Abstract
Genome-wide association studies have revealed that many low-penetrance cancer susceptibility loci are located throughout the genome; however, a very limited number of genes have been identified so far. Using a forward genetics approach to map such loci in a mouse skin cancer model, we previously identified strong genetic loci conferring resistance to chemically induced skin papillomas on chromosome 4 and 7 with a large number of [(FVB/N × MSM/Ms) F1 × FVB/N] backcross mice. In this report, we describe a combination of congenic mapping and allele-specific alteration analysis of the loci on chromosome 4. We used linkage analysis and a congenic mouse strain, FVB.MSM-Stmm3 to refine the location of Stmm3 (Skin tumor modifier of MSM 3) locus within a physical interval of about 34 Mb on distal chromosome 4. In addition, we used patterns of allele-specific imbalances in tumors from N2 and N10 congenic mice to narrow down further the region of Stmm3 locus to a physical distance of about 25 Mb. Furthermore, immunohistochemical analysis showed papillomas from congenic mice had less proliferative activity. These results suggest that Stmm3 responsible genes may have an influence on papilloma formation in the two-stage skin carcinogenesis by regulating papilloma growth rather than development.
Keywords: allelic imbalance, congenic mice, DMBA-TPA, papilloma, tumor susceptibility
Introduction
Identification of the specific genetic variants responsible for increased susceptibility to familial or sporadic cancers remains an important but challenging goal with major implications for the prediction of individual cancer risk, as well as for improved strategies for prevention or targeted therapy [1, 2, 18, 19]. Present approaches to detect low-penetrance tumor-susceptibility alleles in humans involve association studies using DNA samples from hundreds or thousands of cancer patients, and an equal number of well-matched controls. Such studies are plagued by confounding factors such as population heterogeneity, weak effects, and genetic interactions, and require a very large numbers of cases and controls to reach statistical significance [9, 17, 21]. For many complex-trait diseases, including cancer, the total number of significant SNP associations detected can only account for a very small proportion of the total genetic risk [12], leading to considerable discussion of the best ways to discover the majority of disease-causing alleles in human populations.
For these reasons, complementary gene mapping and validation approaches including cross-species comparisons using animal models are required to identify genes that modify disease phenotypes such as skin cancer susceptibility [4, 10, 15]. Exploiting the resistance of M. spretus to the two-stage skin carcinogenesis model involving 7,12-dimethylbenz (a) anthracene (DMBA) initiation and subsequent promotion with 12-O-tetradecanoylphorbol-13-acetate (TPA), 15 skin tumor susceptibility loci, Skts1-15 (Skin tumor susceptibility1-15) were identified in an interspecific [(NIH/Ola × M. spretus) F1 × NIH/Ola] backcross mice using QTL analysis [6, 14]. In addition to Skts series, several other skin tumor modifier loci were identified using commonly used inbred strains or wild-derived strains. Skts-fp1-3 (Skin tumor susceptibility-fvb pwk1-3) were identified in a cross between a wild-derived inbred strain PWK and FVB/N [7]. Skts-fp1 was also identified in a study involving a cross between a wild-derived outbred stock of Mus musculus castaneus and FVB/N [8]. We previously reported mapping of loci conferring resistance to skin tumors, Stmm1 and 2 (Skin tumor modifier of MSM), on mouse chromosome 7 and Stmm3 on chromosome 4 by crossing a resistant Japanese wild-derived inbred strain MSM/Ms with a susceptible strain FVB/N [16]. In the present study, we used an interval-specific congenic mouse strain, FVB.MSM-Stmm3 to refine the location of Stmm3 within a physical interval of about 34 Mb on proximal chromosome 4. In addition, we used patterns of allele-specific imbalances in tumors from N2 and N10 congenic mice to narrow down Stmm3 locus further. We detected the region showing high frequencies of MSM allele loss or FVB allele gain, corresponding to a physical distance of about 25 Mb. These results suggest that Stmm3 responsible genes may have an influence on papilloma growth rather than development in the two-stage skin carcinogenesis.
Materials and Methods
Mice and tumor induction
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The protocol was approved by the Committee on the Ethics of Animal Experiments of Chiba Cancer Center (Permit Number: 13–18). All efforts were made to minimize suffering. FVB/N mice were purchased from Japan Clea. MSM/Ms mice have been maintained in the Experimental Animal Facility at Niigata University and Chiba Cancer Centre for more than 20 years. In a large [(FVB/N × MSM/Ms) F1 × FVB/N] backcross study, papilloma resistance loci were identified by QTL analysis [16]. Resistant [(FVB/N × MSM/Ms) F1 × FVB/N] backcross mice were selected for further backcrossing to FVB/N mice over at least 10 generations, and genotyped by D4SNP508, D4Mit17, D4Mit26, D4Mit255, D4Mit157, D4SNP7-1, and D4Mit254 (Fig. 1A), ultimately leading to congenic lines (4a) and (4b) of FVB.MSM-Stmm3 containing MSM allele of Stmm3 on the FVB/N background (Fig. 1A). These congenic mice were treated following the same skin tumor induction protocol, as reported previously [16]. In short, the mice (8–12 weeks) received a single dose of DMBA (25 µg per mouse with 200 µl of acetone) and, starting 1 week after initiation, the animals were subjected to TPA (200 µl of 0.1 mM solution in acetone) twice weekly for 20 weeks. Papilloma numbers were counted every week until 20 weeks after initiation.
