Abstract
A β-glucosidase producing yeast strain was isolated from Korean traditional rice wine. Based on the sequence of the YCL008c gene and analysis of the fatty acid composition, the isolate was identified as Saccharomyces cerevisiae strain HJ-014. S. cerevisiae HJ-014 produced ginsenoside Rd, F2, and compound K from the ethanol extract of red ginseng. The production was increased by shaking culture, where the bioconversion efficiency was increased 2-fold compared to standing culture. The production of ginsenoside F2 and compound K was time-dependent and thought to proceed by the transformation pathway of: red ginseng extract→Rd→F2→compound K. The optimum incubation time and concentration of red ginseng extract for the production of compound K was 96 hr and 4.5% (w/v), respectively.
Keywords: Bioconversion, Compound K, Red ginseng extract, Saccharomyces cerevisiae HJ-014
Ginseng (Panax ginseng C. A. Meyer) belongs to the Araliaceae family of flowering plants and its' root has been used as a traditional medicine in Asian countries for over 2000 years. The major active components of ginseng are ginsenosides, which have a number of pharmacological effects, including anti-inflammatory [1], anti-cancer [2, 3], anti-aging [4], and antioxidant [5] activities. However, naturally occurring ginsenosides are poorly absorbed by the human intestinal tract [6]. As a result, recent studies have focused on the bioconversion of the ginsenosides to increase their absorption and pharmacological activity [7, 8].
Compound K is the active form of protopanaxadiol saponins and has been reported to be easily absorbed by the human digestive tract [9]. In addition compound K has been shown to exhibit hepatoprotective activity [10], inhibit tumor invasion and metastasis, and induce tumor cell apoptosis [11,12,13]. Since compound K does not exist in natural products, many studies have focused on the development of methods for the production of compound K. Microbial biotransformation with the crude extract of Leuconostoc mesenteroides DC102 [14] and Lactobacillus paralimentarius [15] has been used to prepare compound K from ginsenoside Rb1 via Rd and F2. Enzymatic transformation of gypenoside LXXV by β-glucosidase from Thermus thermophilus [16], has also been used to prepare compound K. β-Glucosidase specifically hydrolyzes two glucose molecules at C-3 and one glucose molecules at C-20 of ginsenoside Rb1 to generate compound K. However, only a few β-glucosidase have reported to be able to produce compound K [17, 18].
So far, the majority of studies have used purified ginsenosides for the production of compound K using bioconversion. In this paper, we isolated and identified a β-glucosidase producing yeast, from the traditional Korean rice wine Makgeolli. Significantly, we have demonstrated that this novel yeast strain is capable of producing compound K from a crude ethanol extraction of red ginseng. This research provides a promising solution for the industrial production of compound K form raw ginseng preparations.
MATERIALS AND METHODS
Chemicals
The ginsenoside standards Rd, F2, and compound K were purchased from Sigma (St. Louis, MO, USA) and Ambo Laboratories (Daejeon, Korea). All other chemicals used were reagent grade.
Screening for microbes producing β-glucosidase
Esculin-MRS agar, containing 0.3% (w/v) esculin and 0.02% (w/v) ferric citrate in MRS agar [19], was used to isolate β-glucosidase producing microorganisms. β-Glucosidase producing-microbes hydrolyze the esculin appearing as colonies surrounded by a reddish-brown to dark brown zone. Potential β-glucosidase producing microbes were subjected to additional two-day incubation at 30℃ in MRS medium.
Isolate identification
The genomic DNA was isolated and purified using a Genomic DNA Purification Kit (Qiagen, Hilden, Germany). PCR amplification was performed using Saccharomyces specific primers YCL008c-U (5'-TTCCGTTGGATGTGCCATCG-3') and YCL008c-L (5'-GGAGCCACCAAGGGATGG-3') [20]. The resulting PCR product was sequenced by Macrogen (Seoul, Korea). Homologous sequences were identified using the BLAST, at the NCBI database. Cells were grown for 3 days at 30℃ on MRS agar and fatty acid methyl esters (FAMEs) were extracted and prepared according to the standard protocol of the Microbial Identification System (MIDI; Microbial ID, Newark, DE, USA). Briefly, whole cells extracts were saponified at 100℃ in 1mL of reagent 1 (15% w/v NaOH in 50% v/v methanol). Fatty acids were then esterified at 80℃ with 2 mL of reagent 2 (3.25 N HCl in 46% v/v methanol) and FAMEs were extracted in 1.25mL of reagent 3 (hexane and methyl-t-butyl ether mixture, 1 : 1 v/v). Finally, the organic extract was washed with 3 mL of 1.2% (w/v) NaOH. The washed extracts were then analyzed on an Agilent 6890N-MIDI Sherlock system 6.0 (Microbial ID).
