Abstract
Resveratrol has received considerable attention as a polyphenol with various biological effects such as anti-inflammatory, anti-oxidant, anti-mutagenic, anti-carcinogenic, and cardioprotective properties. As part of the overall safety assessment of HS-1793, a novel resveratrol analogue free from the restriction of metabolic instability and the high dose requirement of resveratrol, we assessed genotoxicity in three in vitro assays: a bacterial mutation assay, a comet assay, and a chromosomal aberration assay. In the bacterial reverse mutation assay, HS-1793 did not increase revertant colony numbers in S. typhimurium strains (TA98, TA100, TA1535 and TA1537) or an E. coli strain (WP2 uvrA) regardless of metabolic activation. HS-1793 showed no evidence of genotoxic activity such as DNA damage on L5178Y Tk+/− mouse lymphoma cells with or without the S9 mix in the in vitro comet assay. No statistically significant differences in the incidence of chromosomal aberrations following HS-1793 treatment was observed on Chinese hamster lung cells exposed with or without the S9 mix. These results provide additional evidence that HS-1793 is non-genotoxic at the dose tested in three standard tests and further supports the generally recognized as safe determination of HS-1793 during early drug development.
Keywords: Resveratrol, HS-1793, Bacterial mutation assay, Comet assay, Chromosomal aberration assay, Genotoxicity
INTRODUCTION
A variety of vegetable foods and beverages contain several non-flavonoid classes of phenolic compounds synthesized by plants in response to injury or fungal attack. Among them, resveratrol has been identified as the major active compound of stilbene phytoalexins and is presumed to be beneficial to human health. Resveratrol is found in a wide variety of plants, including grapes, peanuts, blueberry, bilberry, cranberry, lingonberry, and Polygonum cuspidatum (1). Resveratrol exhibits various biological effects such as anti-inflammatory, antioxidant, anti-mutagenic, anti-carcinogenic, and cardioprotective properties (2). In addition, resveratrol suppresses proliferation of various tumor cells, including lymphoid and myeloid cancers; breast, colon, pancreas, stomach, prostate, head and neck, ovary, liver, lung and cervical cancers; melanoma; and muscle (3). The chemopreventive and chemotherapeutic properties associated with resveratrol offer promise for designing new chemotherapeutic agents. Nevertheless, exposure to high doses of resveratrol is required to induce apoptosis in cancer cells because resveratrol is not a potent cytotoxic compound when compared with other chemotherapeutic drugs (4). The biological activity of resveratrol is also limited by its photosensitivity and metabolic instability. Thus, development of a potent resveratrol analogue may provide a feasible means of achieving an effective physiologic concentration.
The newly developed resveratrol analogue, HS-1793 (4-(6-hydroxy-2-naphthyl)-1,3-benzenediol) does not contain the unstable double bond found in resveratrol, and the positions of two of the three hydroxyl groups in the HS-1793 aromatic ring are also different (5). These characteristics make HS-1793 more metabolically stable, less photosensitive, and more potent than resveratrol (5-7). Several studies have demonstrated that HS-1793 has potent anti-cancer activity in various cancer cell lines such as human renal clear cell carcinoma Caki-1 cells (6), human leukemic U937 cells (7), murine breast cancer FM3A cells (8), human breast cancer MCF-7 cells (9) and human prostate cancer LNCap cells (10). HS-1793 also enhances anti-tumor immunity by reducing regulatory T cells in a mouse breast tumor model (11,12). HS-1793 may be a potential candidate for cancer treatment, however, it needs more information for efficacy as well as saftety.
The discovery of new drugs requires for safety and efficacy investigations before release into the market. A large number of molecules with great diversity can be rapidly synthesized by combinatorial chemistry and high throughput screening has enormously increased the scope and speed of biological assays for safety evaluations. The primary objective of a drug safety evaluation is to obtain toxicity information, which can be interpreted and/or extended to assess the health risk to the humans. The potential risks and benefits of the drugs under study are carefully considered, so the benefits of using a new drug as a therapeutic agent outweigh the risks and side effects (13). Genotoxicity testing is also an important component of the drug development process and is particularly important for a potential chemopreventive agent if the agent is to be administered for long periods of time. It is generally used for hazard identification with respect to DNA damage and its fixation. This damage can be manifested in the form of gene mutation, structural chromosomal aberration, recombination, and numerical changes. These changes are responsible for heritable effects on germ cells and impose risks to future generations (14). In addition, it has been well documented that somatic mutations also play an important role in malignancy (15). These tests have been used mainly for predicting carcinogenicity and genotoxicity because compounds that are positive in these tests have the potential to be human carcinogens and/or mutagens.
Therefore, the present study evaluated the in vitro genotoxicity potential of HS-1793 with bacterial reverse mutation, chromosomal aberration, and comet assays, which are the simplified standard methods often followed for genotoxicity testing.
MATERIALS AND METHODS
Chemicals and reagents. The clastogens used in this study were: methyl methansulfonate (MMS, CAS# 66-27-3), benzo(A)pryene (BaP, CAS#50-32-8), cyclophosphamide (CPA, CAS# 6055-19-2), mitomycin C (MMC, CAS# 50-07-7), 2-(2-furyl)-3-(5-nitro-2-furyl) acrylamide (AF-2, CAS# 153-78-6), sodium azide (NaN3, CAS# 26628-22-8), 9-aminoacridine (9-AA, CAS# 90-45-9), and 2-aminoanthracene (2-AA, CAS# 613-13-8). All chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA). S9 was obtained from Molecular Toxicology Inc. (Boone, NC, USA) and the cofactor was obtained from Oriental Yeast Co., Ltd. (Tokyo, Japan). MMS, BaP, AF-2, 9-AA, and 2-AA were dissolved in dimethyl sulfoxide (DMSO), whereas while CPA, MMC and NaN3 were dissolved with distilled water. All clastogens were prepared fresh for each experiment. A system for metabolic activation (S9 mix) consisted of rat liver S9 and necessary cofactors.
Preparation of the HS-1793, resveratrol analogue. To obtain HS-1793, the stilbene double bond present in resveratrol was substituted with a naphthalene ring as described previously (9,10) and the synthesis of HS-1793 is summarized in Fig. 1. For the in vitro studies, HS-1793 was freshly prepared in DMSO on the day of use.