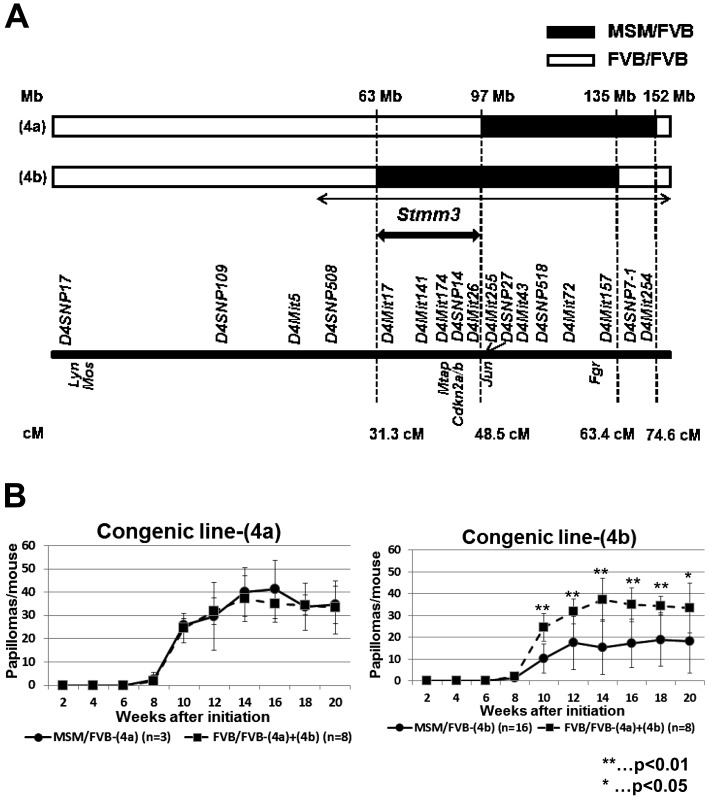
Fig. 1.
Genetic linkage map and papilloma incidence in congenic lines (4a) and (4b) on mouse chromosome 4. (A) A significant linkage peak, Stmm3 was mapped in the previous report [16]. Mouse chromosome 4 is shown horizontally. The black arrowed line indicates previously mapped Stmm3 region by QTL analysis. The bold black arrowed line indicates Stmm3 region refined by congenic mapping. Two black and white bars represents two congenic lines (4a) and (4b). The black bars indicate the heterozygous M/F region, while the white bars indicate the homozygous F/F region. Several well-known genes located on chromosome 4 are indicated with genetic markers. Genetic positions shown are according to the Ensembl database (http://uswest.ensembl.org/index.html), the Mouse Genome Informatics Database (http://www.informatics.jax.org/). (B) Comparison of average papilloma numbers/mouse between a congenic line (4a) and (4b). The panel on the left shows papilloma incidence in a congenic line (4a). The panel on the right shows papilloma incidence in a congenic line (4b). The black lines represent papilloma numbers of M/F heterozygous congenic mice. The dotted lines represent those of F/F heterozygous congenic mice.
DNA preparation, genotyping, and allelotyping using microsatellite markers
DNA was prepared from papillomas and corresponding normal tail tips and kidneys of N2 (n=12) and N10 congenic mice (n=10). Papillomas were selected from N2 mice which were FVB/FVB (F/F) homozygous for Stmm1 and 2 region on chromosome 7 and MSM/FVB (M/F) heterozygous for Stmm3 region on chromosome 4. Microsatellite markers were amplified by standard methods. Each marker’s order and distance were estimated from the Ensembl database (http://uswest.ensembl.org/index.html), the Mouse Genome Informatics Database (http://www.informatics.jax.org/), the NIG Mouse Genome Database (http://molossinus.lab.nig.ac.jp/msmdb/index.jsp), and the Mouse Microsatellite Database of Japan (http://www.shigen.nig.ac.jp/mouse/mmdbj/top.jsp). D4SNP markers are described in [16]. To determine susceptibility in congenic lines, unpaired two-tailed Student’s t-test was used. Differences of 50% or more in the intensity ratios of the two alleles in papilloma DNA relative to the corresponding level in kidney as a normal control were defined as allelic imbalance, as previously described [3, 16]. Percentages of allelic imbalances in different crosses were compared using 2 × 2 Chi square test (Fisher’s test).