Nucleotide sequence accession number
The GenBank accession number for the S. cerevisiae HJ-014 sequence is KM061793.
Assay of enzyme activity
β-Glucosidase activity was determined using p-nitrophenyl-β-D-glucose (pNPG) as a substrate. The reaction mixture, containing 50 µL of 10 mM pNPG, 50 µL of enzyme solution in 50mM acetate buffer (pH 5.0), was incubated at 50℃ for 30 min. The reaction was stopped by adding 100 µL of NaOH-glycine buffer (0.4M, pH 10.8) and measured at 405 nm. One unit of enzyme activity was defined as the amount of enzyme producing 1 µmole of p-nitrophenol per min. Activity staining of β-glucosidase was performed as described previously [21]. After electrophoresis, the gel was incubated in 0.2M acetate buffer containing 0.1% (w/v) esculin and 0.03% (w/v) ferric chloride for 5 min at 50℃. β-Glucosidase appeared as black bands against a transparent background.
Transformation of ethanol extracted red ginseng by S. cerevisiae HJ-014
The 80% ethanol extract of red ginseng was pretreated with α-herbzyme (Korea Enzyme Co., Seoul, Korea). The red ginseng extract (0.5 g) was resolved in 3% α-herbzyme (w/v) in water and incubated at 30℃ with agitation (200 rpm) for 24 hr. MRS broth was inoculated with S. cerevisiae HJ-014 and incubated at 30℃ for 24 hr, the culture broth was then mixed with an equal volume of pretreated red ginseng. The mixture was then incubated at 30℃ with shaking at 200 rpm and a 1.0 mL aliquot was then taken at the indicated time and analyzed by high-performance liquid chromatography (HPLC).
HPLC analysis
The reaction mixtures were analyzed by HPLC (Shimadzu Prominence, Shimadzu, Japan), using a Zorbax Eclipse plus C18 column (4.6 × 250 mm, 5 µm) at 203 nm. The analytes were eluted via gradient elution at 40℃ with acetonitrile/water from 18 : 82 (v/v) to 100 : 0 (v/v) at a flow rate of 1.0mL/min.
RESULTS AND DISCUSSION
The isolation and identification of β-glucosidase producing microbes
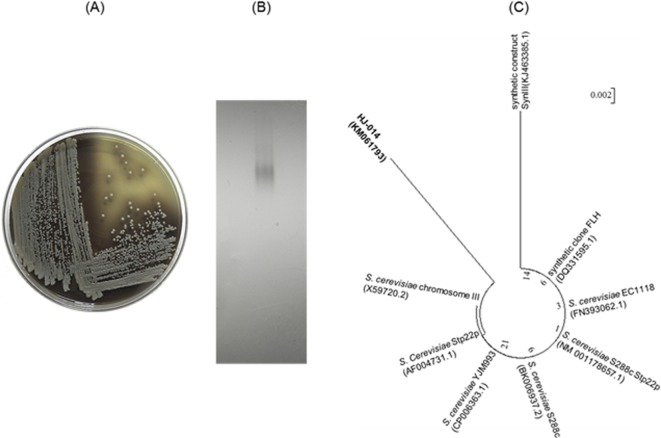
The β-glucosidase producing microbes were screened, using Esculin-MRS agar medium. Black colonies demonstrating β-glucosidase activity were picked and transferred to fresh Esculin-MRS agar medium (Fig. 1A). The production of β-glucosidase from the isolate was confirmed by activity staining (Fig. 1B). The consensus sequence of the Saccharomyces YCL008c gene (1,514 bp) from the isolate was aligned with other S. cerevisiae sequences and the taxonomic relationships were analyzed. Phylogenetic trees based on the aligned sequences were constructed using MEGA ver. 5.1 [22] with neighbor-joining [23], based on 1,000 randomly chosen bootstrap replication and tree/branch style was circle (Fig. 1C). The sequence was 97% homologous to the S. cerevisiae Stp22p genes. The MIDI system was used to analyze the fatty acid composition of the cell membrane from the isolate. The analysis, shown in Table 1, identified palmitoleic acid (16 : 1), oleic acid (18 : 1), palmitic acid (16 : 0), trace amounts of stearic acid (18 : 0) and short chain fatty acids (10 : 0 to 14 : 1). This demonstrated that the isolate contains the same principal fatty acid chains found in S. cerevisiae [24, 25]. Together with the phylogenetic and chemotaxonomic analyses the isolate was identified as S. cerevisiae strain HJ-014.
