Abstract
Polyploidy is common among flowering plants, including the Asteraceae, a relatively recent angiosperm group. EST-SSRs were used to characterize polymorphism among 29 Chrysanthemum and Ajania spp. accessions of various ploidy levels. Most EST-SSR loci were readily transferable between the species, 29 accessions were separated into three groups in terms of the number of fragments. It inferred that the formation from tetraploid to hexaploid and from octoploid to decaploid may be a recent event, while from the diploid to the tetraploid may be an ancient one in the Chrysanthemum lineage. EST-SSR polymorphism was found and some transcripts containing an SSR were transcribed differently in the de novo autotetraploid C. nankingense and C. lavandulifolium than in their progenitor diploid. EST-SSR could provide a potential molecular basis of adaptation during evolution, while whole genome duplication has a major effect on the mutational dynamics of EST-SSR loci, which could also affect gene regulation.
Whole genome duplication (WGD) is a frequent event during the evolution of the angiosperms1, the majority of species now are polyploids or cryptic polyploids2. WGD has been associated with the induction of various alterations to both genome sequence and patterns of gene expression, and some of these changes likely favored evolutionary adaption1,3. It is suggested that changes both in DNA sequence and methylation can result in the differential expression of homoeologous genes, leading to the adaptive phenotypic variation3, however the mechanisms underlying these changes remained poorly understood.
Microsatellites, or simple sequence repeats (SSRs), are ubiquitous throughout both the coding and non-coding regions of all eukaryotic genomes4. SSRs derived from EST (expressed sequence tag) libraries (EST-SSRs) show a higher rate of interspecies transferability than genomic SSRs which reside in the non-coding region of the genome5. SSRs, as highly polymorphic, easily reproducible, codominant markers, have been widely exploited to assess genetic diversity, identify germplasm, and linkage mapping6. New alleles at SSR loci are likely to be generated by a combination of DNA slippage during replication, imperfect mismatch repair, selection and other factors7,8. Although most of the SSR variation are functionally neutral, SSR variation in coding regions may have functional significance, including chromosomal organization, DNA structure, protein binding, gene transcription and translation, etc9,10,11,12,13, which provides the basis for rapid evolution14,15,16,17. Nonetheless, little information about the origin and variation of SSRs in polyploidy is available.
The tribe Anthemideae Cass. (Asteraceae) is one of the largest group of flowering plants. It consists of at least 25,000 species, with a large representation of polyploids18,19. The genus Chrysanthemum has a basic chromosome number of nine, and includes a range of ploidy levels from diploid (2n = 2x = 18) to decaploid (2n = 10x = 90). Based on the EST assembly and gene family statistics for 18 species of Asteraceae, the evolved Asteraceae genomes are thought to have experienced at least three ancient WGD events20. The potential importance of WGD for species diversification21,22 suggests that it has contributed to the ecological and morphological diversity of the Chrysanthemum spp. The diploid species C. nankingense and C. lavandulifolium are native to China, both species contribute to the origin of polyploids of Chrysanthemum22. Although, polyploid is a fundamental biological process, it is relatively underexplored and little is known about duplicate genes or genome changes in WGD. In the previous study, we detected rapid changes at the genetic level and some novel fragments containing microsatellite sequence in Chrysanthemum spp. during allopolyploid formation, suggesting that WGD may be accompanied by the modification of microsatellite sequence23. As no SSR primers were reported before, we then successfully developed over 2,000 EST-SSR primers from C. nankingense EST libraries for Chrysanthemum spp. and some closely related species6. Here we intend to reveal how WGD alters EST-SSR loci employing diploid C. nankingense and C. lavandulifolium and their autotetraploids as well as Chrysanthemum spp. and Ajania spp. of different ploidy levels. The effects of WGD on gene expression were also investigated by comparing the abundance of SSR-containing transcripts in autotetraploid C. nankingense and C. lavandulifolium with that in their diploid progenitors, and whether these changes in DNA sequence could contribute the unidirectional changes in gene expression were discussed, all these findings will further expand our current evolutionary framework.
Results
Polymorphism survey with EST-SSR
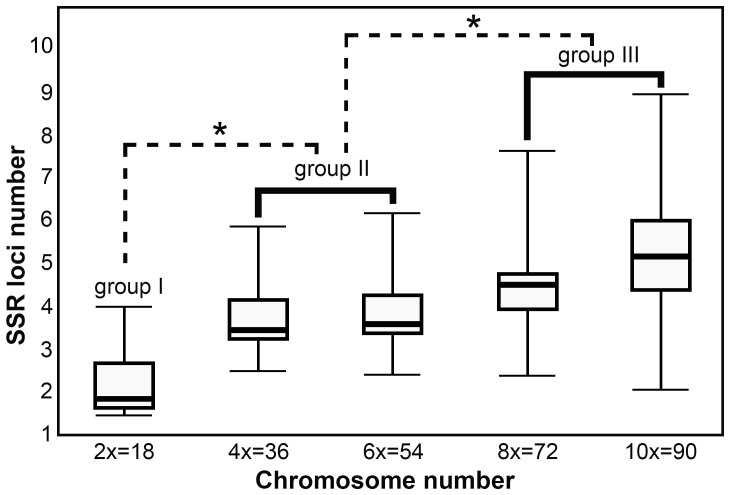
Twenty nine Chrysanthemum sect. accessions including 17 different Chrysanthemum spp. (23 accessions) and 6 Ajania spp. (6 accessions) were analyzed (Table 1). These ploidy levels varied from diploid (2n = 2x = 19) to decaploid (2n = 10x = 90)22,23,24,25,26. In total across the test accessions, 2,807 loci were detected by 20 EST-SSR primer pairs (Table 2, primer pairs #1-#20), and displayed polymorphism between nearly all of the accessions tested. The mean number of fragments amplified from the diploid species was less than from the higher ploidy ones (Fig. 1). For example, primer pair #5 recognized a mean of 1.67 fragments among the diploids, but, 2.56, 2.75, 4.00 and 4.33, respectively, among the tetra-, hexa-, octo- and decaploids (Table 3). Primer pair #8 detected a mean of 3.33 fragments among the diploids, and, 4.22, 5.13, 7.33 and 9.33, respectively, among the higher ploidy species. Similarly, primer pairs #4, #10, #12, and #14 detected lesser fragments in lower ploidy species than in higher ones. But primer pair #17 amplified a larger number of fragments from tetra- and hexaploid templates than from either the octo- or the decaploid ones (Table 3). A statistical analysis suggested 29 accessions fell into three groups in terms of the number of fragments: the diploids (2x species group), the tetra- and hexaploids (4x-6x species group), and the octo- and decaploids (8x-10x species group) according to Tukey's test and the Student's t test (P < 0.05). In general, the number of fragments amplified was highest for high ploidy group (8x-10x) and lowest for the diploids (2x), but there was no significant difference between either the 8x and 10x, or the 4x and 6x ones (Fig. 1).