Immunohistochemistry
Papillomas were fixed with 4% paraformaldehyde at 4°C overnight. The endogenous peroxidase activity in the specimens was blocked by treatment with 0.3% H2O2 and samples were then rinsed with PBS. Sections were incubated with primary antibodies diluted in blocking buffer overnight at 4°C and stained with rat anti-Ki67 (1:100, Dako) antibody and rabbit anti-keratin 14 (1:500, Covance Research). Secondary antibodies were Alexa Fluor 488-conjugated anti-rat antibody (1:100, Molecular Probes, Invitrogen) and Alexa Fluor 568-conjugated anti-rabbit antibody (1:100, Molecular Probes, Invitrogen). Nuclei were counterstained with Hard Set Mounting Medium with DAPI (Vector). All fluorescence images were obtained with a Leica TCS SPE confocal microscope equipped with a DMI4000B (10×/0.40, 20×/0.70, and 40×/1.25 oil immersion objective).
Results
Congenic analysis of Stmm3 (Skin tumor modifier of MSM) located on chromosome 4
We recently identified a series of skin papilloma resistance loci, Stmm (Skin tumor modifier of MSM) loci using [(FVB/N × MSM/Ms) F1 × FVB/N] backcross mice between a wild derived inbred mouse strain MSM/Ms and a susceptible inbred mouse strain FVB/N [16]. We detected two broad LOD score peaks, Stmm1 and 2 on chromosome 7 and one peak, Stmm3 on chromosome 4 [16] (Fig. 1A). To confirm the presence of low-penetrance susceptibility genes in the identified region on chromosome 4, we selected resistant [(FVB/N × MSM/Ms) F1 × FVB/N] backcross mice for further backcrossing to FVB/N mice and generated two N10 congenic mouse lines of FVB.MSM-Stmm3 covering the extended region including Stmm3. These two lines were termed lines (4a) and (4b) (Fig. 1A). First, these congenic lines were subjected to DMBA-TPA skin carcinogenesis experiment, according to the standard protocol and their papilloma development was monitored for a period of 20 weeks (Supplementary Table 1). As previously shown, FVB/N mice are highly susceptible to the two-stage skin chemical carcinogenesis. Similarly, homozygous F/F mice of congenic lines (4a) and (4b) (n=8) were highly susceptible to this carcinogenesis, developing 33.4 ± 11.3 papillomas at 20 weeks after initiation (Fig. 1B and Supplementary Table 1). Heterozygous M/F mice of a congenic line (4a), which were susceptible to papilloma, developed an average of 34.7 ± 8.1 papillomas/mouse at 20 weeks after initiation (n=3) (Fig. 1B and Supplementary Table 1). On the other hand, M/F heterozygous mice of a congenic line (4b) showed the strong suppressive effect on papilloma development. The number was an average of 18.1 ± 14.6 papillomas/mouse at 20 weeks (n=16, compared with control: P=0.0114, by t-test) after initiation (Fig. 1B and Supplementary Table 1). On the basis of skin carcinogenesis experiments with these two lines, the location of the Stmm3 region were narrowed down to an interval of about 34 Mb (indicated by the bold black arrowed line) from 63Mb to 97Mb on chromosome 4 (Fig. 1A) by excluding the region of the negative congenic line (4a) from the region of the positive congenic line (4b) (Fig. 1A and Supplementary Table 1).
