Abstract
Cell therapies utilizing mesenchymal stem cells (MSCs) have a great potential in many research and clinical settings. The mechanisms underlying the therapeutic effects of MSCs have been studied previously and the paracrine effects elicited by their production of various growth factors and cytokines were recognized as being crucial. However, the molecular controls that govern these paracrine effects remain poorly understood. To elucidate the molecular regulators of this process, we performed a global knockdown of microRNAs (miRNAs) in human adipose-derived mesenchymal stem cells (hADSCs) by inhibiting DGCR8, a key protein in miRNA biogenesis. Global disruption of miRNA biogenesis in hADSCs caused dramatic changes in the expression of subsets of growth factors and cytokines. By performing an extensive bioinformatic analysis, we were able to associate numerous putative miRNAs with these genes. Taken together, our results strongly suggest that miRNAs are essential for the production of growth factors and cytokines in hADSCs. [BMB Reports 2014; 47(8): 469-474]
Keywords: Cytokines, DGCR8, Growth factors, Human adipose-derived mesenchymal stem cell, MicroRNA
INTRODUCTION
Cell therapies using adult stem cells, such as mesenchymal stem cells (MSCs), have a great potential for tissue regeneration and wound healing in various diseases (1). MSCs are multipotent stromal cells, which can be readily isolated from various human tissues and have the ability to self-renew and differentiate into multiple cell lineages, including osteoblasts, chondrocytes, and adipocytes (2-4). Originally, the engraftment of MSCs into local tissues and their differentiation was considered a major mechanism underlying tissue repair and wound healing. In particular, local transplantation of high densities of these cells in animal models demonstrated the effects of such engraftment and differentiation (5-7). However, growing evidence suggests that most of the therapeutic effects of MSCs can be explained by their production of various growth factors and cytokines (8). Indeed, the conditioned medium of MSCs has tissue repair effects, which further supports the therapeutic functions of these factors (9-11).
MicroRNAs (miRNAs) are a group of small non-coding RNAs. They negatively regulate the expression of target messenger RNAs (mRNAs) by destabilizing mRNA and/or inhibiting translation (12). The biogenesis of miRNAs involves multiple actions of RNase III enzymes. After transcription by RNA polymerase II, long RNAs containing hairpin structures are first recognized by the RNA-binding protein DGCR8 (DiGeorge syndrome critical region 8), which directs binding of the RNase III enzyme Drosha. This Drosha-DGCR8 complex, known as ‘Microprocessor’, is critical for the initial processing of miRNAs (13-15). After cleavage by Microprocessor, hairpins are transported to the cytoplasm and are then processed by Dicer, another RNase III enzyme (16, 17).
Mounting evidence suggests that miRNAs are key players in diverse biological processes, including the regulation of MSCs (18); however, the global effects of miRNA loss have not been investigated systematically in MSCs. In this study, we examined the roles of miRNAs in the expression of cytokines and growth factors in MSCs by inhibiting the DGCR8 gene in human adipose-derived mesenchymal stem cells (hADSCs). We first confirmed that DGCR8, and hence miRNAs, was functionally depleted in hADSCs by RNA interference (RNAi) technologies. We then demonstrated that groups of cytokines and growth factors were misregulated in DGCR8-knockdown hADSCs. Among the misregulated factors, bone morphogenetic protein (BMP) 2 and insulin-like growth factor 1 (IGF1) genes were significantly upregulated in DGCR8-knockdown hADSCs. Conversely, BMP1, chemokine (C-X-C motif) ligand 12 (CXCL12), and fibroblast growth factor (FGF) 2 mRNAs were downregulated in DGCR8-knockdown hADSCs. By performing an extensive bioinformatic analysis, we were able to associate numerous putative miRNA regulators with these genes. Taken together, our analyses strongly suggest that miRNAs are essential for the production of growth factors and cytokines in hADSCs. Therefore, the manipulation of miRNA expression might be useful for modulating the therapeutic effects of hADSCs.