Table 1. The materials grouped by Chrysanthemum and Ajania on the basis of their traditional classification.
| Ploidy | Taxa | Sample Size | Location |
|---|---|---|---|
| 2 x | Ajania myriantha | 5 | 31°51′N, 102°09′E |
| 2 x | Chrysanthemum nankingense | 5 | 32°07′N, 118°96′E |
| 2 x | Chrysanthemum dichrum | 5 | 37°33′N, 114°41′E |
| 2 x | Chrysanthemum makinoi | 5 | 36°25′N, 138°02′E |
| 2 x | Chrysanthemum boreale | 5 | 36°45′N, 139°05′E |
| 2 x | Chrysanthemum lavandulifolium | 5 | 39°94′N, 116°40′E |
| 4 x | Ajania tenuifolia | 5 | 32°91′N, 102°56′E |
| 4 x | Ajania przewalskii | 5 | 31°95′N, 102°21′E |
| 4 x | Chrysanthemum indicum (NJ) | 5 | 32°21′N, 118°51′E |
| 4 x | Chrysanthemum yoshinaganthum | 5 | 36°20′N, 140°12′E |
| 4 x | Chrysanthemum okiense | 5 | 34°38′N, 132°46′E |
| 4 x | Chrysanthemum zawadskii | 5 | 29°72′N, 118°35′E |
| 4 x | Chrysanthemum japonicum var. wakasaense | 5 | 36°11′N, 140°12′E |
| 4 x | Chrysanthemum chanetii | 5 | 38°74′N, 106°00′E |
| 4 x | Chrysanthemum indicum (HZ) | 5 | 30°27′N, 120°13′E |
| 6 x | Chrysanthemum vestitum (TZS) | 5 | 30°65′N, 116°56′E |
| 6 x | Chrysanthemum vestitum (DBS) | 5 | 31°11′N, 116°17′E |
| 6 x | Chrysanthemum morifolium ‘gongju’ | 5 | 29°78′N, 118°34′E |
| 6 x | Chrysanthemum morifolium ‘jinba’ | 5 | 29°78′N, 118°34′E |
| 6 x | Chrysanthemum japonense | 5 | 35°71′N, 139°73′E |
| 6 x | Chrysanthemum. japonense var. delile | 5 | 34°43′N, 132°49′E |
| 6 x | Chrysanthemum. japonense var. ashizuriense | 5 | 36°12′N, 140°04′E |
| 6 x | Chrysanthemum zawadskii | 5 | 34°40′N, 132°42′E |
| 8 x | Ajania shiwogiku | 5 | 36°11′N, 140°12′E |
| 8 x | Chrysanthemum ornatum | 5 | 33°05′N, 130°99′E |
| 8 x | Ajania × marginatum | 5 | 36°08′N, 140°16′E |
| 10 x | Ajania pacificum | 5 | 35°84′N, 139°91′E |
| 10 x | Chrysanthemum crassum (JD) | 5 | 36°71′N, 136°75′E |
| 10 x | Chrysanthemum crassum (DSN) | 5 | 35°64′N, 139°86′E |
Note. Abbreviations given in parenthesis denote different accessions of the same species.
Table 2. Primer sequences employed to assay variation at 20 EST-SSR loci.
| Primers ID | SSR motifs | Primer sequence (5′-3′) | Polymorphic |
|---|---|---|---|
| #1 | CAT | F: TTCCTCCTATAGCCAAGGCA | |
| R: GGCTGGATCAGTCGTTTCAT | |||
| #2 | ACC | F: AAACCACCAAACCCATCAAA |  |
| R: AACTTTGCCAGCATCGACTT | |||
| #3 | GTG | F: CGACAGTTGAGCAACAGGAA |  |
| R: CCAATCACCGACACATCATC | |||
| #4 | CAC | F: TATCCCACTATGCCCACCAC | |
| R: AAACTGTGGTGCATGGTCAA | |||
| #5 | CACCTG | F: GGCACAGCAACTTCAAGACA | |
| R: ATTTTCTGCGGCATTTTCTG | |||
| #6 | AAGAGG | F: GGAAGAGGAGGAAGAGGACG | |
| R: CGTTTAGGGCTGCTTCTTTG | |||
| #7 | GAATGT | F: ATGGCATTGAGGAGGATGAG | |
| R: AACATTACCCGCACCAACAC | |||
| #8 | GGGTCA | F: CCAACAACCACAACAACAGC | |
| R: ACAGAACCATTATGCGGAGC | |||
| #9 | TGA | F: CAAAAGTGCAAATGGAGGCT | |
| R: TCCCCAAATAACAAACCAACA | |||
| #10 | CCT | F: AACTTACCAACCCCACTCCC | |
| R: AACACTGGTCTCCAATTCCG | |||
| #11 | AAG | F: GTTCACCGTCAATTCACACG | |
| R: ATGTTTCGACAGTCGCAGTG | |||
| #12 | TCA | F: GAGCGCAGGAAGAAGAGAAA | |
| R: GCGCTTCTTCTCACGTCTCT | |||
| #13 | ATC | F: CCCCAATTCCTGTATCATCA | |
| R: ATCAGCTCGGGCTCAATCTA | |||
| #14 | GAA | F: CACCCTCTTCATGTCAAAACC |  |
| R: ACCATTTTCATCCACCCAAT | |||
| #15 | GTG | F: CCGGTGTTCGGTATAAATGG |
 
|
| R: ACAATTCGCTTCGGCTCTAA | |||
| #16 | CAA | F: ATGGGGAGATATGCCCTTTC | |
| R: TAAGTAAATCTGCCCTGGCG | |||
| #17 | GCT | F: GGAATATTCGACCGGGATTT | |
| R: ACTGAATTTGCACAGTCCCC | |||
| #18 | AAG | F: TTCCCCATGAAGAGGAACTG | |
| R: TCTGATGTAGGCATAGATAGCACA | |||
| #19 | GAG | F: TCGTTAAAATGGCGTCATCA | |
| R: TCATTCCTATCGCCAAAACC | |||
| #20 | ATG | F: GACGACGACGACAATGAAGA | |
| R: CTTGATGGGAAACCGTCTGT | |||
| #21 | CAA | F: TGTCGATCATCCCAAGTTTC | |
| R: TTTAAGCCCTGGTACATCGG | |||
