1. Introduction
Despite the promise of human neural stem cells (hNSCs) as an emerging cell source for neural tissue engineering, hNSC applications are hindered by the lack of advanced functional biomaterials that can promote cell adhesion, survival, and differentiation while also integrating neuronal stimulatory cues, specifically electrical stimulation. Electrospun fibrous substrates with controlled fiber architectures provide topographical cues to cells by presenting three dimensional (3-D) geometries that are representative of the extracellular matrix (ECM),1 defined by a high surface-to-volume ratio and porosity, and are thus better suited for differentiation studies of neural stem cells than standard two dimensional substrates. The architecture of ECM is of special importance because it supports 3 -D cellular networks together to form a tissue, allows for the proliferation and growth of cells, and regulates cellular processes capable of enhancing neurite outgrowth and neuronal differentiation of several cell types, including embryonic stem cells and hESC-derived NSCs2-6. Furthermore the inherently high surface to volume ratio of electrospun polymer substrates can facilitate mass transfer of nutrients and waste, promote cell attachment, and enable drug loading, properties that are inherent to bioactive matrix microniches. For stem cells, conductive substrates can promote neuronal maturation by providing electrical shortcuts between developing cells, while also permitting application of electrical stimuli that can mimic the electrophysiological environment experienced by cells in a variety of biological processes, including muscle contraction, wound healing, and synaptic transmission7-12. In addition to enhancing neurite outgrowth and neuronal maturation, applied electrical stimulation may also direct neural stem cell migration, opening the possibility to guide these cells towards injured sites8.
Various methods of delivering electrical stimulation to cells in culture include 2 -dimensional (2-D) substrates such as etched ITO glass13 and conductive polymers1, 14-19 like polypyrrole17. However, the use of conductive polymers alone is impeded by their poor processability, electroactive stability, and mechanical properties after doping20, 21. Another alternative has emerged involving nanocarbon materials such as carbon nanotubes (CNT)22, 23 and graphene24. In particular, single-walled carbon nanotubes (SWNT) have been employed due to their inherently high conductivity and the ability to regulate neuronal behavior both structurally and functionally25. Along with their biocompatibility at low concentrations, SWNT are ideal candidates for biomedical composites26, 27. It has been shown that SWNT interfaced with neural cells can promote neuron growth28-30 and enhance differentiation of NSCs into neurons31, 32. This is likely a result of a combination of topographical cues, enhanced signal transmission from the tight contacts formed between the SWNTs and the neuron membranes, and differential production of ECM proteins that modulate synaptic stability7, 32-34. While multi-walled carbon nanotubes (MWNTs) have been incorporated into electrospun fibers35, the incorporation of SWNTs into fibrous composite substrates that mimic the ECM has proved challenging36. The use of SWNTs during the electrospinning process could be circumvented altogether if the SWNT incorporation or deposition can be designed post facto, thus avoiding any bulk modification of the substrate properties and thereby retaining the SWNT bio-interfacial features. Doing so yields the additional benefit of using insulating polymers, already extensively used for differentiation studies, and thereby providing grounds for the use of highly biocompatible substrates. Other methods to do so have included spraying SWNT onto substrates37, layer-by-layer deposition (LbL)31, 38 and the attachment of SWNT to self assembled monolayers (SAM)39. In the first method, the growth of SWNT on the substrates can leave behind unwanted catalyst particles detrimental to cell viability. Regarding the latter two methods, enhancement of the differentiation kinetics has been reported with mouse embryonic stem cells by the LbL31 approach and with immortalized human neural stem cells via SAM40 approach, neither of which makes use of electrical stimulation.
This study is the first demonstration of electrically actuated SWNT-based composites for differentiation of human neural stem cells. We fabricated extracellular matrix-mimetic, composites by vacuum impregnation of electrospun poly(lactic-co-glycolic acid) (PLGA) membranes with single-walled carbon nanotubes (SWNTs) and investigated the ability of these substrates to enhance differentiation of induced pluripotent stem cell (iPSC)-derived NSCs. The SWNT-polymer substrates are electrically conductive, mechanically robust, and highly biocompatible with human NSC cultures in vitro and showed enhanced levels of electrically responsive cells. Notably, changes in the expression of two major neuronal markers, Neurofilament M (NFM) and microtubule -associated protein-2 (MAP2) of 14-day cultures showed that the composite enhanced neuronal differentiation of NSCs compared to PLGA controls without SWNT. To further utilize the multifunctional nature of SWNT-PLGA to affect neurogenesis, early NSC cultures on SWNT-PLGA were subjected to a 10 minute, 30μA direct current regimen of electrical stimulation. The electrical stimulation markedly increased neuronal differentiation after 14 days. These results highlight the multifunctionality of SWNT-PLGA, which afford a fibrous topography with high surface area to volume ratios to help to organize neuronal networks, along with the ability to exploit electro-conductivity to stimulate neuronal induction and neuronal maturation.