The effect of Stmm3 on papilloma size
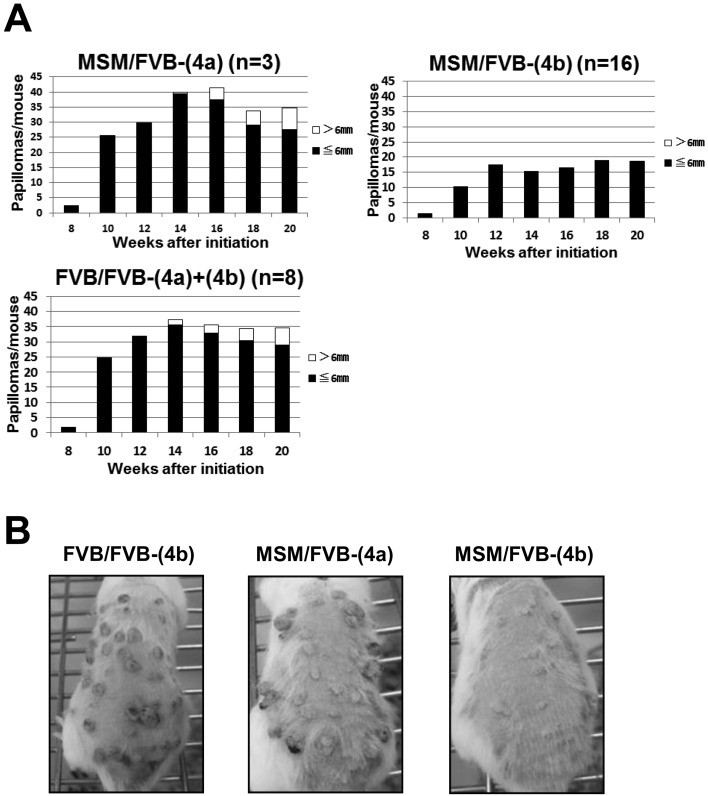
In the original report, we classified papillomas on the basis of their diameter, and carried out linkage analysis for each category. This analysis revealed strong linkage at Stmm3 on chromosome 4 to the number of papillomas > 6 mm. However, this linkage peak at Stmm3 completely disappeared when the analysis was confined to the number of papillomas ≤ 6 mm in diameter. We concluded that Stmm3 genes function only at the late stage of papillomas, but are not involved at the early stage of papillomas, such as papilloma development [16]. To confirm this stage-specific effect of Stmm3, we classified papillomas of congenic mice into two categories on the basis of size. As a result, M/F heterozygous mice of a congenic line (4a) developed 7.0 ± 6.1 papillomas > 6 mm, which was close to the number of papillomas > 6 mm of F/F homozygous mice at 20 weeks after initiation (Figs. 2A, B and Supplementary Table 1). Whereas, M/F heterozygous mice of a congenic line (4b) developed almost no papilloma > 6 mm (n=16; P=0.0000650, by t-test, compared with F/F homozygous mice, Supplementary Table 1). Furthermore, M/F heterozygous mice of a congenic line (4b) developed 18.0 ± 14.6 papillomas ≤ 6 mm, which was less than the number of papillomas ≤ 6 mm of F/F homozygous and M/F heterozygous mice of a line (4a). Although the difference was statistically significant, the suppressive effect on papilloma was much stronger in papillomas > 6 mm, than in ≤ 6 mm (n=16; P=0.0000650 vs 0.0455 by t-test, compared with F/F homozygous mice, Supplementary Table 1). Taken together, these results suggest Stmm3 conferred resistance to larger papillomas and showed a weaker suppressive effect on smaller papillomas. The conclusion in the original report was confirmed by congenic analysis that Stmm3 genes function only at the late stage of papillomas, but are not involved at the early stage of papillomas [16].
Fig. 2.
The effect of Stmm3 on papilloma growth. (A) Papilloma incidence of congenic lines (4a) and (4b) sorted according to size. The Black bars represent the number of papillomas ≤ 6 mm. The white bars represent the number of papillomas > 6 mm in diameter. (C) Photos of representative mice on TPA treatment. Dorsal back skin of a homozygous F/F mouse of a congenic line (4b), M/F heterozygous mice of congenic lines (4a) and (4b) at 20 weeks after initiation from left to right.
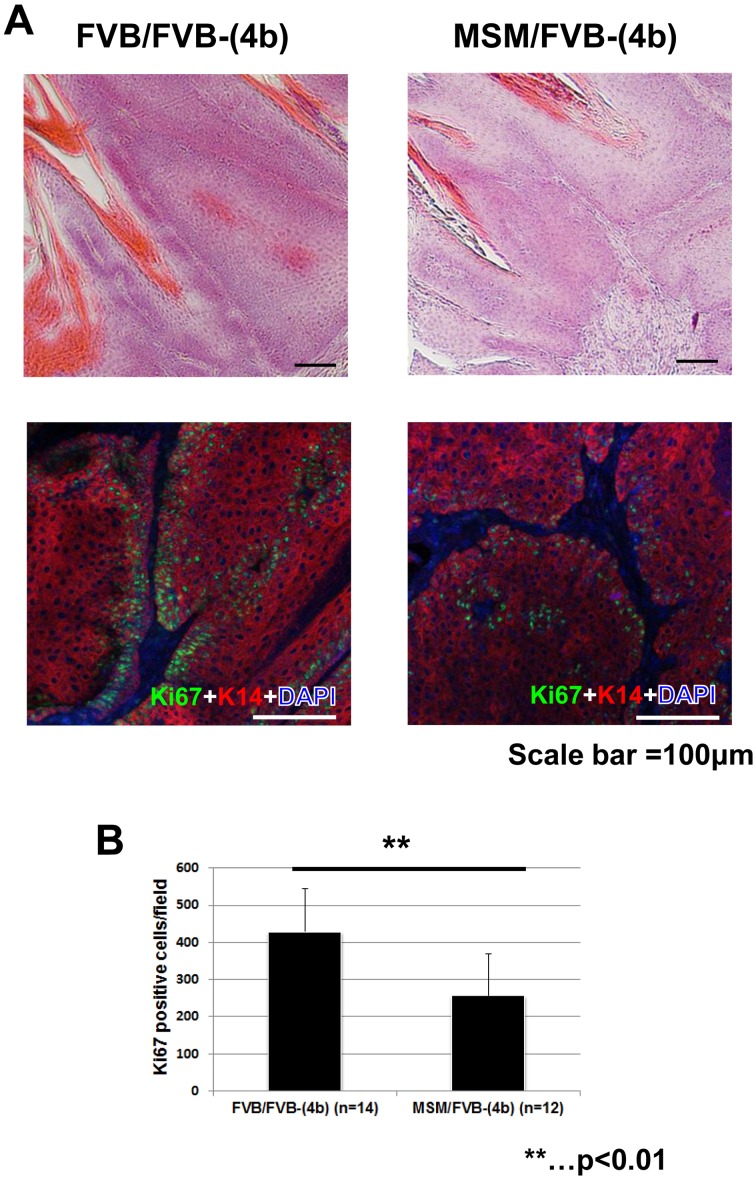
Next, histological analysis was performed using papillomas from congenic mice. HE (hematoxylin and eosin) staining didn’t show significant morphological changes between M/F heterozygous and F/F homozygous mice of a congenic line (4b) (Fig. 3A). We then carried out immunohistochemical analysis with the cell proliferation marker, Ki67. As a result, papillomas from M/F heterozygous mice of a congenic line (4b) showed decreased number of Ki67 positive cells, compared to papillomas from F/F homozygous mice of a congenic line (4b) Figs. 3A, B, P=0.000477, by t-test). These results suggest Stmm3 reduces proliferative cells in papillomas, which is compatible with the results of carcinogenesis experiments that Stmm3 reduces papilloma growth rather than development.
