Abstract
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy CADASIL is caused by more than a hundred NOTCH3 mutations. Virtually all encoded mutant proteins contain an odd number of cysteines. As such, structural changes in NOTCH3 may be the primary molecular abnormality in CADASIL. Thus, we sought evidence for structurally altered NOTCH3 protein in CADASIL tissue. Four antibodies were raised in rabbits against two non-overlapping N-terminal NOTCH3 sequences. These reagents were used in immunohistochemical experiments to detect epitopes in post-mortem CADASIL brains (n=8), control brains, and cells overexpressing NOTCH3. To determine the biochemical nature of NOTCH3 epitopes, we used these antibodies to probe pure NOTCH3-Fc fusion proteins treated with acid, urea, guanidinium, ionic detergents, acrylamide, and thiol- and phosphorus-based reductants. All antibodies avidly stained arteries in 8 of 8 CADASIL brain samples. The most prominent staining was in degenerating media of leptomeningeal arteries and sclerotic penetrating vessels. Normal appearing vessels from control brains were not reactive. Antibodies did not react with cultured cells overexpressing NOTCH3 or with purified NOTCH3-Fc protein. Furthermore, treatment of pure protein with acid, chaotropic denaturants, alkylators, and detergents failed to unmask N-terminal NOTCH3 epitopes. Antibodies, however, recognized novel N-terminal epitopes in purified NOTCH3-Fc protein treated with three different reductants (DTT, beta-mercaptoethanol, and TCEP). We conclude that CADASIL arteries feature latent N-terminal NOTCH3 epitopes, suggesting the first evidence in vivo of NOTCH3 structural alterations.
1. Introduction
In 1996, Joutel and colleagues first described mutations in NOTCH3 in patients with CADASIL, an inherited disorder of smooth muscle cells that causes leukoencephalopathy and manifests as premature stroke and vascular dementia1. Since then, more than one hundred independent NOTCH3 mutations have been published 2, the overwhelming majority of which are missense mutations that change the number of cysteines in the predicted amino acid sequence3. This results in an uneven number of cysteine residues, which likely alters the tertiary structure of Notch3.
The primary sequence of NOTCH3 features a large extracellular domain which is the site of almost all mutations linked to CADASIL3. As in homologous related proteins (NOTCH1, 2, and 4), the NOTCH3 extracellular domain is composed of an array of more than thirty EGF-like domains4. Each of these EGF-like domains contains six conserved cysteine residues. Notch EGF-like structures that have been elucidated at the atomic structural level feature six oxidized cysteine residues that form three pairs of disulfide bonds5, 6.
The conserved disulfide bonding of EGF-like repeats in other extracellular matrix proteins7, 8 suggests that disulfide pairing plays an important role in structural stability of NOTCH3. Introduction or elimination of a cysteine, which hallmarks most mutations seen in CADASIL, is predicted to impair normal disulfide pairing within the NOTCH3 ectodomain and, consequently, may seriously disrupt the structure of the protein. The ectodomain of NOTCH3, which harbors all known CADASIL mutations, accumulates in the blood vessels of patients 9. One potential mechanism of CADASIL pathogenesis, therefore, could be cysteine mutation-driven structural alterations of the protein that drives protein misfolding and accumulation. There is, however, no experimental evidence demonstrating structural alterations of NOTCH3 protein in human tissue.
We describe the discovery of novel N-terminal NOTCH3 epitopes in diseased arteries in human CADASIL brains. We show that these epitopes are undetectable in native NOTCH3 protein, but can be unmasked specifically by disulfide bond disruption. Our studies suggest an important role of reduced cysteine in CADASIL pathogenesis.