2. Results and discussion
2.1. Fabrication of SWNT-Polymer Composites
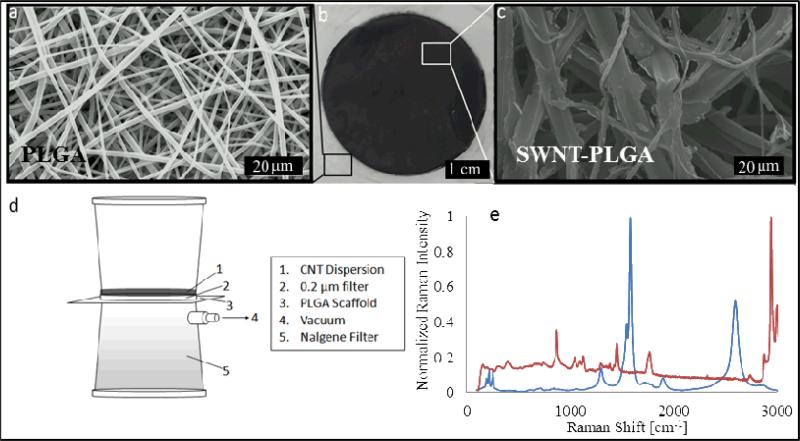
An aqueous dispersion was prepared with SWNTs, bovine serum albumin (BSA) and ascorbate, and was ultrasonicated to disperse the aggregated SWNTs. Fibrous PLGA substrates approximately 10 μm thick were fabricated by electrospinning and characterized visually by scanning electron microscopy (SEM), revealing a uniform fibrous architecture (Figure 1a). SWNTs were deposited onto 5x5 cm substrate fibers by vacuum-driven impregnation (Figure 1d). The SWNT had infiltrated completely through the substrate, appearing as a visible black circle on both sides of the PLGA substrate (Figure 1b). SEM images show that the fiber morphology changes upon impregnation, going from what appears to be smooth overlapping fibers to those with a rougher morphology (Figure 1a&c), indicating the presence of aggregated SWNT strongly adsorbed onto the fibers of the substrate via van der Waals forces41. Although van der Waals forces are typically weak, it is well understood that for SWNT these weak forces arise from the pz orbitals along the longitudinal axis of the SWNT. Given the high aspect ratio of the SWNT (1000:1), the summation of these typically weak forces results in an overall force that is extremely strong, thus explaining why SWNT are so difficult to debundle without the need for chemical modification or the use of surfactants. From Figure 1c, a change in contrast, from light to dark, is associated with the presence of SWNT in the substrates, and demonstrates that the SWNTs remain adsorbed to the substrate surface after drying. The lighter contrast is attributed to the charging associated with the polymeric substrates during SEM imaging, a phenomenon consistent with nonconductive samples. Furthermore a change in fiber diameter is evident that results from the formation of liquid bridges and subsequential coalescence during drying. We have characterized the change in fiber diameter as a function of SWNT dispersion concentration and observe an increase in fiber diameter with an increase in concentration (Supporting Information– Figure S1). Cross sections of the substrates show that an external fibrillar layer located on the polymeric fibers, which based on previous work27, 42 we have identified as SWNT bundles (Supporting Information– Figure S2). The presence of the SWNTs was confirmed using Raman spectroscopy, which shows characteristic peaks located at 1590 and 2700 corresponding to the G and G’ peaks inherent to SWNT ( Figure 1e). The sharpness and lack of broadening of these peaks indicates that the surface is completely coated with SWNT. An additional peak is also prominent at 1350 relating to the characteristic D peak of SWNTs43. The splitting of the G band is the result of a mixture of both metallic and semiconducting SWNT, which is to be expected given that the SWNT were not separated, and the peaks below 500 cm −1 arise from coupling modes between the SWNT and polymer. The conductivity of the substrates was determined to be 227±15 S m−1, which is comparable to previous fiber composites fabricated with agarose[27] or PVA[26], and boron doped silicon, a material commonly used in neural prosthetic device44. In order to verify that our composite materials are indeed safe45, we evaluated the stability of SWNT coated scaffolds using a SWNT release study (Supporting Information– Figure S3). Utilizing SWNTPLGA substrates left in PBS for 3 months, we determined that the SWNT are in fact strongly absorbed within the composite material, indicating little to no desorption from the substrate.
Figure 1. Fabrication and characterization of SWNT-PLGA-Composites.
(a) SEM image of the electrospun PLGA substrate prior to incorporation with SWNT (b) Optical image of the impregnated area (c) SEM image of the substrate after impregnation (Bundles of SWNT can be identified in Figure S2 in the Supporting Information). (d) Vacuum impregnation method used to forcibily drive an aqueous SWNT dispersion into the hydrophobic substrate (e). Raman spectroscopy results confirm the presence of SWNT in substrate after wetting.
2.2. Human Neural Stem Cell Cultures on Composites
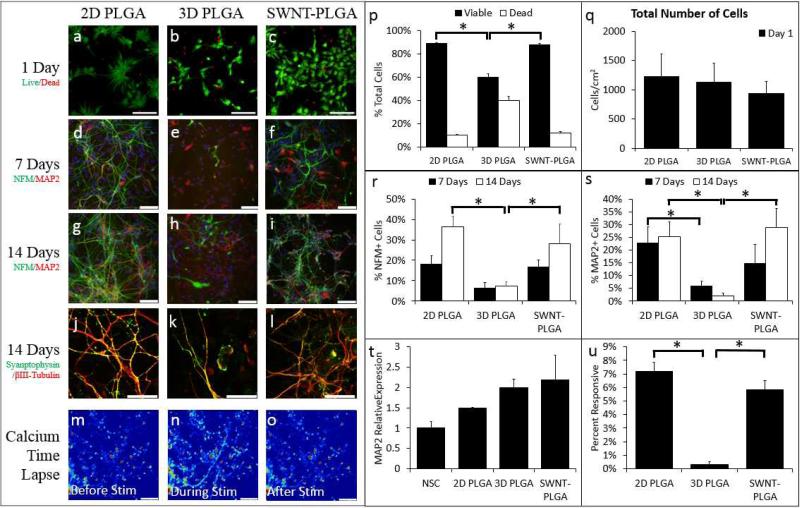
SWNT-PLGA substrates were sterilized with oxygen plasma and pretreated with a minimal amount of poly-D-lysine and laminin to promote attachment and neurite outgrowth and to ensure cellular survival. Two configurations were first tested: SWNT-PLGA and PLGA substrates, with a 2D control of PLGA thin film glass coverslips. These PLGA conditions will hereafter be referred to as 2D PLGA and 3D PLGA. hNSC were primed for neuronal differentiation and seeded onto the substrates. Cell viability was evaluated one day after cell seeding by labeling with calcein AM and propidium iodide (Figure 2a-c). Cell viability on 2D PLGA and SWNT-PLGA was significantly greater than the 3D PLGA case (Figure 2p) with no significant differences in the number of cells per condition (Figure 2q).
