998 |
6446/ 6662 |
6696/ 6938 |
6805/ 7081 |
6167/ 6453 |
5778/ 5857 |
6286/ 6504 |
6337/ 6417 |
7112/ 7172 |
| B | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 0.05/ 0.07 |
0.04/ 0.04 |
0.01/ 0.01 |
0.03/ 0.02 |
0.04/ 0.03 |
0.11/ 0.1 |
0.05/ 0.02 |
0.04/ 0.05 |
0.03/ 0.03 |
0.03/ 0.01 |
0.03/ 0.02 |
0.03/ 0.01 |
0.14/ 0.1 |
| acptr | 0.15/ 0.14 |
0.09/ 0.12 |
0.09/ 0.07 |
0.02/ 0.01 |
0.05/ 0.07 |
0.03/ 0.01 |
0.03/ 0.02 |
0.04/ 0.02 |
0.04/ 0.06 |
0.01/ 0.01 |
0.03/ 0.03 |
0.01/ 0.01 |
0.01/ 0.01 |
Table IX.
Oligoglycine bond-break table for all the hydrogen bonds shorter than 3.0 Å
| A | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 11947/ 12605 |
13296/ 14370 |
12194/ 12759 |
12252/ 13166 |
13200/ 14339 |
12637/ 14084 |
13690/ 14892 |
12902/ 13702 |
12487/ 13437 |
12830/ 13697 |
14526/ 15594 |
15097/ 16065 |
1221/ 1477 |
| acptr | 1098/ 1305 |
11895/ 13886 |
13239/ 15132 |
11888/ 12697 |
11856/ 12972 |
12770/ 13811 |
12901/ 14062 |
13625/ 14904 |
12892/ 13789 |
12510/ 13179 |
12775/ 13829 |
14549/ 15389 |
14970/ 15232 |
| B | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 0.05/ 0.05 |
0.07/ 0.06 |
0.04/ 0.01 |
0.07/ 0.03 |
0.08/ 0.03 |
0.1/ 0.05 |
0.08/ 0.04 |
0.06/ 0.02 |
0.07/ 0.04 |
0.06/ 0.02 |
0.07/ 0.02 |
0.06/ 0.02 |
0.17/ 0.11 |
| acptr | 0.16/ 0.09 |
0.14/ 0.13 |
0.13/ 0.08 |
0.06/ 0.03 |
0.09/ 0.07 |
0.08/ 0.02 |
0.08/ 0.04 |
0.09/ 0.04 |
0.07/ 0.03 |
0.05/ 0.02 |
0.08/ 0.05 |
0.05/ 0.02 |
0.02/ 0.01 |
As for oligoglycine, the bond breaks avoided by H-bond interactions were not very significant. Most of them were less than 10%. This indicates that the oligo-β-alanine structure relies much more on multicenter hydrogen bonds than does the oligoglycine helix.
The same analysis was carried out on oligoalanine:
Overall, the oligo-β-alanine multi-center hydrogen bonds avoid more bond breakage than the hydrogen bonds in oligoalanine, especially in the strongly bonded (<2.5 Å) case. But oligoalanine comes very close in the weak bond (<3.0 Å) case, which suggests i->i+3, i+4 bond switching(not significant in oligoglycine due to its low content of α-helix). All of the evidence above showed that the multicenter hydrogen bonds are more structurally important in oligo-β-alanine than in oligoglycine, or oligoalanine. In spite of the structural flexibility of oligo-β-alanine we found in the previous sections, our data here shows that its hydrogen bonds are quite stable.