2. Results
2.1 Identification of novel N-terminal NOTCH3 epitopes in CADASIL arteries
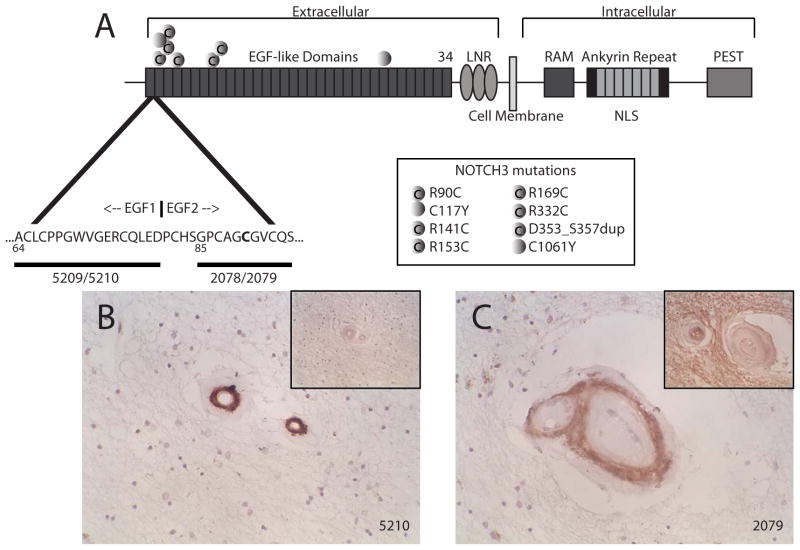
Two synthetic peptides were used to immunize rabbits (Figure 1A). Antisera 5209 and 5210 were raised against the wild type C-terminal end of the first EGF-like domain. Antisera 2078 and 2079 were raised against the N-terminal end of the second EGF-like domain that contained the canonical R90C CADASIL mutation. All four antisera reacted unequivocally at <1:2000 with 1 ug immobilized peptide immunogen.
Figure 1.
Antibodies to human NOTCH3 and reactivity with CADASIL white matter arteries. (A) A schematic of the NOTCH3 protein and expanded view of the N-terminus. Sequences localized to the end of the first EGF-like repeat and the beginning of the second EGF-like repeat, highlighted with horizontal bars, were used as immunogens to generated N-terminal NOTCH3 antibodies 5209, 5210, 2078, and 2079. Gain or loss of cysteine mutations are depicted by shaded circles with or without a “C,” respectively, at the bottom right side of (A). Small penetrating white matter vessels from CADASIL brains were examined by immunohistochemistry using 5210 (B) and 2079 (C). Insets in (B) and (C) show negative vascular staining patterns using preimmune serum. Positive vascular staining was seen with 5209 and 2078, and corresponding preimmune serum failed to react with CADASIL vessels.
Antibodies were used for immunohistochemical analysis of CADASIL (n=8) and control brain samples (n=6). In CADASIL arteries, antisera reacted strongly with thickened white matter arteries (Figure 1B and 1C). The media of CADASIL leptomeningeal arteries also reacted strongly with NOTCH3 antibodies in vessel layers that contain degenerating smooth muscle (Figure 2A and 2C). Morphologically normal arteries of control brains did not react with antibodies (Figures 2B and 2D). Importantly, the 2078 and 2079 antisera, which were raised against a peptide containing the R90C mutation, reacted with the seven CADASIL brains which did not have the R90C mutation. Staining of a brain with the R90C mutations was not stronger than other samples, suggesting that the antibodies react with CADASIL arteries but are not specific for the R90C mutation.
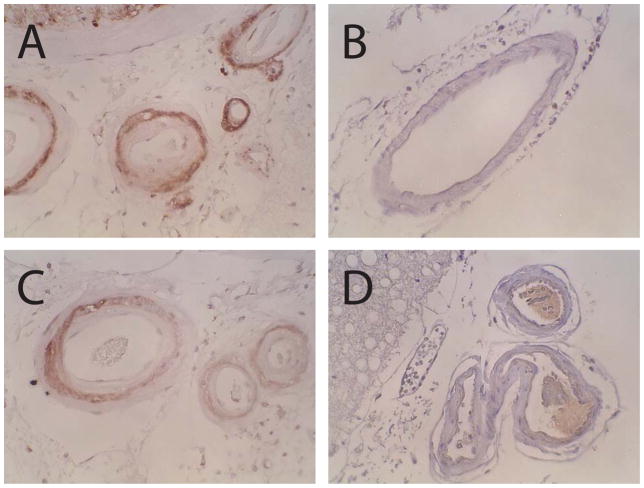
Figure 2.