Fig. 1. Synthesis and chemical structure of resveratrol analogs, HS-1793.
Cell culture. Chinese hamster lung fibroblast cell line (CHL) and L5178Y Tk+/− mouse lymphoma cells were obtained from the American Type Culture Collection (Manassas, VA, USA). CHL cells were maintained in Eagle’s minimum essential medium (GIBCO/Invitrogen, Carlsbad, CA, USA) supplemented with 100 U of penicillin-streptomycin/ml and 10% heat-inactivated fetal bovine serum (GIBCO/Invitrogen) at 37℃ in a 5% CO2 atmosphere. The doubling time was about 13 hr and the modal chromosome number was 25. L5178Y Tk+/− mouse lymphoma cells were cultured in RPMI 1640 medium supplemented with 100 U of penicillin-streptomycin/ml, 200 μg/ml sodium pyruvate, 500 μg/ml pluronic F68, and 10% heat-inactivated fetal bovine serum (GIBCO/Invitrogen) at 37℃ in a 5% CO2 atmosphere.
Bacterial reverse mutation assay. The Ames test was carried out following the recommendations of Maron and Ames (16) and according to the Organization for Economic Co-operation and Development (OECD) guideline number 471 (17), by pre-incubating in the presence or absence of S9 mix. The Salmonella typhimurium strains (TA98, TA100, TA1535, and TA1537) and the Escherichia coli strain (WP2 uvrA) were purchased from Molecular Toxicology Inc. HS-1793 in diluted in DMSO was prepared as test samples, and DMSO was used as a negative control. AF-2, 2-AA, 9-AA and NaN3 were used as the positive controls for the test at 0.01~80 μg/plate. Each sample was assayed in triplicate. To a test tube, 0.5 ml of 0.1 M sodium phosphate buffer (pH 7.4) or 0.5 ml of S9 mix (10% S9 and salt-Cofactor-I solution), and 0.1 ml of bacterial culture were added. After adding 0.1 ml of the test chemical suspension, the solution was incubated for 20 min at 37℃ with shaking (140 rpm). After the incubation, 2.0 ml of top agar was mixed gently with the pre-incubated solution and poured onto a minimal glucose agar plate. The top agar consisted of 0.5% NaCl and 0.6% BactoTM Agar (Becton, Dickinson, Fullerton, CA, USA) in distilled water supplemented with 0.05 mM histidine and 0.05 mM biotin for the test that used S. typhimurium or 0.05 mM tryptophan for the test that used E. coli. Each culture plate was placed in a incubator for 48 hr at 37℃. After the incubation, the number of revertant colonies was counted. Different concentrations of 2-AA for TA98, TA100, TA1535, TA1537, and WP2uvrA were used with or without metabolic activation. Positive controls of each bacterial strain in the presence and absence of metabolic activation are shown in Tables 1 and 2. The test substance was considered positive in the bacterial reverse mutation assay when there was (a) an increase (≥ two-fold) of spontaneous revertants compared with those in the negative control or (b) a dosedependent increase of revertant colonies in at least one of the tester strains without cytotoxicity.
Table 1. Results of bacterial reverse mutation assay by HS-1793 treatment without S9 activation.
| Dose (μg/plate) | Number of reverse mutants/plate (Mean ± SD) | ||||
|---|---|---|---|---|---|
|
| |||||
| Base substitution | Frame shift | ||||
|
|
|
||||
| TA100 | TA1535 | E. coli | TA98 | TA1537 | |
|
| |||||
| 0 | 122 ± 10 | 16 ± 5 | 35 ± 2 | 28 ± 3 | 15 ± 4 |
| 78.1 | 121 ± 8 | 15 ± 2 | 39 ± 3 | 29 ± 2 | 14 ± 2 |
| 156.3 | 117 ± 11 | 17 ± 2 | 33 ± 3 | 27 ± 2 | 17 ± 6 |
| 312.5 | 116 ± 9 | 18 ± 2 | 37 ± 2 | 30 ± 4 | 13 ± 4 |
| 625 | 124 ± 6 | 17 ± 5 | 36 ± 2 | 26 ± 3 | 14 ± 3 |
| 1,250 | 107 ± 12 | 17 ± 2 | 36 ± 3 | 25 ± 2 | 15 ± 6 |
| 2,500 | 108 ± 6 | 18 ± 4 | 32 ± 1 | 28 ± 1 | 18 ± 1 |
| 5,000 | 119 ± 12 | 17 ± 3 | 36 ± 2 | 27 ± 3 | 13 ± 2 |
|
| |||||
| Positive control (μg/plate) | AF-2 (0.01) | NaN3 (0.5) | AF-2 (0.01) | AF-2 (0.1) | 9-AA (80) |
|
| |||||
| 425 ± 23* | 151 ± 17* | 334 ± 23* | 278 ± 20* | 151 ± 11* | |
AF-2: 2-(2-furyl)-3-(5-nitro-2-furyl) acrylamide; NaN3: Sodium azide; 9-AA: 9-aminoacridine.
Three independent assays were performed and SD represents standard deviation. * p<0.05 vs. control.
Table 2. Results of bacterial reverse mutation assay by HS-1793 treatment with S9 activation.
| Dose (μg/plate) | Number of reverse mutants/plate (Mean ± SD) | ||||
|---|---|---|---|---|---|
|
| |||||
| Base substitution | Frame shift | ||||
|
|
|
||||
| TA100 | TA1535 | E. coli | TA98 | TA1537 | |
|
| |||||
| 0 | 114 ± 12 | 20 ± 4 | 41 ± 4 | 36 ± 4 | 17 ± 3 |
| 78.1 | 116 ± 13 | 24 ± 3 | 39 ± 3 | 40 ± 4 | 16 ± 2 |
| 156.3 | 109 ± 8 | 22 ± 3 | 40 ± 2 | 368± 4 | 14 ± 2 |
| 312.5 | 114 ± 4 | 23 ± 4 | 42 ± 3 | 43 ± 8 | 15 ± 5 |
| 625 | 106 ± 19 | 21 ± 4 | 38 ± 3 | 42 ± 4 | 14 ± 2 |
| 1,250 | 108 ± 14 | 21 ± 3 | 37 ± 2 | 43 ± 4 | 14 ± 1 |
| 2,500 | 118 ± 11 | 20 ± 2 | 43 ± 3 | 44 ± 4 | 16 ± 3 |
| 5,000 | 106 ± 9 | 21 ± 6 | 40 ± 2 | 33 ± 2 | 15 ± 2 |
|
| |||||
| Positive control (μg/plate) | 2-AA (1.0) | 2-AA (2.0) | 2-AA (10) | 2-AA (0.5) | 2-AA (2.0) |
|
| |||||
| 598 ± 20* | 156 ± 6* | 428 ± 21* | 232 ± 9* | 118 ± 16* | |
2AA: 2-aminoanthracene.