RESULTS
Characterization of hADSCs
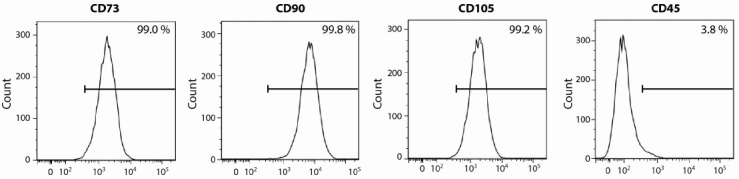
hADSCs were isolated from adipose tissues of four donors and individual cell lines were checked for the expression of various cell surface markers, including CD73, CD90, and CD105. The absence of CD45 expression was also checked. Flow cytometry analysis revealed that the established cell lines showed the characteristic CD marker expression profiles as previously described (Fig. 1) (19). In addition, established hADSCs showed a multipotent differentiation potential in vitro (data not shown), which is another criteria used to define hADSCs (19). Therefore, we used these hADSCs at passage 3-5 for further analysis.
Fig. 1. Fluorescence-activated cell sorting (FACS) analysis of hADSCs. hADSCs were positive for CD73, CD90, and CD105, and negative for CD45. Similar profiles were obtained in four independent hADSC lines.
Disruption of miRNA biogenesis by knocking down the DGCR8 gene
To investigate the functions of miRNAs in hADSCs, we blocked miRNA biogenesis by inhibiting the DGCR8 gene using RNAi, which has been routinely used in various cell lines (20). Quantitative reverse transcription PCR (qRT-PCR) analysis showed that the level of DGCR8 transcripts was significantly reduced at 48 hours post-transfection (Fig. 2A). We then evaluated a group of miRNAs that are known to be expressed in hADSCs by performing miRNA qRT-PCR analysis. The levels of these miRNAs were significantly downregulated (22-32%) (Fig. 2B). hADSCs exhibited severe proliferation defects following depletion of DGCR8 (data not shown), similar to the effects of DGCR8 depletion in mouse embryonic stem cells (21). Taken together, we successfully reduced expression of DGCR8, and hence that of miRNAs, in hADSCs.
Fig. 2. Global miRNA knockdown upon DGCR8 depletion. (A) Quantitative real-time reverse transcription PCR (qRT-PCR) analysis of the DGCR8 gene. Total RNA was prepared from hADSCs 48 hours after transfection of siRNA targeting DGCR8 (siDGCR8) or GFP (siGFP). Error bars denote the standard error of the mean of eight independent experiments performed with four independent samples (*P < 0.0001). (B) Real-time RT-PCR analysis of miRNAs that are expressed in hADSCs. Total RNA was isolated from hADSCs 48 hours after transfection of siDGCR8 or siGFP. Error bars denote the standard error of the mean of four experiments performed with four independent samples (*P < 0.0001). (C) A scatter plot of siGFP- and siDGCR8-transfected hADSCs using the Human Stem Cell RT2 ProfilerTM PCR Array. Black and grey dots represent transcripts that are upregulated and downregulated, respectively, in siDGCR8-transfected hADSCs. Each data point represents a transcript.
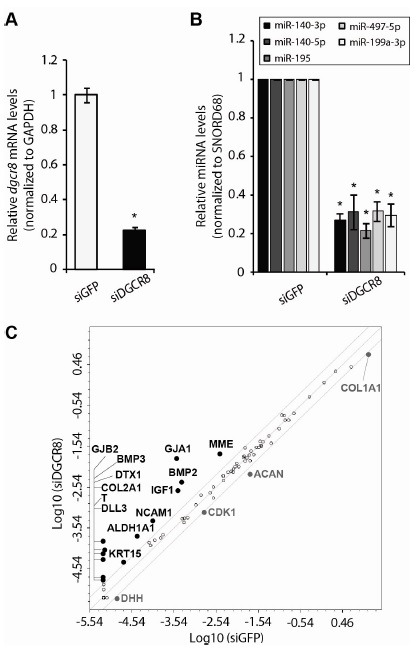
To determine the molecular consequences of DGCR8 depletion in hADSCs, we profiled 84 stem cell-related genes using a PCR array (Fig. 2C). In comparison to hADSCs transfected with control GFP-targeting siRNA (siGFP), 13 genes were upregulated more than 2-fold in hADSCs transfected with DGCR8-targeting siRNA (siDGCR8) and four genes were downregulated. Among the functional groups within the array, expression of various cytokines and growth factors was significantly enhanced upon knockdown of DGCR8, including expression of BMP2 (increased 8.9-fold relative to the control), BMP3 (increased 13.0-fold relative to the control), and IGF1 (increased 6.8-fold relative to the control). Therefore, we characterized these genes further.
