Abstract
Despite the important role of MAPKs in signal transduction, their functions in the cellulase hyper-producing filamentous fungus Hypocrea jecorina haven't been studied except for the Hog1-like Tmk3. In this work, we constructed and explored the features of H. jecorina Δtmk2 to identify the role of this Slt2-homologous Tmk2. It is suggested from the results that Tmk2 is involved in cell wall integrity, sporulation and cellulase production. Although bearing similar roles in cell wall integrity maintenance, Tmk2 and Tmk3 appear to also have distinct functions: Tmk3 participates in high osmolarity resistance while Tmk2 does not; Tmk2 participates in sporulation but not Tmk3; Tmk3 is involved in promoting cellulase production while Tmk2 is involved in repressing cellulase formation. These studies provide the first insight into the function of Tmk2 in H. jecorina and contribute to understanding the signal transduction processes leading to the regulation of cellulase production in this important cellulase hyper-producer.
Hypocrea jecorina (syn. Trichoderma reesei) is one of the most important cellulase-producing industrial filamentous fungal species1. Isolated during World War II, it has received many rounds of improvement over the past 70 years to gain the cellulase hyper-secreting capability: modern industrial H. jecorina strains can usually produce over 20 g/L proteins2. This unusual feature has made H. jecorina one of the model cellulase-producing organisms for the understanding of cellulase production regulation and other prominent physiological processes in filamentous fungi.
The mitogen-activated protein kinase (MAP kinase, MAPK) pathways are among the most well-known signal transduction pathways in eukaryotes3. These pathways feature signal relay cascades involving three kinases: MAP kinase kinase kinase (MAPKKK), MAP kinase kinase (MAPKK) and MAP kinase4. These three kinases collaborate to pass phosphorylation states (the signal) from upstream sensors to MAPKKK, to MAPKK, to MAPK, and eventually to downstream elements such as transcription factors for the regulation of physiological functions. In filamentous fungi, three MAPKs have been identified, respectively homologous to yeast Hog1, Slt2 and Fus35,6.
Investigations on Hog1 homologues in Aspergillus nidulans7, Magnaporthe grisea8, Cryphonectria parasitica9, Neurospora crassa10, Trichoderma harzianum11, and Aspergillus fumigatus12 showed their involvement in adaptation to high osmotic pressure, a conserved role of Hog1 in both filamentous fungi and yeasts13,14. Hog1 homologues were also shown to be involved in other species-specific processes such as sporulation8,9, oxidative stress response12, thermal stress response15, tolerance to heavy metals11 and pigmentation9.
Investigations in filamentous fungi including Beauveria bassiana16, Fusarium graminearum17, N. crassa18, A. fumigatus19, Magnaporthe oryzae20, Claviceps purpurea21, Trichoderma virens22, Mycosphaerella graminicola23, A. nidulans24, and Coniothyrium minitans25 suggested Slt2 homologues primarily participate in cell wall integrity maintenance similarly to the budding yeast Saccharomyces cerevisiae26. Exceptions, however, do exist. For instance, deletion of slt2-like genes in Botrytis cinerea27 and Colletotrichum lagenarium28 didn't lead to weakened cell wall structure. Other pathways in which Slt2-homologues function include virulence16,17,20,29, sporulation17,18,25,27,29, female fertility17,18, secondary metabolism18,29,30, hyphal polarity maintenance18,19,31, surface hydrophobicity maintenance16,22,29, stress response19 and circadian rhythm maintenance32.
The functions of Fus3 homologues in filamentous fungi are quite diverse, out of which the participation in the secretion of extracellular proteins is of particular interest, exemplified in T. virens33,34 and Trichoderma atroviride35.
