Abstract
MicroRNAs (miRNAs) are a large family of post-transcriptional regulators, which are 21-24 nt in length and play a role in a wide variety of biological processes in eukaryotes. The past few years have seen rapid progress in our understanding of miRNA biogenesis and the mechanism of action, which commonly entails a combination of target degradation and translational repression. The target degradation mediated by Argonaute-catalyzed endonucleolytic cleavage exerts a significant repressive effect on target mRNA expression, particularly during rapid developmental transitions. This review outlines the current understanding of the mechanistic aspects of this important process and discusses several different experimental approaches to identify miRNA cleavage targets. [BMB Reports 2014; 47(8): 417-423]
Keywords: Argonaute protein, MicroRNA, MicroRNA-directed cleavage
INTRODUCTION
MicroRNAs (miRNAs) are 21-24 nt long, endogenous non-coding RNAs of rapidly emerging significance of gene regulation in various cellular processes. miRNA biogenesis begins with the transcription of hairpin-structured precursors known as primary miRNAs (pri-miRNAs) by RNA polymerase II (1). The transcript is recognized by the RNase III-type enzyme, Drosha (2) and Dicer in animals (3) or Dicer-Like 1 in plants (4) for a sequential two-step cleavage, converting pri-miRNAs into precursor miRNAs (pre-miRNAs) and finally resulting in the production of the ∼21-24-nt miRNAs. In a miRNA duplex, one strand (the miRNA star strand) is discarded by unwinding or cleavage (5), and the other strand (the miRNA strand) is retained in the RNA-induced silencing complex (RISC) for target recognition, which is governed by nucleotide complementarity between the miRNA and the target mRNA.
Most miRNAs characterized in animals imperfectly base-pair with sequences in the 3'-untranslated region (3'-UTR), with most of the pairing specificity provided by the miRNA 5' proximal “seed” region (positions 2-8). This partial complementarity often facilitates translational repression and/or target degradation via deadenylation of the message (6-8). In plants, however, many miRNAs guide the RISCs to target mRNAs containing perfect or near-perfect complementary sites to direct the cleavage of mRNA through Argonaute (AGO) endonuclease activity, exerting a strong repressive effect on target mRNA expression. Until recently, this type of regulation was restricted to plant miRNAs; however, new findings with transcriptome-wide analyses have indicated that plant-like mechanisms may also operate in animals. Motivated by these recent findings, this review focuses primarily on the present understanding of miRNA-directed cleavage of target mRNAs.
STRUCTURAL FEATURES OF ARGONAUTE PROTEINS
A hallmark of miRNA-directed cleavage pathway is their dependence on Argonaute proteins that have specialized small RNA binding modules and are highly conserved among species. The Argonaute protein was first discovered by screening for mutations that pleiotropically affected general plant architecture in Arabidopsis (9). Many organisms encode a set of Argonaute proteins, ranging from one in S. pombe, 10 in Arabidopsis, and up to 27 in C. elegans (10). The Argonaute protein is defined by the presence of two characteristic domains, PAZ and PIWI (11) and the family can be subdivided into two classes, based on their sequence homology: those that resemble Arabidopsis AGO1 (related to siRNAs and miRNAs) and those that resemble Drosophila PIWI (mainly associates with piRNAs).