Hydrogen Bonding and Structural Relationships
To find out the relationship between hydrogen bonding and helical structure, clustering based on hydrogen bonding and structural RMSD were compared. The RMSD clustering was executed as mentioned above. The hydrogen-bonding clustering was done in the same way, except the RMSD matrix was replaced by the Jaccard distance matrix as mentioned above. The H-bonding matrix used here was the 2.5 Å cutoff, since the information became dispersed among principal components when the 3.0 Å cutoff was tried. After clustering with Jaccard distances, a total of 132 clusters resulted for oligo-β-alanine. The structural data was reclustered with a 10,000 by 10,000 RMSD matrix and 105 clusters resulted. The cumulative probability that one can predict the H-bond clustering with structural clustering is listed in Figure 17.
Figure 17.
The match between Jaccard-distance clustering and RMSD-distance clustering for oligo-β-alanine. The x-axis marks the confidence we can predict a structure’s H-Bonding cluster from its structural cluster.
The overall accuracy when the structure clusters were mapped to the hydrogen bond was 54.56%. This prediction accuracy indicates that there is a non-random relationship between helical structure and hydrogen bonding, but not so significant that one could necessarily infer one from the other.
The predictive accuracy was much less for oligoglycine as shown in Figure 18. The overall predictive accuracy was 36.35%.
Figure 18.
The match between Jaccard-distance clustering and RMSD-distance clustering for oligoglycine.
The predictive accuracy was 31.65% in oligoalanine, which was even less than oligoglycine. The difference between accuracy for oligo-β-alanine, oligoalanine and oligoglycine indicates that oligo-β-alanine has a stronger relationship between its overall dodecamer structure and its hydrogen-bonding pattern. Notice, the clustering is based on the structure of the whole dodecamer, which does not contradict the previous statement that oligo-β-alanine structures are less clearly hydrogen bonded.
Discussion
Dynamics of proteins provide insight into the functional role of components. Helices are not static, but are relatively molten, especially when isolated in solution [40]. Helix packing to form tertiary protein structure is not packing of rigid elements, but rather induced fit of flexible partners. The medicinal chemist desires to mimic the recognition motif of a given biologically active peptide with a semi-rigid structure to optimize affinity by preorganization. Non-peptidic helix mimetics as potential therapeutics have been designed and prepared. Since one seldom knows the exact conformational requirements (both static and dynamic) required for helix recognition, determination of the conformational flexibility available to helices made from different amino acids is a first step to the design of a more rigid non-peptide mimetics. Obviously, static structures only reveal part of the requirements for successful mimicry in molecular recognition. One experimental observation based on analyses of ststic structures regarding relative stability of α- and β-amino helices comes from Seebach’s paradox, the enhanced stability of oligo-β-alanine vesus oligoalanine and oligoglycine helices when the oligo-β-alanine helix should be relatively entropically destabilized.
Conclusions
The fact that hydrogen-bond persistence correlates with helical stability is quite plausible. MD simulations suggest probable explanations for lower activation barriers for hydrogen-bond exchange in less stable helices. In particular, a role of water in bridging donor and acceptors became more apparent with simulation. The observation that intramolecular hydrogen-bond exchange is less likely with oligo-β-alanine from the MD simulations provides a plausible basis for its enhanced stability versus oligoglycine or oligoalanine. Direct correlation with experimental results has proven more difficult, due to limited comparative studies. Our results indicate that oligo-β-alanine does not have a clearly defined structural distribution in water, making it difficult to determine whether the MD simulations fit the experimental NMR results presented by Seebach et al. [18]. But assuming that a 1 Å cutoff is reasonable, our result coincide with the observations by Arvidsson [41, 42] that oligo-β-alanine helical structures are disrupted in neutral pH aqueous environments. Previous work [43, 44] has shown the importance of internal hydrogen bonds in β-peptide helix formation. Given water’s ability to induce external hydrogen bonds, this offers an explanation for why β-peptide helices are less abundant in water.
In hydrogen-bonding analyses, the glycine dodecamer was found to form more transient hydrogen bonds in the 2.5–3.0 Å range than oligo-β-alanine, thus showed that its structure has lower transition barriers for those interactions. The higher hydrogen-bond persistence and density in oligo-β-alanine acceptor+2 diagonal showed that hydrogen bonds were more stable in the oligo-β-alanine helix, but at the same time, the 312 structure was not present in a large amount. This supports our hypothesis that severe fragmentation of 312 helix-like structures occurs in water.
