Abstract
Osteoblasts are specialized mesenchymal cells that are responsible for bone formation. In this study, we examine the role of GATA4 in osteoblast differentiation. GATA4 was abundantly expressed in preosteoblast cells and gradually down-regulated during osteoblast differentiation. Overexpression of GATA4 in osteoblastic cells inhibited alkaline phosphatase activity and nodule formation in osteogenic conditioned cell culture system. In addition, overexpression of GATA4 attenuated expression of osteogenic marker genes, including Runx2, alkaline phosphatase, bone sialoprotein, and osteocalcin, all of which are important for osteoblast differentiation and function. Overexpression of GATA4 attenuated Runx2 promoter activity, whereas silencing of GATA4 increased Runx2 induction. We found that GATA4 interacted with Dlx5 and subsequently decreased Dlx5 binding activity to Runx2 promoter region. Our data suggest that GATA4 acts as a negative regulator in osteoblast differentiation by downregulation of Runx2. [BMB Reports 2014; 47(8): 463-468]
Keywords: GATA4, Transcription factor, Runx2, Dlx5, Osteoblast differentiation
INTRODUCTION
Homeostasis in bone is controlled by a balance between osteoblasts and osteoclasts, which respectively regulate bone formation and bone resorption. Imbalance between osteoblast and osteoclast activity causes abnormal bone density and typical bone diseases.
Runx2 plays a pivotal role in osteoblast differentiation. Runx2 is responsible for mesenchymal condensation, osteoblast differentiation, chondrocyte hypertrophy, and vascular invasion of developing skeletons from mesenchymal stem cells (1, 2). Human defective bone formation disease, cleidocranial dysplasia, results from Runx2 haploinsufficiency (2, 3). Runx2 is controlled by various transcription factors such as TAZ and Sox9 (4, 5). It induces expression of osteoblast marker genes, including alkaline phosphatase (ALP), collagen type I, osteopontin, bone sialoprotein (Bsp) and osteocalcin (OCN), during osteoblast differentiation (6).
Among the positive regulators of bone formation, Dlx5 is one of the important transcription factors in osteoblast differentiation. Dlx5 encodes a member of a homeobox transcription factor gene family, similar to the Drosophila distal-less (Dll) gene. All Dlx genes might play a role in chondrogenesis and/or osteogenesis (7). Dlx5 is expressed at very early stages of bone development (8) and has been proposed to play a central role in bone development and fracture healing (9). Although Dlx5 could be at the same time a downstream target of Runx2 and an upstream regulator of Runx2 type II (9, 10), Dlx5 specifically regulates Runx2 expression by binding to homeodomain-response elements in the Runx2 PI promoter (10). Overexpressed Dlx5 increases OCN expression, which leads to a fully mineralized matrix in cell culture system (11-13).
GATA4 is a member of six GATA family of zinc finger transcription factor and has been investigated its role in cardiac development and adult cardiac hypertrophy. GATAs have consensus DNA-binding sequence (A/T)GATA(A/G) and regulate various biological processes. GATA1, -2, -3 are expressed in hematopoietic stem cells, whereas GATA4, -5, -6 are expressed in mesoderm- and endoderm-derived tissues (14, 15). GATA4 plays various roles through interactions with regulatory proteins such as p300, RXRα, and SRF (16). In the heart, GATA4 interacts with nuclear factor for activated T cells (NFAT), which has been studied in immune and bone cells (17). However, the role of GATA4 in osteoblast differentiation still remains to be determined.
In this present study, we demonstrate how GATA4 regulates the process of osteoblast differentiation. Our data revealed a novel role of GATA4 in modulating Runx2 in osteoblasts.