Although substantial progress has been made in studying the biochemical functions of Argonaute proteins, many questions regarding the details of the mechanism remain unanswered. A breakthrough was first achieved by the crystal and NMR structures of the PAZ domain of Drosophila AGO1 (12) and AGO2 (13, 14), which revealed that the PAZ domain contains an OB (oligonucleotide / oligosaccharide binding) fold, a typical single-stranded nucleic acid binding motif. Further studies have shown that the initial interaction between the 2 nt 3'-overhang of the miRNA strands and the PAZ domain is essential for efficient target silencing (15). A complete crystal structure of a prokaryotic Argonaute protein from the archaeal species Pyrococcus furiosus, first shed light on the overall structural insight for the remaining domains and possible molecular mechanisms (16). The study found that the Nterminal, MID, and PIWI domains formed a crescent-shaped base, with the PAZ domain held above the base by a “stalk”-like region (16). A more recent structural study in Thermus thermophiles Argonaute in complex with a 5'-phosphorylated DNA guide strand enabled the identification of a nucleotide binding channel and a pivot-like conformational change during complex formation (17). The organization indicates a bilobal architecture, with the Nterminal and PAZ domains forming one lobe and the MID and PIWI domains together making up the other (18). Recent structural studies extended to a eukaryotic Argonaute MID domain have showed its role in mediating the interaction with the phosphorylated 5'-end of the guide strand and provided structural evidence for a nucleotide-specific interaction that prefers U or A at the 5'-end of miRNAs in the MID domain (19, 20).
ARGONAUTE AS A SLICER
The PIWI domain, located across the primary groove from the PAZ domain, has a tertiary structure belonging to the RNase H family of enzymes, originally described as being responsible for catalyzing the RNA cleavage of the RNA/DNA hybrids, using a conserved Asp-Glu-Asp-Asp (DEDD) motif for divalent metal ion binding (21, 22). Early biochemical work identified the catalytic triad DD(D/H) of Pyrococcus furiosus Argonaute (PfAGO) (23), which appears to differ from the catalytic tetrad (DEDD) of bacterial RNase H enzymes. A solution to this conundrum came from comparative analyses of the structures of Kluyveromyces polysporus Argonaute (KpAGO) and the NcQDE-2 MID-PIWI lobe (20), which found a significant difference in loop L2 (24). This led Nakanishi et al. to examine whether a conserved glutamate at the tip of this loop was likely to be the fourth catalytic residue of Argonaute.
The release of the 3'-end of the guide strand from the PAZ domain allows passenger-strand unwinding and facilitates the formation of a catalytically competent Argonaute (15). During this process, the loops L1 and L2 undergo a post-rearrangement that refold to form a ‘plugged-in’ conformation, which inserts the invariant glutamate finger into the catalytic pocket that helps to coordinate an active-site metal ion (24). Further mutational analyses suggest that this glutamate indeed constitutes the second residue of the universally conserved RNase H-like DEDD catalytic tetrad that completes the active site of Argonaute (24).
Despite the high sequence conservation of the four human Argonaute proteins (AGOs1-4), a slicing mechanism is only inherent to AGO2 even though AGO3 also has a complete DED(D/H) motif, raising the question as to whether additional determinants other than the presence of the catalytic triad are required for slicer activity. Recent studies provided a vital clue in this regard. They exploited DNA shuffling technology to generate chimeric AGO protein libraries and discovered that two N-terminal motifs are key for the slicing activity in concert with the PIWI domain (25). Interestingly, by swapping the N-terminal motifs and PIWI domains of AGO2 into AGO1, the chimera became an active slicer with activity comparable to wild-type AGO2 (26). Another study found that mutations in the PIWI-domain of AGO1 might misarrange the catalytic triad (27). Recent improvements in these findings help in understanding the additional structural elements that make Argonaute protein an active endonucleolytic enzyme, and solidified the fact that slicing not only requires the catalytic residues but also involves an exquisite interplay between the catalytic residues and more distant regions of the protein.
EXPERIMENTAL VALIDATION OF miRNA-DIRECTED TARGET CLEAVAGES
Understanding the biological function of miRNAs first required identification and characterization of their target mRNAs by a bioinformatics approach, incorporating as many factors as possible that could influence the miRNA and target interaction. In contrast to animal miRNAs, the extensive complementarity between plant miRNAs and their targets allows capturing predicted targets with relatively high confidence, without too many false positives (28). This approach has been in use since the first prediction algorithms, developed in the Bartel laboratory (29), became available and several refinements have been made to improve overall confidence of predictions, such as position-dependent penalty scores that add strong penalties for base mismatches near the 5'-end or the central region of miRNAs; these areas are critical for functional interactions (30). Once miRNA targets are predicted, experimental validation of miRNA-target interaction usually follows. The rest of this review will discuss several experimental techniques for miRNA-directed cleavage target identification.