Three independent assays were performed and SD represents standard deviation. * p<0.05 vs. control.
Cytotoxicity assay. The number of viable cells was determined by the ability of mitochondria to convert MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide, Sigma-Aldrich) to formazan dye. L5178Y Tk+/− mouse lymphoma cells and CHL cells were cultured overnight in 96 well plates, at a density of 1 × 104 cells/200 μl in each well. The next day, L5178Y Tk+/− mouse lymphoma cells and CHL cells were treated with various concentrations of HS-1793 for 24 hr. Following the incubation, the medium was removed and the cells were supplemented with fresh medium containing MTT solution (final 0.5 mg/ml) into each well. Following incubation for another 4 hr at 37℃ in a humidified 5% CO2 atmosphere, the MTT was removed, and cells were lysed with 150 μl DMSO. Absorbance was measured at 550 nm using a microplate reader (VersaMax Molecular Devices, Sunnyvale, CA, USA).
In vitro comet assay. DNA damages were detected by the alkaline version of standard comet assay described by Singh et al. (18) with some modifications (19). The comet assay was performed with L5178Y Tk+/− mouse lymphoma cells. Cells were incubated at a concentration of 1.5 × 105 cells/ml in 2 ml per well in a 12-well plate. The cells were then exposed with or without HS-1793 for 4 hr. DMSO was used as a negative control. The positive clastogens (15 μg/ml MMS without S9 mix, or 35 μg/ml Bap with S9 mix) were treated for 4 hr before harvesting for the comet assay. The cells were collected and mixed with low melting point agarose at 37℃. This mixture was placed on the top of the previous layer of 0.5% normal melting point (NMP) agarose on the slide, then covered with a coverslip and returned to 4℃ until solid. The coverslip was gently removed and some NMP agarose was added onto the slide. The slide was then covered again with a coverslip and placed at 4℃ until the mixture was solid. The slide was placed in chilled lysis buffer (100 mM EDTA, 2.5 M sodium chloride, 10 mM Trizma base and 1% N-lauroylsarcosinate, adjusted to pH 10.0, with 1% Triton X-100) and unwinding buffer (1 mM EDTA and 300 mM sodium hydroxide, pH > 13), respectively, and subjected to electrophoresis. Thereafter, the slides were gently washed with 0.4 M Tris buffer, stained with Gel green DNA dye (Biotium, Inc., Hayward, CA, USA), and visualized and analyzed under a fluorescence microscope (Carl Zeiss, Oberkochen, Germany). The images were captured, and a minimum of 100 comets per slide, in triplicates for a group, were analyzed using the Metafer 4 software (MetaSystems, Carl Zeiss) which gives % DNA in tail, tail length, tail moment (TM), and olive moment (OM) directly. The parameter TM is the product of tail length and % DNA in tail, and OM is the product of the distance between the center of the head and the center of the tail and % DNA in tail.
In vitro chromosomal aberration assay. Chromosome aberration tests were performed on CHL cells. The cells were plated at 1 × 104 cells/dish and incubated for 24 hr. The culture medium was then replaced with fresh medium including a test substance. DMSO was used as a negative control. Preliminary tests for cell growth inhibition and chromosomal aberrations were performed to determine an appropriate concentration range of the test substance for the chromosomal aberrations assay. The concentrations were set within range including 50% inhibition dose. For shortterm treatment, cells were treated with each test substance for 6 hr with either additional culture medium (without metabolic activation) or the S9 mix at a final concentration of 3% (with metabolic activation). After a 6 hr exposure, the cells were washed and incubated in fresh medium for a further 18 hr. For continuous treatment, the cells were treated with each dose for 24 hr. Colcemid was added (10 μg/ml final concentration) 2 hr prior to harvesting. The cells were then harvested, swollen in 0.075 M KCl solution, fixed in methanol: acetic acid, placed on clean glass slides and Giemsa (5%) stained. The images were captured, and one hundred metaphases per slide, were analyzed using the Metafer 4 software. Two hundred metaphases per a group were analyzed for structural chromosomal aberrations (20). The incidences of chromatid/chromosome gaps and numerical aberrations were recorded, but these incidental observations were not classified as chromosomal aberrations. Results were evaluated according to the following criteria (21): negative, < 5%; uncertain, 5~10%; positive, > 10% for structural or numerical chromosomal aberration frequencies. In addition, statistical significance was tested among groups.
Statistical analysis. All data are expressed as mean and standard deviation. Statistical significance was tested using the Statistical Package for the Social Sciences statistical software for Windows, ver. 18.0 (SPSS Inc., Chicago, IL, USA). Data were tested for normality using the Kolmogorov-Smirnov test and for homogeneity of variance using Levene’s test, prior to any statistical analysis. The data were normally distributed, and the variances were homogeneous. Therefore, significant differences between two groups were evaluated by Student’s t-test and significant differences among more than two groups were evaluated by one-way analysis of variance with Dunnett’s post hoc test for multiple comparisons. A difference was considered significant at p < 0.05.