| #22 | CTT | F: CACCGGAATTGTAACCAACC | |
| R: GCAAATCACTTGCTCCAACA | |||
| #23 | TCC | F: CCCTTTTGTCTCAAAGAACACA | |
| R: GCACGTTGTAAAGAATGCGA | |||
| #24 | ACC | F: TCTATCTCACCGGTCACACG | |
| R: GTGCGTCCAAACATTGAGTG | |||
| #25 | TCA | F: AACCATGAATCCAGACACCC | |
| R: ACCAAGCCAGTCGAGTTTTG | |||
| #26 | TTC | F: GGCGATGGATGATGATGATT |  |
| R: GAAAGAGGTGGATCGGATGA | |||
| #27 | AAG | F: CTAGTCGTGCTCCTCAGGCT | |
| R: AGTTCGGCCATGACATTTTC | |||
| #28 | GAATGT | F: CGTTATCCCAATCGTCGTCT | |
| R: AACATTACCCGCACCAACAC | |||
| #29 | GGGTCA | F: CCAACAACCACAACAACAGC | |
| R: ACAGAACCATTATGCGGAGC | |||
| #30 | TCA | F: TCAATGGACATAAAAGAAAGA | |
| R: GGTGGACAATCCGGTCATAC | |||
| #31 | TTC | F: GATTTCCCGATCGAATCAGA |
 
|
| R: TACAACGGACGGTGCATAGA | |||
| #32 | ATC | F: CCCCAATTCCTGTATCATCA | |
| R: ATCAGCTCGGGCTCAATCTA | |||
| #33 | CAC | F: ACCATGGACGCCATATCAAC | |
| R: TGGTGCATATTGCATGGTCT | |||
| #34 | A(G)AGATG | F: CCTAGTATCAAAGCTGCGAACA |
|
| R: CAATCGCGTTATCGTGTACC | |||
| #35 | CCA | F: CTTGACTTCAGCGACCATCA | |
| R: TTTTGGCAATATGGAAAGGC | |||
| #36 | GTAGTG | F: ATCCTATACGACAGCTCCGC | |
| R: TTTGGTGGCCATCTTCTTTC | |||
| #37 | ACA | F: AACTACAACAACAACGGCCA | |
| R: AAGCTTGTGAATTCGTTGCC | |||
| #38 | CAC | F: CACCACCCTTTCCACACTTC | |
| R: AATGAAGACCCTGCACCAAG | |||
| #39 | TTC | F: AGTCCTCCTCCTCCTCCTTG | |
| R: TGGGAGAACAAACAACACCA | |||
| #40 | AAT | F: TCAAAGGATAAGAATTATGGA | |
| R: CTGTTTACGCAGCACTTCCA |
Note: Primer pairs #1-#20 were used for 29 accessions genotyping; Primer pairs #1-#40 were used for diploid C. nankingense and C. lavandulifolium and their autotetraploid;  : Polymorphic between C. nankingense diploid and tetraploid plants;
: Polymorphic between C. nankingense diploid and tetraploid plants;  : Polymorphic between C. lavandulifolium diploid and tetraploid plants.
: Polymorphic between C. lavandulifolium diploid and tetraploid plants.
Figure 1. The relationship between mean SSR allele number and chromosome number in Chrysanthemum spp.
The lines show the grouping of the ploidy levels into (1) diploid, (2) tetra- and hexaploids, and (3) octo- and decaploids according to Tukey's test and the Student's t test (P < 0.05).
Table 3. The relationship between mean SSR fragment number and ploidy level in Chrysanthemum spp.
| Primers IDPloidy level | |||||
|---|---|---|---|---|---|
| Primers ID | 2 x | 4 x | 6 x | 8 x | 10 x |
| #1 | 1.83 ± 0.40 | 3.89 ± 0.61 | 4.00 ± 0.19 | 4.00 ± 1.00 | 4.00 ± 0.00 |
| #2 | 1.83 ± 0.54 | 3.89 ± 0.56 | 5.00 ± 0.19 | 4.33 ± 0.88 | 6.00 ± 0.00 |
| #3 | 3.00 ± 0.45 | 4.44 ± 0.50 | 4.13 ± 0.52 | 4.33 ± 0.33 | 6.00 ± 1.00 |
| #4 | 1.67 ± 0.49 | 3.22 ± 0.36 | 2.88 ± 0.48 | 4.67 ± 0.33 | 5.00 ± 0.00 |
| #5 | 1.67 ± 0.33 | 2.56 ± 0.34 | 2.75 ± 0.31 | 4.00 ± 1.00 | 4.33 ± 0.33 |
| #6 | 1.50 ± 0.22 | 4.11 ± 0.51 | 3.50 ± 0.38 | 3.33 ± 0.67 | 6.00 ± 1.00 |
| #7 | 1.83 ± 0.31 | 3.33 ± 0.55 | 3.38 ± 0.46 | 4.67 ± 1.33 | 4.00 ± 0.00 |
| #8 | 3.33 ± 0.61 | 4.22 ± 0.72 | 5.13 ± 0.55 | 7.33 ± 1.20 | 9.33 ± 0.67 |
| #9 | 1.33 ± 0.21 | 2.44 ± 0.24 | 2.50 ± 0.33 | 2.33 ± 0.88 | 2.33 ± 0.67 |
| #10 | 3.50 ± 0.56 | 5.89 ± 0.59 | 4.88 ± 0.85 | 7.67 ± 0.33 | 9.00 ± 0.00 |
| #11 | 1.50 ± 0.22 | 2.78 ± 0.60 | 2.38 ± 0.42 | 3.67 ± 0.33 | 4.00 ± 1.00 |
| #12 | 1.67 ± 0.21 | 3.44 ± 0.50 | 3.63 ± 0.38 | 4.33 ± 0.33 | 6.00 ± 0.00 |
| #13 | 1.33 ± 0.33 | 3.33 ± 0.53 | 3.88 ± 0.81 | 5.33 ± 0.33 | 5.00 ± 1.00 |
| #14 | 2.67 ± 0.56 | 3.11 ± 0.54 | 3.63 ± 0.42 | 4.33 ± 0.88 | 6.67 ± 1.33 |
| #15 | 2.00 ± 0.26 | 3.22 ± 0.22 | 3.25 ± 0.41 | 3.33 ± 0.33 | 4.67 ± 0.33 |
| #16 | 1.33 ± 0.33 | 3.22 ± 0.22 | 3.38 ± 0.53 | 4.00 ± 1.00 | 5.33 ± 0.33 |
| #17 | 2.17 ± 0.60 | 3.33 ± 0.73 | 3.38 ± 0.60 | 2.33 ± 0.88 | 2.67 ± 0.33 |