The Carrington laboratory pioneered this field through an experimental approach for the validation of putative cleavage targets (31). A modified RNA-ligase-mediated Rapid Amplification of cDNA Ends (5'-RLM-RACE) experiment, the most widely used method to date, has enabled researchers to map the 5'-ends of target mRNAs within the expected cleavage site by taking advantage of the characteristic of Argonaute-mediated cleavage, which leaves ligation-competent 3'-cleavage fragments ending with 5'-monophosphates (Fig. 1A). The plant miRNAs are generally known to induce a site-specific cleavage within the target RNA strand between 10th and 11th nucleotides, relative to the 5'-end of the miRNAs, which is a diagnostic indication for the cleavage.
Fig. 1. Experimental approaches for microRNA-directed cleavage target identification. (A) 5'-RLM-RACE. In this modified protocol, particular steps (dephosphorylation and cap-removal reactions) from the original 5'-RACE protocol are omitted. A 5'-RNA adaptor is directly ligated to the 3'-end of miRNA-directed cleavage products that contain ligation-competent 5'-monophosphate. The ligated RNAs are reverse-transcribed, PCR-amplified, cloned, and sequenced, allowing determination of their mRNA cleavage sites. (B) Northern blot analysis. Shown are typical molecular phenotypes of transient or stable overexpression of miRNAs in plants, which often lead to reduced accumulation of the full-length target RNA and increased accumulation of cleavage product [miRNA (+); left lane on the blot]. miRNA-deficient mutant, such as dcl-1 (right lane on the blot), served as a negative control. (C) qRT-PCR. Shown is a typical qRT-PCR output of relative target mRNA expression in wild-type and miRNA mutants [miRNA (-): knockout / miRNA (+): overexpression]. (D) Reporter assay. Reporter constructs containing WT or a mutant 3'-UTR were cotransfected with a miRNA of interest [miRNA (+)] or a non-cognate miRNA [miRNA (-)]. To support the direct interaction, mutation of the miRNA-binding site is often introduced in the reporter construct, which should eliminate miRNA-target interaction. The level of repression for cleavage targets is usually expected to be higher than that are observed for seed-matched targets. (E) In vitro cleavage assay. The DNA fragment encoding a target RNA is in vitro transcribed under the control of a T7 promoter. This target RNA is cap-radiolabeled and incubated with lysates containing an endogenous miRNA or an exogenously assembled miRNA of interest. When such a target RNA substrate is cleaved, its 5'-end cleavage product remains detectable by autoradiography. A time-course in vitro RNAi reaction is performed and analyzed in a denaturing gel, with the wild-type and target site-mutated substrates as a negative control. (F) Degradome sequencing. Shown is the schematic description of degradome library construction and data analysis. (1) Poly(A)-tailed RNAs are purified from total RNA with oligo-dT magnetic beads. (2) A 5'-adaptor is ligated to the 5'-end of the cleaved molecules with 5'-monophosphate. (3) Reverse transcription and subsequent alkaline hydrolysis of the RNA templates are performed to construct the first-strand cDNA templates. Primer extension is carried out to generate the second-strand cDNAs. (4) Restriction enzyme MmeI cleaves ∼20-nt within the double-stranded cDNAs, which are ligated with a dsDNA 3'-adaptor. (5) The resulting products are PCR-amplified and submitted for high-throughput sequencing. (6) The pre-processed 20-nt sequences are matched to the transcriptome. The miRNAs, extracted from small RNA sequencing data from the same tissue or samples for degradome library construction, are aligned to find miRNA-target transcript base pairing.