We also tested the effect of multicentered-hydrogen bonds on helical structure and hydrogen-bond stability. A reduction of hydrogen-bond cleavage was much more significant in oligo-β-alanine helix than in the oligoglycine and oligoalanine helices, and hydrogen bonds in the 2.5–3.0 Å range were shown to be important for this effect. This showed that the dependency of oligo-β-alanine helix structure on multicenter hydrogen bonds.
The relationship between hydrogen bonding and structural helicity was also tested. A non-random, but only weak relationships, were found. This suggests that interactions between hydrogen bonding and the helicity of longer oligopeptides should be further examined. The capped dodecamer is a minimal model due to fraying at each end because of limited hydrogen bonding. Our study also suggests the hypothesis that the stacking structure is a competing structure with the 31 helix structure in water that dominates in oligo-β-alanine. This problem should be further explored in oligo-γ-alanine.
Supplementary Material
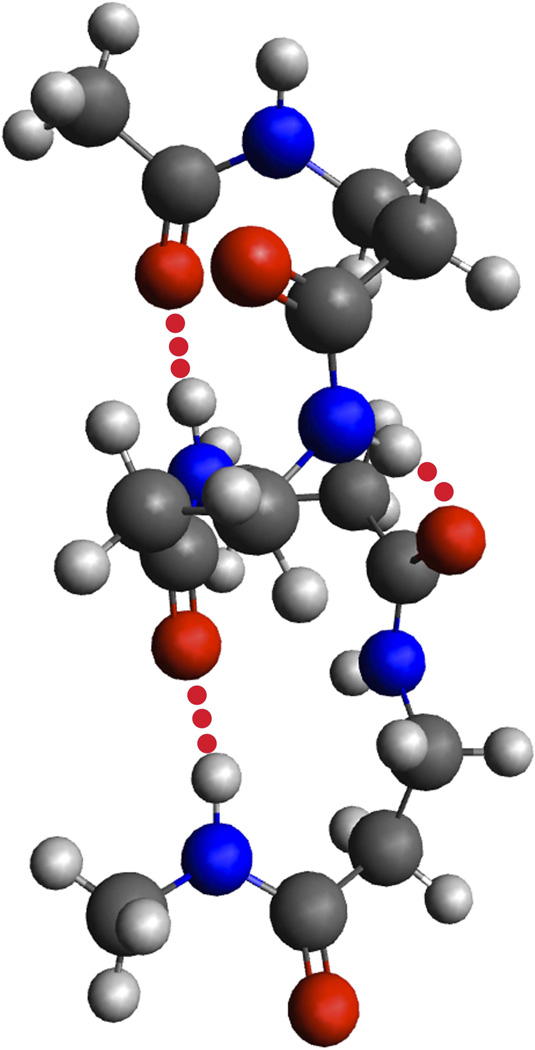
Figure 5.
Hydrogen-bonding patterns seen in oligo-β-alanine.
Figure 19.
The match between Jaccard-distance clustering and RMSD-distance clustering for oligoalanine.
Table I.
Top 10 principal components from the PCA of the RMSD matrix
| Principal Components Number | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Standard Deviation |
60.875 | 28.048 | 21.850 | 18.731 | 14.140 | 12.088 | 9.720 | 6.638 | 6.400 | 5.632 |
| Proportion of Variance |
0.606 | 0.129 | 0.078 | 0.057 | 0.033 | 0.0239 | 0.015 | 0.007 | 0.007 | 0.005 |
| Cumulative Proportion |
0.606 | 0.734 | 0.8122 | 0.870 | 0.902 | 0.926 | 0.942 | 0.949 | 0.955 | 0.961 |
Table II.