RESULTS
Bacterial reverse mutation assay. The results of the bacterial reverse mutation tests with the five bacterial strains for HS-1793 are summarized in Tables 1 and 2. A toxicity test was performed in the presence or absence of metabolic activation with the dosages ranging from 78.1~5,000 μg/plate at a common ratio of approximately 2. Toxicity to all tester strains was observed using the pre-incubation method. Evidence of HS-1793 related toxicity, as indicated by reduced revertant numbers and/or thinning of the background lawn was not observed at 5,000 μg/plate with S. typhimurium strains (TA98, TA100, TA1535, and TA1537) or E. coli strain (WP2 uvrA) in the absence or presence of S9 metabolic activation (data not shown). Based on the toxicity test results, the main test was performed with dose ranges at a common ratio of approximately 2 and the results are shown in Tables 1 and 2. No increases in revertant frequencies were observed at any dose of HS-1793 in any tester strains with or without the S9 mix compared to the concurrent vehicle control cultures (Tables 1 and 2). The positive control chemicals for each tester strain induced at least a two-fold increase in the number of revertant colonies compared to the solvent control. Solvent and positive control values were distributed within the historical control ranges obtained in our laboratory. These results suggest that HS-1793 did not induce genotoxicity during bacterial reverse mutation under the conditions used in this study.
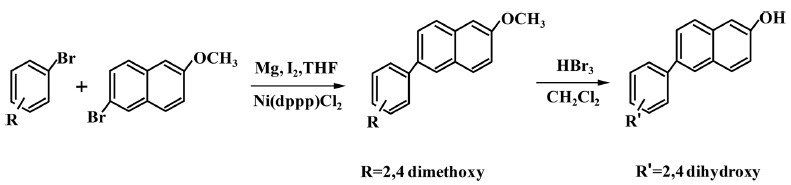
Comet assay. The results of in vitro comet assay with L5178Y Tk+/− mouse lymphoma cells for HS-1793 are summarized in Fig. 2 and Table 3. In the dose-range study for HS-1793, cell viability decreased in dose dependent manner during the 24 hr treatments. To exclude DNA damage by HS-1793 induced cytotoxicity, no reduction of viable cell numbers > 70% was observed at 62.5 μg/ml for 4 hr (Fig. 2A). Based on the cell viability results, the comet assay was performed with the dose ranges at a common ratio of approximately 2, and the results are shown in Table 3. No significant differences in % DNA in the tail, tail length, TM, or OM of L5178Y Tk+/− mouse lymphoma cells were observed between the vehicle and HS-1793 treated groups in the presence or absence of metabolic activation. The comet parameters of cells treated with positive control (MMS without S9 mix and Bap with S9 mix) increased significantly when compared to that of the vehicle (p < 0.05, Table 3 and Fig. 2B). These results indicated that HS-1793 did not induce genotoxicity as DNA damage in L5178Y Tk+/− mouse lymphoma cells under the conditions used in this assay.
Fig. 2. Cytotoxicity and genotoxicity of HS-1793 by the comet assay in L5178Y Tk+/− mouse lymphoma cells. (A) The cells were treated with the indicated concentrations of HS-1793 for 4 hr. Cell viability was determined by the MTT assay. Each percent value in treated cells was calculated with respect to that in the untreated control. All samples were run in triplicate and experiments were repeated three times. Results are expressed as percentages of control, and data are means ± standard deviations of three independent experiments. (B) Photomicrograph (400×) of comet in L5178Y Tk+/− mouse lymphoma cells with or without the S9 mix after a 4 hr exposure to HS-1793.
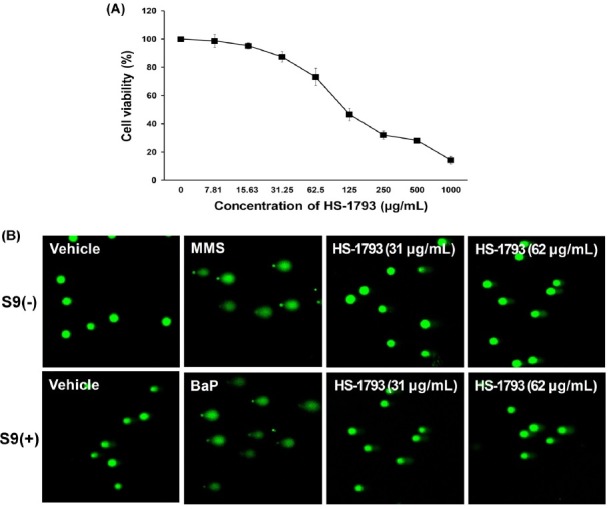
Table 3. Results of in vitro comet assay by HS-1793 treatment without and with S9 activation in L5178Y Tk+/− mouse lymphoma cells.
| S9 Mix | Dose (μg/ml) | % DNA in tail | Tail length (μm) | Tail moment (μm) | Olive moment (μm) |
|---|---|---|---|---|---|
|
| |||||
| (−) | 0 | 1.82 ± 0.21 | 1.91 ± 0.47 | 0.14 ± 0.04 | 0.83 ± 0.21 |
| 3.9 | 0.93 ± 0.21 | 1.49 ± 0.36 | 0.03 ± 0.01 | 0.42 ± 0.03 | |
| 7.8 | 1.09 ± 0.37 | 1.98 ± 0.27 | 0.07 ± 0.02 | 0.62 ± 0.05 | |
| 15.5 | 1.61 ± 0.49 | 2.11 ± 0.41 | 0.13 ± 0.03 | 0.75 ± 0.07 | |
| 31 | 2.13 ± 0.36 | 2.75 ± 0.37 | 0.16 ± 0.03 | 0.98 ± 0.09 | |
| 62 | 2.52 ± 0.34 | 2.82 ± 0.46 | 0.21 ± 0.05 | 1.01 ± 0.07 | |
|
| |||||
| MMS | 15 | 51.29 ± 4.26* | 97.21 ± 6.75* | 58.13 ± 6.12* | 14.68 ± 1.01* |
|
| |||||
| (+) | 0 | 1.34 ± 0.21 | 1.04 ± 0.06 | 0.11 ± 0.03 | 0.64 ± 0.16 |
| 3.9 | 0.98 ± 0.13 | 0.86 ± 0.04 | 0.04 ± 0.01 | 0.41 ± 0.04 | |
| 7.8 | 1.16 ± 0.08 | 0.95 ± 0.13 | 0.06 ± 0.01 | 0.56 ± 0.06 | |
| 15.5 | 1.42 ± 0.16 | 1.04 ± 0.06 | 0.13 ± 0.02 | 0.78 ± 0.12 | |
| 31 | 1.58 ± 0.11 | 1.17 ± 0.07 | 0.09 ± 0.02 | 0.80 ± 0.17 | |
| 62 | 1.65 ± 0.09 | 1.20 ± 0.03 | 0.15 ± 0.01 | 0.85 ± 0.15 | |
| BaP | 35 | 49.14 ± 3.29** | 80.29 ± 6.24** | 42.15 ± 3.98** | 13.74 ± 1.97** |
Postive control: MMS (methyl methansulfonate), BaP (Benzo(A)Pryene).
