Abstract
Colorectal cancer has become the third most common cancer and leads to high mortality worldwide. Although colorectal cancer has been studied widely, the underlying molecular mechanism remains unclear. PER3 is related to tumor differentiation and the progression of colorectal cancer. High expression of miR-103 is associated with poor prognosis in patients with colorectal cancer. However, the relationship between miR-103 and PER3 in CRC cells remains unclear. In this study, we found that PER3 was downregulated in CRC tissues and CRC cell lines, whereas miR-103 was upregulated in CRC cell lines. We also found that PER3 promoted CRC cells apoptosis. These results indicate that PER3 plays a suppressive role in CRC cells. Moreover, we found that PER3 was targeted, at least partially, by miR-103. Taken together, we provide evidence to characterize the role of PER3 in CRC, which may be a new therapeutic target for CRC. [BMB Reports 2014;47(9): 500-505]
Keywords: Circadian clock gene, Colorectal cancer, miR-103, Period3, Vitro
INTRODUCTION
Colorectal cancer has become the third most common cancer and leads to high mortality of worldwide, with high tumor migration and invasion. Although colorectal cancer has been studied widely, the underlying molecular mechanism remains unclear.
Circadian clock genes, which regulate transcription, are associated with circadian rhythms, sleeping, DNA damage/repair, cell proliferation and tumor development (1). The family of Circadian clock genes mainly consist of 8 core genes (2), including Period1 (Per1), Period2 (Per2), Period3 (Per3), Cryptochrome1 (Cry1), Cryptochrome2 (Cry2), Clock, Bmal1 and Casein Kinase Iε (CKIε). Recently, studies indicated that circadian clock genes participated in the tumorgenesis processes.
In colorectal cancer tissue, PER3 mRNA levels were lower in tumor tissues compared to normal tissues, and PER3 expression was related to multiple clinicopathologic factors, including tumor location, differentiation, and stage (3). Matsuo and his colleagues used the core clock genes mutant mice to find that the circadian clock controlled the G2/M transition by regulating the expression of wee1, a major cyclin complex regulating G2/M (4). These results indicated that the PER3 gene might play a role in colon cancer progression. However, the molecular mechanisms underlying PER3 in colorectal cancer cells are still unknown.
Non-coding small RNA negatively regulated a variety of genes. Growing evidence indicates a relationship between cancer and miRNAs. miRNAs act as tumor suppressors or oncogenes, depending on the target genes (5). Among these functional miRNAs, miR-103 increased in multiple cancers. In endometrial cancer cell lines, miR-103 post-transcriptionally reduced the expression of the tumor suppressor tissue inhibitor of metalloproteinase 3 and stimulated growth and invasion (6). MiR-103 showed an inverse correlation with ID2 (repressor of nervous system cancers) during neuroblastoma cell differentiation induced by retinoic acid (7). In colorectal cancer, high expression of miR-103 was associated with metastastic potential of colorectal cancer cell lines and ensuing poor prognosis. Death-associated protein kinase and Krüppel-like factor 4, the known metastasis suppressors in CRC cells, were targeted by miR-103 leading to increased cell motility and decreased cell-cell adhesion and epithelial marker expression (8). All the above data suggested that PER3 gene might play a critical role in colon cancer progression and miR-103 is an oncogene. However, the relationship between miR-103 and PER3 in CRC cells remains unclear.
In the present study, we investigated the precise function of PER3 in colorectal cancer (CRC) cells and the correlation between PER3 and miR-103. The molecular mechanisms underlying PER3 in CRC was explored. Thus it could provide a promising choice therapy for patients with CRC in the future.