Hydrogen bonding shorter than 2.5 Å (before ‘/’) and 3.0 Å (after ‘/’) count matrix for each position for oligo-β-alanine. The larger numbers in the matrix correspond to frequent hydrogen binding between residues (for example, residue i to i+2 and residue i to i+3) and are highlighted by boxes.
| acptr donor |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0/ 2 |
52/ 355 |
503/ 756 |
9/ 33 |
16/ 34 |
4/ 16 |
24/ 24 |
0/ 0 |
0/ 0 |
0/ 2 |
35/ 59 |
0/ 0 |
14/ 16 |
| 2 | 101/ 824 |
0/ 2 |
133/ 459 |
757/ 897 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 3 | 83/ 141 |
225/ 898 |
0/ 2 |
109/ 440 |
649/ 805 |
0/ 0 |
3/ 20 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
1/ 1 |
| 4 | 24/ 142 |
4172/ 4443 |
436/ 1158 |
0/ 1 |
191/ 563 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 5 | 0/ 2 |
167/ 178 |
653/ 747 |
612/ 1722 |
0/ 2 |
142/ 584 |
250/ 348 |
0/ 0 |
0/ 2 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 6 | 53/ 58 |
34/ 82 |
0/ 2 |
2255/ 2436 |
207/ 1004 |
0/ 0 |
211/ 736 |
1121/ 1478 |
106/ 168 |
0/ 1 |
0/ 0 |
0/ 0 |
0/ 0 |
| 7 | 0/ 0 |
0/ 0 |
0/ 0 |
111/ 135 |
998/ 1058 |
379/ 1338 |
0/ 2 |
51/ 423 |
508/ 670 |
0/ 0 |
0/ 0 |
0/ 1 |
0/ 0 |
| 8 | 0/ 0 |
0/ 0 |
0/ 0 |
69 114 |
7/ 15 |
1450/ 1561 |
449/ 1558 |
0/ 2 |
108/ 446 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 9 | 0/ 0 |
0/ 1 |
0/ 0 |
0/ 0 |
3/ 5 |
132/ 183 |
2261/ 2383 |
486/ 1222 |
0/ 1 |
258/ 914 |
1891/ 2136 |
0/ 1 |
0/ 0 |
| 10 | 0/ 0 |
0/ 0 |
0/ 0 |
0/ 6 |
0/ 1 |
62/ 107 |
97/ 108 |
1497/ 1611 |
548/ 1510 |
0/ 0 |
47/ 231 |
263/ 352 |
13/ 57 |
| 11 | 0/ 1 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
1/ 9 |
2829/ 2996 |
119/ 965 |
0/ 1 |
4/ 116 |
264/ 306 |
| 12 | 0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
221/ 252 |
2433/ 2544 |
141/ 766 |
0/ 3 |
114/ 496 |
| 13 | 0/ 0 |
0/ 2 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 9 |
168/ 271 |
0/ 0 |
65/ 93 |
61/ 166 |
2559/ 3004 |
238/ 1053 |
0/ 3 |
Table IV.
Hydrogen bonding shorter than 2.5 Å (before ‘/’) and 3.0 Å (after ‘/’) count matrix for each position for oligoalanine. The larger numbers in the matrix indicating frequent hydrogen bonds between residues are highlighted by boxes.