RESULTS
Expression of GATA4 was down-regulated during osteoblast differentiation
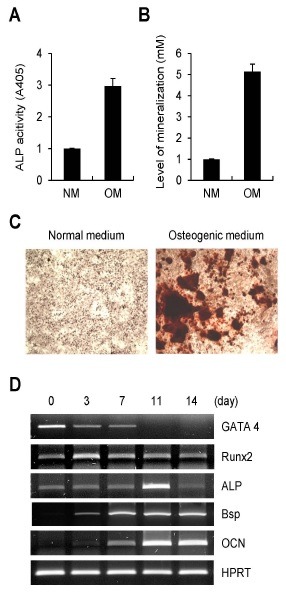
To investigate the role of GATA4 in osteoblasts, we examined the expression pattern of GATA4 during osteoblast differentiation. Consistent with previous findings (18), ALP activity and nodule formation were strongly increased, when primary calvarial cells were cultured in osteogenic media (Fig. 1A-C). In RT-PCR analysis, the expressions of well-known osteogenic maker genes, including Runx2, ALP, Bsp, OCN were strongly induced during osteoblast differentiation. In contrast, GATA4 was abundantly expressed in preosteoblast cells and gradually decreased in time-dependent manner (Fig. 1D), suggesting that GATA4 might play a role in osteoblast differentiation.
Fig. 1. Expressions of GATA4 and osteogenic marker genes during osteoblast differentiation. Primary calvarial osteoblast precursor cells were incubated with normal medium (NM) or osteogenic medium (OM) containing ascorbic acid and β-glycerophosphate. (A) After 7 days of culture, alkaline phosphatase (ALP) activity was measured at 405 nm using alkaline phosphatase yellow (pNPP) liquid substrate system. (B, C) After 14 days of culture, nodule formation was assayed using Alizarin red S. (B) Stained cells were extracted using cetylpyridinium chloride, and mineralization level was quantified by measuring its absorbance at 562 nm. (C) The mineral nodule deposition was visualized by alizarin red S staining. (D) Total RNA was collected at each time point. RT-PCR was performed for GATA4 and osteogenic marker genes, including Runx2, alkaline phosphatase (ALP), bone sialoprotein (Bsp), osteocalcin (OCN), and hypoxanthine-guanine phosphoribosyltransferase (HPRT) for control.
Overexpression of GATA4 down-regulates ALP activity and nodule formation
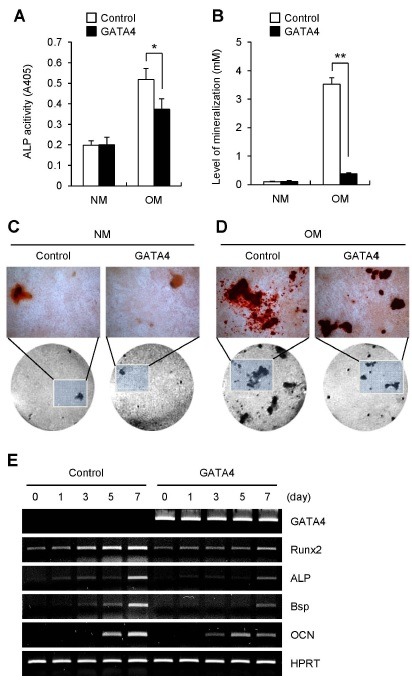
To investigate the effect of GATA4 on osteoblast differentiation, we overexpressed GATA4 in primary preosteoblast cells using a retroviral vector. Transduced cells were cultured in normal medium or osteogenic medium. Exogenous overexpression of GATA4 strongly attenuated induction of ALP activity (Fig. 2A) and bone nodule formation under osteogenic conditions (Fig. 2B-D). Even though GATA4 expression was suppressed during osteoblast differentiation, exogenous GATA4 could inhibit osteoblast differentiation in an osteogenic cell culture model, suggesting that GATA4 is a negative regulator during osteoblast differentiation.