RESULTS
PER3 was frequently downregulated in colorectal cancer tissues and cell lines, but miR-103 was upregulated in colorectal cancer cell lines
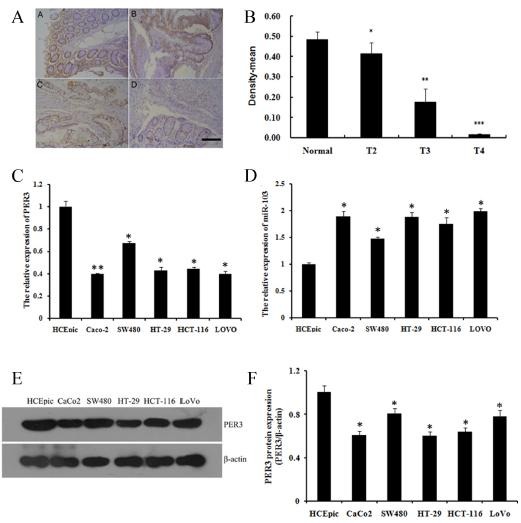
We performed tissue assay analysis to detect the expression of PER3 in colorectal cancer tissues. In most of the 70 cases of colorectal cancer tissues and normal colorectal tissues, we found that PER3 was significantly decreased in different stages of colorectal cancer tissues compared with that of in the normal tissues (Fig. 1A, B). We studied five human colorectal cancer cell lines to study the expression of PER3 and miR-103. All five cell lines showed a notable down-regulation of PER3 in mRNA (Fig. 1C) and protein level (Fig. 1E, F), whereas the miR-103 expression was high (Fig. 1D).
Fig. 1. expression of per3 and mir-103 in colorectal cancer tissues and human colorectal cancer cell lines. (A) representative images of immunohistochemistry staining of per3 in normal and colorectal cancer tissue in different stages. A) normal colorectal tissue, B) T2 stage, C) T3 stage, D) T4 stage. the images were shown at 100 × magnifications, brown color represents per3 staining; (B) optical density analysis of per3 immunohistochemistry staining; (C, D) expression of per3 and mir-103 mrna in different colorectal cancer cell lines; (E, F) expression of per3 protein in different colorectal cancer cell lines and quantification. data shown are means ± S.D, *P < 0.05, **P < 0.01, ***P < 0.001 versus control.
Overexpression of PER3 repressed proliferation and invasion, and regulated the expression of P53, cyclin B1, CDC2, Bcl2, Bid, cleaved-caspase3, and cleaved-caspase8 in SW480 and HCT-116 cells
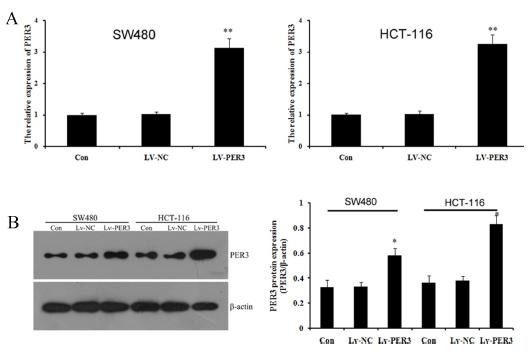
In this study, we selected SW480 and HCT-116 cells to study the function of PER3 in colorectal cancer cells. We overexpressed PER3 using lentiviral transfection. After transfection with lentiviral recombinant vectors, qRT-PCR and western blot were performed to analyze the expression of PER3 mRNA and protein. As shown in Fig. 2A, the expression of PER3 mRNA increased significantly in transfected LV-PER3 cells compared with transfected LV-NC vector and control cells. Western blot showed a similar and statistically significant increase (Fig. 2B). The results indicated that PER3 gene was over-expressed effectively by the LV-PER3 transfected in SW480 and HCT-116 cell lines cells.
Fig. 2. PER3 was over-expressed in SW480 and HCT-116 cell lines. (A) qPCR detected the expression of PER3 after transfecting with Lv-PER3 or Lv-NC; (B) western blot detected and quatified the expression of PER3. Data shown are means ± S.D, *P < 0.05, **P < 0.01 versus control.