Despite the importance of H. jecorina in the understanding of fungal physiology, experimental investigations on its signal transduction pathways are quite limited5. In particular, signal transduction pathways transmitting external signals for the regulation of cellulase production, the most important physiological process in this cellulase hyper-producing species, are yet to be well understood. Previous in silico reconstruction of signal transduction pathways of H. jecorina suggested the presence of three MAPKs: Tmk1 that is homologues to yeast Fus3, Tmk2 that is homologues to yeast Slt2 and Tmk3 that is homologous to yeast Hog11. The functions of these MAPKs in H. jecorina were unknown before research carried out in our laboratory showed Tmk3 functions in cell wall integrity maintenance, high osmolarity resistance, biosynthesis and cellulase production regulation5. The unusual role of Tmk3 in cell wall integrity maintenance and the interesting participation of Tmk3 in cellulase production regulation lead to three questions: 1) since the Hog1-like Tmk3 acts similarity to Slt2-type MAPK in cell wall integrity maintenance, what is the role of the Slt2-like Tmk2? 2) Is Tmk2 simply a redundancy to Tmk3? 3) Does Tmk2 also regulate cellulase production and how? In this study, with these questions in mind, we attempted to identify the role of Tmk2 in H. jecorina, and to compare the functions of Tmk2 and Tmk3 in this very important cellulase-producing model organism.
Results
Construction, growth and sporulation of H. jecorina Δtmk2
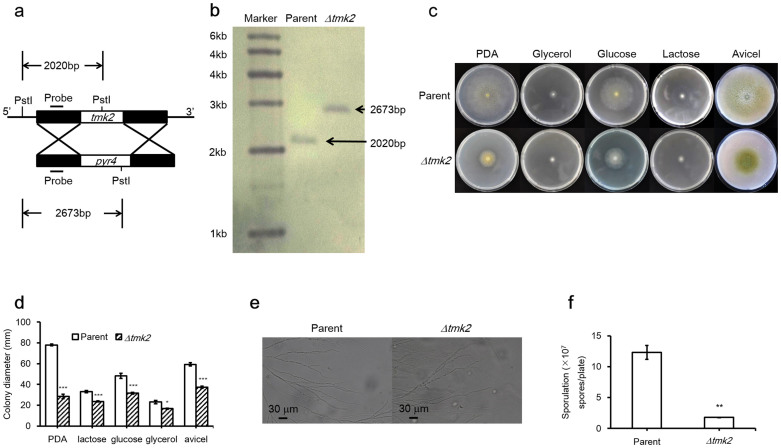
Sequence comparison between Slt2 from S. cerevisiae and Tmk2 from H. jecorina showed 69% sequence identity, supporting the previous suggestion that Tmk2 encodes a Slt2-type MAPK5 (see Supplementary Information 1 online for sequence alignment). H. jecorina Δtmk2 was constructed by homologous recombination using H. jecorina Δku70 as the parent strain, whose non-homologous end joining pathway was disrupted (Fig 1a–b). Both H. jecorina parent and Δtmk2 strains were grown on PDA plates, minimal media plates containing glucose, glycerol, or lactose as carbon sources, as well as Avicel double layer plates. A reduction of growth on agar plates was observed for H. jecorina Δtmk2 in comparison to the parent strain (Fig. 1c–d). To examine whether deletion of tmk2 leads to phenotypic changes of hyphae, spores of both H. jecorina parent and Δtmk2 strains were inoculated on glass slides on which a thin layer of PDA agar media was overlaid. The slides were incubated for two days to allow hyphal growth and were subsequently observed with a microscope. As is shown in Figure 1e, no apparent changes of hyphae morphology were observed, suggesting deletion of tmk2 doesn't substantially impact hyphae development in H. jecorina.
Figure 1. Growth of H. jecorina parent and Δtmk2 strains on plates, sporulation and microscopic observations of hyphae.