Fig. 2. The effect of GATA4 on osteoblast differentiation. Primary calvarial osteoblasts were transduced with control (pMX-IRES-EGFP) or GATA4 retrovirus. Transduced cells were cultured with normal medium (NM) or osteogenic medium (OM) containing ascorbic acid and β-glycerophosphate. (A) After 7 days of culture, alkaline phosphatase (ALP) activity was measured at 405 nm using alkaline phosphatase yellow (pNPP) liquid substrate system. (B-D) After 21 days of culture, nodule formation was assayed using Alizarin red S. (B) The stained cells were extracted using cetylpyridinium chloride, and mineralization level was quantified by measuring its absorbance at 562 nm. (C, D) The mineral nodule deposition cultured with NM (C) or OM (D) was visualized by alizarin red S staining. Original magnification, top panels, X100; bottom panels, X40. Data are presented as mean ± SD. *P < 0.05, **P < 0.01 versus control. (E) Primary calvarial osteoblasts were transduced with control (pMX-IRES-EGFP) or GATA4 retrovirus. Transduced cells were cultured with osteogenic medium containing ascorbic acid, β-glycerophosphate, and bone morphogenetic protein 2 (BMP-2). Total RNA was collected from each time point. RT-PCR was performed for expression of GATA4, Runx2, ALP, Bsp, OCN and HPRT.
Overexpression of GATA4 down-regulates the expression of osteogenic marker genes during osteoblast differentiation
To understand how GATA4 regulates osteoblast differentiation, we investigated the expression patterns of various osteogenic marker genes. Osteoblasts transduced with control retrovirus were cultured in osteogenic media containing ascorbic acid, β-glycerophosphate, and bone morphogenetic protein 2 (BMP-2). Subsequently, the expression of osteogenic marker genes such as Runx2, ALP, Bsp and OCN were strongly induced. However, overexpression of GATA4 strongly diminished induction of theses osteoblastic marker genes, which are known to be important modulators of osteogenic development (Fig. 2E). This finding indicates that GATA4 is involved in regulating osteoblast differentiation through directly or indirectly regulating the expression of osteoblastogenic marker genes, including Runx2.
GATA4 attenuates Dlx5-mediated Runx2 induction
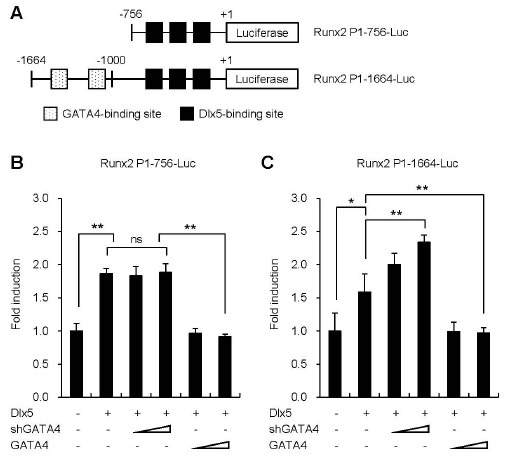
As mentioned above, Dlx5 had been reported to regulate Runx2 expression in osteoblast differentiation (10, 11). Since overexpression of GATA4 down-regulated Runx2 expression in osteoblasts, we tested whether GATA4 regulates Dlx5-mediated Runx2 induction. To investigate the mechanism by which GATA4 regulates Dlx5-induced Runx2 expression, we performed transient transfection assays using Runx2 P1 promoter plasmids: Runx2 P1-756-Luc, which contains only three putative Dlx5-binding sites, and Runx2 P1-1664-Luc, which contains three putative Dlx5-binding sites and two putative GATA4-binding sites (Fig. 3A). Dlx5 significantly enhanced the activities of Runx2 P1 promoters (Fig. 3B, C). Dlx5-mediated Runx2 promoter activity was further enhanced by knockdown of GATA4 using a shGATA4 construct in Runx2 P1-1664-Luc (Fig. 3C), which contains putative GATA4-binding sites. However, Runx2 P1-756 promoter without GATA4-binding elements demonstrated no significant increase in activity from GATA4 knockdown (Fig. 3B). In contrast, GATA4 overexpression abrogated Dlx5 transactivation effect on both Runx2 P1 promoters (Fig. 3B, C). Our data imply that putative GATA4 binding sites exist between -1664 and -756 bp and that GATA4 effectively abrogates Dlx5 effect on Runx2 P1 promoter.