Three independent assays were performed and SD represents standard deviation. *, ** p < 0.05 and p < 0.01, respectively vs. control.
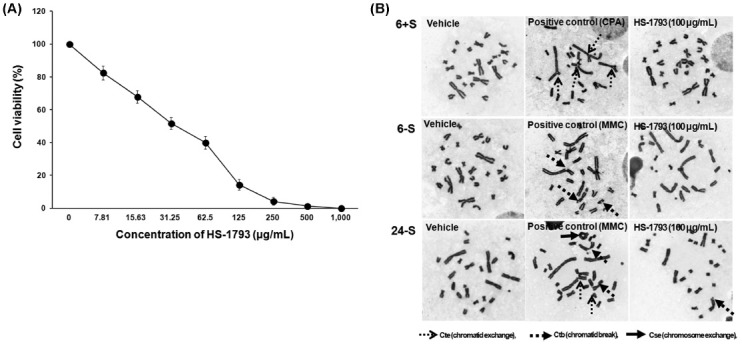
Chromosomal aberration assay. Results of the HS-1793 in vitro chromosomal aberration test are shown in Tables 4 and 5. HS-1793 was tested at concentrations ranging from 7.81 to 1,000 μg/ml and cell viability decreased in dose dependent manner during the 24 hr treatments (Fig. 3A). The IC50 was calculated as 118.7 μg/ml for the 24 hr treatment and the chromosomal aberration test was performed at a common ratio of approximately 2 based on the IC50. As shown Table 4, HS-1793 did not induce a significant increase in the incidence of structural or numerical chromosomal aberrations after short term treatment for 6 hr at doses of 6.25, 12.5, 25, 50 and 100 μg/ml in the presence or absence of the S9 mix. In addition, HS-1793 did not induce a significant increase in the incidence of structural or numerical chromosomal aberrations after continuous treatment for 24 hr at concentrations of 6.25, 12.5, 25, 50 and 100 μg/ml in the absence of the S9 mix (Table 5 and Fig. 3B). Both the positive controls, containing either MMC for treatment without metabolic activation or CPA for treatment with metabolic activation, resulted in an increase in the frequency of cells with > 10% chromosomal aberration, supporting the validity of the study. Our data show that HS-1793 did not induce chromosomal aberrations in CHL cells under the conditions of the study.
Table 4. Results of the chromosome aberration assay by short term treatment of HS-1793 treatment in CHL cells.
| S9 Mix/Treatment time | Dose (μg/ml) | Growth index (%) | Structural aberrations | Numerical aberrations | Gap | Number of total cell | Total frequency (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Chromosome type | Chromatid type | Others | PP + ER | (−) Gap | (+) Gap | |||||||
|
|
|
|||||||||||
| csb | cse | ctb | cte | |||||||||
|
| ||||||||||||
| (−)/6~18 hr | 0 | 100 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 200 | 0.5 | 1 |
| 6.25 | 98 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 200 | 1 | 1.5 | |
| 12.5 | 90 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 200 | 1 | 2 | |
| 25 | 84 | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 200 | 1 | 2 | |
| 50 | 72 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 200 | 1 | 1.5 | |
| 100 | 65 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 200 | 0.5 | 1 | |
| MMC | 0.5 | - | 5 | 3 | 53 | 96 | 0 | 0 | 22 | 200 | 78.5* | 89.5* |
| (+)/6~18 hr | 0 | 100 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 200 | 1 | 2 |
| 6.25 | 98 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 200 | 1 | 1.5 | |
| 12.5 | 95 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 200 | 0.5 | 1 | |
| 25 | 89 | 0 | 0 | 2 | 3 | 0 | 0 | 2 | 200 | 2.5 | 3.5 | |
| 50 | 2 | 0 | 0 | 3 | 2 | 0 | 0 | 1 | 200 | 2.5 | 3 | |
| 100 | 71 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 200 | 0.5 | 1.5 | |
| CPA | 10 | - | 1 | 5 | 59 | 166 | 0 | 0 | 33 | 200 | 116** | 132** |
Postive control: MMC (mitomycin C), CPA (cyclophosphamide).
ctb: chromatid break; cte: chromatid exchange; csb; chromosome break; cse: chromosome exchange;: pol: polyploidy; end: endo-reduplication.
Three independent assays were performed and SD represents standard deviation. *, ** p < 0.05 and p < 0.01, respectively vs. control.
Table 5. Results of the chromosome aberration assay by continuous treatment of HS-1793 in CHL cells.
| S9 Mix/Treatment time | Dose (μg/ml) | Growth index (%) | Structural aberrations | Numerical aberrations | Gap | Number of total cell | Total frequency (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Chromosome type | Chromatid type | Others | PP + ER | (−) Gap | (+) Gap | |||||||
|
|
|
|||||||||||
| csb | cse | ctb | cte | |||||||||
|
| ||||||||||||
| (−)/24~0 hr | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 200 | 0 | 0 |
| 6.25 | 93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 200 | 0 | 0 | |
| 12.5 | 85 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 200 | 1 | 1 | |
| 25 | 79 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 200 | 0.5 | 0.5 | |
| 50 | 72 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 200 | 1.5 | 2 | |
| 100 | 58 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 200 | 2.5 | 1.5 | |
| MMC | 0.5 | - | 3 | 12 | 15 | 181 | 0 | 0 | 8 | 200 | 106* | 109.5* |
Postive control: MMC (mitomycin C).
ctb: chromatid break; cte: chromatid exchange; csb: chromosome break; cse: chromosome exchange; pol: polyploidy; end: endo-reduplication.