Panel a, schematic representation of tmk2 deletion. Panel b, Southern blotting analysis of H. jecorina parent and Δtmk2 strains. Panel c, H. jecorina parent and Δtmk2 strains grown on PDA plates, Avicel double layer plates and minimal media plates containing various carbon sources. Gly, glycerol; Glu, glucose; Lac, lactose; Avi, Avicel. Panel d, colony diameters of H. jecorina parent and Δtmk2 strains grown on PDA plates, Avicel double layer plates and minimal media plates containing various carbon sources. Panel e, microscopic images of hyphae from H. jecorina parent and Δtmk2 strains. Panel f, sporulation of H. jecorina parent and Δtmk2 strains grown on PDA plates. * P<0.05; ** P<0.01; *** P<0.001.
Sporulation levels of H. jecorina parent and Δtmk2 strains were assayed by inoculating the same amount of spores (1 × 105) on PDA plates and comparing the level of spore production after 6 days of growth. A significant smaller amount of spores was produced by H. jecorina Δtmk2 (1.78 ± 0.04 × 107/plate, mean ± s.d., n = 9) than the parent strain (1.23 ± 0.11 × 108/plate, mean ± s.d., n = 9, P = 0.0038, Fig. 1f). These results lead to the suggestion that deletion of tmk2 results in reduced sporulation in H. jecorina.
Involvement of Tmk2 in cell wall integrity maintenance
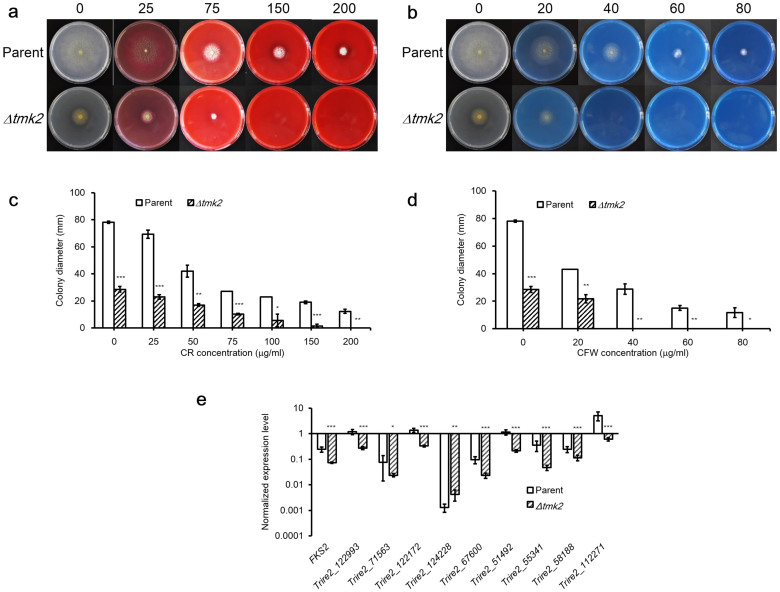
The sensitivity of H. jecorina parent and Δtmk2 strains to cell wall interfering substances Congo Red (CR) and Calcofluor White (CFW) was tested, in order to find out if deletion of tmk2 leads to defective cell wall. The parent strain could tolerate at least 200 μg/ml CR or 80 μg/ml CFW, while the Δtmk2 strain was unable to grow on plates containing 150 μg/ml CR or 40 μg/ml CFW (Fig. 2a–2d). These results lead to the suggestion that deletion of tmk2 results in increased sensitivity to cell wall interfering substances.
Figure 2. Involvement of H. jecorina parent and Δtmk2 strains in cell wall integrity maintenance.
Panel a, growth of H. jecorina parent and Δtmk2 strains on PDA plates containing various concentrations of CR. Numbers above the plates indicate concentrations of CR in each plate (in μg/ml). Panel b, growth of H. jecorina parent and Δtmk2 strains on PDA plates containing various concentrations of CFW. Numbers above the plates indicate concentrations of CFW in each plate (in μg/ml). Panel c, colony diameters of H. jecorina parent and Δtmk2 strains grown on PDA plates containing various concentrations of CR. Panel d, colony diameters of H. jecorina parent and Δtmk2 strains grown on PDA plates containing various concentrations of CFW. Panel e, normalized expression levels of chitin synthase- and β-1,3-glucan synthase-coding genes. The expression levels of each gene were compared to the expression levels of Actin-coding gene for normalization. * P<0.05; ** P<0.01; *** P<0.001.