Fig. 1.
A, Production of β-glucosidase from isolated yeast detected in Esculin-MRS agar medium; B, Activity staining to detect β-glucosidase; C, Phylogenetic tree based on YCL008c gene sequences.
Table 1.
Cellular fatty acid composition (%) of Saccharomyces cerevisiae HJ-014
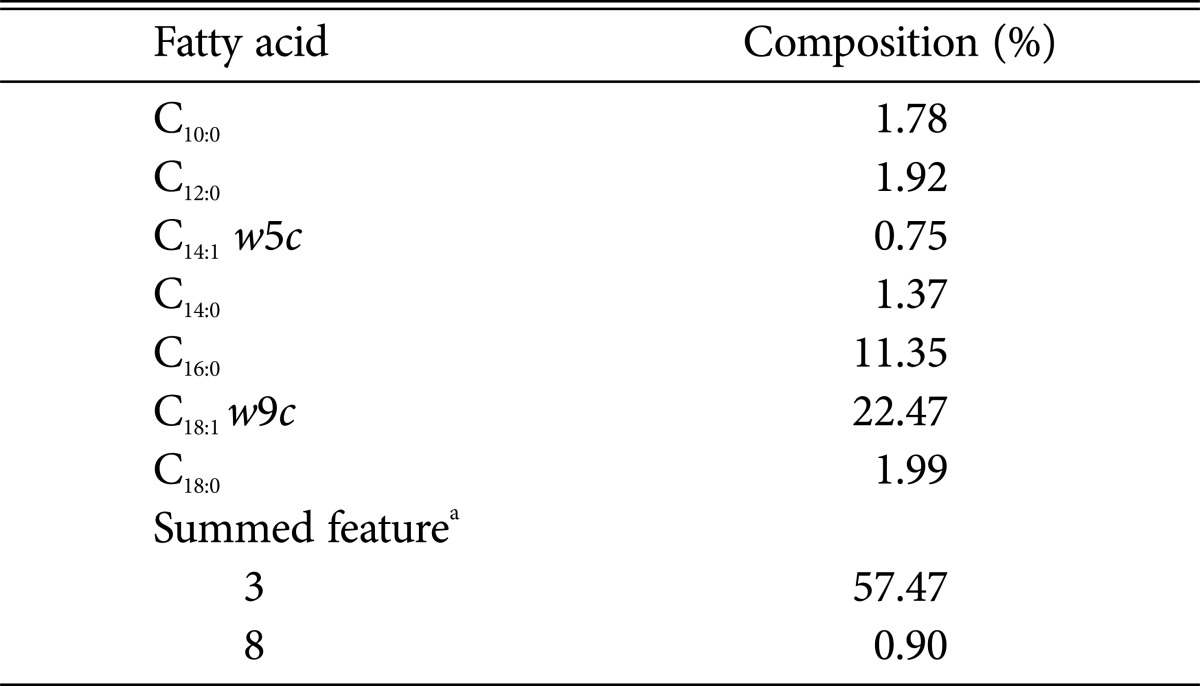
aSummed features represent groups of two or more fatty acids that could not be separated using the MIDI system. Summed feature 3 comprised C16:1 w7c/C16:1 w6c and/or C16:1 w6c/C16:1 w7c and summed feature 8 comprised C18:1 w7c and/or C18:1 w6c.