Fig. 3. GATA4 attenuates Dlx5 transactivation on Runx2 promoter. (A) Deletion constructs of the Runx2 P1 promoter: Runx2 P1-756-Luc which contains only three putative Dlx5-binding sites, and Runx2 P1-1664-Luc, which contains three putative Dlx5-binding sites and two putative GATA4-binding sites. (B, C) C2C12 cells were co-transfected with Runx2 P1-756-Luc (B) or Runx2 P1-1664-Luc (C) luciferase reporter together with Dlx5, and shRNA-GATA4 or GATA4 (100 ng or 400 ng) as indicated. Each well was also co-transfected with 20 ng Renilla expression vector to control for transfection efficiency. Luciferase activity was normalized to Renilla activity as expressed by the co-transfected plasmid. Data are presented as mean ± SD of triplicate samples. Results are representative of at least 3 independent sets of similar experiments. *P < 0.05, **P < 0.01 versus control, ns; not significant.
GATA4 attenuates Dlx5 binding ability to Runx2 promoter by interaction with Dlx5
To investigate the inhibitory mechanism of GATA4 on Dlx5 transcriptional activity, we used an immunoprecipitation assay to determine the interaction between GATA4 and Dlx5. Human embryonic kidney (HEK) 293T cells were co-transfected with Flag-GATA4 and HA-Dlx5. This portion of the experiment revealed that GATA4 could directly interact with Dlx5 (Fig. 4A). This finding suggests that GATA4 might inhibit Dlx5-mediated Runx2 induction through interacting with Dlx5.
Fig. 4. GATA4 blocks Dlx5 binding to Runx2 promoter region. (A) HEK 293T cells were transfected with Flag-GATA4 and HA-Dlx5. Lysates were immunoprecipitated (IP) with anti-Flag antibody. Immunoprecipitated samples (upper panel) or whole cell lysates (WCL; lower panel) were subjected to Western blotting for detection of GATA4 (Flag) and Dlx5 (HA). (B) ChIP assay of GATA4 binding to the Runx2 promoter region. C2C12 cells were transfected with pMX-IRES-EGFP (C) or pMX-IRES-GATA4-EGFP (G4) plasmid. After 48 hours of transfection, samples were immunoprecipitated with control IgG or with anti-Dlx5 antibodies and were subjected to PCR amplification with primers specific for the Dlx5 binding sites of Runx2 P1 promoter region.
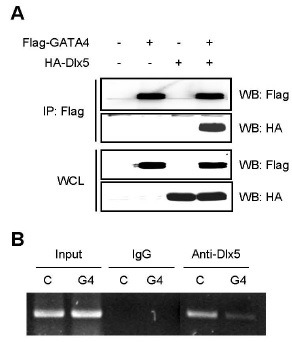
Next, to determine whether GATA4 proteins can modulate the binding of Dlx5 to Dlx-binding sites in Runx2 P1 promoter region, we carried out the chromatin immunoprecipitation (ChIP) assay. GATA4 was transfected in C2C12 cell lines and immunoprecipitated with anti-Dlx5. Consistent with luciferase data, ChIP assay revealed that overexpression of GATA4 strongly attenuated Dlx-5 binding to Runx2 P1 promoter region, compared to the control (Fig. 4B). Collectively, these results suggest that GATA4 decreases Dlx5 binding effect to Dlx-binding sites in the Runx2 P1 promoter region.
DISCUSSION
GATA4 had been found to colocalize with nuclear Runx2 in the trabecular bone near the growth plate and to regulate estrogen receptor-α-mediated osteoblast transcription (19). Morphological expression assay using ALP and nodule formation revealed significant down-regulation in osteogenic induction by GATA4 overexpression. In addition, we showed that GATA4 down-regulated osteogenic marker genes such as Runx2, ALP, Bsp, and OCN. Collectively, the results suggest that GATA4 plays a negative role in osteoblastogenesis by down-regulating Runx2 and its downstream target genes.