We further analyzed the transcription of chitin synthase coding genes in H. jecorina parent and Δtmk2 strains to see if Tmk2 control the synthesis of chitin, a key component of cell wall. From the sequenced genome of H. jecorina, we were able to identify 9 chitin synthase coding genes (Trire2_112271, Trire2_58188, Trire2_55341, Trire2_51492, Trire2_124228, Trire2_122172, Trire2_71563, Trire2_67600, Trire2_122993) and one β-1,3-glucan synthase coding gene (Trire2_78176, fks), respectively involved in the synthesis of chitin and β-1,3-glucan, two major components of fungal cell wall. Real-time PCR analysis of the transcriptional levels of these genes in both H. jecorina parent and Δtmk2 strains showed they are clearly and significantly downregulated in H. jecorina Δtmk2 (Fig. 2e) except for Trire2_124228. Respectively, Trire2_112271, Trire2_58188, Trire2_55341, Trire2_51492, Trire2_122172, Trire2_71563, Trire2_67600, Trire2_122993 and fks were downregulated by 8.6 (P = 1.1 × 10−4, n = 9), 2.1 (P = 2.2 × 10−4, n = 9), 7.6 (P = 3.0 × 10−4, n = 9), 5.4 (P = 5.2 × 10−6, n = 9), 4.2 (P = 1.8 × 10−6, n = 9), 3.2 (P = 0.035, n = 9), 4.2 (P = 8.6 × 10−5, n = 9), 4.3 (P = 6.6 × 10−6, n = 9) and 3.3 (P = 9.8 × 10−6, n = 9) folds.
Involvement of Tmk2 in cellulase production
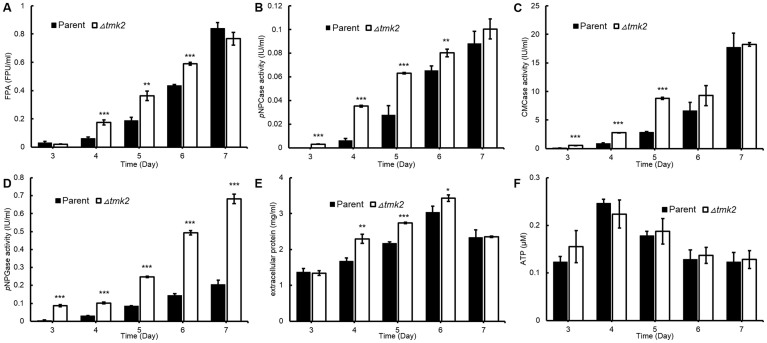
We assayed extracellular cellulase production in both H. jecorina parent and Δtmk2 strains. The cellobiohydrolase, endoglucanase and β-glucosidase activities were respectively measured by abilities to hydrolyze p-nitrophenyl-β-D-cellobioside (pNPC), carboxymethylcellulose (CMC) and p-nitrophenyl-β-D-glucopyranoside (pNPG). The overall cellulase activity was measured by the Filter Paperase Activity (FPA). As is shown in Figure 3a–e, higher cellulase production levels could be identified in the Δtmk2 strain: out of all 25 data sets (5 days ×5 activities/concentrations), 18 are statistically significantly different (n = 3, P<0.05) between H. jecorina parent and Δtmk2 strain. We further measured the ATP level in cultures according to previous suggestions and used it to represent the cellular biomass in the cultures36. As is shown in Figure 3f, the ATP accumulation in H. jecorina parent and Δtmk2 is not statistically different in submerged cultures used for cellulase production analysis, therefore leading to the suggestion that H. jecorina Δtmk2 produces more cellulase per unit of biomass than the parent strain.
Figure 3. Cellulase production and biomass accumulation of H. jecorina parent and Δtmk2 strains.