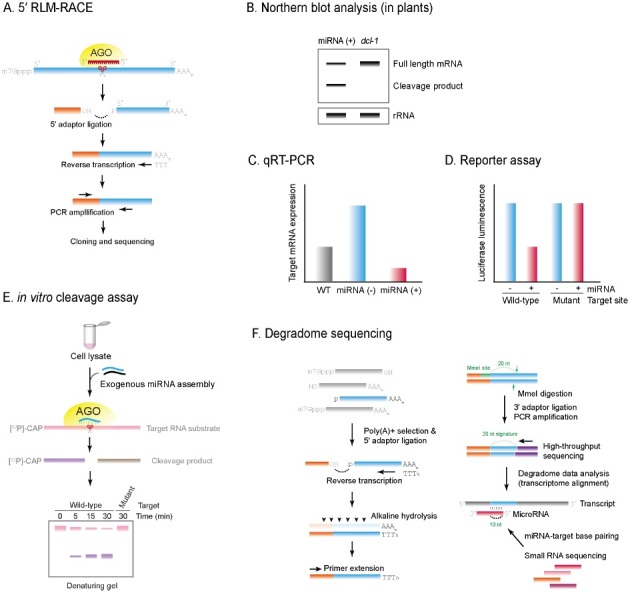
Well-established techniques such as Northern blot analysis (Fig. 1B) and qRT-PCR (Fig. 1C), can be used and are especially useful when miRNA-defective mutants are available. Of these techniques, Northern blot analysis most closely reflects a steady state level of mRNA following miRNA action − as two populations, of cleaved and uncleaved transcripts, are detected so the extent of miRNA-directed cleavage can be estimated (Fig. 1B). The accumulation of fast-migrating 3'-cleavage fragments indicates a high degree of cleavage. On some occasions, a substantial level of full-length target mRNAs is detectable, suggesting that only a minor fraction of targets are under the control of miRNA-directed cleavage. miRNA-mediated inhibition at the protein level is generally evaluated by a reporter assay (or Western blot analysis) with miRNA target sequence inserted into the 3'-UTR after the luciferase ORF, but this type of method can also be used to validate miRNA-directed cleavage because the downregulation of mRNA levels is often accompanied by a reduction in protein levels (Fig. 1D).
Biochemical approaches using cell lysates or purified Argonaute proteins have been used widely for the faithful and effective validation of miRNA-directed cleavage in vitro (Fig. 1E). This cell-free system was first developed by Tuschl and colleagues (32). The lysates prepared from Drosophila melanogaster embryos effectively recapitulate in vitro RNA interference triggered by miRNAs and allow the identification of 5'-cleavage products because a cap-radiolabeled target RNA without a poly (A) tail is stable in this lysates for several hours. Other cell lysates, including HeLa cell cytosol extract, wheat germ extract, and HEK 293T cells stably or transiently overexpressing hAGO2, have been used successfully for in vitro cleavage assays (Please see figure legends for detailed information about these experimental techniques).
However, these methods are specific for a single miRNA at a time and are labor-intensive. Furthermore, a certain experiment, such as 5'-RLM-RACE, is a qualitative but not a quantitative method, which does not reflect the extent of miRNA-directed cleavage in target gene regulation. Inspired by these appreciations, the recent advent of high-throughput sequencing of RNA cleaved fragments (33-35), since named “degradome sequencing” (also referred to as “parallel analysis of RNA ends,” PARE) opens up a new avenue for a large-scale experimental validation of miRNA-directed cleavage (Fig. 1F). This has been successfully applied for global target identification and validation in plant species, including Arabidopsis (33, 34), rice (36), tomato (37), grapevine (38), and a few others (39).