Misregulation of cytokines and growth factors in DGCR8-knockdown hADSCs
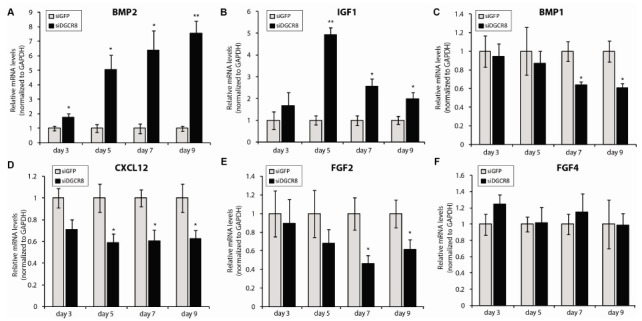
Using qRT-PCR analysis, we next validated the PCR array results in expanded pools of siRNA-transfected hADSCs. We focused on the panel of cytokines and growth factors identified from the PCR array, which included BMP1, BMP2, BMP3, CXCL12, fibroblast growth factor (FGF) 1, FGF2, FGF3, FGF4, growth differentiation factor 2 and 3, IGF1, and jagged 1. After testing the efficiencies of the customized primers (Supplementary Table 1) for the respective genes, we further characterized the expression patterns of BMP1, BMP2, CXCL12, FGF2, FGF4, and IGF1. The primers for these six genes exhibited consistent amplification efficiencies in hADSCs transfected with siRNAs. Consistent with our PCR array results, BMP2 and IGF1 were significantly upregulated upon DGCR8 knockdown up to 9 days after siRNA transfection (Fig. 3A, B). By contrast, BMP1, CXCL12, and FGF2 mRNAs were significantly downregulated in siDGCR8-transfected hADSCs, in comparison to siGFP-transfected control hADSCs (Fig. 3C-E). mRNA expression of FGF4 was similar in siGFP- and siDGCR8-transfected hADSCs (Fig. 3F).
Fig. 3. Effects of DGCR8 depletion on the expression of various cytokines and growth factors. Quantitative real-time reverse transcription PCR (qRT-PCR) analysis of (A) BMP2, (B) IGF1, (C) BMP1, (D) CXCL12, (E) FGF2, and (F) FGF4. Total RNA was isolated at the indicated time points after siRNA transfection. Error bars denote the standard error of the mean of at least five experiments (*P<0.05; **P<0.001).
Numerous miRNAs are predicted to target each tested cytokine and growth factor
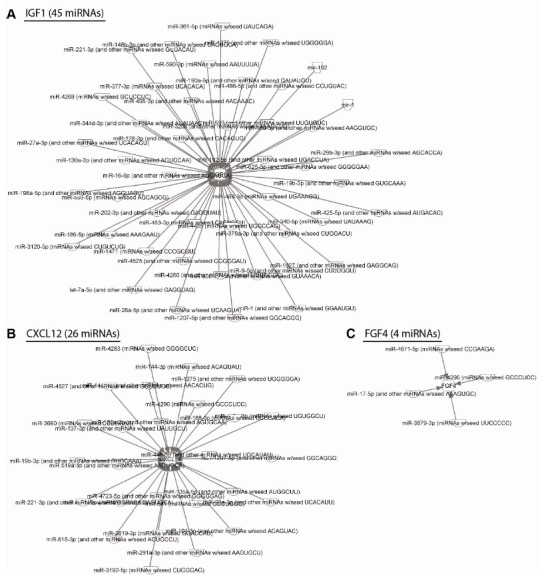
To associate putative miRNA regulators with the tested genes, TargetScan 6.0 (22) and Ingenuity Pathway Analysis (IPA) software was used. In this analysis, we applied an miRNA-mRNA interaction filter on the respective cytokines and growth factors. We next prioritized the filter to include targets that are experimentally validated and/or highly predicted in the IPA software. Finally, the application of direct interactions revealed numerous putative miRNA regulators of each gene. A network map of miRNA-mRNA interactions was then created with the IPA software (Fig. 4). One of the genes that was upregulated upon DGCR8 knockdown, IGF1, had 45 predicted miRNA regulators (Fig. 4A). Twenty-six miRNAs were predicted to be upstream regulators of CXCL12 (Fig. 4B). Notably, only four direct miRNA interactions were detected for FGF4 (Fig. 4C), which exhibited minimal expression changes upon DGCR8 depletion (Fig. 3F).