| acptr/ donor |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0/ 6 |
1719/ 5025 |
0/ 2 |
59/ 80 |
0/ 0 |
0/ 0 |
0/ 0 |
1/ 7 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 2 | 59/ 392 |
0/ 4 |
1846/ 5361 |
0/ 4 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 3 | 84/ 487 |
232/ 770 |
0/ 4 |
2005/ 5140 |
0/ 1 |
0/ 5 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 4 | 852/ 942 |
57/ 593 |
35/ 346 |
0/ 11 |
1340/ 3749 |
0/ 2 |
0/ 3 |
0/ 0 |
636/ 734 |
6/ 40 |
0/ 0 |
0/ 0 |
0/ 0 |
| 5 | 0/ 0 |
1315/ 1562 |
208/ 1115 |
69/ 356 |
0/ 9 |
1877/ 4512 |
1/ 3 |
0/ 0 |
0/ 1 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 6 | 0/ 0 |
0/ 0 |
1756/ 2129 |
257/ 1433 |
33/ 370 |
0/ 5 |
1206/ 3804 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 7 | 280/ 437 |
0/ 0 |
1/ 1 |
2038/ 2517 |
386/ 1969 |
112/ 586 |
0/ 3 |
1194/ 3124 |
0/ 0 |
0/ 0 |
0/ 0 |
142/ 144 |
0/ 0 |
| 8 | 500/ 552 |
0/ 0 |
0/ 0 |
28/ 81 |
2579/ 3293 |
590/ 2465 |
43/ 357 |
0/ 9 |
1922/ 5468 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
| 9 | 58/ 111 |
0/ 0 |
0/ 0 |
0/ 1 |
2/ 29 |
2786/ 3536 |
467/ 2279 |
103/ 438 |
0/ 2 |
1538/ 4963 |
0/ 0 |
0/ 0 |
5/ 15 |
| 10 | 0/ 0 |
0/ 0 |
44/ 48 |
0/ 0 |
0/ 0 |
1/ 10 |
2191/ 3011 |
442/ 1818 |
99/ 762 |
0/ 3 |
1841/ 5120 |
0/ 0 |
0/ 0 |
| 11 | 0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 2 |
1264/ 1687 |
207/ 1005 |
131/ 698 |
0/ 4 |
1933/ 5847 |
0/ 2 |
| 12 | 0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
0/ 0 |
653/ 830 |
185/ 596 |
113/ 501 |
0/ 4 |
1964/ 6018 |
| 13 | 0/ 0 |
0/ 0 |
0/ 0 |
0/ 1 |
0/ 0 |
0/ 0 |
0/ 0 |
7/ 25 |
40/ 70 |
503/ 747 |
96/ 446 |
62/ 417 |
0/ 2 |
Table X.
Oligoalanine bond-break table for all the hydrogen bonds shorter than 2.5 Å.
| A | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 5678/ 5708 |
3974/ 4018 |
5736/ 5870 |
6540/ 7131 |
6668/ 7646 |
6070/ 7083 |
7155/ 8447 |
7058/ 8655 |
6079/ 7508 |
7942/ 9165 |
7143/ 8291 |
7761/ 8445 |
1069/ 1265 |
| acptr | 1486/ 2267 |
7665/ 8356 |
5599/ 6495 |
6669/ 7685 |
7338/ 8504 |
6888/ 8110 |
6342/ 7491 |
6993/ 7851 |
6537/ 7255 |
5131/ 5330 |
6822/ 7027 |
6107/ 6206 |
6605/ 6655 |
| B | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 0.01/ 0.01 |
0.01/ 0.01 |
0.02/ 0.02 |
0.08/ 0.05 |
0.13/ 0.05 |
0.14/ 0.05 |
0.15/ 0.11 |
0.18/ 0.14 |
0.19/ 0.08 |
0.13/ 0.05 |
0.14/ 0.07 |
0.08/ 0.06 |
0.15/ 0.09 |
| acptr | 0.34/ 0.05 |
0.08/ 0.04 |
0.14/ 0.05 |
0.13/ 0.07 |
0.14/ 0.1 |
0.15/ 0.11 |
0.15/ 0.06 |
0.11/ 0.06 |
0.1/ 0.04 |
0.04/ 0.03 |
0.03/ 0.03 |
0.02/ 0.01 |
0.01/ 0.01 |
Table XI.