Figure 2. SWNT-PGLA substrates elicit enhanced neuronal differentiation of hNSCs.
hNSCs were cultured on 2D PLGA, 3D PLGA and SWNT-PLGA to determine the effect of SWNT vacuum impregnation on hNSC differentiation and viability. The cells were labeled as live (green) or dead (red) after 1 day of substrate growth (a-c). The cells were significantly more viable in 2D PLGA (p<1e-6) and SWNT-PLGA (p<1e-6) versus 3D PLGA culture platforms (p-q). These cells were labeled for antibodies NFM (green) and MAP2 (red) at 7 and 14 days in situ (d-i) and for Synaptophysin (green) and βIII-tubulin (red) at 14 days in the substrate (j-l). Mature, punctate Synaptophysin can be seen in the neurites and cell bodies of the cells of both conditions at day 14. The percentage of cells positive for NFM (r) increased with time on SWNT-PLGA and showed greater enhancement (p<0.01) compared to 3D PLGA after 14 days of culture. Percentage of MAP2 (s) positive cells increased through14 days after plating on SWNT-PLGA, with greater enhancement (p<2e-4) compared to 3D PLGA. PCR was performed to assay the fold gene expression of MAP2(t). Time lapse of calcium imaging, taken after 14 days of culture, (m-o) shows calcium levels in cells before, during, and after brief electrical stimulation. There is a significant increase in percentage of cells responsive to electrical stimulation via calcium influx on SWNT-PLGA (p<1e-5) compared to 3D PLGA (u). All statistics were carried out via ANOVA with Bonferroni-Holm correction. Scale bars on Live/Dead images are 10μm (a-c); those on NFM/MAP2 and Synaptophysin images are 50μm (d-l); and those on Calcium Time Lapse images are 1μm (m-o). *Significantly Different from 3D PLGA
After 1, 7, and 14 days of differentiation, neuronal differentiation was characterized by evaluating the expression of Neurofilament M (NFM) and microtubule-associated protein 2 (MAP2), two proteins commonly found in soma and dendrites of neurons, respectively ( Figure 2d-i). These biomarkers have been shown to play a critical role in the maintenance of the neuronal architecture, cellular differentiation, and structural and functional plasticity46, 47. After 7 days of differentiation, there were 17% NFM+ cells and 15% MAP2+ cells observed in SWNT-PLGA, compared to 6% NFM+ cells and 5% MAP2+ cells in 3D PLGA, which indicates a significant difference for the MAP2+ cells (Figure 2r-s). These numbers increased steadily after 14 days of culture to 28% NFM+ cells and 29% MAP2+ cells in SWNT-PLGA, compared to 8% NFM+ cells and 2% MAP2+ cells in 3D PLGA (Figure 2r-s). Thus, clear differences emerge in the fraction of neuronal marker-positive cells between SWNT-containing and SWNT-lacking substrates for both markers. Further, after 14 days, hNSCs in both 3D conditions expressed mature neuronal marker synaptophysin, a synaptic vesicle protein (Figure 2j-l). qRT-PCR also revealed increased expression of the MAP2 gene at day 14 of differentiation in SWNT-PLGA relative to PLGA controls (Figure 2t). Additionally, neuronal functionality was assessed by calcium imaging, where electrically active cells are defined as those that show an increase in fluorescence due to calcium influx in response to an external electrical stimulation. We observed a significant increase in the number of electrically responsive cells in SWNT-PLGA versus 3D PLGA conditions, 0.3% to 5.9% (Figure 2u). Treatment with 1μM tetrodotoxin (TTX), a voltage-gated sodium channel blocker, abolished most calcium activity, suggesting that these temporal increases in intracellular calcium are due to elicited action potentials 48.
The enhancement of neuronal differentiation kinetics within SWNT-PLGA relative to 3D PLGA can be attributed to several factors including a change in electrical conductivity, substrate topography, and substrate surface chemistry. The individual and concerted contributions of these factors remain to be elucidated. The increase in conductivity is reported to facilitate communication between neuronal cells, by allowing the cells to form “electrical shortcuts”, or transient electrical connections without being synapsed with one another34. Enhanced neuronal network connectivity has been proposed as the primary mechanism by which SWNT increase the frequency of postsynaptic currents 7, 34, 49. In addition, cell behavior has been shown to be markedly altered by substrate topography.50 In our study, the impregnation of the PLGA scaffolds with the SWNT dispersion resulted in a marked enhancement in the fiber diameter (Supporting Information – Table 6 & Figure S1). Increased fiber diameters yield increased interfacial areas and a reduction in void size between fibers, supporting increased local neuronal spreading and outgrowth. Similarly, the increased viability evidenced in the SWNT-PLGA versus uncoated PLGA fibers could be due to the altered topography of the SWNT -PLGA substrate that is less porous and this can translate into a higher degree of cell-cell contact compared to the PLGA substrate, which is advantageous for hNSC survival. Previous studies implicate the combination of nanoscale grooves and hydrophobic environments to elicit increased adsorption of proteins and growth factors essential for neuronal differentiation32. We observed that the contact angle of the aqueous SWNT dispersion changes markedly after impregnation, from a hydrophobic surface, 117°, to a hydrophilic one, 80°, (Supporting Information – Figure S4). Our preliminary studies suggest that the levels of protein adsorption are not significantly altered (see Supplementary Information), which could result from the opposing effects of increased interfacial area and surfaces with diminished hydrophobicity, however further protein conformation and dynamics studies will be necessary to dissect the underlying mechanisms.