As the data shows, we observed that overexpression of PER3 significantly decreased cell proliferation (Fig. 3A, B), and inhibited cell invasion ability (Fig. 3C) in SW480 and HCT-116 cells compared to control cells. Besides, we observed that overexpression of PER3 upregulated significantly apoptosis in SW480 and HCT-116 cells compared to control (Fig. 3D). It is suggested that up-regulation of PER3 inhibited the proliferation and invasion of colorectal cancer cells.
Fig. 3. Effects of PER3 overexpression in SW480 and HCT-116 cells. (A) MTT assay was used to evaluate cell growth. The data are presented as 570 nm optical density in different groups. (B) Colony formation assay was used to determinate cell proliferation. (C) Transwell assay was used to determinate cell invasion. Representative images of crystal violet staining are shown. The data are presented as 570 nm optical density in different groups. (D) Flow cytometric analysis of apoptosis with Annexin-V/PI double staining; the data of histogram shows the percentage of early and late apoptotic cells. (E) After PER3 overexpression, expression of P53, cyclin B1, CDC2, Bcl2, Bid, cleaved-caspase3, and cleaved -caspase8 were determined and quantified by western blot. Data shown are means ± S.D, *P < 0.05, **P < 0.01 versus the control, #P < 0.05, ##P < 0.01 versus LV-NC groups.

To explore potential molecular mechanisms underlying PER3-induced cell apoptosis, we tested the expression of apoptosis-related genes including tumor suppressor P53, cell-cycle regulation genes cyclin B1 and CDC2, anti-apoptosis gene Bcl2, pro-apoptosis gene Bid, caspase3, and caspase8. Overexpression of PER3 significantly increased the expression of P53, cyclin B1, CDC2, Bid, cleaved-caspase3, and cleaved-caspase8 proteins compared to that of control cells, whereas significantly decreased the expression of Bcl2 (Fig. 3E). The results indicated that overexpression of PER3 could inhibit colorectal cancer cell proliferation. Taken together, our results suggested that PER3 plays a critical role in the apoptotic pathway.
PER3 was directly targeted by miR-103
To investigate if the predicted binding site of miR-103 directly targeted 3'UTR of PER3, we cloned the 3'UTR of PER3 downstream to a luciferase reporter gene (wt-PER3), its mutant version (mut-PER3) by the binding site mutagenesis was also constructed. wt-PER3 vectors were co-transfected with miR-103 mimics or inhibitor into SW480 and HCT-116 cells. The luciferase activity of cells transfected with miR-103 was significantly decreased compared with inhibitor control cells (Fig. 4A). Moreover, miR-103-mediated repression of luciferase activity was abolished by the mutant putative binding site (Fig. 4A). QPCR showed that enhanced or repressed miR-103 significantly decreased or increased PER3 mRNA levels compared to cells transfected with pre-scramble control in SW480 and HCT-116 cells, respectively (Fig. 4B, C). These results suggested that miR-103 inhibited PER3 expression on the transcriptional level.
Fig. 4. PER3 3'UTR was directly targeted by miR-103 and miR-103 regulated proliferation and migration of SW480 and HCT-116 cells. (A) The repression of luciferase activity by PER3 3'UTR was dependent on miR-103. Mutated PER3 3'UTR abrogated miR-103 mediated repression luciferase activity. (B) qPCR detected the expression of miR-103 after transfecting with pre-miR-103, pre-scramble, anti-miR-103, or anti- scramble. (C) qPCR detected the expression of PER3 mRNA after transfecting with pre-miR-103, pre-scramble, anti-miR-103, or anti- scramble. (D) Colony formation evaluated the proliferation of SW480 and HCT-116 cells treated with pre-miR-103, pre-scramble, anti-miR-103, or anti- scramble. (E, F) MTT assay measured the proliferation of SW480 and HCT-116 cells treated with pre-miR-103, pre-scramble, anti-miR-103, or anti- scramble. (G, H) Transwell assay measured the migration of SW480 and HCT-116 cells treated with pre-miR-103, pre-scramble, anti-miR-103, or anti- scramble. Data shown are means ± S.D, #P < 0.05, *P < 0.05, **P < 0.01 versus control or pre-scramble.