TRANSCRIPTOME-WIDE IDENTIFICATION OF CLEAVAGE TARGETS IN ANIMALS
miRNA-directed target RNA cleavage has been recognized as a plant-specific mechanism, but the first observation in animals, where miR-196-directed cleavage of HOXB8 transcript was detected (40), raised the possibility that some fraction of mammalian miRNA targets might also be regulated by such a cleavage mechanism. Motivated by these recent advances, degradome sequencing was also employed to directly detect endogenous miRNA cleavage targets in mammalian cells (41, 42) and in C. elegans (43). The results from degradome sequencing in mammalian cells suggested that only extensively paired sites could impart robust cleavage in cells (41). Park et al. found that miR-249 directed the cleavage of the ZK637.6 transcript with extensive and evolutionarily conserved complementarity in a nematode. Such miRNA-mediated cleavage, however, did not appear to be a prevalent mechanism in C. elegans, which would be expected to be rarer than in mammals, because cleavage signals in degradome data were not significantly associated with even near-perfect sites of miRNAs, suggesting that extensive pairing did not guarantee the contribution of miRNA-mediated cleavage in C. elegans. Nevertheless, the first reported example in C. elegans raises the possibility that there may be other miRNA-directed cleavages in worms faced with environmental or physical stresses, such as overcrowding, limited food, or elevated temperatures.
Recently, an interesting observation emerged from a large-scale investigation of the biological activity of miRNAs in the sea anemone Nematostella vectensis, one of the non-bilaterian animals (44). Unlike bilaterian animals, Nematostella miRNAs frequently mediate the cleavage of their target mRNAs via near-perfect complementarity (44). Given that (i) non-bilaterian animals have been shown to carry a homolog of HYL1, once considered to be a plant-specific miRNA biogenesis factor (45), (ii) a large proportion of their miRNAs are methylated, presumably by HEN1 (44), and (iii) some of their miRNAs are highly conserved with plant miRNAs (44), the miRNAs in plants and animals may not have evolved independently and those miRNAs in the common ancestor of the two kingdoms might also share an ancestral mechanism of miRNA-directed cleavage.
Most examples of miRNA-directed cleavage observed so far in bilaterian animals are generally found to have a significantly low number of degradome sequencing reads. The direct detection of 5'-monophosphorylated cleavage fragments, as a result of Argonaute-mediated cleavage, is theoretically challenging if they are degraded or recapped. This seems, however, unlikely to happen since previous studies revealed that the knockdown of xrn-1 (or xrn-4 in plants), a primary 5'-3' exonuclease, did not markedly increase the detection of Argonaute-mediated cleavage (34, 41). Likewise, a genome-wide analysis of 5'-capped RNAs from various mouse tissues suggested that the recapping of cleaved fragment is unlikely to occur (46), indicating that the low level of sequencing reads is probably not because of their rapid degradation or recapping. Thus, the infrequency of these events may instead suggest that miRNA-directed cleavage might have biological significance for a very limited subset of genes, raising the essential question as to why target mRNAs in bilaterian animals might have been under selective pressure to escape miRNA-directed cleavage. It may be that target cleavage, which is an irreversible form of repression, is deleterious to essential mRNAs and less efficient for gene regulation, which should be highly flexible and respond dynamically to internal and external stimuli.
CONCLUSIONS AND PROSPECT
Research in the RNA silencing field has profited greatly from recent developments in high-throughput sequencing technologies that allow an unprecedented level of small RNA profiling, which seems to have reached a plateau in our capacity to functionally or mechanistically characterize these molecules. Degradome sequencing has certainly contributed to overcome these limitations by providing a global survey of uncapped RNA molecules and parallel validation of miRNA targets, which has been especially successful in plants where miRNAs predominantly operate through target cleavage pathways. Although this is a powerful strategy, it overlooked an important point that translational inhibition is more widespread than was previously appreciated in plants (47, 48). Some controversy has emerged due to several apparent discrepancies where the abundance of target mRNA was unaffected by miRNA overexpression, whereas the protein was significantly down-regulated (49-51). More direct evidence came from the results of a forward genetic screen, showing that translational inhibition can be genetically uncoupled from target cleavage (47). It was demonstrated that mutations in several genes, including microtubule-severing KATANIN and P-body component VARICOSE proteins, impaired miRNA-mediated repression at the protein level, but not at the transcript level (47).