In our osteogenic induction model for osteoblast differentiation, all GATA family members were expressed during osteoblast differentiation, even though each expression patterns of GATAs was slightly different (data not shown). Among GATAs, we found that only GATA4 expression was decreased whereas well-known osteogenic marker genes gradually increased, during osteoblast differentiation. Because GATA family members share a highly conserved DNA-binding domain, there is apparent redundancy in DNA-binding properties to target GATA sequences (15). Therefore, we could not exclude the possibility that other GATAs can also modulate osteoblast differentiation. However, in contrast to in vitro redundant property of GATAs, GATAs often show nonredundant roles in vivo. The specificity of GATA action is governed, in part, through protein-protein interactions with other transcriptional partners. Indeed, various factors have been known to cooperate with GATAs to control tissue-specific transcription in the hematopoietic system, the heart, and many other tissues (15). Further studies are needed to elucidate the role of GATAs in bone cells using animal models.
Previously, Lee et al. reported that Dlx5 enhances Runx2 P1 promoter activity through binding Dlx5-reponse elements, which are locate between -756 and -342 bp from the Runx2 transcription start site (10). P1 promoter deletion analysis indicated a strong increase in reporter activity between P1-342 and P1-756. Interestingly, the stimulatory activity of Dlx5 disappeared for longer constructs (Runx2 P1-1664 and Runx2 P1-2782) (10). These findings imply that repressor-binding site(s) might be located between -1664 and -756 bp from the Runx2 transcription start site. We found that putative GATA4 binding sites are located in this region of Runx2 P1 promoter. In order to demonstrate the effect of putative GATA4-reponse elements on Runx2 promoter, we used the 3 different sizes of the Runx2 P1 promoter (P1-2782, P1-1664, P1-756 bp). As a result, down-regulation of GATA4 further enhanced Dlx5-mediated transactivation of Runx2 for longer promoters, P1-2782 and P1-1664 (Fig. 3C and data not shown for Runx2 P1-2782 promoter), whereas this enhancement was not observed for the shorter P1-756 promoter (Fig. 3B). This result suggests that GATA4 might inhibit Dlx5-mediated Runx2 induction through binding to putative GATA-response elements of Runx2 P1 promoter. Although we believe that GATA4 acts as a negative regulator, in part, by Dlx5 sequestration from Dlx5-binding sites in the Runx2 promoter region, we could not rule out the possibility that GATA4 itself might suppress Runx2 expression by binding to GATA-binding sites.
GATA4 is not only essential for maintenance of postnatal cardiac function and protection from stress-induced heart failure but is also responsible for pathological cardiac hypertrophy that is characterized by increased cardiomyocyte size, increased protein synthesis, and altered gene expression, all of which induce hypertrophic agonists such as artrial natriuretic factor (ANF), brain natriuretic peptide (BNP), skeletal α-actin, α-myosin heavy chain (α-MHC), and β-myosin heavy chain (β-MHC) (14,16,20,21). GATA4 has been shown to act as a cardiac-specific transcriptional activator; however, our study demonstrates that GATA4 can act as a repressor in osteoblasts.
In summary, GATA4 might play a negative role in osteoblast differentiation by regulating Dlx5 binding ability to Runx2 promoter. Therefore, the detailed mechanism of GATA4 gene regulation should be further elucidated to provide additional therapeutic approaches to various bone diseases.
MATERIALS AND METHODS
Reagents
Alkaline phosphatase yellow (pNPP) liquid substrate system, Alizarin Red S, ascorbic acid, and β-glycerophosphate were purchased from Sigma-Aldrich (St Louis, MO); recombinant bone morphogenetic protein-2 was acquired from Korea Bone Bank (Seoul, Korea). Antibodies against for Flag and hemagglutinin (HA) were obtained from Sigma-Aldrich (St Louis, MO) and Roche Applied Sciences (Indianapolis, IN), respectively. Anti-Dlx5 was purchased from Santa Cruz Biotechnology, INC (Dallas, TX). Dynabead protein G for immunoprecipitation was obtained from Novex Life technologies (Carlsbad, CA).