Panel a, extracellular protein secretion levels. Panel b, FPA levels. Panel c, abilities to hydrolyze pNPC (pNPCase activity levels). Panel d, abilities to hydrolyze CMC (CMCase activity levels). Panel e, abilities to hydrolyze pNPG (pNPGase activity levels). Panel f, biomass measured by ATP concentration in cultures. * P<0.05; ** P<0.01; *** P<0.001.
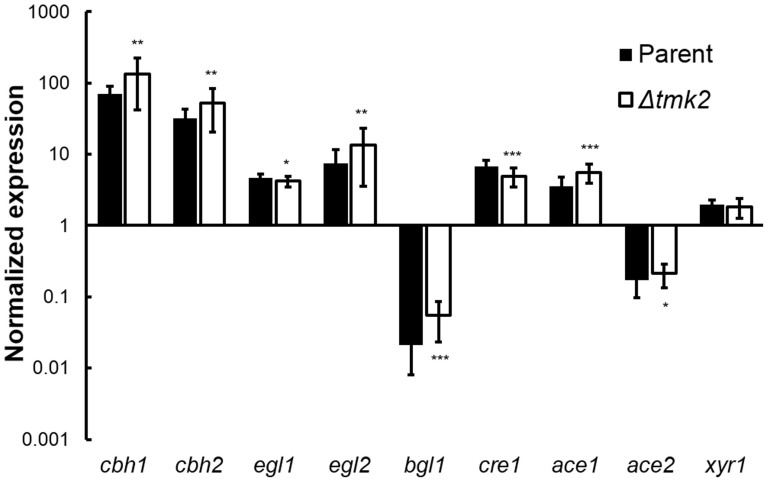
The transcriptional levels of cbh1, cbh2, egl1, egl2, bgl1, cre1, ace1, ace2 and xyr1, respectively coding cellobiohydrolase I, cellobiohydrolase II, endoglucanase I, endoglucanase II, β-glucosidase I, and known transcription factors that regulate cellulase expression were analyzed using qPCR in both H. jecorina parent and Δtmk2 strains. Although statistical tests suggested the upregulation or downregulation of all these genes but xyr1 in H. jecorina Δtmk2 in comparison with the parent strain, the difference between the two strains is too minor to conclude a biologically significant difference (Fig. 4). A suggestion can therefore be made that the modulation of Tmk2 on cellulase production is not on the transcriptional level.
Figure 4. Transcriptional abundance of cellulase- and transcription factor-coding genes.
The expression levels of each gene were compared to the expression levels of Actin-coding gene for normalization. * P<0.05; ** P<0.01; *** P<0.001.
Tmk2 is not involved in high osmolarity resistance or oxidative stress response
Both H. jecorina parent and Δtmk2 strains were grown on PDA plates containing NaCl or H2O2 (Fig. 5). Both strains could grow on all the plates containing NaCl. The ratios of colony diameters between H. jecorina Δtmk2 and parent strain didn't decrease when NaCl concentrations increased: the ratios were respectively 0.37, 0.51, 0.62, 0.71, 0.79 and 0.81 for plates containing 0 M, 0.2 M, 0.4 M, 0.6 M, 0.8 M and 1 M NaCl. These results suggested there is no apparent increase of sensitivity to higher contents of NaCl following tmk2 deletion, and further suggested Tmk2 is not involved in combating high osmolarity. Both H. jecorina parent and Δtmk2 strains could grow on plates containing 1 mM or 2 mM H2O2, but neither strains could grow on plates containing 3 mM or 4 mM H2O2 (Fig. 5). Similarly to NaCl-containing plates, the ratios of colony diameters between H. jecorina Δtmk2 and parent strains didn't decrease when H2O2 concentrations increased: 0.37 for plates containing 0 mM H2O2, 0.45 for plates containing 1 mM H2O2, and 0.48 for plates containing 2 mM H2O2, suggesting the sensitivity to H2O2 didn't increase after tmk2 deletion and that Tmk2 is not involved in combating oxidative stress.