2.3. Electrical Stimulation of hNSC on SWNT-PLGA Substrates
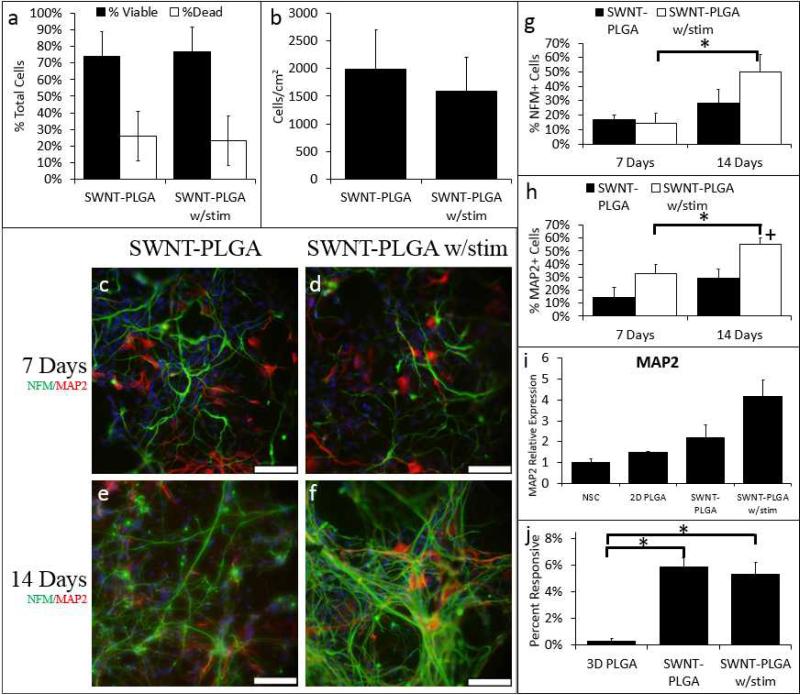
While passive cell culture on conductive substrates has been previously shown to enhance neuronal differentiation, applied electrical stimulation can also modulate cell and tissue growth both in nerve cultures24 and more recently for the differentiation of myoblasts51. However, a combination of the electrospun substrates possessing high surface area/volume fibrous topography with applied electrical stimulation has yet to be explored for hNSCs. We hypothesized that our SWNT-PLGA could deliver an electrical stimulus to cultured hNSCs, and that this electrical stimulation would further enhance neuronal differentiation. On day 3 of NSC differentiation in SWNT-PLGA, a 30μA direct current was applied for 10 minutes. This mode of electrical stimulation is similar to pre viously reported methods that indicate that short periods of electrical stimuli has the potential to alter cellular behavior17 by opening Na+ and K+ channels within the cells through depolarization. Electrical stimuli were delivered to substrates via bridging current through etched conductive ITO-coated glass. (Figure S5). Tests on cultures on day 4, or 1 day after electrical stimulation, show that the electrical stimulation had a negligible effect on cell viability and the total cell number ( Figure 3a-b). It should be noted that in the latter panels some cells are still undergoing mitosis, which we attribute to the change in cell density. NSC differentiation was then evaluated after 7 and 14 days in situ for samples with and without electrical stimulation. We found that electrical stimulation increased %NFM+ cells at day 14 compared to 3D PLGA and %MAP2+ cells after days 7 and 14 compared to 3D PLGA (Figure 3g-h). This remains true when compared to 2D PLGA and SWNT-PLGA without stimulation at day 14. Changes in MAP2 gene expression are shown in Figure 3i. For comparison purposes we also performed stimulation on the bare PLGA substrates in the NSC medium (Supporting Information -Figures S6 & S7) observing that there is no significant effect from the medium. Previous reports have demonstrated in vivo that electrical stimulation can increase MAP2 expression, which can in turn increase dendritic and synaptic plasticity52. It is interesting to note that the change in MAP2 and NFM expression relative to non-stimulated samples shows a somewhat delayed but sustained effect on the expression of these cytoskeletal proteins. While the exact mechanisms are not understood, it is likely that the stimulation activates a pathway far upstream of MAP2 or dendrite formation, or that it induces differences in secondary remodeling phenomena, which results in longer term effects on neuronal differentiation. Notably, similar to trends seen in Figure 2, we have also observed that the network connectivity in electrically stimulated SWNT-PLGA supported an elevated level of calcium influx responsiveness compared to PLGA scaffolds (Figure 3j). However, electrically stimulated SWNTPLGA scaffolds did not elicit detectable “field-wide” enhancements in levels of electrical activity relative to 2D or unstimulated SWNT-PLGA conditions, possibly due to the non-uniform levels of cell density in these different conditions. We hypothesize that cell density, in addition to neuronal differentiation and substrate conditions, may play a regulatory role in promoting neuronal maturation. The electrical activity in the stimulated conditions may reflect the slightly diminished cell numbers in these conditions. Nonetheless, we have achieved similar levels of electrically active cells in our SWNT-PLGA conditions to non-transplantable 2D controls, which is a key step towards establishing transplantable functional neuronal cells in vivo within SWNT-PLGA composite scaffolds.
Figure 3. Electrical stimulation further enhances neuronal differentiation of hNSCs in SWNT PLGA substrates.
hNSCs were cultured on SWNT-PLGA, and at day 3, selected substrates were electrically stimulated for ten minutes at 30μA. On day 4, one day after stimulation, cell viability testing determined that there was no significant decrease in cell viability or number due to electrical stimulation (a & b). After 7 and 14 days, the cells were fixed and all conditions were labeled with antibodies for NFM (green) and MAP2 (red) (c-f). The percentage of NFM and MAP2 cells for each condition are shown (g & h). On day 7 there is a significant increase in the percentage of MAP2 positive cells on the electrically stimulated SWNT-PLGA substrates versus 2D PLGA (p<5e-4). On day 14, electrical stimulation significantly increased percentage MAP2 positive cells compared to 2D PLGA (p=0.008), 3D PLGA (p<1e-8), and SWNT-PLGA alone (p= 0.02). PCR analysis of MAP2 gene fold expression (i) showed responsiveness following electrical stimulation. The percentage of cells responsive to electrical stimulation via calcium influx was quantified (j); Cells on SWNT-PLGA with (p<1e-3) and without (p<2e-6) stimulation are significantly more responsive compared to 2D PLGA. A comparison study for PLGA substrates containing no SWNT but electrically stimulated can be found in the supplementary information. *Significantly Different from 3D PLGA +Significantly different from 3D PLGA and SWNT-PLGA (ANOVA with Bonferroni-Holm Correction). Scale bars on NFM/MAP2 images are 50μm (c-f);
In summary, the SWNT-PLGA composite can elicit enhanced kinetics of human NSC neuronal differentiation and maturation, which is further accelerated in a sustained manner by a simple electrical stimulation regimen. Such substrates could be vital to expanding neuronal cells derived from, say, patient-derived induced pluripotent stem cells for in vitro applications such as drug screening or disease modeling. In addition, interfacing human neuronal cells with SWNT composites opens up a wide range of applications to testing the plasticity, functionality, and subtype specificity of neurons following programmed regimen of electrical stimulation. Such a system could also be used to transplant human neuronal cells within biomaterial constructs into models of spinal cord or neurodegeneration in the brain and serve to accelerate neuronal differentiation, improve integration of exogenous cells within host tissue, and provide the ability to deliver or monitor electrical signals to or from the transplanted constructs.