Our understanding of miRNA biology is still very limited by our poor appreciation of their cell-type specific and spatio-temporal regulation. Many miRNAs exhibit discrete expression patterns and their regulation depends largely on the availability of particular AGO proteins and distinct target mRNAs within a given cell type. Although many exhaustive studies have identified more than a million small RNAs, the biological function of only a few miRNAs have been identified. The combinatorial analysis of small RNA sequencing and accurate expression data for target mRNA/proteins and various miRNA processing enzymes, could ultimately provide us with a global snapshot of miRNA regulation in both plants and animals.
Acknowledgments
We thank lab members for comments on the manuscript. We also thank Dr. Byeong-ha Lee for critical reading of the manuscript. This work was supported by the National Research Foundation of Korea (NRF) grant, funded by the Korea government (MEST; No. 2012R1A2A2A01045528) and the Next-Generation BioGreen 21 Program (No. PJ00820603 and No. PJ00801101), Rural Development Administration, Republic of Korea.
References
- 1.Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., Kim V. N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. (2004);23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V. N. The nuclear RNase III Drosha initiates microRNA processing. Nature. (2003);425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. (2010);11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. (2009);136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 5.Shin C. Cleavage of the star strand facilitates assembly of some microRNAs into Ago2-containing silencing complexes in mammals. Mol. Cells. (2008);26:308–313. [PubMed] [Google Scholar]
- 6.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. (2011);12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 7.Subtelny A. O., Eichhorn S. W., Chen G. R., Sive H., Bartel D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. (2014);508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazzini A. A., Lee M. T., Giraldez A. J. Ribosome Profiling Shows That miR-430 Reduces Translation Before Causing mRNA Decay in Zebrafish. Science. (2012);336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. (1998);17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ender C., Meister G. Argonaute proteins at a glance. J. Cell Sci. (2010);123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- 11.Cerutti L., Mian N., Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends. Biochem. Sci. (2000);25:481–482. doi: 10.1016/S0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 12.Yan K. S., Yan S., Farooq A., Han A., Zeng L., Zhou M. M. Structure and conserved RNA binding of the PAZ domain. Nature. (2003);426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 13.Lingel A., Simon B., Izaurralde E., Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. (2003);426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 14.Song J. J., Liu J., Tolia N. H., Schneiderman J., Smith S. K., Martienssen R. A., Hannon G. J., Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. (2003);10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Juranek S., Li H., Sheng G., Wardle G. S., Tuschl T., Patel D. J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. (2009);461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song J. J., Smith S. K., Hannon G. J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. (2004);305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Sheng G., Juranek S., Tuschl T., Patel D. J. Structure of the guide-strand-containing argonaute silencing complex. Nature. (2008);456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M., Doudna J. A. A three-dimensional view of the molecular machinery of RNA interference. Nature. (2009);457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 19.Frank F., Sonenberg N., Nagar B. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. (2010);465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 20.Boland A., Huntzinger E., Schmidt S., Izaurralde E., Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl. Acad. Sci. U. S. A. (2011);108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowotny M., Gaidamakov S. A., Crouch R. J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. (2005);121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 22.Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. (1990);347:306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- 23.Rivas F. V., Tolia N. H., Song J. J., Aragon J. P., Liu J., Hannon G. J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. (2005);12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi K., Weinberg D. E., Bartel D. P., Patel D. J. Structure of yeast Argonaute with guide RNA. Nature. (2012);486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schurmann N., Trabuco L. G., Bender C., Russell R. B., Grimm D. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol. (2013);20:818–826. doi: 10.1038/nsmb.2607. [DOI] [PubMed] [Google Scholar]