Fig. 4. miRNA-mRNA target networks in hADSCs. (A) Using IPA software, miRNAs that might directly regulate IGF1 are shown. A confidence level filter was adopted that prioritized experimentally validated and/or highly predicted targets. This analysis revealed 45 miRNAs that directly regulate IGF1. (B) Using the same miRNA analysis filter, CXCL12 had 26 miRNA regulators. (C) FGF4 had direct interactions with only four miRNAs.
DISCUSSION
Our results highlight the critical roles of miRNAs in the regulation of genes encoding cytokines and growth factors. We globally removed canonical miRNAs by knocking down DGCR8, an RNA-binding protein required for miRNA processing. Consistent with previous reports in numerous model systems (reviewed in (23)), loss of DGCR8 resulted in differential expression profiles of genes involved in stem cell biology (Fig. 2). Notably, analysis of siDGCR8-transfected hADSCs indicated the misregulation of genes related to the functions of mesenchymal cells, cell-cell communication, the Notch signaling pathway, and cytokines/growth factors. Indeed, qRT-PCR analysis of subsets of cytokines and growth factors confirmed the PCR array results. Expression of BMP1, CXCL12, and FGF2 was reduced in DGCR8-knockdown hADSCs according to qRT-PCR analysis (Fig. 3C-E), which was not detected by the PCR array. This is likely due to the use of different primer sets in the two assays, which might affect the detection sensitivity.
To date, the essential roles of miRNAs in human MSCs have been studied mostly by dissecting the functions of individual miRNAs (reviewed in (18)). Considering that a single miRNA can regulate hundreds of target mRNAs simultaneously (24-26), it is important to evaluate the consequences of global miRNA depletion. However, the systematic effects of global miRNA loss in hADSCs have not been previously reported. In the present study, we demonstrated the critical effects of miRNAs on the expression of cytokines and growth factors, which are considered to be key factors in the therapeutic effects of human MSCs. Many cytokines and growth factors exert their functions by affecting the proliferation, differentiation, and migration of human MSCs (10, 27). Therefore, it is imperative to tightly regulate the expression of these molecules during the development of human MSCs. Based on the results of the current study as well as previous reports, we postulate that miRNAs are essential upstream regulators of cytokines and growth factors. In addition, our miRNA-mRNA network data suggest that multiple distinct miRNAs target a single common factor (Fig. 4). Interestingly, the 3' untranslated region of FGF4 is relatively short (280 nucleotides in length) and has a small number of predicted miRNA regulators (Fig. 4C), which might partly explain the DGCR8-independent expression pattern of this gene.
In summary, our data demonstrate that global miRNA loss can affect genes involved in stem cell development in general, highlighting the essential functions of miRNAs in somatic stem cell biology.
MATERIALS AND METHODS
Isolation and culture of hADSCs
This study was carried out according to the guidelines and with the approval of the Institutional Review Board of Asan Medical Center. hADSCs were isolated from human adipose tissue of four donors and cultured as previously described (28, 29). Adipose tissues were washed extensively with phosphate-buffered saline (Gibco) and incubated with 0.75% type I collagenase (Sigma) at 37℃ for 1 hour. After neutralization with Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), the suspension was centrifuged at 1,200 rpm (400 g) for 10 min. The pellet was resuspended in red blood cell lysis buffer (Sigma) and centrifuged at 1,200 rpm (300 g) for 10 min. The collected pellet was filtered through a 100-μm cell strainer (BD Falcon) to obtain a uniform cell suspension, which was incubated overnight at 37℃ and in an atmosphere of 5% CO2 in Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco). The medium was replaced every 3 days and the cells were used at passage 3-5 for all experiments.