Microbial bioconversion of ethanol extracted red ginseng to compound K
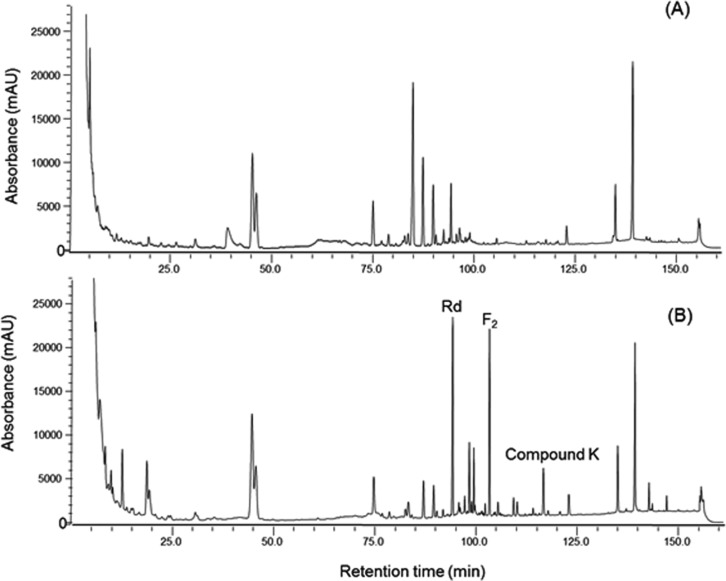
The culture broth of S. cerevisiae HJ-014 was mixed with an equal volume of pretreated red ginseng extract and incubated at 30℃ for 24 hr at 200 rpm. The reaction mixture was analyzed by HPLC, with standard ginsenosides including Rd, F2, and compound K. Ginsenoside F2 and compound K were not detected in the absence of S. cerevisiae HJ-014 treatment (Fig. 2A). Three ginsenosides were detected in the reaction mixture incubated with isolate and the retention times of Rd, F2, and compound K were approximately 94, 103, and 116.5 min, respectively (Fig. 2B). It has been shown that Sphingomonas echinoides GP50 can completely convert ginsenoside Rb1 to ginsenoside Rd, but it produces a limited amount of compound K [26]. In Lactobacillus pentosus DC101, over 97% of ginsenoside Rd is degraded and converted into compound K [27]. However, only one report has been published on the production of compound K from raw ginseng extract [28].
Fig. 2.
Production of ginsenoside Rd, F2 and compound K by Saccharomyces cerevisiae HJ-014. A, The reaction mixture was incubated at 30℃ for 24 hr at 200 rpm without S. cerevisiae HJ-014; B, The reaction mixture was incubated with S. cerevisiae HJ-014 at same conditions.
The effects of agitation on the production of compound K
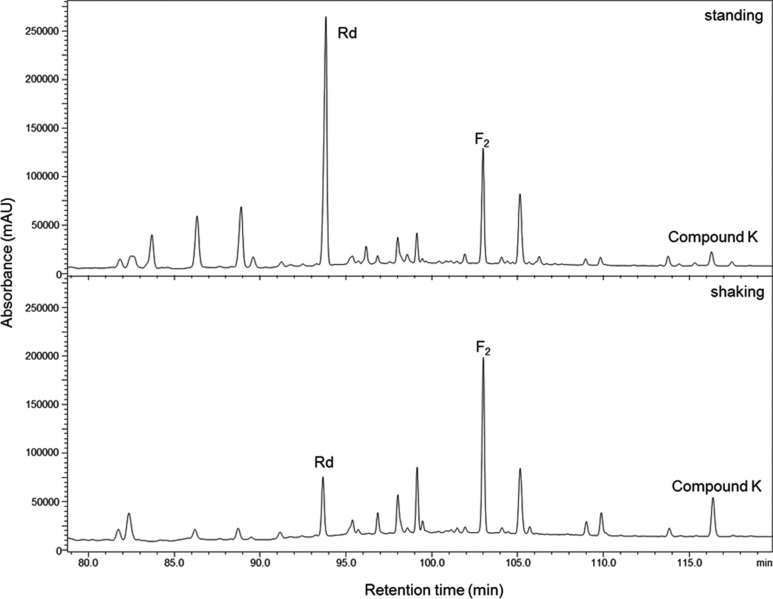
To investigate the effect of agitation on the production of compound K, we incubated the reaction mixture either with or without agitation at 30℃ for 48 hr. As shown in Fig. 3 ginsenoside Rd was the major product detected in the standing culture, while compound K was only detected in trace amounts, compared to the shaking culture. The bioconversion pathway of ginsenoside Rd by L. pentosus DC101 has been reported; where Rd is initially converted to F2, followed by its conversion into compound K [27]. The bioconversion efficiency of the standing culture was lower than that of the shaking culture possibly because sufficient degradation of Rd had not occurred in in the standing culture.