Effect of miR-103 on SW480 and HCT-116 cells growth and migration
To validate whether miR-103 regulates colorectal cancer cells growth or not, Colony formation and MTT assay were performed in SW480 and HCT-116 cells transfected with miR-103 mimics, anti-miR-103 or scramble control. It showed that the up-regulation of miR-103 induced significant promotion of cell growth from 48 h to 72 h after transfection; on the contrary, the down-regulation of miR-103 induced significant inhibition of cell growth from 48 h to 72 h after transfection (Fig. 4D-F). Cell migration of transfected cells was also measured by transwell assays. As shown in Fig. 4G and H, compared to the scramble control, cells transfected with pre-miR-103 exhibited significant increase of migratory ability; on the contrary, the cells transfected with anti-miR-103 exhibited significant decrease of migratory ability.
DISCUSSION
PER3 gene is one of the circadian rhythm-controlled genes. There was a hypothesis that if cell cycle and differentiation controlled by PER3 were disrupted, the tumorigenesis process would probably be started (9). Some epidemiologic evidences supported this hypothesis. For example, abnormal expression of circadian genes was observed in breast and prostate cancers (10). In human gingival cancer CA9-22 cells, PER3 was identified to play a pro-apoptotic role (11). In CRC, there was a positive correlation between poor survival rates and low expression of PER3 in the tumor tissue (12). Tumor invasion or metastasis was demonstrated to be promoted (13) or suppressed (14) by some miRNAs, which provided useful potential therapeutic targets for cancers. Generally, miRNAs effected cancer development by post-transcriptionally regulating the target genes. MiR-137 inhibited invasion of CRC cell by targeting Cdc42 (15). And miR-21 has been demonstrated to enhance cellular invasion by suppressing PDCD4 gene expression in CRC (16). According to previous findings, miR-103 might be a potential minimally-invasive biomarker for the diagnosis of mesothelioma (17). Chen et al. showed that miR-103/107 promotes metastasis in CRC by targeting tumor suppressors DAPK and KLF4 (12). Thus, we speculated PER3 and miR-103 might be involved in CRC tumorigenesis processes.
In this present study, we confirmed that the expression of PER3 in colorectal cancer tissues was decreased significantly compared with that in normal tissue, consistent with previous studies (18). In addition, we found that expression of PER3 was associated with tumor differentiation, with a stage-dependent manner. Moreover, we tested extendedly the expression of PER3 and miR-103 in five CRC cell lines. We found that the expression of miR-103 in CRC cell lines was increased compared with that of normal cells, which had an inverse relationship with that of PER3. These results indicated that PER3 and miR-103 play roles in CRC development.
To investigate the precise function of PER3 in CRC cells, lentiviral transfection was used to overexpress PER3 gene in SW480 and HCT-116 cells. The results showed that overexpression of PER3 significantly inhibited cell proliferation and invasion, and promoted apoptosis in SW480 and HCT-116 cells. The mitochondrial pathway is the one of the ways to induce apoptosis. The Bcl-2 family is the central regulator of the mitochondrial pathway, including both anti-apoptotic members and pro-apoptotic members (19). In our data, the expression of Bid, one of Bcl-2 pro-apoptotic members, was increased, and that of anti-apoptotic member-Bcl2 was decreased. Moreover, we found that the expressions of cyclin B1 and cell division cycle 2 (CDC2) were dramatically increased; Cyclin B1/ CDC2 complex plays a critical role on the regulation of cell-cycle progression from G2/M phase (20). p53 has been widely identified as a tumor suppressor protein that regulates apoptosis and cell cycle arrest in cancer cells (21). Many studies suggested that p53 was involved in caspases activation and cell death (22). In our present study, the expression of p53, cleaved-caspase3 and cleaved-caspase8 was increased significantly in CRC cells transfected with lentivirus. Additionally, we verified that miR-103 directly target PER3 gene by binding to a specific complementary site within its 3' UTR. Our results newly found a vital molecular relationship between miR-103 and PER3 in CRC cell lines. Moreover, the proliferation and migration of CRC cells were regulated by miR-103.