Oligoalanine bond-break table for all the hydrogen bonds shorter than 3.0 Å.
| A | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 4904/ 4981 |
5083/ 5340 |
8370/ 9034 |
7211/ 10528 |
7208/ 11410 |
6169/ 10217 |
6710 10903 |
6738/ 10876 |
5763/ 9624 |
7346/ 10960 |
7394/ 10298 |
8627/ 11160 |
2737/ 3382 |
| acptr | 1529/ 4212 |
6227/ 10153 |
5602/ 9575 |
6295/ 10566 |
6644/ 10780 |
7438/ 11458 |
6437/ 9732 |
7439/ 10019 |
7114/ 9054 |
6798/ 7907 |
8174/ 9373 |
7550/ 8309 |
7481/ 7575 |
| B | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| segs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| type | |||||||||||||
| donor | 0.02/ 0.02 |
0.05/ 0.01 |
0.07/ 0.02 |
0.32/ 0.09 |
0.37/ 0.16 |
0.4/ 0.14 |
0.38/ 0.21 |
0.38/ 0.18 |
0.4/ 0.16 |
0.33/ 0.13 |
0.28/ 0.09 |
0.23/ 0.08 |
0.19/ 0.14 |
| acptr | 0.64/ 0.11 |
0.39/ 0.13 |
0.41/ 0.17 |
0.4/ 0.15 |
0.38/ 0.16 |
0.35/ 0.15 |
0.34/ 0.08 |
0.26/ 0.06 |
0.21/ 0.08 |
0.14/ 0.07 |
0.13/ 0.04 |
0.09/ 0.02 |
0.01/ 0.01 |
Acknowledgements
G.R.M. acknowledge support by DOD (contract HDTRA1-08-C-0015) and NIH (R01 GM106974-01). J.W.P. acknowledge support from NSF (CHE-0535675) and NIH (R01-GM6955302).
References
- 1.Ponder JW, et al. Current status of the AMOEBA polarizable force field. J Phys Chem B. 2010;114(8):2549–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appella DH, et al. Residue-based control of helix shape in beta-peptide oligomers. Nature. 1997;387(6631):381–384. doi: 10.1038/387381a0. [DOI] [PubMed] [Google Scholar]
- 3.Kritzer JA, et al. Helical beta-peptide inhibitors of the p53-hDM2 interaction. J Am Chem Soc. 2004;126(31):9468–9469. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- 4.Seebach D, Gardiner J. Beta-peptidic peptidomimetics. Acc Chem Res. 2008;41(10):1366–1375. doi: 10.1021/ar700263g. [DOI] [PubMed] [Google Scholar]
- 5.Johnson LM, Gellman SH. alpha-Helix mimicry with alpha/beta-peptides. Methods Enzymol. 2013;523:407–429. doi: 10.1016/B978-0-12-394292-0.00019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby E. Biphenyls as potential mimetics of protein alpha-helix. Bioorg Med Chem Lett. 2002;12(6):891–893. doi: 10.1016/s0960-894x(02)00031-8. [DOI] [PubMed] [Google Scholar]
- 7.Orner BP, Ernst JT, Hamilton AD. Toward proteomimetics: terphenyl derivatives as structural and functional mimics of extended regions of an alpha-helix. J Am Chem Soc. 2001;123(22):5382–5383. doi: 10.1021/ja0025548. [DOI] [PubMed] [Google Scholar]
- 8.Ernst JT, et al. Design and application of an alpha-helix-mimetic scaffold based on an oligoamide-foldamer strategy: antagonism of the Bak BH3/Bcl-xL complex. Angew Chem Int Ed Engl. 2003;42(5):535–539. doi: 10.1002/anie.200390154. [DOI] [PubMed] [Google Scholar]
- 9.Bourne GT, Kuster DJ, Marshall GR. Bipyridal-Based α-Helical Mimetics. Chemistry Eur J. 2010;16(28):8439–8445. doi: 10.1002/chem.201000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Che Y, Brooks BR, Marshall GR. Protein recognition motifs: design of peptidomimetics of helix surfaces. Biopolymers. 2007;86(4):288–297. doi: 10.1002/bip.20744. [DOI] [PubMed] [Google Scholar]