NOTCH3 N-terminal epitopes in leptomeningeal arteries in CADASIL and controls. Antibodies 5210 (A, B) and 2079 (C, D) stained leptomeningeal arteries of a CADASIL brain (A, C) but not those of a control brain (B, D). Staining in CADASIL was observed in the medial layer of arteries that showed intense thickening due to intimal hyperplasia and medial fibrosis in CADASIL. Antibodies 5209 and 2078 resulted in similar staining patterns.
Antibodies were also tested on cell lines overexpressing wild type or mutant NOTCH3 (Figure 3) that were fixed and embedded in the same manner as brain tissues. Immunohistochemistry performed on these samples using all four antibodies to the N-terminal sequences of NOTCH3 did not result in signal. This suggested that antibodies reacted with NOTCH3 epitopes generated by post-translational processes in vessels in CADASIL.
Figure 3.
Immunohistochemistry of Notch3 N-terminal epitopes in 293 cell lines. overexpressing wild type or mutant NOTCH3. Human 293 cell lines were collected, fixed, and paraffin embedded prior to analysis by immunohistochemistry. (A) Key showing antibodies used to analyze cell lines. N3 MO1, a monoclonal against the N-terminus of NOTCH3, was used as a positive control to verify NOTCH3 expression. Antibodies were each applied to vector negative control 293 (B), wild type NOTCH3 (C), and R90C NOTCH3 (D) transfected cell lines. N-terminal antibodies 5210 and 2079 did not stain cells overexpressing NOTCH3.
2.2 Unmasking of latent NOTCH3 N-terminal epitopes by protein reduction
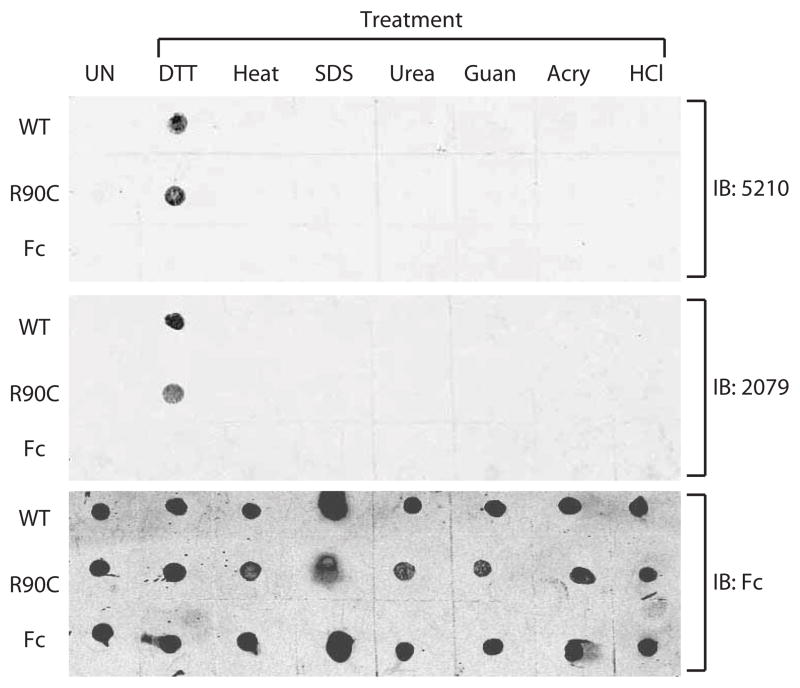
We purified recombinant NOTCH3 ectodomain (EGF domains 1–27) fused to immunoglobulin Fc to apparent homogeneity using protein A chromatography. A wild type protein and the R90C point mutation-containing protein were analyzed after adsorbing proteins to nitrocellulose. None of the four antibodies recognized native or mutant NOTCH3 ectodomain (UN in Figure 4).
Figure 4.
Unmasking of NOTCH3 N-terminal epitopes in recombinant NOTCH3 protein. Recombinant fusions of NOTCH3 ectodomain to mouse Fc were purified to homogeneity from 293 cell lines. Fifty ng of wild type NOTCH3 (WT), R90C NOTCH3 (R90C), or mouse Fc (C) were treated with chemicals indicated for 60 minutes at 37°C (or at 65°C without chemicals) and then spotted on nitrocellulose. 5210 or 2079 were used to detect N-terminal NOTCH3 epitopes. Untreated protein (UN) was applied on the left-most column. Conditions included 10 mM dithiothreitol (DTT), heat, 1% SDS, 6M urea, 6M guanidinium HCl (Guan), 0.3% acrylamide (Acry), and 0.5M hydrochloric acid (HCl). To verify presence of proteins, blots were also probed against Fc (bottom panel).