| #18 | 1.83 ± 0.31 | 4.11 ± 0.26 | 3.63 ± 0.42 | 5.00 ± 0.58 | 5.33 ± 0.33 |
| #19 | 2.83 ± 0.31 | 4.44 ± 0.47 | 4.00 ± 0.33 | 4.67 ± 0.33 | 5.00 ± 0.00 |
| #20 | 2.50 ± 0.34 | 4.44 ± 0.34 | 4.25 ± 0.31 | 5.00 ± 1.53 | 7.33 ± 0.66 |
The effect on EST-SSR genotype of autopolyploidization
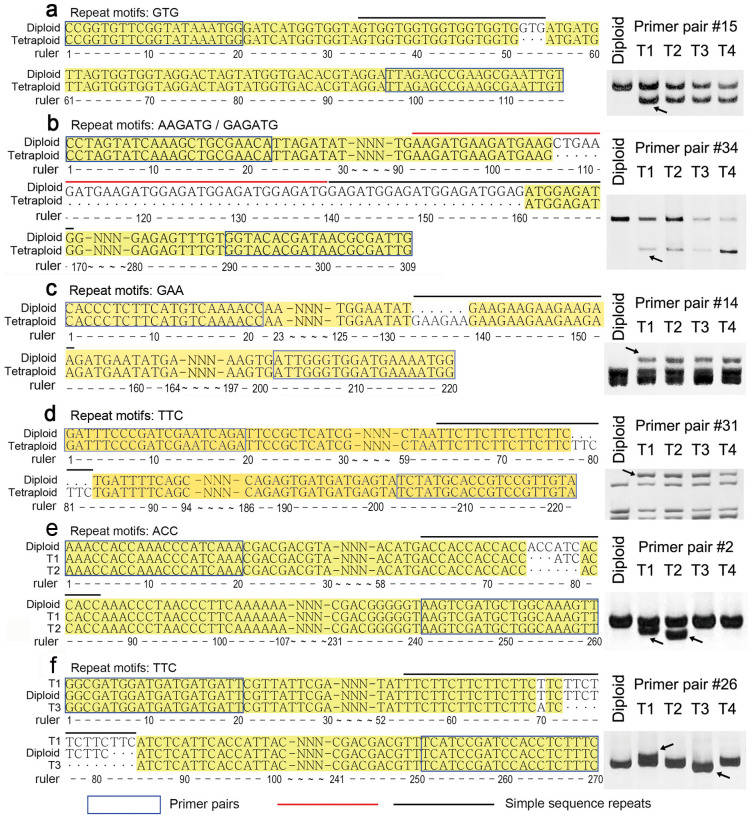
In the previous study, we detected rapid genomic SSRs variation in Chrysanthemum spp. during allopolyploid formation via amplified fragment length polymorphism markers, indicating that mutation of SSR sequence might be a rapid process in the nascent allopolyploid23. In present study, we aimed to address SSRs changes in autopolyploidy using four individual plants of autotetraploid C. nankingense (referred to as T1-T4)27, and six individuals of autotetraploid C. lavandulifolium. The changes were compared between autotetraploid and their progenitor diploids. For PCR amplification, genome DNA was used as template, 20 EST-SSR primer pairs (Table 2, primer pairs #1-#20) used for 29 accessions genotyping were employed. Only four primer pairs (#2, #3, #14 and #15) out of above 20 EST-SSR primer pairs were polymorphic. To generate more polymorphic fragments, additional 20 SSRs primer pairs were employed (Table 2, primer pairs #21-#40), as a result, three additional polymorphic primer pairs #26, #31 and #34 were screened. The PCR amplicons were electrophoresed through a 6% denaturing polyacrylamide gel. The expectation was that autopolyploidization would have no effect on the SSR genotype of polyploid, since their parent was a homozygous autotetraploid. However, six out of 40 EST-SSR loci detected showed polymorphic variation between C. nankingense diploid and its tetraploid plants. For example, primer pair #15 produced a 116 bp fragment including seven GTG repeats in diploid and tetraploid T1-T4 lines, in addition, a fragment including six repeats (113 bp) in tetraploids was detected (Fig. 2a). Primer pair #34 amplified a 309 bp fragment from both diploid and tetraploids, and additionally a 255 bp fragment from the tetraploid lines (Fig. 2b). Similarly, primer pairs #14 and #31 detected five GAA/TTC repeats in the diploid, but seven in the tetraploid (Fig. 2c, d). Primer pair #2 amplified a 260 bp fragment from the diploid, tetraploid T3 and T4 lines, but one more 257 bp fragment (one fewer ACC repeat) from tetraploid T1 line and one more 254 bp fragment (loss of ACCATC) from tetraploid T2 line (Fig. 2e). Primer pair #26 produced identical fragment among diploid and tetraploid T2 and T4 lines, however, the tetraploid T1contained an extra TTC repeat and the T3 amplicon included three TTC repeats delete compared with that in diploid (Fig. 2f). Although, a similar polymorphic variation was also found between C. lavandulifolium diploid and tetraploid plants, for example, primer pair #3 amplified a shorter fragment in three out of six tetraploid lines, while primer pair #15, #31 and #34 all produced an additional fragment in all tetraploid lines (Fig. 3).
Figure 2. Variation in SSR sequence among diploid and autotetraploid C. nankingense plants.
(a) A GTG repeat absent from the tetraploid amplicon; (b) #34 nine A(G)AGATG repeat absent from the tetraploid amplicon; (c–d) Two GAA (c) and TTC (d) repeat present in the tetraploid amplicons; (e) an ACC repeat absent from the T4 amplicon and an ACCATC repeat from the T1 amplicon; (f) an additional TTC repeat in the T1 amplicon, and the loss of three TCT repeats in the T3 amplicon. a: primer pair #15, b: primer pair #34, c: primer pair #14, d: primer pair #31, e: primer pair #2, f: primer pair #26. The cropped gels have been run under the same experimental conditions. Full-length gels are presented in Supplementary Fig. S1.