Fig. 3.
The effect of agitation on the production of ginsenoside Rd, F2 and compound K. The reaction mixture was incubated the either with (200 rpm) or without agitation at 30℃ for 48 hr.
Time-dependent production of compound K
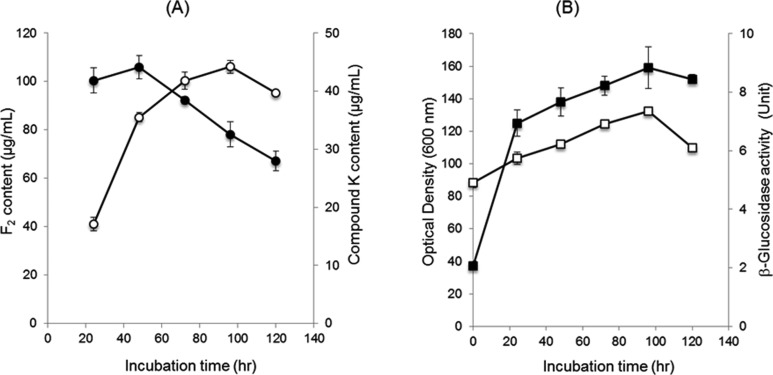
The production rate of F2 and compound K was time-dependent [27]. The ginsenoside F2 peak reached a maximum level (106 µg/mL) (Fig. 4A) after 48 hr incubation and then decreased gradually (Fig. 4A). The compound K peak also increased continuously until 96 hr, when the amount of compound K produced reached its maximum (44 µg/mL) (Fig. 4A). The growth rate and β-glucosidase activity was increased gradually up to 96 hr and then decreased (Fig. 4B). Our results show that the transformation pathway of red ginseng extract by S. cerevisiae HJ-014 is red ginseng extract→Rd→F2→compound K and β-glucosidase may play an important role in compound K production.
Fig. 4.
The time-dependent production of ginsenoside F2 and compound K. A, Time-dependent change in the concentrations of F2 and compound K. The closed and open symbols represent the concentration of F2 and compound K, respectively; B, The growth (closed square) and the production of β-glucosidase (open square) according to the incubation time.
The effects of red ginseng extract concentration on the production of compound K
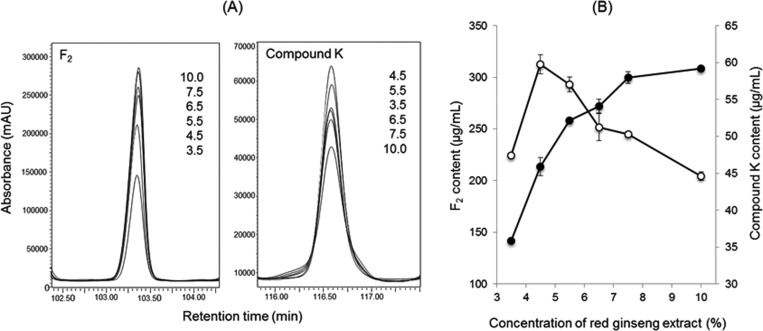
It have been demonstrated that a high concentration of ginseng extract inhibits microbial growth [28]. The optimum concentration of red ginseng extract was determined for the production of the ginsenoside F2 and compound K in S. cerevisiae HJ-014. HPLC was used to monitor the production of F2 and compound K produced from various concentrations of red ginseng extract. The amount of F2increased in proportion to the concentration of red ginseng extract treated (Fig. 5A) and reached a maximum (308 µg/mL, Fig. 5B), at 10% (w/v). However, a large amount of compound K was produced at low concentrations of red ginseng extract, the optimum concentration being 4.5% (w/v) (Fig. 5).
Fig. 5.
The effect of red ginseng extract concentration on the production of ginsenoside F2 and compound K. A, Enlarged HPLC chromatogram of F2 and compound K. The numbers in the figure indicate the concentration of red ginseng extract (%, w/v); B, Effect of red ginseng extract on the production of F2 and compound K. The closed and open symbols represent the concentration of F2 and compound K, respectively.