Three independent assays were performed and SD represents standard deviation. * p < 0.05 vs. control.
Fig. 3. Cytotoxicity and genotoxicity of HS-1793 by chromosomal aberration assay in CHL cells. (A) The cells were treated with the indicated concentrations of HS-1793 for 24 hr. Cell viability was determined by the MTT assay. Each percent value in treated cells was calculated with respect to that in the untreated control. All samples were run in triplicate and experiments were repeated three times. Results are expressed as percentages of control, and data are means ± standard deviations of three independent experiments. (B) Photomicrograph (400×) of chromosomal aberration in CHL cells with or without the S9 mix after a 6 or 24 hr exposure to HS-1793.
DISCUSSION
We report the findings of genotoxicity testing conducted on the novel resveratrol analogue HS-1793 using a bacterial mutation assay, in vitro comet assay, and in vitro chromosomal aberration assay. HS-1793 showed negative results in the three established genotoxicity tests.
The bacterial reverse mutation assay (Ames assay) is primarily used to determine the mutagenic potential of various compounds in a prokaryote organism and whether the reactive metabolite is a product of metabolic activation by mammalian enzymes (22). In the present study, the mutagenic effect of HS-1793 was assessed in the TA98, TA100, TA1535 and TA1537 S. typhimurium tester strains, and E. coli WP2 uvrA in the presence and absence of S9 metabolic activation. Toxicity was evaluated based on a decrease in the number of revertant colonies and/or thinning of the bacterial lawn. HS-1793 did not produce an increase in the number of revertant colonies tested with or without the S9 mix in S. typhimurium strains TA98 and TA1537, which are strains indicative of a frame shift mutation. In addition, HS-1793 produced negative results in the TA100, TA1535 and WP2uvrA strains that specifically detect base pair point mutations. The concurrent positive controls demonstrated the sensitivity of the assay, and the mean revertant colony counts for the vehicle controls were within 99% confidence limits of the current historical control range of our laboratory. These results were consistent with the study results of Matsuoka et al. (21), who assessed the genotoxic potential of resveratrol. Resveratrol was negative in the bacterial reverse mutation assay with S. typhimurium strain TA100 and TA98, as well as the E. coli strain WP2uvrA strain tested in the presence and absence of the S9 mix at doses of 0.02~5000 μg per plate (23). Our results suggest no evidence of bacterial reverse mutagenicity of HS-1793 at concentrations up to 5000 μg/plate.
Although negative results were observed in the Ames test, further testing was conducted using in vitro mammalian assays to confirm these results. DNA damage considered relevant to mammalian cells cannot be adequately measured in bacteria and should be evaluated in mammalian cells for investigations on cytogenetic effects. Several mammalian cell systems are used to detect both DNA damage and gross chromosomal damage in in vitro test systems (24). Certain chemical substances induce the risk of other diseases or the individual chemical is even demonstrated to be carcinogenic or to cause damage to DNA (25). The genetic damage that a cell can incur, the damage response mechanisms available to cells and organisms, and the potential consequences of such damage can be referred to as cell genotoxicity. Genotoxic agents or genotoxins are those substances causing genotoxicity in a cell.
The comet assay has high sensitivity and low specificity for the association between genotoxicity test findings and rodent carcinogenicity outcomes. Genotoxicity with special reference to DNA damage in a cell shows comet formation after mini-gel electrophoresis. DNA fragments form a comet as a result of DNA damage in a cell (26). When viewed under a microscope, the cell has the appearance of a comet with a head, i.e., the nuclear region and the tail contain DNA fragments in strands (27). The formation of comets indicates primary DNA lesion leading to genetic damage in the cell. In particular, this low specificity of the comet assay is associated with cytotoxicity (28). Thus, to exclude the possibility of HS-1793 induced DNA damage resulting from cytotoxicity, viability was determined using the MTT assay. In this study, we determined the concentrations of HS-1793 that did not affect cell survival in L5178Y Tk+/− mouse lymphoma cells for the comet assay. The results demonstrate no significant induction of DNA damage after a 4 hr exposure to HS-1793 with or without the S9 mix. The concurrent positive controls (BaP with S9 mix and MMS without S9 mix) showed significantly higher values in the comet parameters (p < 0.05). Our data suggest that HS-1793 is not responsible for DNA strand breakage capacity in mammalian cells under tested conditions.
Resveratrol acts as an antimutagenic/anticarcinogenic agent by preventing oxidative DNA damage which plays a pivotal role in the carcinogenic activity of many genotoxic agents (29). Cellular oxidative stress generates free radicals, including reactive oxygen species (ROS) and these interact with critical macromolecules leading to DNA damage. DNA damage resulting from attack by ROS includes base modifications, sugar damage, strand breaks, abasic sites and DNA protein-crosslinks and it has been implicated in mutagenesis and carcinogenesis (30). Resveratrol has effective antioxidant activity in different in vitro assays including: total antioxidant activity, reducing power, DPPH•, ABTS•+, DMPD•+ and O2• radical scavenging, hydrogen peroxide scavenging and metal chelating activities (31). It is also a potent inhibitor of cellular ROS production in various cells (32). Similarly, our previous data demonstrate that HS-1793 effectively scavenges free radicals and inhibits radiationinduced plasmid DNA strand breaks an in vitro assay. In addition, HS-1793 significantly decreased ROS and cellular DNA damage in 2 Gy irradiated Chinese hamster ovary (CHO)-K1 cells (33). Indeed, HS-1793 showed no evidence of mutagenic activity when assessed for genotoxicity in the comet assay and it may be involved in the anti oxidant activity of HS-1793.