Figure 3. Variation in SSR sequence among diploid and autotetraploid C. lavandulifolium plants and pattern of variation in SSR sequence after polyploidization.
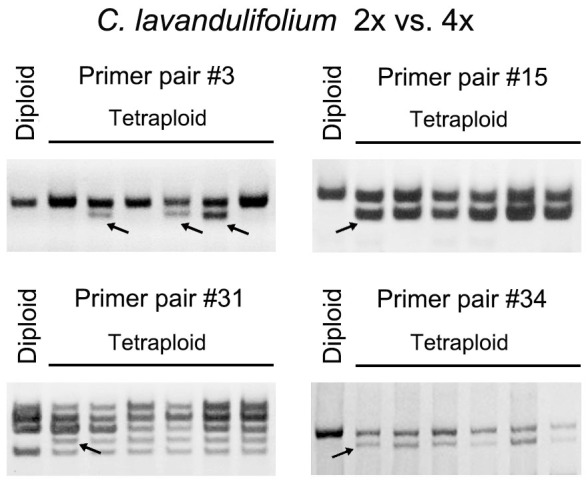
Four polymorphic variation EST-SSR loci were shown. The cropped gels have been run under the same experimental conditions. Full-length gels are presented in Supplementary Fig. S2.
To test whether changes in DNA sequence may contribute the unidirectional changes in gene expression and function, we isolated total RNA from leaves of diploid and autotetraploid C. nankingense and C. lavandulifolium, RT-PCR was conducted to compare whether these SSRs allelic variations were being expressed in tetraploid plants. RT-PCR based on the seven polymorphic primer pairs described above showed that no amplicon was produced from diploid and tetraploid T1-T4 lines using primer pairs #14 or #15 (Fig. 4a, b)(cDNA positive control and negative control data not shown), which suggested that loci #14 and #15 had the special temporal or spatial expression patterns. Primer pairs #2 and #31 of C. nankingense, and #31 and #15 of C. lavandulifolium all produced the same profile from all five templates (Fig. 4c, d). However, primer pair #26 in C. nankingense amplified an additional fragment from the T1 template and a shorter fragment from the T3 template and primer pair #3 in C. lavandulifolium amplified an additional shorter fragment in three tetraploid lines. Finally, primer pair #34 amplified a 309/297 bp fragment from the diploid C. nankingense/C. lavandulifolium template, but a 255/273 bp one from their tetraploid template (Fig. 4a, b), which mean that the genomic SSR variation could be transcribed in tetraploid plants.
Figure 4. Changed transcription patterns of SSR-containing genes upon autopolyploidization.
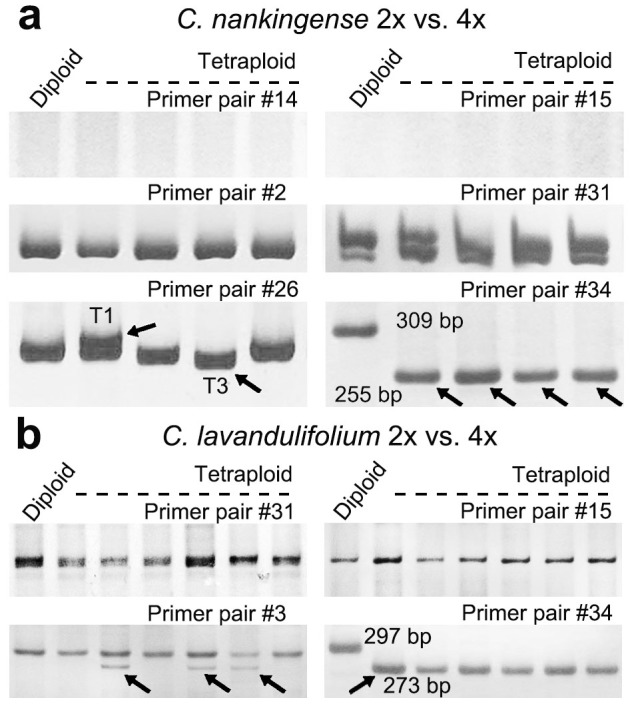
No amplicons were produced from either the diploid or any of the four tetraploid C. nankingense cDNA templates using primer pairs #14 or #15, while primer pairs #2 and #31 in autotetraploid C. nankingense and primer pairs #31 and #15 in autotetraploid C. lavandulifolium produced an identical amplicon from all five templates. However, primer pair #26 detected transcription changes in two (T1 and T3) of the four tetraploid C. nankingense plants and primer pair #3 detected transcription changes in three of six autotetraploid C. lavandulifolium plants. Primer pair #34 detected transcription in all autotetraploid but not in the diploid. The cropped gels have been run under the same experimental conditions. Full-length gels are presented in Supplementary Fig. S3.
Discussion
Understanding the evolution is one of the greatest questions and challenges in contemporary science28. WGD is considered as a prominent driver of evolution among the angiosperms. As polyploidy within the Asteraceae family is more frequent than among flowering plants in general, thus the Chrysanthemum spp. provide a particularly suitable group of plants for investigating the adaptation process resulting from polyploidization29,30,31.
The impact of evolution on genomic sequence is particularly well reflected in the behavior of SSR loci13. EST-SSRs resided in the open reading frame or any of the 5′- or 3′-untranslated regions are also an important part of genomic sequence, which may acquire a new function (i.e. neofunctionalization) or evolve towards complementation functions (i.e. subfunctionalization) via increasing or decreasing the length of homopolymeric amino acid stretches or regulating gene expression especially in the promoter region, so the lability of an SSR can vary the polyploids for local adaptation promoting long-term persistence15. The present data showed a positive correlation between ploidy level and EST-SSR fragment number (2x species group VS. 4-6x species group VS. 8-10x species group) in Chrysanthemum spp. due to the long-term evolution and development (Fig. 1). Traditionally, microsatellite mutation rates range from 10−3 to 10−4 per generation and microsatellite variation appears to be a complex phenomenon that is influenced by DNA slippage, mismatch-repair efficiency, selection and other factors7,8. However, when two different/same genomes are combined into a single cell, they must respond to the consequences of genome duplication, especially duplicate copies of genes with similar or redundant ones. Increased gene or genome dosage may induce disease syndromes and abnormal development32. This resembles the “genomic shock” phenomenon proposed by McClintock32. The genomic shock occurs rapidly in allo- or auto-polyploids, resulting in mutations, sequence eliminations, chromosomal rearrangements and large-scale transcriptional changes. Although, the evolutionary model of EST-SSR loci and the full range of factors underlying their sequence mutation rate remain uncertain following WGD, it seems to have been a measurable induction of EST-SSR sequence alteration in Chrysanthemum, since we detected 6 and 4 alleles at SSR loci (loci # 2, #14, #15, #26, #31 and #34 in C. nankingense and # 3, #15, #31 and #34 in C. lavandulifolium) shown a rapid variability, just as there was in de novo amphidiploids derived from the wide cross C. morifolium x Leucanthemum paludosum (Asteraceae)23. The assumption is that these changes are driven by genomic shock3, which may also accelerate SSR sequence variation and that some of the novel alleles induced provides the polyploid cell with a degree of genetic flexibility, as shown also with respect to repeat-induced point mutations in duplicated sequences of Neurospora crassa33.