Flow cytometry
hADSCs were labeled with phycoerythrin (PE)-conjugated mouse anti-human CD73, PE-conjugated mouse anti-human CD90, PE-conjugated mouse anti-human CD105, and PE-conjugated mouse anti-human CD45 antibodies (BD Biosciences). Cells were then analyzed on a FACSCanto II (BD Biosciences) as described in the manufacturer’s protocol. A total of 10,000 events were acquired and the cells were gated properly for analysis.
Cell transfection
hADSCs were plated at a density of 5-6 × 104/ml in 6-well plates containing complete medium. The next day, hADSCs were transfected with siGFP or siDGCR8 (ST Pharm) at a final concentration of 32 nM using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The sequence of siDGCR8 was 5'-CAUCGGACAAGAGUGUGAUdTdT-3', as previously described (20). The cells were harvested on the indicated number of days after transfection.
qRT-PCR and PCR array
For qRT-PCR analysis of mRNAs, total RNA was isolated from cultured cells using TRIzol reagent (Life technologies) according to the manufacturer’s protocol. Reverse transcription was then performed using random hexamers and SuperScript III reverse transcriptase (Life technologies). Quantitative PCR was performed on a 7900 Real Time PCR system (Applied Biosystems) using Power SYBR Green PCR master mix (Applied Biosystems). The primers used for qRT-PCR are provided in Supplementary Table 1. mRNA expression was normalized against that of GAPDH. For qRT-PCR analysis of miRNAs, total RNA was isolated from cultured cells using TRIzol reagent (Life Technologies) or a miRVana miRNA Isolation Kit (Life Technologies) according to the manufacturer’s protocol. miRNAs were reverse-transcribed using a miScript II RT Kit and their expression was then quantified using a miScript SYBR Green PCR Kit (Qiagen) according to the manufacturer’s instructions. miRNA expression was normalized against that of SNORD68. Each reaction was performed in triplicate and repeated with at least three different samples.
The Human Stem Cell RT2 ProfilerTM PCR Array (PAHS-405Z) (Qiagen) was used according to the manufacturer’s protocol to profile the expression of 84 genes related to stem cell development. Data were analyzed using online analysis software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php) (Qiagen). The relative expression levels of the genes were plotted in a scatter plot.
Target prediction and functional analysis
The associations of mRNAs with particular miRNAs were identified using TargetScan 6.0 and IPA software (Qiagen). For each mRNA, a filter with the relationship type of RNA-RNA interactions of miRNA targeting was applied. Next, the confidence levels were adapted to targets that are experimentally observed and/or highly predicted in the IPA software. Finally, only direct interactions were selected to build miRNA-mRNA networks.
Statistical analysis
Statistical significance was examined using two-tailed Students t-test. Results with P values less than 0.05 were considered significant.
Acknowledgments
We thank Professor Euiyoung Bae for critical comments on the manuscript. We acknowledge E-J Kim and HY Lee for their technical help. This work was supported by the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea (2012- 517), the Basic Science Research Program through the National Research Foundation of Korea (NRF) from the Ministry of Education, Science, and Technology (2012R1A1A3013555), the Next-Generation BioGreen 21 Program from the Rural Development Administration (PJ009097), and the Korean Health Technology R&D Project from Ministry of Health and Welfare, Republic of Korea (A121957).