3. Conclusion
We have introduced an integrated platform for growth and electrical stimulation of human neural stem cells combining three salient features: (i) First the SWNT-PLGA substrates can be easily fabricated based on vacuum-driven impregnation of an electrospun fibrous substrate with a SWNT dispersion. (ii) The resulting substrates combine the salient features of highly conductive SWNTs with the topography of electrospun substrates, and were found to be highly biocompatible for the support of hNSCs derived from iPSC's. (iii) The application of electrical stimulation to the conductive substrates was found to elicit an enhanced effect on the hNSC differentiation process. Thus, the composite substrates are promising candidates for probing neurogenesis and neural activity, given the high level of biocompatibility, 3-D geometries, extracellular matrix-mimetic topography, and tunable differentiation/maturation cues. In addition, a conductive polymer scaffold may improve cell survival and functional integration after transplantation in vivo by providing structural support for transplanted cells and facilitating synaptogenesis with host cells. It is also anticipated that the novel SWNT composite fabrication technique presented here can be extended to similar electrospun scaffolds fabricated from different polymers, which would permit tuning of scaffold material properties for specific in vivo applications. Similarly, the amount of SWNT that can be deposited within the substrate can be controlled, allowing future studies into the effects of SWNT concentration and topography on differentiation. This will allow for future studies to determine the optimal substrate and stimulation conditions to maximally promote neuronal differentiation and maturation, which may lead to transplantable cell-scaffold constructs for CNS regeneration.
4. Experimental
Preparation of SWNT dispersion: All chemicals were of reagent grade or higher. An aqueous dispersion of SWNT was prepared containing 0.4 (wt% by vol) of SWNT (Unidym ), bovine serum albumin (BSA, Sigma-Aldrich) and ascorbic acid (Sigma-Aldrich). The dispersion was mixed with a horn tip sonicator (Mixsonix S400) for 10 minutes at a pulsed rate of one second on and one second off at 40 amperes.
Fabrication of PLGA scaffolds
Fibrous PLGA scaffolds were made by dissolving 15% weight/volume PLGA (Sigma-Aldrich, 85:15 PLA:PGA) in 1,1,1,3,3,3-Hexafluoro-2-propanol (Sigma-Aldrich, HFIP) with gentle agitation. This solution was electrospun with a 23 gauge needle and a potential difference of +18kV at a distance of 18cm onto a grounded flat plate collector.
Impregnation of PLGA scaffolds with SWNT dispersions
A 5 cm by 5 cm PLGA scaffold was placed on a Nalgene 0.2 μm pore filter with a 150 mL capacity. The scaffold was sealed in place by the upper cup of the filter. A volume of 3 mL of the dispersion was then placed onto the scaffold and pulled through the scaffold by vacuum. The conductivity of the substrates was determined from resistance measurements shown in the supplementary information and compared with the conductivity of the dried SWNT and the PLGA scaffolds without SWNT. A microBCA assay (Pierce Biotechnology) was performed to assess any differences in adsorbed level of protein (laminin), as elaborated in the Supplementary Information.
Scanning Electron Microscopy
All images were imaged using a Hitachi S-4500 Field emission SEM. PLGA and PLGA-SWNT substrates were sputtercoated with platinum prior to imaging.
Fabrication of PLGA Thin Film Coverslips
A 1% (w/v) PLGA polymer solution was prepared by dissolving the polymer in HFIP overnight at room temperature. The polymer solution was then spin-coated onto 12 mm glass coverslips. The thin films were dried for at least 24 h under vacuum.
iPSC to NSC Differentiation
Induced pluripotent stem cells (HFF1-iPSCs, gift from the Rutgers University Cell and DNA Repository, RUCDR) were differentiated to neural stem cells (hNSCs), using Neurobasal Media (Life Technologies) with 1x N2 (Life Technologies), 1x B27 without vitamin A (Life Technologies), 1x ITS (Life Technologies), and 2mM L-glutamine (Invitrogen) supplemented with 500ng/mL Noggin (Peprotech). hNSCs were further propagated in proliferation media consisting of 50% DMEM/F12 (Life Technologies) with 2mM L-glutamine (Invitrogen) and 50% Neurobasal Media (Life Technologies) supplemented with 0.5× N2 supplement (Life Technologies), 0.5× B27 supplement without Vitamin A (Life Technologies), and 20 ng/ml bFGF (Peprotech).
hNSC Priming for Differentiation
In order to prime hNSCs for differentiation, they were cultured in proliferation media with bFGF withdrawn once the cells reached 70% confluence, where the proliferation media consisted of Neurobasal Media (Life Technologies), DMEM/F12 (Life Technologies), 0.5× N2 (Life Technologies), and B27 without vitamin A (Life Technologies) without any additional growth factors. The hNSCs would subsequently be switched to differentiation media consisting of Neurobasal Media (Life Technologies), 1x B27 (Life Technologies), and 1x Glutamax (Invitrogen) with 10ng/mL BDNF (Peprotech).
hNSC Neuronal Differentiation Analysis
Neuronal differentiation was evaluated using immunocytochemistry and qRT-PCR. The presence of MAP2 and Neurofilament M (NFM) were evaluated using primary antibodies for those proteins, anti-MAP2 (Becton Dickinson) and anti-NFM (Invitrogen), linked to secondary antibodies labeled at 594nm and 488nm respectively. Cell nuclei were labeled with DAPI. Images were taken with an Olympus IX81 Inverted Fluorescent Microscope. Each marker's presence per cell was quantified using the ImageJ software. For the qRT -PCR analysis MAP2 (TaqMan Hs 00258900_m1) was evaluated for undifferentiated NSCs and for each experimental condition after 14 days of differentiation. Undifferentiated NSCs were used as the reference sample. For NFM and MAP2 percentages the average number of cells per data point was 6,165 cells with a minimum of 1,429 and a maximum of 10,777. For electrical activity >1,000 cells were used per data point.