Fig. 3.
Stmm3 congenic mice show a decrease of proliferative cells in papillomas. (A) Histological analysis of papillomas from congenic mice. HE staining patterns of papillomas from F/F homozygous (left) and M/F heterozygous mice (right) of a congenic line (4b) are shown on the top. Double-immunostaining patterns of Ki67 (green) and Keratin 14 (K14) (red) in papillomas from F/F homozygous (left) and M/F heterozygous (right) mice of a congenic line (4b) are shown on the bottom. (B) The number of Ki67 positive cells in papillomas. The black bars represent the number of Ki67 positive cells in papillomas from F/F homozygous (left) and M/F heterozygous (right) mice of a congenic line (4b). The P-value was calculated for Ki67 positive cell number by t-test. Error bar represents standard deviation (S.D.). Scale bars;100 µm.
Allelic imbalances in favor of FVB/N were found around Stmm3 locus in skin tumors from N2 and N10 congenic mice
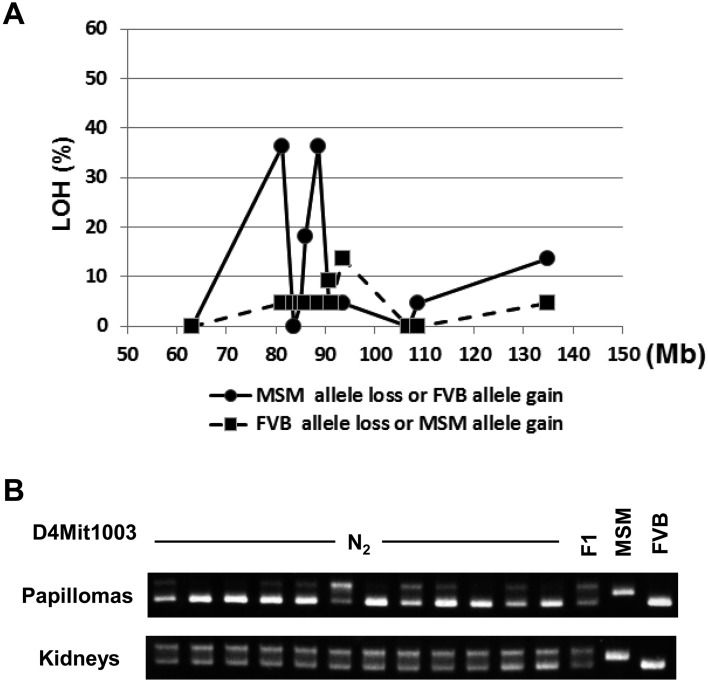
We carried out a detailed investigation of allelic imbalance on chromosome 4 to determine whether somatic change would allow more specific localization of Stmm3. This allelic imbalance analysis was performed using twelve informative microsatellite markers (for detailed information, see Supplementary Table 2). Fig. 4A shows combined results of allelic imbalance analysis of papillomas from N2 (n=12) and N10 congenic mice (n=10) around Stmm3 locus. An example of allelic imbalance analysis of papillomas from N2 mice (n=12) by a representative marker, D4Mit1003 is shown in Fig. 4B. Four papillomas showed losses of MSM alleles across the whole region, but others exhibited regional losses involving smaller chromosome fragments (data not shown). Maximal allele-specific imbalance was detected using marker D4Mit1003 and D4Mit26 located at approximately 81.0 Mb and 88.7 Mb, respectively. These markers are mapped within the minimal candidate region of 34 Mb identified with the two congenic lines of Stmm3 (Fig. 4A and Supplementary Table 2). The somatic mapping data allowed us to refine the interval containing Stmm3 to the shorter region. Two allelic imbalance peaks are detected (Fig. 4A and Supplementary Table 2). The proximal peak is the region from D4Mit17 (63.0 Mb) D4Mit1003 (81.3 Mb) D4Mit328 (83.6 Mb), the physical size is about 20.6 Mb. The distal peak is the region from D4Mit327 (86.1 Mb) D4Mit26 (88.7 Mb) D4Mit165 (90.7 Mb), the physical size is about 4.6 Mb (Fig. 4A and Supplementary Table 2). When these two regions are combined, the total physical size of the candidate region can be cut down to about 25 Mb. This interval still contains a large number of genes. Further congenic and somatic mapping will refine the candidate region for future gene identification step.