In conclusion, we confirmed that PER3 was decreased in CRC tissues and first described precisely the function of PER3 gene as a tumor suppressor in CRC cells. PER3, at least partially, was targeted by miRNA-103, which might affect cells apoptosis in G2/M phase by modulating apoptosis-related gene in p53 pathway. As a result, restoration of PER3 or inhibition of miRNA-103 expression would be an important target for the clinical treatment of CRC.
MATERIALS AND METHODS
Tissue samples and cell lines
All patients signed an informed consent, approved by the Independent Ethical Committee of Central South University. The colorectal cancer and the adjacent normal colorectal epithelial tissues used in the study were collected from 70 patients from 2010 to 2013. Tumor tissue were representative of the different pathologic stages of colorectal carcinoma, including normal tissue (n=10), T2 stage (n=10), T3 stage (n=20), T4 stage (n=34); T1 stage (n=1) and 5 samples with unclear classification were excluded in further analysis. All samples were embedded in paraffin and stored at 4℃ until tissue section.
Five human colorectal cancer (CRC) cell lines were used, including Caco-2, SW480, HT-29, HCT-116, LOVO; and HCEpic cells were used as normal control. Cells were grown routinely in RPMI-1640 medium (Life technology, USA) containing 10% fetal bovine serum (Gibco, USA) and cultured under a 37℃ humidified air of 5% CO2.
Antibodies
Antibodies of Per3 were obtained from Immunoway (Immunoway, USA). Antibodies of CDC2, Cyclin B1, and p53 were purchased from Santa Cruz Biotechnology (Santa Cruz, USA). Antibodies of β-actin, caspase-3, caspase-8, Bcl-2, and Bid were purchased from Abzoom (Abzoom, USA).
Tissue microarray
A tissue microarray (Auragene Bioscience Co., Changsha, China), containing 64 cases of colorectal cancer (including 10 cases of T2, 20 cases of T3, 34 cases of T4) and 10 cases of matched adjacent normal tissue, was used for immunohistochemistry analysis. Briefly, tissue array was performed with citrate-based antigen retrieval using 10 mM sodium citrate buffer (pH 6.0). The array was then incubated with PER3 antibody, and stained using a DAB kit counter-staining with hematoxylin. Staining integral density was measured and quantified with a Leica Q550 image analysis system Leica Quin Software.
Quantitative PCR analysis
Total RNA was extracted from the indicated cells using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. Expressions of Per3 mRNA were detected by SYBR green qPCR assay (BioRad, USA), and β-actin was used as an endogenous control. The specific primers were as follows: PER3, F: GCAGGTCTATGCCAGTGTGA, R: TGCCTT GTGGTTCTGTTTGT; β-actin, F: AGGGGCCGGACTCGTCA TACT, R: GGCGGCACCACCATGTACCCT. The relative expressions of miR-103 were measured using All-in-OneTM miRNA qRT-PCR Detection Kit (GeneCopoeia). The specific primer sets for miRNA-103 and U6 and the PCR mix were purchased from GeneCopoeia. Expression of U6 was used as an endogenous control. Data were processed using 2-ΔΔCT method.
Western blotting
Total cellular extracts (50 μg) were prepared from each group and separated on 10% SDS-polyacrylamide gels for Per3, cleaved-caspase-3, cleaved-caspase-8, Bid, Bcl-2, CDC2, Cyclin B1, p53 and β-actin detection and transferred to nitrocellulose membranes. Membranes were incubated with the indicated antibodies overnight at 4℃. Then the membranes were incubated with HRP-conjugated secondary antibody. Signals were visualized using ECL Substrates (Millipore, USA). β-actin was taken as an endogenous protein for normalization.