- 11.Marshall GR. Limiting assumptions in structure-based design: binding entropy. J Comput Aided Mol Des. 2012;26(1):3–8. doi: 10.1007/s10822-011-9494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seebach D, Beck AK, Bierbaum DJ. The world of beta- and gamma-peptides comprised of homologated proteinogenic amino acids and other components. Chem Biodivers. 2004;1(8):1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 13.Elliot A. In: Poly-a-Amino Acids. Fasman GD, editor. New York: Dekker; 1967. pp. 48–57. [Google Scholar]
- 14.Henzler Wildman KA, et al. Determination of the conformation and stability of simple homopolypeptides using solid-state NMR. Solid State Nucl Magn Reson. 2003;24(2–3):94–109. doi: 10.1016/s0926-2040(03)00048-1. [DOI] [PubMed] [Google Scholar]
- 15.Itoh SG, Okumura H. Length Dependence of Polyglycine Conformations in Vacuum. Journal of the Physical Society of Japan. 80(9):094801. [Google Scholar]
- 16.Bykov S, Asher S. Raman studies of solution polyglycine conformations. J Phys Chem B. 2010;114(19):6636–6641. doi: 10.1021/jp100082n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumas AM, Bode JW. Synthesis of acyltrifluoroborates. Org Lett. 2012;14(8):2138–2141. doi: 10.1021/ol300668m. [DOI] [PubMed] [Google Scholar]
- 18.Seebach D, Hook DF, Glattli A. Helices and other secondary structures of beta- and gamma-peptides. Biopolymers. 2006;84(1):23–37. doi: 10.1002/bip.20391. [DOI] [PubMed] [Google Scholar]
- 19.Cheng RP, Gellman SH, DeGrado WF. beta-Peptides: from structure to function. Chem Rev. 2001;101(10):3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 20.Glättli A, Seebach D, van Gunsteren WF. Do Valine Side Chains Have an Influence on the Folding Behavior of β-Substituted β-Peptides? Helvetica Chimica Acta. 2004;87(10):2487–2506. [Google Scholar]
- 21.Dean JC, Buchanan EG, Zwier TS. Mixed 14/16 helices in the gas phase: conformation-specific spectroscopy of Z-(Gly)n, n = 1, 3, 5. J Am Chem Soc. 2012;134(41):17186–17201. doi: 10.1021/ja306652c. [DOI] [PubMed] [Google Scholar]
- 22.Baldauf C, Gunther R, Hofmann HJ. Mixed helices--a general folding pattern in homologous peptides? Angew Chem Int Ed Engl. 2004;43(12):1594–1597. doi: 10.1002/anie.200353249. [DOI] [PubMed] [Google Scholar]
- 23.Ohnishi S, et al. Conformational preference of polyglycine in solution to elongated structure. J Am Chem Soc. 2006;128(50):16338–16344. doi: 10.1021/ja066008b. [DOI] [PubMed] [Google Scholar]
- 24.Rutledge SE, Volkman HM, Schepartz A. Molecular recognition of protein surfaces: high affinity ligands for the CBP KIX domain. J Am Chem Soc. 2003;125(47):14336–14347. doi: 10.1021/ja034508o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, et al. p53 alpha-Helix mimetics antagonize p53/MDM2 interaction and activate p53. Mol Cancer Ther. 2005;4(6):1019–1025. doi: 10.1158/1535-7163.MCT-04-0342. [DOI] [PubMed] [Google Scholar]
- 26.Cummings CG, et al. Synthesis and biological evaluation of a 5-6-5 imidazole-phenyl-thiazole based alpha-helix mimetic. Org Lett. 2009;11(1):25–28. doi: 10.1021/ol8022962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill DJ, et al. A field guide to foldamers. Chem Rev. 2001;101(12):3893–4012. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 28.Chin JW, Schepartz A. Design and Evolution of a Miniature Bcl-2 Binding Protein. Angew Chem Int Ed Engl. 2001;40(20):3806–3809. doi: 10.1002/1521-3773(20011015)40:20<3806::AID-ANIE3806>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 29.Park JS, et al. Accommodation of alpha-substituted residues in the beta-peptide 12-helix: expanding the range of substitution patterns available to a foldamer scaffold. J Am Chem Soc. 2003;125(28):8539–8545. doi: 10.1021/ja034180z. [DOI] [PubMed] [Google Scholar]