We hypothesized that NOTCH3 could be modified from its native state to reveal latent epitopes in the N-terminus. To test this possibility, we chemically treated purified wildtype or R90C NOTCH3 fusions prior to probing with antibodies. Of the six biochemical treatments initially tested, only DTT converted wild type or mutant NOTCH3 to a form that was recognized by antibodies (Figure 4).
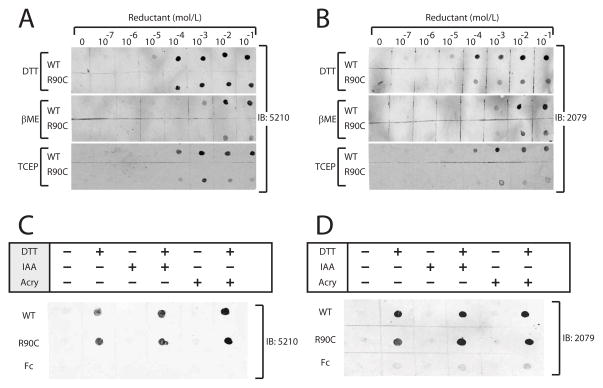
We then tested the effects of increasing concentrations of DTT, beta-mercaptoethanol (a thiol reductant) and TCEP (a phosphorus-based reductant) on NOTCH3 protein. All reductants efficiently converted both wildtype and R90C NOTCH3 to a form recognizable by all four antibodies (Figure 5).
Figure 5.
Effect of reducing reagents on N-terminal epitopes in NOTCH3. As in Figure 4, purified recombinant NOTCH3 ectodomain fused to mouse Fc were treated prior to application to nitrocellulose filters. Proteins were treated with increasing concentrations of indicated reducing agents for 60 minutes at 37°C. Antibody binding to wild type NOTCH3-Fc (WT) and R90C NOTCH3-Fc (R90C) protein is shown. Similar results were obtained with the 5210 (A) and 2079 (B) antibodies. Chemical used to treat protein include thiol reductants dithiothreitol (DTT) and beta-mercaptoethanol (βME) and the phosphorus-based reductant tris(2-carboxyethyl)phosphine (TCEP). (C–D) After reduction with 10mM DTT, NOTCH3 protein was alkylated with either iodoacetic acid (IAA; 100mM) or acrylamide (acry; 0.3%) and applied to nitrocellulose filters. Filters were then probed with 5210 (C) or 2079 (D). Alkylation had no effect on epitope recognition by either N-terminal NOTCH3 polyclonal antiserum.
Since antibodies only bound to reduced NOTCH3, it was possible that they interacted with thiol groups of NOTCH3. If this were the case, we would expect that reduction followed by alkylation of the protein would reduce antibody recognition. However, addition of alkylating agents acrylamide or iodoacetate after protein reduction did not affect antibody reactivity (Figure 6), suggesting that epitope recognition did not require protein thiols.
3. Discussion
CADASIL has been extensively studied because, in part, it is the most common small vessel disorder with an identified monogenic cause2. Stereotyped cysteine mutations in NOTCH3 and accumulation of NOTCH3 protein in vessels have suggested a proteinopathy involving NOTCH3, but changes in the protein structure of NOTCH3 have yet to be explored. Here, we present the first evidence of molecular changes in the NOTCH3 protein in CADASIL brain tissue.
Characteristics of NOTCH3 N-terminal epitopes
The molecular changes in NOTCH3 described herein were only found after protein reduction. Two classes of molecular changes could account for induction of latent epitopes by protein reduction: 1) an accessible (surface-exposed) region of the protein shifts conformation and 2) an inaccessible region of the protein moves to the surface of the protein and becomes accessible to antibodies. The antibodies we used were polyclonal and generated against peptides that likely adopt many conformations. It would be unlikely that all four polyclonal antibodies fail to bind accessible regions of the protein, particularly after exposure of the protein to multiple denaturing treatments (Figure 4). The absence of polyclonal antibody binding to native NOTCH3 is thus unlikely to result solely from a conformational change in an antibody-accessible N-terminal target region.