Figure 5. Growth of H. jecorina parent and Δtmk2 strains on plates containing NaCl and H2O2.
Panel a, growth on plates containing NaCl. Panel b, growth on plates containing H2O2.
Discussion
The improved sensitivity of H. jecorina Δtmk2 to cell wall interfering substances, a typical phenomenon in cell wall defective strains, as well as the hampered transcription of chitin synthases in H. jecorina Δtmk2 lead to the suggestion that Tmk2 is involved in cell wall integrity maintenance in H. jecorina. This physiological role is not a surprise, because Slt2 homologues have been found to participate in this process in many other filamentous fungi such as T. virens22 and A. fumigatus19. The observation in this (Fig. 2) and previous work5 that both Tmk2 (Slt2-type MAPK) and Tmk3 (Hog1-type MAPK) are involved in cell wall integrity pathway, however, was novel in filamentous fungi. Our results cannot lead to the suggestion of the relationship between the two MAPKs in cell wall integrity pathway, but it is not uncommon that two different MAPKs play similar roles in the same organism. For instance, both Fus3 and Slt2 homologues in Cochliobolus heterostrophus are implicated to participate in conidiation, surface hydrophobicity maintenance and virulence29,37. Possible explanations to this phenomenon include the redundancy of signal transduction pathways to key physiological functions or the ability to receive the same upstream phosphorylation signals for structurally similar proteins (Tmk2 and Tmk3 share 43% sequence identity, see Supplementary Information 2 online for sequence alignment). It cannot be ruled out, however, that these MAPKs sharing similar roles play different important roles in the same pathway. All these possibilities need to be examined in more future in-depth investigations to further clarify the relationship between different MAPKs.
Cellulase production is a complicated process that is influenced by many external factors. Carbohydrate signals have a profound impact on cellulase production: quite a few di- and poly-saccharides such as cellulose, lactose and sophorose induce cellulase production38,39, while glucose represses cellulase production38. Light was shown to stimulate cellulase production in the model organism H. jecorina40. Investigations on signal transduction pathways that transmit these external signals to nucleus have helped to identify several key factors. For instance, G proteins, a PAS/LOV domain protein ENVOY and protein kinase A have been shown to be involved in light-regulated cellulase production41,42,43,44. It is a reasonable proposal that these external signals eventually determine phosphorylation states of transcription factors that regulate cellulase transcription and eventually lead to the modulation of cellulase production. Quite a few transcription factors have been shown to regulate the transcription of cellulase-coding genes. The most well studied transcription factors include activators Xyr145, XlnR46, ACEII47, Clr-148, Clr-248, as well as repressors ACEI49 and CreA50. Out of these transcription factors, Xyr1, ACEI, ACEII and CreA (termed Cre1 in H. jecorina) are present and have been investigated in H. jecorina. Interestingly, our previous prediction of phosphorylation sites in these proteins suggested the presence of MAPK phosphorylation sites in all of these transcription factors5, leading to the proposal that MAPKs are involved in the regulation of cellulase-coding genes, but which MAPK is involved in regulating the transcription of cellulase-coding genes is unknown. Our investigations on the role of Tmk2 in cellulase production lead to the suggestion that Tmk2 is involved in repressing cellulase production but not transcription (Fig. 3–4). This is in contrast to our previous research on Tmk35 in two aspects. For one, Tmk3 is involved in promoting cellulase production while Tmk2 is involved in repressing cellulase production. For two, Tmk3 regulates cellulase production on the transcription level but not Tmk2. From these results, we can exclude the possibility that Tmk2 phosphorylates these transcription factors. The mechanism by which Tmk2 regulates cellulase production is unknown and requires further investigation, but we can raise a preliminary hypothesis that Tmk2 is involved in post-transcriptional modulation and/or protein synthesis and secretion. More specifically, it is reasonable and promising to propose that the weakened cell wall benefits cellulase secretion in H. jecorina Δtmk2, a phenomenon also observed in H. jecorina Δtmk3 during growth on solid-state media5.