In general, the higher ploidy Chrysanthemum species tended to be more polymorphic and have more amplified fragments than the diploids (Fig. 1). This pattern is quite general among Chrysanthemum. EST-SSR loci become duplicated upon WGD, but there are also instances of sub-genome level duplications involving whole chromosomes or segments within a chromosome. We also note that the variation of an allele or locus may occur due to single (or multiple) nucleotide substitutions, insertions or deletions occurring in flanking regions or SSR regions (even the eliminating DNA sequence) of different polyploid individuals, like loci #2 and 26 in C. nankingense and #3 in C. lavandulifolium, which may in time become fixed in the population as a result of “genomic shock” and a lack of further fragment accumulation in higher ploidy levels, however, all species which share similar randomly selected homologous polymorphic microsatellite systems will have fragment frequency spectra with the same mean size34,35. While comparisons involving a broad spectrum of species have shown that estimates of the timing of a whole genome (or other) duplication event is reasonably robust across the species set2,36. This means these species which have the similar polymorphic microsatellite systems will perform a more concentrated divergence time of WGD. The present statistical analysis suggested that although there was a difference in allele number between the 2x and 4x-6x species group, and between the 4x-6x and 8x-10x species group, there was no significant difference between either the 8x and 10x, or the 4x and 6x ones. Based on this observation, the formation time from tetraploid to hexaploid and from octoploid to decaploid may well have been a close event, but that from the diploid to the tetra-hexaploid group or to octo-decaploid group are more likely to have been a widely separated one in the Chrysanthemum lineage (Fig. 1). This scenario fits well with the observation that meiotic chromosome pairing is much looser in diploid than in polyploid ones, and that hybridization barriers are only serious in diploid x polyploid combinations37,38,39,40,41,42,43.
It is of interest to note that, although many SSR loci are genetically neutral44, strong evidence shows that some EST-SSRs can provide the molecular basis for rapid adaptation to a new environment especially for these tri-nucleotide and tri-nucleotide-based repeats SSR loci45. For example, rapidly evolved triplet repeats in plant may be the main reason for modulation of transcription factor activity and thus result in subtle or overt genomic effects9. These repeated codon changes in the number can increase or decrease the length of homopolymeric amino acid stretches which in turn may affect such properties as protein flexibility and binding affinity9. The number of repeats in different SSRs also can affect gene function, particularly vulnerable to mutation by virtue of having SSRs in its own coding regions7,8. Here, we have shown that WGD induced EST-SSR polymorphism in C. nankingense and C. lavandulifolium, and that certain genes (containing an SSR) were transcribed differently in de novo autotetraploids compared with that in their progenitor diploid (loci #26 and #34 in C. nankingense, loci #3 and #34 in C. lavandulifolium). In particular, the gene targeted by primer pair #34 produced an additional shorter version of the transcript in both C. nankingense and C. lavandulifolium autotetraploid compared with diploid (Fig. 4a, b), the cSSR variation completely replaced the origin one in the autotetraploid compared with diploid. Transcription polymorphism was also shown by the gene targeted by primer pair #26 and #3 (Fig. 4), where the autotetraploid plants generated an additional (Fig. 4a, b) and shorter transcript (Fig. 4a) than the one generated by the diploid.
To a certain degree, these changes of microsatellites may reflect the genetic variation after WGD. We cannot at the present exclude the possibility that the EST-SSR polymorphism induced in the C. nankingense and C. lavandulifolium autotetraploid modifies the function and/or efficiency of the gene products, possibly in an advantageous manner (Of course, the probability is too little). Plant genomes contain very large numbers of EST-SSRs, a proportion of which can contribute to the process of species adaptation during evolution9,46. In the previous study, de novo autotetraploids of C. nankingense is fertilisable and shows an improved level of adaptation to cold, drought and salinity stress47, here, EST-SSR variation was found almost immediately after WGD. Considering the potential function of EST-SSRs variation, we deduced that these EST-SSR variation may be a potential molecular basis for adaptation to new environments of Chrysanthemum spp.
Methods
Plant material and EST-SSR polymorphism survey
All materials have been maintained by the Chrysanthemum Germplasm Resource Preserving Centre, Nanjing Agricultural University, China. These ploidy levels varied from diploid (2n = 2x = 19) to decaploid (2n = 10x = 90)22,23,24,25,26. Fully expanded fourth and fifth leaves of plants from each accession were harvested and frozen in liquid nitrogen. Each accession consists of 5 individual plants. Genomic DNA was extracted using a modified CTAB method48. DNA extract was treated with RNase to remove any contamination of RNA, then dissolved in 50-μl TE (pH 8.0). The concentration and purity of the DNA preparations were measured using NanoDropND-1000 spectrophotometer (Nanodrop Technologies, USA), and the ratio of OD260/OD280 was calculated.