Construct
GATA4 was prepared by reverse transcription polymerase chain reaction (RT-PCR) using RNA from C57BL/6 heart. The primer sequences for RT-PCR are as follows: GATA4 sense: 5’-CGGGATCCACCATGTACCAAAGCCTGGCCATGG-3’ and GATA4 antisense: 5’-CCGCTCGAGCGCGGTGAT TATGTCCC CATGAC-3’. The amplified PCR fragments were cloned into the pMX-IRES-EGFP vector.
Retroviral infection
To generate retroviral stocks, retroviral vectors were transfected into the packaging cell line Plat E, using TransIT-2020 (Mirus, Madison, WI). Viral supernatant was collected from cultured media 48 hours after transfection. Osteoblast cells were incubated with viral supernatant for 8 hours in the presence of polybrene (10 μg/ml). After removing the viral supernatant, osteoblast cells were cultured with ascorbic acid (50 μg/ml) and β-glycerophosphate (10 mM) for the indicated times.
Cell culture and assay of ALP and nodule formation
Primary osteoblast precursor cells were isolated from the calvarial bone of newborn mice by enzymatic digestion with α-MEM containing 0.1% collagenase (Invitrogen) and 0.2% dispase II (Roche Applied Sciences) (18). Shortly, enzymes were removed, and cells were cultured in α-MEM, containing 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. Osteoblast differentiation was promoted in osteogenic media containing ascorbic acid (50 μg/ml) and β-glycerophosphate (10 mM) for 1-3 weeks and culture medium was replaced every 3 days. In some experiments, BMP-2 (100 ng/ml) was added to osteogenic media, as indicated. To access ALP activity assay, cells were either stained with ALP or lysed at day 4 of culture with osteoblast lysis buffer [50 mM Tris-HCl (pH 7.4), 1% Triton X-100, 150 mM NaCl, 1 mM EDTA]. Following this, cell lysates were incubated with p-nitrophenyl phosphate (Sigma), and ALP activity was measured using a spectrophotometer at 405 nm. For ALP staining, cultured cells were fixed in 10% formalin for 10 min. The cells were permeabilized for 30 min with 0.1% Triton-100 in phosphate buffered saline (PBS), and treated with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate for 10-30 min. To measure the level of calcium deposition in the extracellular matrix, the preosteoblast cells were seeded in 48-well tissue culture plates and cultured for 21 days under osteogenic media contained ascorbic acid and β-glycerophosphate. Cultured cells were washed twice with PBS and fixed in 70% ethyl alcohol for 1 h. After three times washes with PBS, they were then stained with 40 mM Alizarin red solution (pH 4.2) for 10 min to stain the calcium deposits. Cultured cells were then washed five times with distilled water, followed by PBS for 15 min to remove non-specific stained cells. To quantify the degree of mineralization, they were extracted using 10% (w/v) cetylpyridinium chloride in 10 mM sodium phosphate (pH 7.0). The concentration was evaluated by measuring the absorbance at 562 nm on a multiplate reader using an Alizairn red S standard curve in the same solution. All values are expressed as fold changes over the control, which were preosteoblast cells treated with standard osteogenic media.
Semiquantitative RT-PCR
To determine the expression of various genes, total RNA was extracted from cultured cells using TRIzol (invitrogen, Carlsbad, CA). First-strand cDNA was transcribed from 1 μg RNA using Superscript RT (Invitrogen) following the manufacturer’s protocol. Primers used were the following: 5’-ALP, 5’-AACCCAGACACAAGCATTCC-3’; 3’-ALP, 5’-AAAACGTGG GAATGATCAGC-3’; 5’-Runx2, 5’-CCCAGCCACCTTTACCTA CA-3’; 3’-Runx2, 5’-CAGCGTCAACACCATCATTC-3’; 5’-osteocalcin, 5’-GCGCTCTGTCTCTCTGACCT-3’; 3’-osteocalcin, 5’-A CCTTATTGCCCTCCTGCTT-3’; 5’-bone sialoprotein, 5’-AAAGT GAAGGAAAGCGACGA-3’; 3’-bone sialoprotein, 5’-ACTCAAC GGTGCTGCTTTTT-3’; 5’-GATA4, 5’-TCTCACTATGGGCACAG CAG-3’; 3’-GATA4, 5’-GCGATGTCTGAGTGACAGGA-3’; 5’-HPRT, 5’-GTAATGATCAGTCAACGGGGGAC-3’; 3’-HPRT, 5’-CCAGCAAGCTTGCAACCTTAACCA-3’.