Although the comet assay is a sensitive and rapid method for detecting DNA single-strand breaks and alkali-labile sites in individual cells, it does not detect aneuploidy, chromosome rearrangement, DNA–DNA cross-linking, or DNA adducts. The in vitro chromosome aberration test is primarily used to identify the potential of a chemical to induce structural chromosome aberrations in cultured mammalian cells (14). Thus, the in vitro chromosome aberration test was performed using CHL cells to investigate the potency of HS-1793. As a result, no structural chromosome aberrations were caused by HS-1793 in the absence or presence of the S9 mix for either the short or continuous exposure periods at doses of 6.25~100 μg/ml. Resveratrol induces sister chromatid exchange dose-dependently at up to 10 μg/ml and shows weak aneuploidy induction in a Chinese hamster lung cell line (34). In addition, some analogues of resveratrol differing in number and position of hydroxy groupsclear positive responses in a concentration-dependent manner in chromosomal aberration and sister chromatid exchange (35). These results indicate that HS-1793 is non clastogenic at the tested concentration of 100 μg/ml.
In summary, the potential genotoxic effects of HS-1793 were assessed in an in vitro standard test battery. HS-1793 was non-genotoxic in a bacterial mutation assay, chromosome aberration assay, and the comet assay in vitro test. These data suggest that the risk of HS-1793 having genotoxic activity is low. Some studies on the beneficial characteristics of HS-1793, including a reduction in cancer risk have been reported but should be further assessed in human clinical studies. Therefore, many investigation of HS-1793-induced genotoxicity including in vivo assays and elucidation of the mechanism of genotoxicity are needed to clarify the potential risk for humans and to develop a new drug.
CONFLICT OF INTETEST
The authors declare that there are no conflicts of interest.
Acknowledgments
This research was supported by 2014 national R&D Program through the Dong-nam Institute of Radiological & Medical Sciences (DIRAMS) funded by the Ministry of Education, Science and Technology (50493-2014 and 50592-2014).
References
- 1.Gusman J., Malonne H., Atassi G. A reappraisal of the potential chemopreventive and chemotherapeutic properties of resveratrol. Carcinogenesis. (2001);22:1111–1117. doi: 10.1093/carcin/22.8.1111. [DOI] [PubMed] [Google Scholar]
- 2.Pervaiz S., Holme A.L. Resveratrol: its biologic targets and functional activity. Antioxid. Redox. Signaling. (2009);11:2851–2897. doi: 10.1089/ars.2008.2412. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal B.B., Bhardwaj A., Aggarwal R.S., Seeram N.P., Shishodia S., Takada Y. Role of resveratrol in prevention and therapy of cancer: Preclinical and clinical studies. Anticancer Res. (2004);24:2783–2840. [PubMed] [Google Scholar]
- 4.Park J.W., Choi Y.J., Suh S.I., Baek W.K., Suh M.H., Jin I.N., Min D.S., Woo J.H., Chang J.S., Passaniti A., Lee Y.H., Kwon T.K. Bcl-2 overexpression attenuates resveratrol-induced apoptosis in U937 cells by inhibition of caspase-3 activity. Carcinogenesis. (2001);22:1633–1639. doi: 10.1093/carcin/22.10.1633. [DOI] [PubMed] [Google Scholar]
- 5.Saiko P., Szakmary A., Jaeger W., Szekeres T. Resveratrol and its analogs: defense against cancer, coronary disease and neurodegenerative maladies or just a fad? Mutat. Res. (2008);658:68–94. doi: 10.1016/j.mrrev.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Jeong S.H., Lee J.S., Jeong N.Y., Kim T.H., Yoo K.S., Song S., Suh H., Kwon T.K., Park B.S., Yoo Y.H. A novel resveratrol analogue HS-1793 treatment overcomes the resistance conferred by Bcl-2 and is associated with the formation of mature PML nuclear bodies in renal clear cell carcinoma Caki-1 cells. Int. J. Oncol. (2009);35:1353–1360. doi: 10.3892/ijo_00000453. [DOI] [PubMed] [Google Scholar]
- 7.Jeong S.H., Jo W.S., Song S., Suh H., Seo S.Y., Lee S.H., Kwon T.K., Yoo Y.H. A novel resveratrol derivative, HS1793, overcomes the resistance conferred by Bcl-2 in human leukemic U937 cells. Biochem. Pharmacol. (2009);77:1337–1347. doi: 10.1016/j.bcp.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Kim H.J., Yang K.M., Park Y.S., Choi Y.J., Yun J.H., Son C.H., Suh H.S., Jeong M.H., Jo W.S. The novel resveratrol analogue HS-1793 induces apoptosis via the mitochondrial pathway in murine breast cancer cells. Int. J. Oncol. (2012);41:1628–1634. doi: 10.3892/ijo.2012.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong S.H., Song I.S., Kim H.K., Lee S.R., Song S., Suh H., Yoon Y.G., Yoo Y.H., Kim N., Rhee B.D., Ko K.S., Han J. An alogue of resveratrol HS-1793 exhibits anticancer activity against MCF-7 Cells via Inhibition of mitochondrial biogenesis bene expression. Mol. Cells. (2012);34:357–365. doi: 10.1007/s10059-012-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong N.Y., Yoon Y.G., Rho J.H., Lee J.S., Lee S.Y., Yoo K.S., Song S., Suh H., Choi Y.H., Yoo Y.H. The novel resveratrol analog HS-1793-induced polyploid LNCaP prostate cancer cells are vulnerable to downregulation of BclxL. Int. J. Oncol. (2011);38:1597–1604. doi: 10.3892/ijo.2011.979. [DOI] [PubMed] [Google Scholar]