- 26.Faehnle C. R., Elkayam E., Haase A. D., Hannon G. J., Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell Reports. (2013);3:1901–1909. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauptmann J., Dueck A., Harlander S., Pfaff J., Merkl R., Meister G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. (2013);20:814–817. doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- 28.Jones-Rhoades M. W., Bartel D. P., Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant. Biol. (2006);57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 29.Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., Bartel D. P. Prediction of plant microRNA targets. Cell. (2002);110:513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 30.Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M., Weigel D. Specific effects of MicroRNAs on the plant transcriptome. Dev. Cell. (2005);8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Llave C., Xie Z., Kasschau K. D., Carrington J. C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. (2002);297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 32.Tuschl T., Zamore P. D., Lehmann R., Bartel D. P., Sharp P. A. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. (1999);13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Addo-Quaye C., Eshoo T. W., Bartel D. P., Axtell M. J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. (2008);18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.German M. A., Pillay M., Jeong D. H., Hetawal A., Luo S., Janardhanan P., Kannan V., Rymarquis L. A., Nobuta K., German R., De Paoli E., Lu C., Schroth G., Meyers B. C., Green P. J. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. (2008);26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 35.Gregory B. D., O R. C., Lister R., Urich M. A., Tonti-Filippini J., Chen H., Millar A. H., Ecker J. R. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. (2008);14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Li Y. F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., Axtell M. J., Zhang W. X., Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. (2010);62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 37.Karlova R., van Haarst J. C., Maliepaard C., van de Geest H., Bovy A. G., Lammers M., Angenent G. C., de Maagd R. A. Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J. Exp. Bot. (2013);64:1863–1878. doi: 10.1093/jxb/ert049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pantaleo V., Szittya G., Moxon S., Miozzi L., Moulton V., Dalmay T., Burgyan J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. (2010);62:960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 39.Jeong D. H., Schmidt S. A., Rymarquis L. A., Park S., Ganssmann M., German M. A., Accerbi M., Zhai J., Fahlgren N., Fox S. E., Garvin D. F., Mockler T. C., Carrington J. C., Meyers B. C., Green P. J. Parallel analysis of RNA ends enhances global investigation of microRNAs and target RNAs of Brachypodium distachyon. Genome Biol. (2013);14:R145. doi: 10.1186/gb-2013-14-12-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yekta S., Shih I. H., Bartel D. P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. (2004);304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 41.Shin C., Nam J. W., Farh K. K., Chiang H. R., Shkumatava A., Bartel D. P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. (2010);38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karginov F. V., Cheloufi S., Chong M. M., Stark A., Smith A. D., Hannon G. J. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. (2010);38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J. H., Ahn S., Kim S., Lee J., Nam J. W., Shin C. Degradome sequencing reveals an endogenous microRNA target in C. elegans. FEBS Letters. (2013);587:964–969. doi: 10.1016/j.febslet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 44.Moran Y., Fredman D., Praher D., Li X. Z., Wee L. M., Rentzsch F., Zamore P. D., Technau U., Seitz H. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. (2014);24:651–663. doi: 10.1101/gr.162503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran Y., Praher D., Fredman D., Technau U. The evolution of microRNA pathway protein components in Cnidaria. Mol. Biol. Evol. (2013);30:2541–2552. doi: 10.1093/molbev/mst159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer T. R., Dinger M. E., Bracken C. P., Kolle G., Szubert J. M., Korbie D. J., Askarian-Amiri M. E., Gardiner B. B., Goodall G. J., Grimmond S. M., Mattick J. S. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. (2010);20:1639–1650. doi: 10.1101/gr.112128.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y. Y., Sieburth L., Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. (2008);320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 48.Iwakawa H., Tomari Y. Molecular Insights into microRNA-Mediated Translational Repression in Plants. Mol. Cell. (2013);52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 49.Aukerman M. J., Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. (2003);15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bari R., Datt Pant B., Stitt M., Scheible W. R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. (2006);141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gandikota M., Birkenbihl R. P., Hohmann S., Cardon G. H., Saedler H., Huijser P. The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. (2007);49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]