Immunoprecipitation assay and Western blot analysis
293T cells were transfected with Flag-GATA4 and HA-Dlx5 for 48 hours, and then washed with chilled PBS and lysed in extraction buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, PMSF, and protease inhibitors]. Cell lysates were immunoprecipitated with monoclonal anti-Flag antibody. Cell lysates and immunoprecipitated samples were subsequently separated by SDS-PAGE and transferred to PVDF membrane (Milipore). The membrane was blocked with TBS-T [10 mM Tri-HCl (pH 7.6), 150 mM NaCl, 0.1% Tween 20] containing 5% skim milk and was probed with horseradish peroxidase (HRP)-conjugated antibodies including anti-Flag-HRP (Sigma) and anti-HA-HRP (Sigma). Signals were detected with enhanced chemiluminescence (ECL) and analyzed by LAS3000 luminescent image analyzer (Fuji Photo Film).
Promoter assay
For transfection of reporter plasmids, C2C12 cells were plated on 24-well plates at a density of 2 × 104 cells/well one day before the transfection. Plasmid DNA was mixed with TransIT-2020 (Mirus, Madison, WI) and transfected into the cells following the manufacturer’s protocol. After 48 h of transfection, the cells were washed twice with PBS and then lysed in reporter lysis buffer (Promega, Madison, WI). Luciferase activity was measured with a luciferase assay system (Promega) according to the manufacturer’s instructions. Luciferase activity was measured in triplicate, averaged, and then normalized to β-galactosidase activity using o-nitrophenyl-β-D-galactopyranoside (Sigma-Aldrich) as a substrate.
ChIP assay
A chromatin immunoprecipitation (ChIP) assay was performed with a ChIP kit (Upstate Biotechnology, Lake Placid. NY), according to the manufacturer’s instructions, using antibodies against Dlx5 or control IgG (Santa Cruz Biotechnology, Santa Cruz, CA). The precipitated DNA was subjected to PCR amplification with specific primers for the Runx2 P1 promoter region containing Dlx5-binding sites. The following primers were used for PCR: Runx2 P1 sense, 5’-AAGGCAAACAGA AGGAAGCA-3’; Runx2 P1 antisense, 5’-AGGAGCCCACACT CCTGTAA-3’.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant (MRC for Gene Regulation, 2011-0030132), which is funded by the Korea government (MSIP), and by the Basic Science Research Program through National Research Foundation of Korea (NRF) at the Ministry of Education, Science, and Technology (NRF 2011-0023311). We thank H.M. Ryoo for Runx2 P1 promoters and T. Kitamura for Plat-E cells.
