Abstract
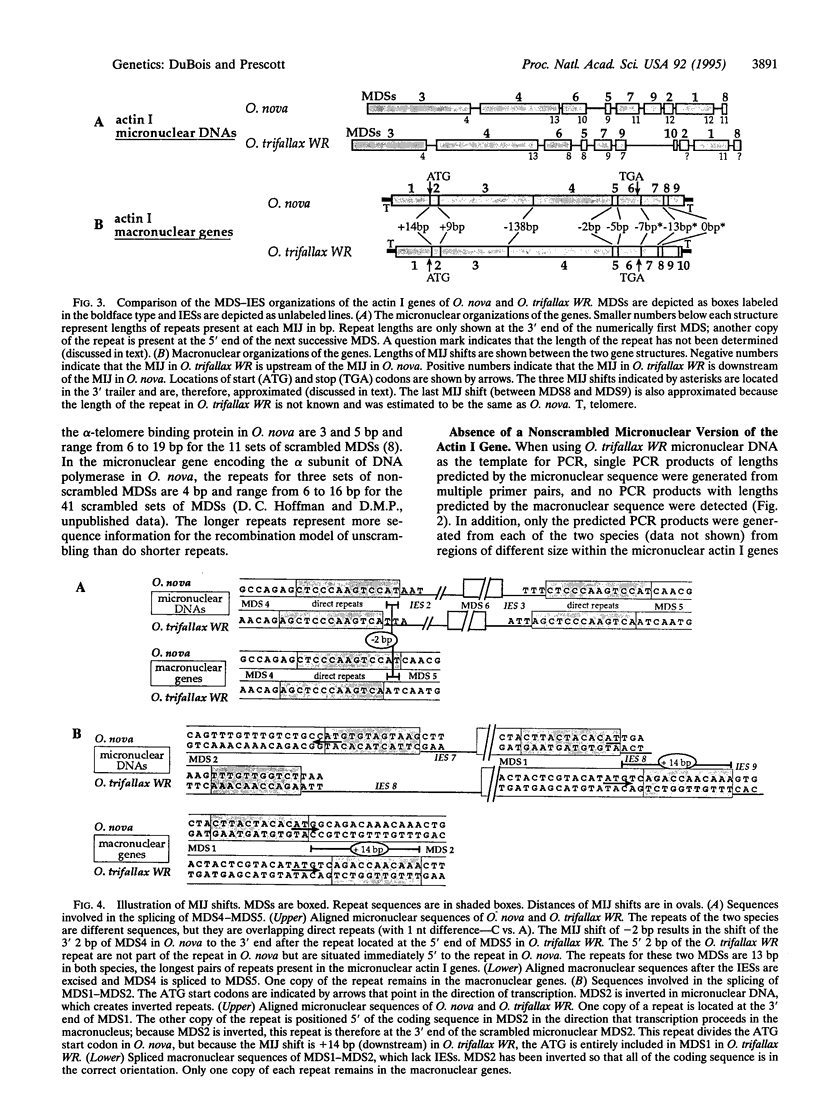
The DNA in a germ-line nucleus (a micronucleus) undergoes extensive processing when it develops into a somatic nucleus (a macronucleus) after cell mating in hypotrichous ciliates. Processing includes destruction of a large amount of spacer DNA between genes and excision of gene-sized molecules from chromosomes. Before processing, micronuclear genes are interrupted by numerous noncoding segments called internal eliminated sequences (IESs). The IESs are excised and destroyed, and the retained macro-nuclear-destined sequences (MDSs) are spliced. MDSs in some micronuclear genes are not in proper order and must be reordered during processing to create functional gene-sized molecules for the macronucleus. Here we report that the micronuclear actin I gene in Oxytricha trifallax WR consists of 10 MDSs and 9 IESs compared to the previously reported 9 MDSs and 8 IESs in the micronuclear actin I gene of Oxytricha nova. The MDSs in the actin I gene are scrambled in a similar pattern in the two species, but the positions of MDS-IES junctions are shifted by up to 14 bp for scrambled and 138 bp for the nonscrambled MDSs. The shifts in MDS-IES junctions create differences in the repeat sequences that are believed to guide MDS splicing. Also, the sizes and sequences of IESs in the micronuclear actin I genes are different in the two Oxytricha species. These observations give insight about the possible origins of IES insertion and MDS scrambling in evolution and show the extraordinary malleability of the germ-line DNA in hypotrichs.