Fig. 4.
Allelic imbalance analysis of Stmm3 region on chromosome 4. (A) Frequency of allelic imbalance detected by SSLP (Simple Sequence Length Polymorphisms) analysis on distal chromosome 4. Data are derived from papillomas of N2 and N10 congenic mice (N2, n=12; N10, n=10). The percentages of chromosome imbalances are plotted for different microsatellite markers (for detailed information, see Supplementary Table 2). The black line represents MSM allele loss or FVB allele gain. The dotted line represents FVB allele loss or MSM allele gain. (B) SSLP analysis results for a representative microsatellite marker, D4Mit1003 of normal genomic (kidney) DNA and of DNA from twelve independent papillomas from different N2 mice. The upper bands represent the MSM allele, whereas the lower band represents the FVB allele.
Discussion
In this study, the location of Stmm3 was refined within a physical interval of about 34 Mb on chromosome 4 by using two congenic lines. By allelic alteration analysis of papillomas from N2 and N10 congenic mice, we further narrowed down the physical interval to about 25 Mb. Furthermore, we carried out immunohistochemical analysis by a proliferation marker, Ki67 using papillomas from congenic lines containing Stmm3. Interestingly, we observed a decrease of Ki67 positive cells papillomas in a congenic strain that is M/F heterozygous for Stmm3 on chromosome 4. Taken together, these results suggest that Stmm3 modifies papilloma growth rather than development.
As previously shown, we screened stage-specific papilloma modifier loci on the basis of the size. Stmm3 were identified on chromosome 4 as modifier loci conferring resistance to papillomas of larger size [16]. They were mapped on chromosome 4 and the effect was confirmed by congenic analysis in this study. As previously shown, mouse chromosome 4 contains several tumor modifier loci including Skts7 and Skts-fp1 mapped at 40–60 cM on chromosome 4 for chemically induced skin papilloma susceptibility by the analysis of NIH and Mus spretus cross, and FVB and PWK cross, respectively [7, 8, 14]. Common genes might be responsible for the susceptibility of these two and our crosses. PWK, Mus spretus and MSM/Ms, are wild derived inbred strains that could share common haplotypes. The haplotype analysis would help to identify a candidate gene in the near future. This region contains many QTLs for susceptibility to other types of tumors [5]. Pctr1 and 2 (Plasmacytoma resistance 1 and 2) were mapped as loci for susceptibility to pristane induced plasmacytoma [13, 20], and Cdkn2a has been suggested to be a corresponding gene for Pctr1 [22, 23]. Cdkn2a is also a strong candidate gene for Papg1 (Pulmonary adenoma progression 1), QTL for progression of urethane induced lung tumors [24]. Allelic imbalance analysis in this study showed Cdkn2a is localized in the peak region (around 88 Mb) showing loss of MSM allele (Fig. 4A). This could be a good candidate gene for late stage papilloma on chromosome 4 in our future study.