Lentiviral stable transfection
The Lv-Per3 and Lv-NC lentiviral suspensions were purchased from GeneChem (Shanghai, China), and transfected into SW480 and HCT-116 cells according to the manufacturer's instructions. qPCR and western blotting were performed to determinate the mRNA and protein levels of Per3 in the SW480 and HCT-116 cells after lentiviral transfection for 6 days. The stably transfected cells were used for the following further analysis.
MTT assay
SW480 or HCT-116 cells were seeded at 10000 cells per well in 96-well plates after transfection. MTT assay was performed to test cell viability at 0, 24, 48, and 72 hours, and the absorbance was measured at 570 nm using an enzyme immunoassay analyzer.
Colony formation assay
One hundred cells pretreated with Lv-Per3 were prepared and plated into 6-well plates. Cells were grown in high glucose DMEM medium with 10% FBS. When the cell clones can be visible to the naked eye, colon was stopped. The supernatant was discarded. Cells were washed, and fixed by 4% paraformaldehyde for 15 min, then stained by GIMSA (Solarbio, Beijing, China). The total cellular clones were counted using an inverted microscope (Nikon, Japan). Colony-forming efficiency was calculated as the number of total clones divided by the number of plated cells.
Transwell assay
Briefly, 5×104 SW480 or HCT-116 cells resuspended in serum-free media were plated into the upper chamber for migration assay after glucose treatment and transfection, and media supplemented with 10% FBS was filled into the lower chamber. After incubation for 8 h, the cells that had migrated through the membrane to the lower surface were fixed, stained with crystal violet, and then quantified by measuring the optical density (OD) of crystal violet at 570 nm using an enzyme immunoassay analyzer.
Flow cytometric analysis of apoptosis with Annexin-V/PI double staining
SW480 and HCT-116 cells pretreated with Lv-Per3 were trypsinized, collected, and resuspended. About 2.5 × 105 cells were harvested and washed twice with cold phosphate buffer saline, then resuspended in 500 μl 1 ×binding buffer. 5 μl Annexin V-FITC and 5 μl Propidium Iodide were added to the solution and mixed well. After incubation for 15 min at room temperature in the dark, the cells were analyzed using flow cytometric analysis (BD Biosciences, USA).
Dual luciferase reporter assay
A fragment of wild type (Wt) 3'UTR of Per3 containing the putative miR-103 binding site was amplified and recombined into a psiCHECK-2 vector (Promega) downstream to the luciferase gene sequence. A psiCHECK-2 construct containing 3'UTR of Per3 with a mutant sequence of miR-103 was synthesized. Wt-PER3 and Mut-PER3 primers were purchased from FulenGen corporation(FulenGen, Guangzhou, China). SW480 and HCT-116 cells were plated in 96-well plates, then Wt-PER3-3' UTR-psi-CHECK2 or Mut-PER3-3' UTR-psi-CHECK2 were co-transfected with pre-miR-103, anti-miR-103, pre-scramble, and anti-scramble, respectively. after transfection for 48 h luciferase activity was detected using a dual-luciferase reporter assay system (Promega) and was normalized to Renilla activity.
Statistical analysis
All data were expressed as mean ± SD. SPSS 13.0 statistical software was used to process data. Student’s t test was used if only two groups were applied. P values of <0.05 were considered statistically significant.