References
- 1.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. (1997);89:755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 2.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. (1997);89:765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 3.Lee B., Thirunavukkarasu K., Zhou L., Pastore L., Baldini A., Hecht J., Geoffroy V., Ducy P., Karsenty G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. (1997);16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 4.Hong J. H., Hwang E. S., McManus M. T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B. M., Sharp P. A., Hopkins N., Yaffe M. B. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. (2005);309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 5.Zhou G., Zheng Q., Engin F., Munivez E., Chen Y., Sebald E., Krakow D., Lee B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc. Natl. Acad. Sci. U. S. A. (2006);103:19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marie P. J. Transcription factors controlling osteoblastogenesis. Arch. Biochem. Biophys. (2008);473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Samee N., de Vernejoul M. C., Levi G. Role of DLX regulatory proteins in osteogenesis and chondrogenesis. Crit. Rev. Eukaryot. Gene Expr. (2007);17:173–186. doi: 10.1615/CritRevEukarGeneExpr.v17.i3.10. [DOI] [PubMed] [Google Scholar]
- 8.Zhao G. Q., Zhao S., Zhou X., Eberspaecher H., Solursh M., de Crombrugghe B. rDlx, a novel distal-less-like homeoprotein is expressed in developing cartilages and discrete neuronal tissues. Dev. Biol. (1994);164:37–51. doi: 10.1006/dbio.1994.1178. [DOI] [PubMed] [Google Scholar]
- 9.Holleville N., Mateos S., Bontoux M., Bollerot K., Monsoro-Burq A. H. Dlx5 drives Runx2 expression and osteogenic differentiation in developing cranial suture mesenchyme. Dev. Biol. (2007);304:860–874. doi: 10.1016/j.ydbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Lee M. H., Kim Y. J., Yoon W. J., Kim J. I., Kim B. G., Hwang Y. S., Wozney J. M., Chi X. Z., Bae S. C., Choi K. Y., Cho J. Y., Choi J. Y., Ryoo H. M. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J. Biol. Chem. (2005);280:35579–35587. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- 11.Lee M. H., Kim Y. J., Kim H. J., Park H. D., Kang A. R., Kyung H. M., Sung J. H., Wozney J. M., Kim H. J., Ryoo H. M. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. (2003);278:34387–34394. doi: 10.1074/jbc.M211386200. [DOI] [PubMed] [Google Scholar]
- 12.Miyama K., Yamada G., Yamamoto T. S., Takagi C., Miyado K., Sakai M., Ueno N., Shibuya H. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev. Biol. (1999);208:123–133. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- 13.Tadic T., Dodig M., Erceg I., Marijanovic I., Mina M., Kalajzic Z., Velonis D., Kronenberg M. S., Kosher R. A., Ferrari D., Lichtler A. C. Overexpression of Dlx5 in chicken calvarial cells accelerates osteoblastic differentiation. J. Bone Miner. Res. (2002);17:1008–1014. doi: 10.1359/jbmr.2002.17.6.1008. [DOI] [PubMed] [Google Scholar]
- 14.Molkentin J. D. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. (2000);275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 15.Viger R. S., Guittot S. M., Anttonen M., Wilson D. B., Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol. Endocrinol. (2008);22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pikkarainen S., Tokola H., Kerkela R., Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. (2004);63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 17.Molkentin J. D., Lu J. R., Antos C. L., Markham B., Richardson J., Robbins J., Grant S. R., Olson E. N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. (1998);93:215–228. doi: 10.1016/S0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song I., Kim B. S., Kim C. S., Im G. I. Effects of BMP-2 and vitamin D3 on the osteogenic differentiation of adipose stem cells. Biochem. Biophys. Res. Commun. (2011);408:126–131. doi: 10.1016/j.bbrc.2011.03.135. [DOI] [PubMed] [Google Scholar]
- 19.Miranda-Carboni G. A., Guemes M., Bailey S., Anaya E., Corselli M., Peault B., Krum S. A. GATA4 regulates estrogen receptor-alpha-mediated osteoblast transcription. Mol. Endocrinol. (2011);25:1126–1136. doi: 10.1210/me.2010-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisping E., Ikeda S., Kong S. W., Tarnavski O., Bodyak N., McMullen J. R., Rajagopal S., Son J. K., Ma Q., Springer Z., Kang P. M., Izumo S., Pu W. T. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc. Natl. Acad. Sci. U. S. A. (2006);103:14471–14476. doi: 10.1073/pnas.0602543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz A. M. The cardiomyopathy of overload: an unnatural growth response. Eur. Heart J. (1995);16(Suppl O):110–114. doi: 10.1093/eurheartj/16.suppl_O.110. [DOI] [PubMed] [Google Scholar]