- 11.Choi Y.J., Yang K.M., Kim S.D., Yoo Y.H., Lee S.W., Seo S.Y., Suh H., Yee S.T., Jeong M.H., Jo W.S. Resveratrol analogue HS-1793 induces the modulation of tumorderived T cells. Exp. Ther. Med. (2012);3:592–598. doi: 10.3892/etm.2012.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong M.H., Yang K.M., Choi Y.J., Kim S.D., Yoo Y.H., Seo S.Y., Lee S.H., Ryu S.R., Lee C.M., Suh H.S., Jo W.S. Resveratrol analog, HS-1793 enhance anti-tumor immunity by reducing the CD4+ CD25+ regulatory T cells in FM3A tumor bearing mice. Int. Immunopharmacol. (2012);14:328–333. doi: 10.1016/j.intimp.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Nath J., Krishna G. Safety screening of drugs in cancer therapy. Acta Haematol. (1998);99:138–147. doi: 10.1159/000040828. [DOI] [PubMed] [Google Scholar]
- 14.Madle S., Korte A., Ball R. Experience with mutagenicity testing of new drugs: view point of a regulatory agency. Mutat. Res. (1987);182:187–192. doi: 10.1016/0165-1161(87)90060-4. [DOI] [PubMed] [Google Scholar]
- 15.Tennant R.W., Spalding J., Stasiewicz S., Ashby J. Prediction of the outcome of rodent carcinogenicity bioassays currently being conducted on 44 chemicals by the national toxicology program. Mutagenesis. (1990);5:3–14. doi: 10.1093/mutage/5.1.3. [DOI] [PubMed] [Google Scholar]
- 16.Maron D.M., Ames B.N. Revised methods for the Salmonella mutagenicity test. Mutat. Res. (1983);113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 17.Organization for Economic Co-operation and Development (OECD). OECD guideline for testing of chemicals, no. 471,Genetic Toxicology: Bacterial Reverse Mutation Test. (1997). pp. 1–11. [Google Scholar]
- 18.Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. (1988);175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y.J., Kim M.S., Jeong H.K., Ryu J.C. Genotoxicity study on Khal, a halocidin derivative, in bacterial and mammalian cells. Mol. Cell. Toxicol. (2006);2:151–158. [Google Scholar]
- 20.Organization for Economic Co-operation and Development (OECD). OECD guideline for testing of chemicals, no. 473, Genetic Toxicology: In vitro mammalian chromosome aberration test. (1997). pp. 1–16. [Google Scholar]
- 21.Matsuoka A., Sofuni T., Miyata N., Ishidate M. Jr. Clastogenicity of 1-nitropyrene, dinitropyrenes, fluorine and mononitrofluorenes in cultured Chinease Hamster cells. Mutat. Res. (1991);259:103–110. doi: 10.1016/0165-1218(91)90114-2. [DOI] [PubMed] [Google Scholar]
- 22.Ames B.N., Lee F.D., Durston W.E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc. Natl. Acad. Sci. USA. (1973);70:782–786. doi: 10.1073/pnas.70.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams L.D., Burdock G.A., Edwards J.A., Beck M., Bausch J. Safety studies conducted on high-purity trans-resveratrol in experimental animals. Food Chem. Toxicol. (2009);47:2170–2182. doi: 10.1016/j.fct.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Kirkland D. Chromosome aberration testing in genetic toxicology-past, present and future. Mutat. Res. (1998);404:173–185. doi: 10.1016/S0027-5107(98)00111-0. [DOI] [PubMed] [Google Scholar]
- 25.Buschini A., Carboni P., Frigerio S., Furlini M., Marabini L., Monarca S., Poli P., Radice S., Rossi C. Genotoxicity and cytotoxicity assessment in lake drinking water produced in a treatment plant. Mutagenesis. (2004);19:341–347. doi: 10.1093/mutage/geh040. [DOI] [PubMed] [Google Scholar]
- 26.Lee M., Kwon J., Chung M.K. Enhanced prediction of potential rodent carcinogenicity by utilizing comet assay and apoptotic assay in combination. Mutat. Res. (2003);541:9–19. doi: 10.1016/S1383-5718(03)00175-X. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann A., Agurell E., Beevers C., Brendler-Schwaab S., Burlinson B., Clay P., Collins A., Smith A., Speit G., Thybaud V., Tice R.R. Recommendation for conducting the in vivo alkaline comet assay. Mutagenesis. (2003);18:45–51. doi: 10.1093/mutage/18.1.45. [DOI] [PubMed] [Google Scholar]
- 28.Henderson L., Wolfreys A., Fedyk J., Bourner C., Windebank S. The ability of the comet assay to discriminate between genotoxins and cytotoxins. Mutagenesis. (1998);13:89–94. doi: 10.1093/mutage/13.1.89. [DOI] [PubMed] [Google Scholar]
- 29.Jang M., Cai L., Udeani G.O., Slowing K.V., Thomas C.F., Beecher C.W., Fong H.H., Farnsworth N.R., Kinghorn A.D., Mehta R.G., Moon R.C., Pezzuto J.M. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. (1997);275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 30.Wang D., Kreutzer D.A., Essigmann J.M. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res. (1998);400:99–115. doi: 10.1016/S0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 31.Gülçin I. Antioxidant properties of resveratrol: A structure-activity insight. Innovative Food Sci. Emerging Technol. (2010);11:210–218. doi: 10.1016/j.ifset.2009.07.002. [DOI] [Google Scholar]
- 32.Leonard S.S., Xia C., Jiang B.H., Stinefelt B., Klandorf H., Harris G.K., Shi X. Resveratrol scavenges reactive oxygen species and effects radical-induced cellular responses. Biochem. Biophys. Res. Commun. (2003);309:1017–1026. doi: 10.1016/j.bbrc.2003.08.105. [DOI] [PubMed] [Google Scholar]
- 33.Jeong M.H., Yang K.M., Jeong D.H., Lee C.G., Oh S.J., Jeong S.K., Lee K.W., Jo Y.R., Jo W.S. Protective activity of a novel resveratrol analogue, HS-1793, against DNA damage in 137Cs-irradiated CHO-K1 Cells. J. Radiat. Res. (2014);55:464–475. doi: 10.1093/jrr/rrt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka A., Furuta A., Ozaki M., Fukuhara K., Miyata N. Resveratrol, a naturally occurring polyphenol, induces sister chromatid exchanges in a Chinese hamster lung (CHL) cell line. Mutat. Res. (2001);494:107–113. doi: 10.1016/S1383-5718(01)00184-X. [DOI] [PubMed] [Google Scholar]
- 35.Matsuoka A., Takeshita K., Furuta A., Ozaki M., Fukuhara K., Miyata N. The 4-hydroxy group is responsible for the in vitro cytogenetic activity of resveratrol. Mutat. Res. (2002);521:29–35. doi: 10.1016/S1383-5718(02)00211-5. [DOI] [PubMed] [Google Scholar]