A more likely possibility is that epitopes are shielded in the native disulfide bonded protein and that reduction of disulfide bonds frees the molecule from structural constraints, allowing antibodies access to epitopes previously buried in the N-terminus. As cysteine alkylation failed to block antibody binding, the epitopes identified here do not interact with antibodies via reduced cysteines. Because the two peptides used to generate the antibodies are very close within the primary sequence, we further hypothesize that the end of the first EGF repeat and the beginning of the second EGF repeat are both sequestered, unavailable to antibodies, and may be in spatial proximity.
Potential mechanisms of N-terminal NOTCH3 epitope unmasking
The NOTCH3 N-terminal epitopes are abundant in CADASIL tissues but not normal-appearing control vessels or cells overexpressing native NOTCH3. Moreover, antibodies raised against N-terminal sequences of NOTCH3 bind strongly to protein treated with reductants but not to native protein or denatured NOTCH3 that remains oxidized. Together, these findings suggest that vascular redox changes in NOTCH3 may be one of the potential mechanisms for N-terminal epitope unmasking in CADASIL. This possibility is further strengthened by the stereotyped molecular genetics of CADASIL, which support a critical role of NOTCH3 cysteine imbalance. If reducing conditions indeed regulate latency of NOTCH3 epitopes, two experimental findings need additional explanation.
First, why do both wildtype and mutant NOTCH3 require chemical reduction for recognition by antibodies? The most basic explanation is that a single cysteine mutation is insufficient to cause CADASIL N-terminal NOTCH3 epitope generation. Instead, multiple reduced cysteines (as in the case of chemically reduced proteins) must occur to generate CADASIL N-terminal NOTCH3 epitopes. In CADASIL, a single mutation may predispose the protein to further reduction which consequently results in unmasking of N-terminal NOTCH3 epitopes.
A second question raised by our results is: how does one explain the finding that the same N-terminal NOTCH3 epitopes are unmasked in CADASIL samples that include a wide range of cysteine mutations dispersed throughout NOTCH3? Any molecular mechanism must account for how multiple different NOTCH3 mutants each results in changes within this focal area of the protein which, in some cases, is far removed from the site of the mutation. In vessels, it is possible that disulfides throughout NOTCH3 may slowly isomerize by exchange of cysteine disulfide partners. Many secreted proteins (e.g. RNase, BPTI, TSP) are known to shuffle disulfide pairing. When mutant NOTCH3 containing an odd number of cysteines undergoes disulfide isomerization, reduced cysteines may migrate to wide regions of the protein13. A series of isomerizations could yield the reduction of a critical cysteine important for generation of CADASIL NOTCH3 N-terminal epitopes, even if the CADASIL mutation is not at the N-terminus. Of note, remote effects of NOTCH3 mutation have been reported before; separate mutations in NOTCH3 have previously been observed to affect glycosylation at a single site14.
The model that CADASIL NOTCH3 N-terminal epitope unmasking is gated by a critical cysteine is experimentally testable; using the described antibodies, it may be feasible to perform mutagenesis experiments to identify the cysteine residue(s) required for generation of N-terminal structural changes. Another possibility is that other factors (e.g. Notch-binding matrix proteins10, 15–18), not active in our expression system, bind specifically to NOTCH3 with a reduced cysteine. Subsequently, this factor could facilitate the unmasking of N-terminal epitopes in mutant NOTCH3 protein. The antibodies to NOTCH3 described herein could be utilized in biochemical assays to identify such factors.
Eventual identification of molecular pathways leading to exposure of new epitopes in NOTCH3 could provide insight into biochemical pathways involved in small vessel pathology. As such, our novel observations provide a launch point to investigating structural alterations of NOTCH3 as a key target in cerebral small vessel disease.