EST-SSR screening and polymorphism survey
A set of 20 EST-SSR primer pairs (synthesized by Invitrogen, Shanghai, China) was used to amplify each of the 29 templates (Table 2). All primers were developed from C. nankingense EST libraries (NCBI SRA accession: SRP041330; Dataset name: SRS595330-SRS595332), which could be used for Chrysanthemum spp. and some closely related species due to their cross-species transferability6. The PCR followed the three primer amplification protocol described elsewhere49 with minor modification. The PCR regime comprised an initial denaturation of 95°C/5 min, followed by seven cycles of 94°C/45 s, 68°C/45 s (decreasing by 2°C per cycle), 72°C/60 s, then 30 cycles of 94°C/45 s, 54°C/45 s, 72°C/60 s, with an extension step of 72°C/5 min. Each forward primer carried an M13 tail (5′-CACGACGTTGTAAAACGAC-3′) to enable fluorescent tailing, the fluorescent labeled amplicons were separated using an ABI3730xl DNA analyzer (Applied Biosystems, foster City, CA, USA) following the manufacturer's instructions, and amplicon sizes were estimated from ABI GeneScan LIZ500 size standards using GeneMapper v3.7 software (Applied Biosystems). The scored DNA-SSR fragments were transformed into a binary character matrix, where “1” indicated the presence and “0” showed the absence of a fragment at a particular position. Monomorphic fragments were excluded from the data sets. The used fragments were those fixed in all the 5 individuals of a species or variant, or at least with > 80% occurrence.
Alterations in EST-SSR sequences after WGD
Autotetraploid C. nankingense and C. lavandulifolium were induced following our previous report27. Briefly, nodal segments were immersed in 600 mg/L colchicine for 48 h, then rinsed three times in sterile water and placed on hormone-free MS medium41. 30 days later, the lateral buds had developed, and were transferred onto a rooting medium, until they had developed roots of length 1 cm, after which they were potted into a 2:2:1 (v/v/v) mixture of perlite, vermiculite and leaf mould, and grown in a greenhouse under natural light for two years.
Four individual plants of autotetraploid C. nankingense (referred to as T1-T4) and six individual autotetraploid C. lavandulifolium were genotyped, along with the progenitor diploid, 40 EST-SSR primer pairs were used (Table 2), those which proved to be polymorphic are shown in Table 2 (primer pairs # 2, #14, #15, #26 and #31 for C. nankingense and # 3, #15, #31 and #34 for C. lavandulifolium). The same PCR conditions were imposed as described above. The amplicons were electrophoresed at 200 V for 2 h through a 6% denaturing polyacrylamide gel in 1 × TBE buffer, and visualized by silver staining. Some polymorphic DNA fragments presented in diploid but absent in autotetraploid or those presented in autotetraploid while absent in diploid were excised directly from the gel using a razor blade, submerged in 40 μl ddH2O and boiled for 10 min, then cooled slowly to room temperature, and centrifuged at 17,000 g for 15 min. The DNA dissolved in the liquid phase was re-amplified using the same PCR conditions, and the resulting amplicon was visualized by electrophoresis (1.0% agarose gels) and inserted into the pMD19 TA vector (Takara, Japan) for sequencing confirmation (ABI3730xl DNA analyzer)50.
RNA extraction and RT-PCR analysis
Total RNA was isolated from leaves of diploid and autotetraploid C. nankingense and C. lavandulifolium using the TRIzol reagent (Takara), treated with RNase-free DNase I (Takara) at 37°C for 30 min according to the manufacturer's instructions to remove any contamination of DNA. The concentration of the extracted RNA was measured using a NanoDropND-1000 spectrophotometer. Total RNA samples (2 µg) with an OD260/OD280 ratio of >2.0 and an OD260/OD230 ratio of >1.8 were used to synthesize the first cDNA strand based on random priming and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Prior to RT–PCR analysis, the synthesized cDNA was diluted to 1/5 by adding sterilized ddH2O. A Chrysanthemum GAPDH gene (Forward primer sequence: 5′–CTGCTTCTTTCAACATCATTCC–3′, Reverse primer: 5′–CTGCTCATAGGTAGCCTTCTTC–3′; GenBank accession No. CMF007119) was used as the internal control for ensuring equal cDNA loading. To investigate the polymorphic fragments, the resulting cDNA was amplified using Pfu DNA polymerase (Takara) using the seven (of 40) informative EST-SSR primer pairs (Table 2), applying the same PCR conditions as above. Pure water was used as negative control. The amplicons were electrophoresed through a 6% denaturing polyacrylamide gel, visualized by silver staining. The scored cDNA-SSR fragments were transformed into a binary character matrix, where “1” indicated the presence and “0” showed the absence of a fragment at a particular position. The polymorphic cDNA fragments presented in diploid but absent in autotetraploid or those presented in autotetraploid while absent in diploid were isolated and sequenced as above.
Author Contributions
Conceived and designed the experiments: H.W., F.C., R.G., J.J., S.C., Performed the experiments: H.W., F.C., R.G., X.Q., Analyzed the data: H.W., B.D., Y.L. Contributed reagents/materials/analysis tools: W.F., Z.G., J.W. Wrote the paper: H.W., R.G., J.J., S.C. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
The study was supported by the National Natural Science Foundation of China (31372100, 31272203, 31272202, 31272196, 31171987), the Program for New Century Excellent Talents in University of the Chinese Ministry of Education (NCET-10-0492, NCET-12-0890), the Natural Science Fund of Jiangsu Province (BK2012773, BK2011641), the Fundamental Research Funds for the Central Universities” (KYZ201147), a scholarship granted to Dr. Sumei Chen (File No. 201306855010, CSC, Ministry of Education, China).