- 30.Trzesniak D, et al. Interpreting NMR data for beta-peptides using molecular dynamics simulations. J Am Chem Soc. 2005;127(41):14320–14329. doi: 10.1021/ja044285h. [DOI] [PubMed] [Google Scholar]
- 31.Raguse TL, et al. Toward beta-peptide tertiary structure: self-association of an amphiphilic 14-helix in aqueous solution. Org Lett. 2001;3(24):3963–3966. doi: 10.1021/ol016868r. [DOI] [PubMed] [Google Scholar]
- 32.Rutledge SE, Chin JW, Schepartz A. A view to a kill: ligands for Bcl-2 family proteins. Curr Opin Chem Biol. 2002;6(4):479–485. doi: 10.1016/s1367-5931(02)00352-6. [DOI] [PubMed] [Google Scholar]
- 33.Zheng X, et al. Molecular dynamics of beta-hairpin models of epigenetic recognition motifs. J Am Chem Soc. 2012;134(38):15970–15978. doi: 10.1021/ja306803v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 35.Patriksson A, van der Spoel D. A temperature predictor for parallel tempering simulations. Phys Chem Chem Phys. 2008;10(15):2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- 36.Penev ES, Lampoudi S, Shea J-E. TiReX: Replica-exchange molecular dynamics using Tinker. Computer Physics Communications. 2009;180(10):2013–2019. [Google Scholar]
- 37.Levandowsky M, Winter D. Distance between Sets. Nature. 1971;234(5323):34–35. [Google Scholar]
- 38.Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data via the EM Algorithm. Journal of the Royal Statistical Society, Series B. 1977;39(1):1–38. [Google Scholar]
- 39.Wu YD, Wang DP. Theoretical studies of beta-peptide models. J Am Chem Soc. 1998;120(51):13485–13493. [Google Scholar]
- 40.Smythe ML, Huston SE, Marshall GR. The molten helix: effects of solvation on the α- to 310-transition. J Amer Chem Soc. 1995;117(20):5445–5452. [Google Scholar]
- 41.Arvidsson PI, et al. On the antimicrobial and hemolytic activities of amphiphilic beta-peptides. Chembiochem. 2001;2(10):771–773. doi: 10.1002/1439-7633(20011001)2:10<771::aid-cbic771>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 42.Arvidsson PI, et al. Exploring the antibacterial and hemolytic activity of shorter- and longer-chain beta-, alpha, beta-, and gamma-peptides, and of beta-peptides from beta2-3-aza- and beta3-2-methylidene-amino acids bearing proteinogenic side chains--a survey. Chem Biodivers. 2005;2(3):401–420. doi: 10.1002/cbdv.200590020. [DOI] [PubMed] [Google Scholar]
- 43.Seebach D, et al. Beta-depsipeptides--the effect of a missing and a weakened hydrogen bond on the stability of the beta-peptidic 3(14)-helix. Chem Commun (Camb) 2002;(15):1598–1599. doi: 10.1039/b204187c. [DOI] [PubMed] [Google Scholar]
- 44.Gattin Z, et al. Simulation of beta-depsipeptides: the effect of missing hydrogen-bond donors on their folding equilibria. Biopolymers. 2007;85(4):318–332. doi: 10.1002/bip.20658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.