Immunostaining
The NSCs in substrates were processed for immunocytochemistry by fixing the live cells with 0.4% paraformaldehyde (paraformaldehyde) for 30 minutes at room temperature. The sample was then washed 3 times with phosphate buffered saline (PBS). To block and permeabolize the sample it was immersed in a solution PBS based solution of 10% normal goat serum (Fisher), 1% bovine serum albumin (Sigma Aldrich), and 0.1% Triton X (Sigma Aldrich) f or one hour at room temperature. All antibody and isotype control solutions were made in 10% normal goat serum and 1% bovine serum albumin and were incubated on the samples for one hour at room temperature. Three washes with PBS, minimum of 15 minutes each, cleared the sample for one hour at room temperature with the secondary, fluorescent antibody. Three washes with PBS followed secondary antibody incubation. DAPI was used to mark the nuclei of each cell and was added at a final concentration of 1μg/mL for 5 minutes to the samples before three PBS washes. Each sample was then mounted onto a glass coverslip using ProLong® Gold Antifade Reagent (Invitrogen) for imaging. The samples were not sectioned. All imaging was done on either an Olympus IX81 Inverted Fluorescent Microscope. For the immunocytochemistry imaging 4 images were taken of each sample in random locations on the substrate or coverslip. Each image was quantified until a minimum of 3 images and 150 cells were counted. Quantification is shown as the mean ± standard error of 3 independent experiments, where at least four frames from each of two biological replicates were analyzed for each experiment. DAPI staining was used to quantify how many cells were present. Electrical Stimulation Set-up: A circuit board was constructed allowing for the application of external electrical stimuli. Each electrode starts as an indium tin oxide (ITO) covered glass slide. The middle portion is etched with solution of 20% HCl and 5% HNO3 to create a non-conductive gap, forming two electrodes38. This ensures that the applied current travels through the SWNT-PLGA and not around a path of least resistance. SWNT-PLGA are fitted with cell culture rings to house the cells and growth media. The power source consists of a 2.5 volt watch battery. The ITO glass slides had a resistance of 200 Ω (+/−50 Ω) whereas the substrate contained a resistance of 4.4 kΩ (+/− 6.5 kΩ) on the top and 10.7 kΩ (+/−9.3 kΩ) on the bottom. Electrical stimulation of the cells was applied for 10 minutes on the third day of differentiation under the experimental conditions.
Cell Viability Analysis
To determine cell viability, cells were incubated for 30 minutes at 37°C with 4μM calcein AM and 1μg/mL propidium iodide (both from Life Technologies), which labeled live and dead cells, respectively. Cells were then imaged by acquiring z-stacks using a Leica TCS SP2 confocal microscope. This stacking, combined with a higher magnification allows for a more accurate percentages of NFM+ and MAP2+ cells in each condition. For each condition there was a minimum of two biological replicates in each of the three experiments. Images were taken with a 40x objective and quantified for cell number and percentage of NFM or MAP2 positive cells using custom ImageJ macros. Maximum intensity projections of each stack were constructed and used for quantification of cell viability. Each cell in one frame was counted as either live or dead using ImageJ software (NIH).
Quantitative Real Time Polymerase Chain Reaction
Total RNA was extracted from hNSCs on day 14 in each condition using the RNEasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions, including removal of genomic DNA using Rnasefree DNase. A high -capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) containing random primers was used to reverse transcribe RNA from each sample to cDNA. TaqMan gene expression master mix and TaqMan gene expression assays (Applied Biosystems) were used for template amplification of 10 ng cDNA/reaction. qRT-PCR was carried out on a 7500 Fast Real-Time PCR instrument (Applied Biosystems). Relative gene expression was calculated using the ΔΔCT method, normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the endogenous control and undifferentiated hNSCs in standard 2-D culture. Graphs represent mean ± standard deviation. The TaqMan gene expression assays MAP2 (Hs00258900_m1) and GAPDH (4326317E) (Applied Biosystems) were used in these studies.
Calcium Imaging
Electrical activity was assessed using a calcium indicator dye which reflects changes intensity in response to changes in calcium levels. 3μM Fluo-4 AM (Invitrogen) was loaded into the cells in an imaging solution which consisted of 140mM NaCl, 5mM KCl, 2mM CaCl2, 2mM MgCl2, 10mM HEPES, and 10mM Glucose, supplemented with 0.2% pluronic F127 (Invitrogen). The chamber consisted of a 2 well LabTek (Thermo Scientific) modified to include a platinum wire on either side of the well, acting as electrodes. To suspend the immersed substrate for image capture, a coverslip was altered to include 4 plastic pillars, each approximately 1mm high. A second coverslip was placed on top of the pillars, forming a containment space for the sample. Each sample was excited using a 10V pulse train with 10ms pulse width and 100ms pulse spacing. The dye's response was captured at 1Hz on a Leica SP2 Confocal Microscope. Stacks were made from the images in ImageJ and shifting in the sample was corrected using normalized correlation coefficient template matching. Each cell in the image was selected as a region of interest, and the cells’ average fluorescence at each frame exported to MATLAB. A MATLAB program detected fluorescence spikes in terms of the change in fluorescence over initial fluorescence (dF/F0) due to the stimulation with a threshold of a 1dF/F0 rise over a 10 second window and calculated the percentage of cells in the frame that showed this activity which was manually checked for each series of images.
Statistics
One way ANOVA was used to test for statistical significance between conditions at each individual time-point with a Bonferroni-Holm's post-hoc test. Samples with a p value below 0.05 were considered significant.
Supplementary Material
Acknowledgements
This work was funded in part by NIH EB 001046 (PVM), NSF IGERT DGE 0801620 (PVM), NJSCR Exploratory Research Grant (PVM), a GAANN award from the Department of Education in pharmaceutical engineering and NIH R01 EB007467 (AVN). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or DoE. The authors would like to thank Valery Starovoytov for expert guidance and assistance with SEM. The authors would like to thank Jen Moore and the RUCDR facility at Rutgers University for the gift of the iPSC cell line. JL would like to thank Nedjma Bendiab for helpful discussion relating to Raman measurements. Finally we would like to thank Prof. Asefa and Yuying Meng for help with UV-Vis measurements.