The bioconversion of raw red ginseng extract into compound K by S. cerevisiae HJ-014 has a potential application in industry. Further studies concerning the purification, characterization, and application of this strain and its enzyme system to the production of compound K is required.
ACKNOWLEDGEMENTS
This work was supported by the RIC program of MOTIE (Ministry of Trade, Industry and Energy) in Daejeon University.
References
- 1.Wang L, Zhang Y, Chen J, Li S, Wang Y, Hu L, Wang L, Wu Y. Immunosuppressive effects of ginsenoside-Rd on skin allograft rejection in rats. J Surg Res. 2011;176:267–274. doi: 10.1016/j.jss.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 2.Sato K, Mochizuki M, Saiki I, Yoo YC, Samukawa K, Azuma I. Inhibition of tumor angiogenesis and metastasis by a saponin of Panax ginseng, ginsenoside-Rb2. Biol Pharm Bull. 1994;17:635–639. doi: 10.1248/bpb.17.635. [DOI] [PubMed] [Google Scholar]
- 3.Mochizuki M, Yoo YC, Matsuzawa K, Sato K, Saiki I, Tonooka S, Samukawa K, Azuma I. Inhibitory effect of tumor metastasis in mice by saponins, ginsenoside-Rb2, 20(R)- and 20(S)-ginsenoside-Rg3, of red ginseng. Biol Pharm Bull. 1995;18:1197–1202. doi: 10.1248/bpb.18.1197. [DOI] [PubMed] [Google Scholar]
- 4.Wang BX, Cui JC, Liu AJ, Wu SK. Studies on the anti-fatigue effect of the saponins of stems and leaves of Panax ginseng (SSLG) J Tradit Chin Med. 1983;3:89–94. [PubMed] [Google Scholar]
- 5.Cho WC, Chung WS, Lee SK, Leung AW, Cheng CH, Yue KK. Ginsenoside Re of Panax ginseng possesses significant antioxidant and antihyperlipidemic efficacies in streptozotocin-induced diabetic rats. Eur J Pharmacol. 2006;550:173–179. doi: 10.1016/j.ejphar.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 6.Odani T, Tanizawa H, Takino Y. Studies on the absorption, distribution, excretion and metabolism of ginseng saponins. III. The absorption, distribution and excretion of ginsenoside Rb1 in the rat. Chem Pharm Bull (Tokyo) 1983;31:1059–1066. doi: 10.1248/cpb.31.1059. [DOI] [PubMed] [Google Scholar]
- 7.Park SY, Bae EA, Sung JH, Lee SK, Kim DH. Purification and characterization of ginsenoside Rb1-metabolizing β-glucosidase from Fusobacterium K-60, a human intestinal anaerobic bacterium. Biosci Biotechnol Biochem. 2001;65:1163–1169. doi: 10.1271/bbb.65.1163. [DOI] [PubMed] [Google Scholar]
- 8.Kim MK, Lee JW, Lee KY, Yang DC. Microbial conversion of major ginsenoside Rb1 to pharmaceutically active minor ginsenoside Rd. J Microbiol. 2005;43:456–462. [PubMed] [Google Scholar]
- 9.Akao T, Kanaoka M, Kobashi K. Appearance of compound K, a major metabolite of ginsenoside Rb1 by intestinal bacteria, in rat plasma after oral administration - measurement of compound K by enzyme immunoassay. Biol Pharm Bull. 1998;21:245–249. doi: 10.1248/bpb.21.245. [DOI] [PubMed] [Google Scholar]
- 10.Lee HU, Bae EA, Han MJ, Kim NJ, Kim DH. Hepatoprotective effect of ginsenoside Rb1 and compound K on tert-butyl hyroperoxide-induced liver injury. Liver Int. 2005;25:1069–1073. doi: 10.1111/j.1478-3231.2005.01068.x. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa H, Uchiyama M. Antimetastatic efficacy of orally administered ginsenoside Rb1 in dependence on intestinal bacterial hydrolyzing potential and significance of treatment with an active bacterial metabolite. Planta Med. 1998;64:696–700. doi: 10.1055/s-2006-957560. [DOI] [PubMed] [Google Scholar]