References
- 1.Gery S., Koeffler H. P. The role of circadian regulation in cancer. Cold. Spring. Harb. Symp. Quant. Biol. (2007);72:459–464. doi: 10.1101/sqb.2007.72.004. [DOI] [PubMed] [Google Scholar]
- 2.Buhr E. D., Takahashi J. S. Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. (2013);217:3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Yan D., Teng M., Fan J., Zhou C., Li D., Qiu G., Sun X., Li T., Xing T., Tang H., Peng X., Peng Z. Reduced expression of PER3 is associated with incidence and development of colon cancer. Ann. Surg. Oncol. (2012);19:3081–3088. doi: 10.1245/s10434-012-2279-5. [DOI] [PubMed] [Google Scholar]
- 4.Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. (2003);302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 5.Calin G. A., Croce C. M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. (2006);6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Yu D., Zhou H., Xun Q., Xu X., Ling J., Hu Y. microRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3. Oncol. Lett. (2012);3:1221–1226. doi: 10.3892/ol.2012.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annibali D., Gioia U., Savino M., Laneve P., Caffarelli E., Nasi S. A new module in neural differentiation control: two microRNAs upregulated by retinoic acid, miR-9 and -103, target the differentiation inhibitor ID2. PLoS One. (2012);7:e40269. doi: 10.1371/journal.pone.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H. Y., Lin Y. M., Chung H. C., Lang Y. D., Lin C. J., Huang J., Wang W. C., Lin F. M., Chen Z., Huang H. D., Shyy J. Y., Liang J. T., Chen R. H. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. (2012);72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 9.Sahar S., Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer. (2009);9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Y., Stevens R. G., Hoffman A. E., Fitzgerald L. M., Kwon E. M., Ostrander E. A., Davis S., Zheng T., Stanford J. L. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. (2009);69:9315–9322. doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato F., Wu Y., Bhawal U. K., Liu Y., Imaizumi T., Morohashi S., Kato Y., Kijima H. PERIOD1 (PER1) has anti-apoptotic effects, and PER3 has pro-apoptotic effects during cisplatin (CDDP) treatment in human gingival cancer CA9-22 cells. Eur. J. Cancer. (2011);47:1747–1758. doi: 10.1016/j.ejca.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 12.Mazzoccoli G., Panza A., Valvano M. R., Palumbo O., Carella M., Pazienza V., Biscaglia G., Tavano F., Di Sebastiano P., Andriulli A., Piepoli A. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol. Int. (2011);28:841–851. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 13.Gaziel-Sovran A., Segura M. F., Di Micco R., Collins M. K., Hanniford D., Vega-Saenz D. M. E., Rakus J. F., Dankert J. F., Shang S., Kerbel R. S., Bhardwaj N., Shao Y., Darvishian F., Zavadil J., Erlebacher A., Mahal L. K., Osman I., Hernando E. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. (2011);20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y., Zhao F., Wang Z., Song Y., Luo Y., Zhang X., Jiang L., Sun Z., Miao Z., Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. (2012);31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M., Lang N., Qiu M., Xu F., Li Q., Tang Q., Chen J., Chen X., Zhang S., Liu Z., Zhou J., Zhu Y., Deng Y., Zheng Y., Bi F. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int. J. Cancer. (2011);128:1269–1279. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- 16.Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. (2008);27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 17.Weber D. G., Johnen G., Bryk O., Jockel K. H., Bruning T. Identification of miRNA-103 in the cellular fraction of human peripheral blood as a potential biomarker for malignant mesothelioma--a pilot study. PLoS One. (2012);7:e30221. doi: 10.1371/journal.pone.0030221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niquet J., Wasterlain C. G. Bim, Bad, and Bax: a deadly combination in epileptic seizures. J. Clin. Invest. (2004);113:960–962. doi: 10.1172/JCI21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danial N. N., Korsmeyer S. J. Cell death: critical control points. Cell. (2004);116:205–219. doi: 10.1016/S0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro G. I. Cyclin-dependent kinase pathways as targets for cancer treatment. J. Clin. Oncol. (2006);24:1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y. Induction of genetic instability by gain-offunction p53 cancer mutants. Oncogene. (2008);27:3501–3507. doi: 10.1038/sj.onc.1211023. [DOI] [PubMed] [Google Scholar]
- 22.Vousden K. H., Lane D. P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. (2007);8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]