Footnotes
Supporting Information
Supporting Information is available online from the Wiley Online Library or from the author.
References
- 1.Xie JW, MacEwan MR, Willerth SM, Li XR, Moran DW, Sakiyama-Elbert SE, Xia YN. Advanced Functional Materials. 2009;19(14):2312–2318. doi: 10.1002/adfm.200801904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherry JF, Carlson AL, Benarba FL, Sommerfeld SD, Verma D, Loers G, Kohn J, Schachner M, Moghe PV. Biointerphases. 2012;7(1-4):16. doi: 10.1007/s13758-012-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu HF, Lim SX, Leong MF, Narayanan K, Toh RPK, Gao SJ, Wan ACA. Biomaterials. 2012;33(36):9179–9187. doi: 10.1016/j.biomaterials.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Kim H, Cooke MJ, Shoichet MS. Trends Biotechnol. 2012;30(1):55–63. doi: 10.1016/j.tibtech.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Christopherson GT, Song H, Mao HQ. Biomaterials. 2009;30(4):556–564. doi: 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Xie JW, Willerth SM, Li XR, Macewan MR, Rader A, Sakiyama-Elbert SE, Xia YN. Biomaterials. 2009;30(3):354–362. doi: 10.1016/j.biomaterials.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cellot G, Toma FM, Varley ZK, Laishram J, Villari A, Quintana M, Cipollone S, Prato M, Ballerini L. Journal of Neuroscience. 2011;31(36):12945–12953. doi: 10.1523/JNEUROSCI.1332-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCaig CD, Rajnicek AM, Song B, Zhao M. Physiological Reviews. 2005;85(3):943–978. doi: 10.1152/physrev.00020.2004. [DOI] [PubMed] [Google Scholar]
- 9.Graves MS, Hassell T, Beier BL, Albors GO, Irazoqui PP. Annals of Biomedical Engineering. 2011;39(6):1759–1767. doi: 10.1007/s10439-011-0259-8. [DOI] [PubMed] [Google Scholar]
- 10.Geremia NM, Gordon T, Brushart TM, Al-Majed AA, Verge VMK. Experimental Neurology. 2007;205(2):347–359. doi: 10.1016/j.expneurol.2007.01.040. [DOI] [PubMed] [Google Scholar]
- 11.Cho YN, Ben Borgens R. Journal of Biomedical Materials Research Part A. 2010;95A(2):510–517. doi: 10.1002/jbm.a.32841. [DOI] [PubMed] [Google Scholar]
- 12.Luo XL, Weaver CL, Zhou DD, Greenberg R, Cui XYT. Biomaterials. 2011;32(24):5551–5557. doi: 10.1016/j.biomaterials.2011.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura K, Yanagida Y, Haruyama T, Kobatake E, Aizawa M. Journal of Biotechnology. 1998;63(1):55–65. doi: 10.1016/s0168-1656(98)00075-3. [DOI] [PubMed] [Google Scholar]
- 14.Ghasemi-Mobarakeh L, Prabhakaran MP, Morshed M, Nasr-Esfahani MH, Ramakrishna S. Tissue Engineering Part A. 2009;15(11):3605–3619. doi: 10.1089/ten.TEA.2008.0689. [DOI] [PubMed] [Google Scholar]
- 15.Huang LH, Zhuang XL, Hu J, Lang L, Zhang PB, Wang YS, Chen XS, Wei Y, Jing XB. Biomacromolecules. 2008;9(3):850–858. doi: 10.1021/bm7011828. [DOI] [PubMed] [Google Scholar]
- 16.Lee JY, Lee JW, Schmidt CE. Journal of the Royal Society Interface. 2009;6(38):801–810. doi: 10.1098/rsif.2008.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt CE, Shastri VR, Vacanti JP, Langer R. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(17):8948–8953. doi: 10.1073/pnas.94.17.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson BC, Richardson RT, Moulton SE, Evans AJ, O'Leary S, Clark GM, Wallace GG. Journal of Controlled Release. 2010;141(2):161–167. doi: 10.1016/j.jconrel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Rouabhia M, Wang ZX, Roberge C, Shi GX, Roche P, Li JM, Dao LH. Artificial Organs. 2007;31(1):13–22. doi: 10.1111/j.1525-1594.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 20.Bakhshi AK, Bhalla G. Journal of Scientific & Industrial Research. 2004;63(9):715–728. [Google Scholar]
- 21.Ghasemi-Mobarakeh L, Prabhakaran MP, Morshed M, Nasr-Esfahani MH, Baharvand H, Kiani S, Al-Deyab SS, Ramakrishna S. J Tissue Eng Regen Med. 2011 doi: 10.1002/term.383. [DOI] [PubMed] [Google Scholar]
- 22.Huang YJ, Wu HC, Tai NH, Wang TW. Small. 2012;8(18):2869–2877. doi: 10.1002/smll.201200715. [DOI] [PubMed] [Google Scholar]
- 23.Mattson MP, Haddon RC, Rao AM. Journal of Molecular Neuroscience. 2000;14(3):175–182. doi: 10.1385/JMN:14:3:175. [DOI] [PubMed] [Google Scholar]
- 24.Park SY, Park J, Sim SH, Sung MG, Kim KS, Hong BH, Hong S. Advanced Materials. 2011;23(36):H263–+. doi: 10.1002/adma.201101503. [DOI] [PubMed] [Google Scholar]
- 25.Fabbro A, Bosi S, Ballerini L, Prato M. Acs Chemical Neuroscience. 2012;3(8):611–618. doi: 10.1021/cn300048q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubin RA, Callegari GC, Kohn J, Neimark AV. Ieee Transactions on Nanobioscience. 2008;7(1):11–14. doi: 10.1109/TNB.2008.2000144. [DOI] [PubMed] [Google Scholar]