Koning et al. (2007) showed that low-penetrance susceptibility genes, even when present in a heterozygous state in congenic mice, can influence somatic genetic changes in tumors and that these alterations can be exploited for the rapid fine mapping of putative susceptibility loci [3]. On the basis of this hypothesis, to narrow down Stmm3 candidate region,we carried out allelic imbalance analysis of papillomas from [(FVB/N × MSM/Ms) F1 × FVB/N] backcross and M/F heterozygous mice of N10 congenic line (4b). As a result, we detected allelic imbalance showing the highest frequency in Stmm3 region (Fig. 4A). This clearly shows that one or more low-penetrance susceptibility genes in Stmm3 locus can affect somatic genetic change in tumors. Allelic alterations with the highest rate in favor of FVB alleles were detected within the minimal-overlapping region of Stmm3 congenic lines using the markers D4Mit1003 and D4Mit26, which are located at 81.3 and 88.7 Mb, respectively. The physical size of this region showing allelic alterations is about 25 Mb. However this region still contains a large number of genes, some of which (e.g. Cdkn2a, Mtap, Nfib, Cer1) are interesting candidates for future study.
Several lines of evidence come from studies on mouse skin tumor development where the concept that slowly dividing LRCs (Labell Retaining Cells) rather than rapidly proliferating TA (Transit Amplifying) cells are capable to expand during skin tumor promotion is long established [11]. On the basis of these concepts, we carried out a long term BrdU-LRC analysis using congenic mice containing Stmm1 on chromosome 7 and Stmm3 on chromosome 4 (unpublished data). Interestingly, Stmm1 congenic mice exhibited a significant reduction of BrdU-LRCs in the bulge of hair follicles, whereas Stmm3 congenic mice didn’t show any change. These results suggest Stmm1 gene could suppress papilloma formation by altering the behavior of adult epidermal quiescent stem cells in hair follicles. However, Ki67 staining of papilloms from Stmm3 congenic mice showed a significant reduction of proliferative cells. Taken together, these results indicate Stmm3 responsible genes confer resistance to late stage papillomas by regulating papilloma growth rather than development in the two-stage skin carcinogenesis.
In this report, we showed a combination of congenic mapping and allele-specific alteration analysis of Stmm3 on chromosome 4. We used linkage analysis and congenic mouse strains to refine the location of Stmm3 locus within a physical interval of about 34 Mb on distal chromosome 4. In addition, the region of Stmm3 locus was further narrowed down to a physical distance of about 25 Mb by using patterns of allele-specific imbalances in papillomas from N2 and N10 congenic mice. Combination of DMBA-TPA carcinogenesis and allelic imbalance analysis using congenic mice could facilitate gene identification and functional characterization of the gene responsible for Stmm3.
The authors declare no conflict of interest associated with this manuscript.
Supplementary Table
Acknowledgments
We would like to thank Dr. Minh To for critical reading of the manuscript. This work was supported by MEXT’s program “Promotion of Environmental Improvement for Independence of Young Researchers” under the Special Coordination Funds for Promoting Science and Technology at Niigata University, and Grants-in-Aid for Scientific Research (23790435) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- 1.Balmain A.2002. Cancer as a complex genetic trait: tumor susceptibility in humans and mouse models. Cell 108: 145–152. doi: 10.1016/S0092-8674(02)00622-0 [DOI] [PubMed] [Google Scholar]
- 2.Balmain A., Gray J.,, Ponder B.2003. The genetics and genomics of cancer. Nat. Genet. 33: (Suppl): 238–244. doi: 10.1038/ng1107 [DOI] [PubMed] [Google Scholar]
- 3.de Koning J.P., Wakabayashi Y., Nagase H., Mao J.H., Balmain A.2007. Convergence of congenic mapping and allele-specific alterations in tumors for the resolution of the Skts1 skin tumor susceptibility locus. Oncogene 26: 4171–4178. doi: 10.1038/sj.onc.1210206 [DOI] [PubMed] [Google Scholar]
- 4.DiGiovanni J., Imamoto A., Naito M., Walker S.E., Beltrán L., Chenicek K.J., Skow L.1992. Further genetic analyses of skin tumor promoter susceptibility using inbred and recombinant inbred mice. Carcinogenesis 13: 525–531. doi: 10.1093/carcin/13.4.525 [DOI] [PubMed] [Google Scholar]
- 5.Dragani T.A.2003. 10 years of mouse cancer modifier loci: human relevance. Cancer Res. 63: 3011–3018. [PubMed] [Google Scholar]