References
- 1.Bianco P., Cao X., Frenette P. S., Mao J. J., Robey P. G., Simmons P. J., Wang C. Y. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat. Med. (2013);19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.da Silva Meirelles L., Chagastelles P. C., Nardi N. B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. (2006);119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 3.Brighton C. T., Hunt R. M. Early histological and ultrastructural changes in medullary fracture callus. J. Bone Joint Surg. Am. (1991);73:832–847. [PubMed] [Google Scholar]
- 4.Brighton C. T., Hunt R. M. Early histologic and ultrastructural changes in microvessels of periosteal callus. J. Orthop. Trauma. (1997);11:244–253. doi: 10.1097/00005131-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Kopen G. C., Prockop D. J., Phinney D. G. Marrow stromal cells migrate throughout forebrain and cerebellum, and they differentiate into astrocytes after injection into neonatal mouse brains. Proc. Natl. Acad. Sci. U. S. A. (1999);96:10711–10716. doi: 10.1073/pnas.96.19.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackenzie T. C., Flake A. W. Human mesenchymal stem cells persist, demonstrate site-specific multipotential differentiation, and are present in sites of wound healing and tissue regeneration after transplantation into fetal sheep. Blood Cells Mol. Dis. (2001);27:601–604. doi: 10.1006/bcmd.2001.0424. [DOI] [PubMed] [Google Scholar]
- 7.Prockop D. J. Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Mol. Ther. (2009);17:939–946. doi: 10.1038/mt.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caplan A. I. Why are MSCs therapeutic? New data: new insight. J. Pathol. (2009);217:318–324. doi: 10.1002/path.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salgado A. J., Reis R. L., Sousa N. J., Gimble J. M. Adipose tissue derived stem cells secretome: soluble factors and their roles in regenerative medicine. Curr. Stem Cell Res. Ther. (2010);5:103–110. doi: 10.2174/157488810791268564. [DOI] [PubMed] [Google Scholar]
- 10.Chen L., Tredget E. E., Wu P. Y., Wu Y. Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One. (2008);3:e1886. doi: 10.1371/journal.pone.0001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinnaird T., Stabile E., Burnett M. S., Lee C. W., Barr S., Fuchs S., Epstein S. E. Marrow-derived stromal cells express genes encoding a broad spectrum of arteriogenic cytokines and promote in vitro and in vivo arteriogenesis through paracrine mechanisms. Circ. Res. (2004);94:678–685. doi: 10.1161/01.RES.0000118601.37875.AC. [DOI] [PubMed] [Google Scholar]
- 12.Fabian M. R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. (2012);19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 13.Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of micro RNAs. Nature. (2004);432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 14.Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J. Processing of primary microRNAs by the Microprocessor complex. Nature. (2004);432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 15.Han J., Lee Y., Yeom K. H., Kim Y. K., Jin H., Kim V. N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. (2004);18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. (2001);409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 17.Hutvagner G., McLachlan J., Pasquinelli A. E., Balint E., Tuschl T., Zamore P. D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. (2001);293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 18.Guo L., Zhao R. C., Wu Y. The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. Exp. Hematol. (2011);39:608–616. doi: 10.1016/j.exphem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. (2006);8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 20.Han J., Pedersen J. S., Kwon S. C., Belair C. D., Kim Y. K., Yeom K. H., Yang W. Y., Haussler D., Blelloch R., Kim V. N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. (2009);136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. (2007);39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. Prediction of mammalian microRNA targets. Cell. (2003);115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 23.Suh N., Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. (2011);138:1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baek D., Villen J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. The impact of microRNAs on protein output. Nature. (2008);455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. (2008);455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 26.Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. (2010);466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haider H., Jiang S., Idris N. M., Ashraf M. IGF-1-overexpressing mesenchymal stem cells accelerate bone marrow stem cell mobilization via paracrine activation of SDF-1alpha/CXCR4 signaling to promote myocardial repair. Circ. Res. (2008);103:1300–1308. doi: 10.1161/CIRCRESAHA.108.186742. [DOI] [PubMed] [Google Scholar]
- 28.Zuk P. A., Zhu M., Mizuno H., Huang J., Futrell J. W., Katz A. J., Benhaim P., Lorenz H. P., Hedrick M. H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. (2001);7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 29.Zuk P. A., Zhu M., Ashjian P., De Ugarte D. A., Huang J. I., Mizuno H., Alfonso Z. C., Fraser J. K., Benhaim P., Hedrick M. H. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell. (2002);13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]