- 12.Wakabayashi C, Murakami K, Hasegawa H, Murata J, Saiki I. An intestinal bacterial metabolite of ginseng protopanaxadiol saponins has the ability to induce apoptosis in tumor cells. Biochem Biophys Res Commun. 1998;246:725–730. doi: 10.1006/bbrc.1998.8690. [DOI] [PubMed] [Google Scholar]
- 13.Oh SH, Yin HQ, Lee BH. Role of the Fas/Fas ligand death receptor pathway in ginseng saponin metabolite-induced apoptosis in HepG2 cells. Arch Pharm Res. 2004;27:402–406. doi: 10.1007/BF02980081. [DOI] [PubMed] [Google Scholar]
- 14.Quan LH, Piao JY, Min JW, Kim HB, Kim SR, Yang DU, Yang DC. Biotransformation of ginsenoside Rb1 to prosapogenins, gypenoside XVII, ginsenoside Rd, ginsenoside F2, and compound K by Leuconostoc mesenteroides DC102. J Ginseng Res. 2011;35:344–351. doi: 10.5142/jgr.2011.35.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan LH, Kim YJ, Li GH, Choi KT, Yang DC. Microbial transformation of ginsenoside Rb1 to compound K by Lactobacillus paralimentarius. World J Microbiol Biotechnol. 2013;29:1001–1007. doi: 10.1007/s11274-013-1260-1. [DOI] [PubMed] [Google Scholar]
- 16.Shin KC, Seo MJ, Oh HJ, Oh DK. Highly selective hydrolysis for the outer glucose at the C-20 position in ginsenosides by β-glucosidase from Thermus thermophilus and its application to the production of ginsenoside F2 from gypenoside XVII. Biotechnol Lett. 2014;36:1287–1293. doi: 10.1007/s10529-014-1472-y. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y, Luan H, Hao D, Xiao H, Yang S, Yang L. Purification and characterization of a novel ginsenoside-hydrolyzing β-D-glucosidase from China white jade snail (Achatina fulica) Enzyme Microb Technol. 2007;40:1358–1366. [Google Scholar]
- 18.Yan Q, Zhou XW, Zhou W, Li XW, Feng MQ, Zhou P. Purification and properties of a novel β-glucosidase, hydrolyzing ginsenoside Rb1 to CK, from Paecilomyces Bainier. J Microbiol Biotechnol. 2008;18:1081–1089. [PubMed] [Google Scholar]
- 19.De Man JD, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Microbiol. 1960;23:130–135. [Google Scholar]
- 20.Casaregola S, Nguyen HV, Lapathitis G, Kotyk A, Gaillardin C. Analysis the constitution of beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int J Syst Evol Microbiol. 2001;51(Pt 4):1607–1618. doi: 10.1099/00207713-51-4-1607. [DOI] [PubMed] [Google Scholar]
- 21.Kwon KS, Lee J, Kang HG, Hah YC. Detection of β-glucosidase activity in polyacrylamide gels with esculin as substrate. Appl Environ Microbiol. 1994;60:4584–4586. doi: 10.1128/aem.60.12.4584-4586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Cottrell M, Viljoen BC, Kock JL, Lategan PM. The long-chain fatty acid compositions of species representing the genera Saccharomyces, Schwanniomyces and Lipomyces. J Gen Microbiol. 1986;132:2401–2403. [Google Scholar]
- 25.Van der Rest ME, Kamminga AH, Nakano A, Anraku Y, Poolman B, Konings WN. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev. 1995;59:304–322. doi: 10.1128/mr.59.2.304-322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MK, Im WT, Ohta H, Lee M, Lee ST. Sphingopyxis granuli sp. nov., a β-glucosidase-producing bacterium in the family Sphingomanadaceae in α-4 subclass of the Proteobacteria. J Microbiol. 2005;43:152–157. [PubMed] [Google Scholar]
- 27.Quan LH, Cheng LQ, Kim HB, Kim JH, Son NR, Kim SY, Jin HO, Yang DC. Bioconversion of ginsenoside Rd into compound K by Lactobacillus pentosus DC101 isolated from Kimchi. J Ginseng Res. 2010;34:288–295. [Google Scholar]
- 28.Doh ES, Chang JP, Lee KH, Seong NS. Ginsenoside change and antioxidation activity of fermented ginseng. Korean J Med Crop Sci. 2010;18:255–265. [Google Scholar]