- 6.Ewart-Toland A., Briassouli P., de Koning J.P., Mao J.H., Yuan J., Chan F., MacCarthy-Morrogh L., Ponder B.A., Nagase H., Burn J., Ball S., Almeida M., Linardopoulos S., Balmain A.2003. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat. Genet. 34: 403–412. doi: 10.1038/ng1220 [DOI] [PubMed] [Google Scholar]
- 7.Fujiwara K., Igarashi J., Irahara N., Kimura M., Nagase H.2007. New chemically induced skin tumour susceptibility loci identified in a mouse backcross between FVB and dominant resistant PWK. BMC Genet. 8: 39. doi: 10.1186/1471-2156-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujiwara K., Wie B., Elliott R., Nagase H.2010. New outbred colony derived from Mus musculus castaneus to identify skin tumor susceptibility loci. Mol. Carcinog. 49: 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvan A., Ioannidis J.P., Dragani T.A.2010. Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends Genet. 26: 132–141. doi: 10.1016/j.tig.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter K.W., Crawford N.P.2008. The future of mouse QTL mapping to diagnose disease in mice in the age of whole-genome association studies. Annu. Rev. Genet. 42: 131–141. doi: 10.1146/annurev.genet.42.110807.091659 [DOI] [PubMed] [Google Scholar]
- 11.Kangsamaksin T., Park H.J., Trempus C.S., Morris R.J.2007. A perspective on murine keratinocyte stem cells as targets of chemically induced skin cancer. Mol. Carcinog. 46: 579–584. doi: 10.1002/mc.20355 [DOI] [PubMed] [Google Scholar]
- 12.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., Cho J.H., Guttmacher A.E., Kong A., Kruglyak L., Mardis E., Rotimi C.N., Slatkin M., Valle D., Whittemore A.S., Boehnke M., Clark A.G., Eichler E.E., Gibson G., Haines J.L., Mackay T.F., McCarroll S.A., Visscher P.M.2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. doi: 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mock B.A., Hartley J., Le Tissier P., Wax J.S., Potter M.1997. The plasmacytoma resistance gene, Pctr2, delays the onset of tumorigenesis and resides in the telomeric region of chromosome 4. Blood 90: 4092–4098. [PubMed] [Google Scholar]
- 14.Nagase H., Bryson S., Cordell H., Kemp C.J., Fee F., Balmain A.1995. Distinct genetic loci control development of benign and malignant skin tumours in mice. Nat. Genet. 10: 424–429. doi: 10.1038/ng0895-424 [DOI] [PubMed] [Google Scholar]
- 15.Nagase H., Mao J.H., de Koning J.P., Minami T., Balmain A.2001. Epistatic interactions between skin tumor modifier loci in interspecific (spretus/musculus) backcross mice. Cancer Res. 61: 1305–1308. [PubMed] [Google Scholar]
- 16.Okumura K., Sato M., Saito M., Miura I., Wakana S., Mao J.H., Miyasaka Y., Kominami R., Wakabayashi Y.2012. Independent genetic control of early and late stages of chemically induced skin tumors in a cross of a Japanese wild-derived inbred mouse strain, MSM/Ms. Carcinogenesis 33: 2260–2268. doi: 10.1093/carcin/bgs250 [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Losada J., Castellanos-Martín A., Mao J.H.2011. Cancer evolution and individual susceptibility. Integr. Biol. Camb. 3: 316–328. doi: 10.1039/c0ib00094a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peto J.2001. Cancer epidemiology in the last century and the next decade. Nature 411: 390–395. doi: 10.1038/35077256 [DOI] [PubMed] [Google Scholar]
- 19.Ponder B.A.2001. Cancer genetics. Nature 411: 336–341. doi: 10.1038/35077207 [DOI] [PubMed] [Google Scholar]
- 20.Potter M., Mushinski E.B., Wax J.S., Hartley J., Mock B.A.1994. Identification of two genes on chromosome 4 that determine resistance to plasmacytoma induction in mice. Cancer Res. 54: 969–975. [PubMed] [Google Scholar]
- 21.Stadler Z.K., Thom P., Robson M.E., Weitzel J.N., Kauff N.D., Hurley K.E., Devlin V., Gold B., Klein R.J., Offit K.2010. Genome-wide association studies of cancer. J. Clin. Oncol. 28: 4255–4267. doi: 10.1200/JCO.2009.25.7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S., Qian X., Redman C., Bliskovski V., Ramsay E.S., Lowy D.R., Mock B.A.2003. p16 INK4a gene promoter variation and differential binding of a repressor, the ras-responsive zinc-finger transcription factor, RREB. Oncogene 22: 2285–2295. doi: 10.1038/sj.onc.1206257 [DOI] [PubMed] [Google Scholar]
- 23.Zhang S., Ramsay E.S., Mock B.A.1998. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc. Natl. Acad. Sci. USA 95: 2429–2434. doi: 10.1073/pnas.95.5.2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Wang Y., Herzog C.R., Liu G., Lee H.W., DePinho R.A., You M.2002. A strong candidate gene for the Papg1 locus on mouse chromosome 4 affecting lung tumor progression. Oncogene 21: 5960–5966. doi: 10.1038/sj.onc.1205725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.