References
- Adams K. L. & Wendel J. F. Novel patterns of gene expression in polyploid plants. Trends Genet 21, 539–543 (2005). [DOI] [PubMed] [Google Scholar]
- Jiao Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011). [DOI] [PubMed] [Google Scholar]
- Chen Z. J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58, 377–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell W., Machray G. C. & Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1, 215–222 (1996). [Google Scholar]
- Zwenger S. R., Alsaggaf R. & Basu C. Does an expressed sequence tag (EST) library of Salsola iberica (tumbleweed) help to understand plant responses to environmental stresses? Plant Signal Behav 5, 1330–1335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Next-generation sequencing of the Chrysanthemum nankingense (Asteraceae) transcriptome permits large-scale unigene assembly and SSR marker discovery. PLoS One 8, e62293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. L. & Wong C. Mutation of human short tandem repeats. Hum Mol Genet 2, 1123–1128 (1993). [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet 16, 551–558 (2000). [DOI] [PubMed] [Google Scholar]
- Li Y. C., Korol A. B., Fahima T., Beiles A. & Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11, 2453–2465 (2002). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Microsatellite diversity correlated with ecological-edaphic and genetic factors in three microsites of wild emmer wheat in North Israel. Mol Biol Evol 17, 851–862 (2000). [DOI] [PubMed] [Google Scholar]
- Gur-Arie R. et al. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res 10, 62–71 (2000). [PMC free article] [PubMed] [Google Scholar]
- Hammock E. A. & Young L. J. Functional microsatellite polymorphism associated with divergent social structure in vole species. Mol Biol Evol 21, 1057–1063 (2004). [DOI] [PubMed] [Google Scholar]
- Li Y. C., Korol A. B., Fahima T. & Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21, 991–1007 (2004). [DOI] [PubMed] [Google Scholar]
- Caporale L. H. Natural selection and the emergence of a mutation phenotype: an update of the evolutionary synthesis considering mechanisms that affect genome variation. Microbiology 57, 467 (2003). [DOI] [PubMed] [Google Scholar]
- Kashi Y. & King D. G. Simple sequence repeats as advantageous mutators in evolution. Trends Genet 22, 253–259 (2006). [DOI] [PubMed] [Google Scholar]
- Parisod C., Holderegger R. & Brochmann C. Evolutionary consequences of autopolyploidy. New Phytol 186, 5–17 (2010). [DOI] [PubMed] [Google Scholar]
- Soltis D. E. et al. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56, 13–30 (2007). [Google Scholar]
- Bremer K. & Humphries C. J. Generic monograph of the Asteraceae-Anthemideae. Bulletin of the Natural History Museum. Botany series 23, 71–177 (1993). [Google Scholar]
- Lim K. Y. et al. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS One 3, e3353 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M. S. et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol 25, 2445–2455 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk V. A. et al. Everywhere but Antarctica: Using a supertree to understand the diversity and distribution of the Compositae Kgl. Biol Skr 55, 343–374 (2005). [Google Scholar]
- Liu P. L., Wan Q., Guo Y. P., Yang J. & Rao G. Y. Phylogeny of the genus Chrysanthemum L.: evidence from single-copy nuclear gene and chloroplast DNA sequences. PLoS One 7, e48970 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Rapid genetic and epigenetic alterations under intergeneric genomic shock in newly synthesized Chrysanthemum morifolium× Leucanthemum paludosum hybrids (Asteraceae). Genom Biol Evol 6, 247–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Rapid genomic and transcriptomic alterations induced by wide hybridization: Chrysanthemum nankingense x Tanacetum vulgare and C. crassum x Crossostephium chinense (Asteraceae). BMC genomics 14, 902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. B., Li C., Tang F. P., Chen F. D. & Chen S. M. Chromosome numbers and morphology of eighteen Anthemideae (Asteraceae) taxa from China and their systematic implications. Caryologia 62, 288–302 (2009). [Google Scholar]
- Zhao H. B., Chen F. D., Chen S. M., Wu G. S. & Guo W. M. Molecular phylogeny of Chrysanthemum, Ajania and its allies (Anthemideae, Asteraceae) as inferred from nuclear ribosomal ITS and chloroplast trnL-F IGS sequences. Plant Syst Evol 284, 153–169 (2010). [Google Scholar]
- Liu S. Y. et al. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. shows an improved level of abiotic stress tolerance. Sci Hortic 127, 411–419 (2011). [Google Scholar]
- Pennisi E. et al. So much more to know. Science 309, 78–102 (2005). [DOI] [PubMed] [Google Scholar]
- Yang J., Huang J., Gu H., Zhong Y. & Yang Z. Duplication and adaptive evolution of the chalcone synthase genes of Dendranthema (Asteraceae). Mol Biol Evol 19, 1752–1759 (2002). [DOI] [PubMed] [Google Scholar]
- Kane N. C., Barker M. S., Zhan S. H. & Rieseberg L. H. Molecular evolution across the Asteraceae: micro- and macroevolutionary processes. Mol Biol Evol 28, 3225–3235 (2011). [DOI] [PubMed] [Google Scholar]
- Beest T. M. et al. The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot 109, 19–45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science 226, 792–801 (1984). [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet 24, 579–613 (1990). [DOI] [PubMed] [Google Scholar]
- Chambers G. K. & MacAvoy E. S. Microsatellites: consensus and controversy. Comp Biochem Phys B 126, 455–476 (2000). [DOI] [PubMed] [Google Scholar]
- Amos W. A comparative approach to the study of microsatellite evolution. Microsatellites: Evol Appl 66, 79 (1999). [Google Scholar]
- Bowcock A. M. et al. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368, 455–457 (1994). [DOI] [PubMed] [Google Scholar]
- Dowrick G. The chromosomes of Chrysanthemum. III. Meiosis in C. atratum. Heredity 7, 219–226 (1953). [Google Scholar]
- Watanabe K. Studies on the control of diploid-like meiosis in polyploid taxa of Chrysanthemum. Theor Appl Genet (TAG) 66, 9–14 (1983). [DOI] [PubMed] [Google Scholar]
- Naxin C., Fadi C., Hongbo Z., Weimin F. & Zhouming W. Research on meiosis of some Dendranthema species and their hybrids. Acta Hortic Sinica 33, 1033 (2006). [Google Scholar]
- Li X., Chen F. & Zhao H. Daisy species hybridization compatibility study. Horticulture (in chinese) 35, 257–262 (2008). [Google Scholar]
- Watanabe K. The control of diploid-like meiosis in polyploid taxa of Chrysanthemum (Compositae). J Genetics (in japanese) 52, 125–131 (1977). [DOI] [PubMed] [Google Scholar]
- Langton F. Inheritance in Chrysanthemum morifolium Ramat. Heredity 62 (1989). [Google Scholar]
- Ronald W. & Ascher P. Self compatibility in garden chrysanthemum: occurrence, inheritance and breeding potential. Theor Appl Genet (TAG) 46, 45–54 (1975). [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P. & Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220 (1994). [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. The tuning function of tandemly repeating sequences: a molecular device for fast adaptation. In: Solomon, P. W. ed, Evolutionary Theory and Processes: Modern Horizons. Springer Netherlands, pp 115–138 (2004). [Google Scholar]
- Fondon J. W. 3rd & Garner H. R. Molecular origins of rapid and continuous morphological evolution. P Natl Acad Sci USA 101, 18058–18063 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T. & Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum 15, 473–497 (1962). [Google Scholar]
- Attitalla I. H. Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J Biol Sci 14, 998–999 (2011). [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnol 18, 233–234 (2000). [DOI] [PubMed] [Google Scholar]
- Ennis P. D., Zemmour J., Salter R. D. & Parham P. Rapid cloning of HLA-A,B cDNA by using the polymerase chain reaction: frequency and nature of errors produced in amplification. P Natl Acad Sci USA 87, 2833–2837 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information