4. Materials and Methods
Generation of polyclonal antibodies
Two peptide sequences from human NOTCH3 were used to immunize rabbits (Figure 1A). Immunization of amino acids 64–80 (ACLCPPGWVGERCQLED) resulted in antisera 5209 and 5210. Immunization of amino acids 85–95 with the R90C substitution (GPCAGCGVCQS) resulted in antisera 2078 and 2079. For some studies, 5209 and 5210 were pooled and affinity purified against the immunizing peptide. 2078 and 2079 were tested separately and both exhibited the same specificity for CADASIL arteries and for reduced purified NOTCH3 protein. Since both 2078 and 2079 reacted against CADASIL tissues with diverse mutations and to wild type and R90C proteins, we reasoned that the antisera was not specific for the R90C mutant. These antibodies were used as crude sera in experiments, unless specified.
The specificity of antisera was determined in multiple experiments, described in Results. First, all sera were found to react avidly against CADASIL but not against control tissues. Second, all sera had significantly greater reactivity with reduced NOTCH3-Fc compared to Fc control protein. Third, no specific reactivity with vessels or purified protein was observed with pre-immune sera. Fourth, reactivity of 5209 and 5210 was more than 30- and 50-fold higher against reduced NOTCH3 compared to NOTCH1 and NOTCH2. 2078 and 2079 reacted two- and six-fold higher against NOTCH3 compared to NOTCH1 and NOTCH2. Thus, 5209 and 5210 were significantly more specific than 2078 and 2079 for NOTCH3.
Immunohistochemistry
Brain tissue and genetic mutations in NOTCH3 from six CADASIL patients have been described before10. The study was granted an exemption from IRB review. We examined two additional patients with the mutations R153C and R141C. Locations of mutations of these patients are shown in Figure 1A. Control patients have been described elsewhere11. Tissue was processed into formalin-fixed, paraffin embedded sections followed by citrate mediated antigen retrieval. Control stains were performed on tissue sections with anti-H2 (sc-59467; Santa Cruz Biotechnology) to validate antigenicity of brain proteins in archival material. Cell lines stably expressing NOTCH3 were derive from human embryonic kidney 293 (293) cells stably transfected with plasmids directing the expression of wildt ype or mutant human NOTCH3 and have been previously described 12. Cells were collected by centrifugation, fixed in formalin, and then processed using the same procedure used for brain tissue. All staining was performed using Vector Elite ABC reagents.
Epitope characterization
The production and purification of recombinant NOTCH3 has been described elsewhere 12. The protein contains EGF-like repeats 1–27 from human NOTCH3 fused to mouse Fc. Briefly, 293 cell lines stably transfected with NOTCH3-Fc constructs were shifted to serum free media. Media was collected and then passed over a protein A agarose column. Fusion protein was eluted at with acidic buffer, immediately neutralized with Tris pH 7.4, and then extensively dialyzed. Mouse Fc was purchased from R&D Systems (Minneapolis, MN). Chemicals were purchased from Sigma-Aldrich.
To investigate determinants that unmask NOTCH3 epitopes, chemicals that exert predicted effects on proteins were mixed with NOTCH3 protein. For alkylation experiments, proteins were mixed with reductants for 60 minutes at 37°C, and then a 10-fold molar excess of alkylating agent (compared to thiol reductants) was added for an additional 60 minutes. Protein (50ng) was then spotted on nitrocellulose. Protein spots were allowed to dry completely, and the filter was then rinsed extensively to remove small molecules. Primary antibodies were added at 1:100 overnight, followed by detection with infrared labeled secondary antibodies which were detected on a flatbed scanner (Licor Biosciences, Lincoln, NE).
Highlights.
Four NOTCH3 antibodies specifically recognize degenerating arteries in CADASIL.
These antibodies recognize epitopes in chemically reduced, but not denatured, NOTCH3 protein.
This is the first evidence of structural changes in NOTCH3 in small vessel disease in human brain.
Acknowledgments
Funding: NS054724 (NIH), NS062816 (NIH), and BX000375 (VA). The Michigan ADRC is supported by grant P50-AG08671 and supplied control tissues for studies. Other human tissues were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, contract HHSN275200900011C, Ref. No. NO1-HD-9-0011.