Full text
PDF



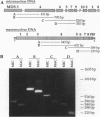
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boswell R. E., Klobutcher L. A., Prescott D. M. Inverted terminal repeats are added to genes during macronuclear development in Oxytricha nova. Proc Natl Acad Sci U S A. 1982 May;79(10):3255–3259. doi: 10.1073/pnas.79.10.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greslin A. F., Prescott D. M., Oka Y., Loukin S. H., Chappell J. C. Reordering of nine exons is necessary to form a functional actin gene in Oxytricha nova. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6264–6268. doi: 10.1073/pnas.86.16.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick G., Cartinhour S. W., Williams K. R., Kotter K. P. Multiple sequence versions of the Oxytricha fallax 81-MAC alternate processing family. J Protozool. 1987 Nov;34(4):429–434. doi: 10.1111/j.1550-7408.1987.tb03207.x. [DOI] [PubMed] [Google Scholar]
- Jahn C. L., Krikau M. F., Shyman S. Developmentally coordinated en masse excision of a highly repetitive element in E. crassus. Cell. 1989 Dec 22;59(6):1009–1018. doi: 10.1016/0092-8674(89)90757-5. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Jahn C. L. Developmentally controlled genomic rearrangements in ciliated protozoa. Curr Opin Genet Dev. 1991 Oct;1(3):397–403. doi: 10.1016/s0959-437x(05)80306-5. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Jahn C. L., Prescott D. M. Internal sequences are eliminated from genes during macronuclear development in the ciliated protozoan Oxytricha nova. Cell. 1984 Apr;36(4):1045–1055. doi: 10.1016/0092-8674(84)90054-0. [DOI] [PubMed] [Google Scholar]
- Mitcham J. L., Lynn A. J., Prescott D. M. Analysis of a scrambled gene: the gene encoding alpha-telomere-binding protein in Oxytricha nova. Genes Dev. 1992 May;6(5):788–800. doi: 10.1101/gad.6.5.788. [DOI] [PubMed] [Google Scholar]
- Mitcham J. L., Prescott D. M., Miller M. K. The micronuclear gene encoding beta-telomere binding protein in Oxytricha nova. J Eukaryot Microbiol. 1994 Sep-Oct;41(5):478–480. doi: 10.1111/j.1550-7408.1994.tb06045.x. [DOI] [PubMed] [Google Scholar]
- Prescott D. M., Greslin A. F. Scrambled actin I gene in the micronucleus of Oxytricha nova. Dev Genet. 1992;13(1):66–74. doi: 10.1002/dvg.1020130111. [DOI] [PubMed] [Google Scholar]
- Prescott D. M. The DNA of ciliated protozoa. Microbiol Rev. 1994 Jun;58(2):233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas-Aparicio R. M., Sparkowski J. J., Proulx A. E., Mitchell J. D., Klobutcher L. A. Nucleic acid splicing events occur frequently during macronuclear development in the protozoan Oxytricha nova and involve the elimination of unique DNA. Genes Dev. 1987 Jun;1(4):323–336. doi: 10.1101/gad.1.4.323. [DOI] [PubMed] [Google Scholar]
- Roth M., Prescott D. M. DNA intermediates and telomere addition during genome reorganization in Euplotes crassus. Cell. 1985 Jun;41(2):411–417. doi: 10.1016/s0092-8674(85)80014-3. [DOI] [PubMed] [Google Scholar]
- Tausta S. L., Klobutcher L. A. Detection of circular forms of eliminated DNA during macronuclear development in E. crassus. Cell. 1989 Dec 22;59(6):1019–1026. doi: 10.1016/0092-8674(89)90758-7. [DOI] [PubMed] [Google Scholar]
- Weber K., Kabsch W. Intron positions in actin genes seem unrelated to the secondary structure of the protein. EMBO J. 1994 Mar 15;13(6):1280–1286. doi: 10.1002/j.1460-2075.1994.tb06380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]