- 27.Lewitus DY, Landers J, Branch JR, Smith KL, Callegari G, Kohn J, Neimark AV. Adv. Funct. Mater. 2011;21(14):2624–2632. doi: 10.1002/adfm.201002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander JK, Fuss B, Colello RJ. Neuron Glia Biology. 2006;2:93–103. doi: 10.1017/S1740925X0600010X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon T, Udina E, Verge VMK, de Chaves EIP. Motor Control. 2009;13(4):412–441. doi: 10.1123/mcj.13.4.412. [DOI] [PubMed] [Google Scholar]
- 30.Wood MD, Willits RK. Journal of Neural Engineering. 2009;6(4):8. doi: 10.1088/1741-2560/6/4/046003. [DOI] [PubMed] [Google Scholar]
- 31.Jan E, Kotov NA. Nano Lett. 2007;7(5):1123–1128. doi: 10.1021/nl0620132. [DOI] [PubMed] [Google Scholar]
- 32.Chao TI, Xiang SH, Chen CS, Chin WC, Nelson AJ, Wang CC, Lu J. Biochemical and Biophysical Research Communications. 2009;384(4):426–430. doi: 10.1016/j.bbrc.2009.04.157. [DOI] [PubMed] [Google Scholar]
- 33.Lovat V, Pantarotto D, Lagostena L, Cacciari B, Grandolfo M, Righi M, Spalluto G, Prato M, Ballerini L. Nano Letters. 2005;5(6):1107–1110. doi: 10.1021/nl050637m. [DOI] [PubMed] [Google Scholar]
- 34.Cellot G, Cilia E, Cipollone S, Rancic V, Sucapane A, Giordani S, Gambazzi L, Markram H, Grandolfo M, Scaini D, Gelain F, Casalis L, Prato M, Giugliano M, Ballerini L. Nature Nanotechnology. 2009;4(2):126–133. doi: 10.1038/nnano.2008.374. [DOI] [PubMed] [Google Scholar]
- 35.Liao HH, Qi RL, Shen MW, Cao XY, Guo R, Zhang YZ, Shi XY. Colloids and Surfaces B-Biointerfaces. 2011;84(2):528–535. doi: 10.1016/j.colsurfb.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Yeo LY, Friend JR. Journal of Experimental Nanoscience. 2006;1(2):177–209. [Google Scholar]
- 37.Balani K, Anderson R, Laha T, Andara M, Tercero J, Crumpler E, Agarwal A. Biomaterials. 2007;28(4):618–624. doi: 10.1016/j.biomaterials.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Gheith MK, Pappas TC, Liopo AV, Sinani VA, Shim BS, Motamedi M, Wicksted JR, Kotov NA. Advanced Materials. 2006;18(22):2975–+. [Google Scholar]
- 39.Uchida T, Yagi A, Oda Y, Nakada Y, Goto S. Chem. Pharm. Bull. 1996;44(1):235–236. doi: 10.1248/cpb.44.235. [DOI] [PubMed] [Google Scholar]
- 40.Park SY, Choi DS, Jin HJ, Park J, Byun KE, Lee KB, Hong S. Acs Nano. 2011;5(6):4704–4711. doi: 10.1021/nn2006128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleis J, Hyldgaard P, Schroder E. Computational Materials Science. 2005;33(1-3):192–199. [Google Scholar]
- 42.Neimark AV, Ruetsch S, Kornev KG, Ravikovitch PI. Nano Lett. 2003;3(3):419–423. [Google Scholar]
- 43.Dresselhaus MS, Dresselhaus G, Saito R, Jorio A. Physics Reports-Review Section of Physics Letters. 2005;409(2):47–99. [Google Scholar]
- 44.HajjHassan M, Chodavarapu V, Musallam S. Sensors. 2008;8(10):6704–6726. doi: 10.3390/s8106704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen EJ, Zhang L, Mattison NT, O'Carroll DM, Whelton AJ, Uddin N, Tinh N, Huang Q, Henry TB, Holbrook RD, Chen KL. Environmental Science & Technology. 2011;45(23):9837–9856. doi: 10.1021/es201579y. [DOI] [PubMed] [Google Scholar]
- 46.Xiong Y, Zeng YS, Zeng CG, Du BL, He LM, Quan DP, Zhang W, Wang JM, Wu JL, Li Y, Li J. Biomaterials. 2009;30(22):3711–3722. doi: 10.1016/j.biomaterials.2009.03.046. [DOI] [PubMed] [Google Scholar]
- 47.Zhang N, Kang TG, Xia Y, Wen QP, Zhang XD, Li HY, Hu Y, Hao HG, Zhao D, Sun D, Yan YP, Zhang GX, Yang JX. European Journal of Pharmacology. 2012;697(1-3):32–39. doi: 10.1016/j.ejphar.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 48.Shimada H, Okada Y, Ibata K, Ebise H, Ota S, Tomioka I, Nomura T, Maeda T, Kohda K, Yuzaki M, Sasaki E, Nakamura M, Okano H. PLoS One. 2012;7(11):e49469. doi: 10.1371/journal.pone.0049469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazzatenta A, Giugliano M, Campidelli S, Gambazzi L, Businaro L, Markram H, Prato M, Ballerini L. Journal of Neuroscience. 2007;27(26):6931–6936. doi: 10.1523/JNEUROSCI.1051-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fioretta ES, Fledderus JO, Burakowska-Meise EA, Baaijens FPT, Verhaar MC, Bouten CVC. Macromolecular Bioscience. 2012;12(5):577–590. doi: 10.1002/mabi.201100315. [DOI] [PubMed] [Google Scholar]
- 51.Quigley AF, Razal JM, Kita M, Jalili R, Gelmi A, Penington A, Ovalle-Robles R, Baughman RH, Clark GM, Wallace GG, Kapsa RMI. Advanced Healthcare Materials. 2012;1(6):801–808. doi: 10.1002/adhm.201200102. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Q, Zhang Q, Zhao XQ, Duan YYW, Lu Y, Li CY, Li T. Brain Research. 2010;1311:148–157. doi: 10.1016/j.brainres.2009.11.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.