Dr. Andrew Lieberman of the Michigan Alzheimer’s Disease Research Center (ADRC) provided control patient samples. Additional control samples were also obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, contract HHSN275200900011C, Ref. No N01-HD-9-0011. This study would not be possible without patients and families who provided important tissues resources for this study. Alan Burgess, Gia Straw, and Ano Nwokoye (UM Comprehensive Cancer Center) and Drs. Phillip Ursell and Melike Pakmezci (UCSF) provided important support for this study. The Michigan ADRC was supported by grant P50-AG08671.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, et al. Notch3 mutations in cadasil, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 2.Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG. Cadasil. Lancet Neurol. 2009;8:643–653. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- 3.Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C, et al. Strong clustering and stereotyped nature of notch3 mutations in cadasil patients. Lancet. 1997;350:1511–1515. doi: 10.1016/S0140-6736(97)08083-5. [DOI] [PubMed] [Google Scholar]
- 4.Kopan R, Ilagan MX. The canonical notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiruma-Shimizu K, Hosoguchi K, Liu Y, Fujitani N, Ohta T, Hinou H, et al. Chemical synthesis, folding, and structural insights into o-fucosylated epidermal growth factor-like repeat 12 of mouse notch-1 receptor. J Am Chem Soc. 2010;132:14857–14865. doi: 10.1021/ja105216u. [DOI] [PubMed] [Google Scholar]
- 6.Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, et al. Structural and functional properties of the human notch-1 ligand binding region. Structure. 2004;12:2173–2183. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Knott V, Downing AK, Cardy CM, Handford P. Calcium binding properties of an epidermal growth factor-like domain pair from human fibrillin-1. J Mol Biol. 1996;255:22–27. doi: 10.1006/jmbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 8.Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, Stuart D. The structure of a ca(2+)-binding epidermal growth factor-like domain: Its role in protein-protein interactions. Cell. 1995;82:131–141. doi: 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 9.Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, et al. The ectodomain of the notch3 receptor accumulates within the cerebrovasculature of cadasil patients. J Clin Invest. 2000;105:597–605. doi: 10.1172/JCI8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Meng H, Blaivas M, Rushing EJ, Moore BE, Schwartz J, et al. Von willebrand factor permeates small vessels in cadasil and inhibits smooth muscle gene expression. Transl Stroke Res. 2012;3:138–145. doi: 10.1007/s12975-011-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong H, Ding H, Young K, Blaivas M, Christensen PJ, Wang MM. Advanced intimal hyperplasia without luminal narrowing of leptomeningeal arteries in cadasil. Stroke. 2013;44:1456–1458. doi: 10.1161/STROKEAHA.111.000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng H, Zhang X, Yu G, Lee SJ, Chen YE, Prudovsky I, et al. Biochemical characterization and cellular effects of cadasil mutants of notch3. PLoS One. 2012;7:e44964. doi: 10.1371/journal.pone.0044964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang EM, Detwiler TC, Milev Y, Essex DW. Thiol-disulfide isomerization in thrombospondin: Effects of conformation and protein disulfide isomerase. Blood. 1997;89:3205–3212. [PubMed] [Google Scholar]
- 14.Arboleda-Velasquez JF, Rampal R, Fung E, Darland DC, Liu M, Martinez MC, et al. Cadasil mutations impair notch3 glycosylation by fringe. Hum Mol Genet. 2005;14:1631–1639. doi: 10.1093/hmg/ddi171. [DOI] [PubMed] [Google Scholar]
- 15.Meng H, Zhang X, Hankenson KD, Wang MM. Thrombospondin 2 potentiates notch3/jagged1 signaling. J Biol Chem. 2009;284:7866–7874. doi: 10.1074/jbc.M803650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng H, Zhang X, Lee SJ, Strickland DK, Lawrence DA, Wang MM. Low density lipoprotein receptor-related protein-1 (lrp1) regulates thrombospondin-2 (tsp2) enhancement of notch3 signaling. J Biol Chem. 2010;285:23047–23055. doi: 10.1074/jbc.M110.144634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monet-Lepretre M, Haddad I, Baron-Menguy C, Fouillot-Panchal M, Riani M, Domenga-Denier V, et al. Abnormal recruitment of extracellular matrix proteins by excess notch3 ecd: A new pathomechanism in cadasil. Brain. 2013;136:1830–1845. doi: 10.1093/brain/awt092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Meng H, Wang MM. Collagen represses canonical notch signaling and binds to notch ectodomain. Int J Biochem Cell Biol. 2013;45:1274–1280. doi: 10.1016/j.biocel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]