There have been many previous reports on factors involved in cellulase formation. Notably most of these identified factors are involved in promoting cellulase formation in filamentous fungi5,51, rather than suppressing cellulase formation. This is not a surprise because: 1) the cellulase production is a complicated process and therefore requires the involvement of many factors; 2) promoting, rather than repressing cellulase formation is beneficial to filamentous fungi except for in certain circumstances. One of these circumstances is when ‘easier' substances such as glucose are abundant. Under these circumstances, a process termed carbon catabolite repression is triggered, resulting in the inhibition of cellulase formation52. The well-known repressor of cellulase-coding gene transcription CreA is involved in this process. Reported in this work is another factor that's shown to repress cellulase production, which is different from previously identified factors on that the regulation doesn't happen on the transcriptional level, therefore providing an additional possibility for engineering H. jercorina to improve cellulase production.
Research carried out on Tmk3 in H. jecorina revealed one interesting phenomenon: the Δtmk3 strain grew very poorly on minimal media but only slightly worse than the parent strain on complete media (PDA)5. A preliminary hypothesis was made that Tmk3 could be involved in the biosynthesis of certain critical compounds that is not available in minimal media but available in PDA5. Unlike H. jecorina Δtmk3, although H. jecorina Δtmk2 doesn't grow as well as the parent strain on both minimal media plates and PDA plates, the growth on minimal media plates was not substantially worse than on PDA plates (Fig. 1c–d), suggesting the involvement of Tmk2 in the same biosynthetic pathway as Tmk3 is unlikely.
Another aspect of fungal physiology in which Tmk2 and Tmk3 show different participation is the resistance to high osmolarity. H. jecorina Δtmk3 was apparently more sensitive to elevated levels of NaCl in media, suggesting involvement of Tmk3 in resistance of high osmolarity. Results obtained in this work show a different response of H. jecorina Δtmk2 to higher osmolarity. If H. jecorina Δtmk2 is more sensitive to high osmolarity, we should be able to see a progressively decreased diameter ratio between H. jecorina Δtmk2 and parent strains following increased NaCl concentration in media. As is shown in Supplementary Information 3 online and Fig. 5a, what we actually observed is that the diameter ratio between H. jecorina Δtmk2 and parent strains stayed approximately constant between 0.2 to 1 M NaCl supplemented in the media, therefore suggesting H. jecorina Δtmk2 is not more sensitive to higher osmolarity in media and Tmk2 is not involved in high osmolarity resistance. The higher diameter ratio between H. jecorina Δtmk2 and parent strains when media is supplemented with 0.2 M NaCl than 0 M NaCl is an interesting finding. The intracellular osmolarity is similar to that of 0.2 M NaCl. When the media is not supplemented with additional osmotic stabilizers, the cells require the support of intact cell walls to prevent from too much water intake53. It can be proposed from these observations that the compromised cell wall in H. jecorina Δtmk2 couldn't provide the support cells needed to fully survive a low osmolarity environment, and therefore grew more poorly on plates containing no NaCl in comparison to growth on plates containing at least 0.2 M NaCl. This finding is in consistence with, and also provides additional evidence to, our suggestion that Tmk2 is involved in cell wall integrity maintenance.
Besides high osmolarity resistance and biosynthesis in which Tmk3 but not Tmk2 participates in, identified in this work is also one aspect of fungal physiology in which Tmk2 but not Tmk3 is involved. The decreased sporulation level in H. jecorina Δtmk2 in comparison with the parent strain suggests Tmk2 is involved in sporulation (Fig. 1f). This phenomenon was not observed in H. jecorina Δtmk3. Although these results cannot lead to the proposal of specified mechanism by which Tmk2 participates in the sporulation pathway, they are a clear indication that Tmk2 and Tmk3 function differently in this important physiological process.
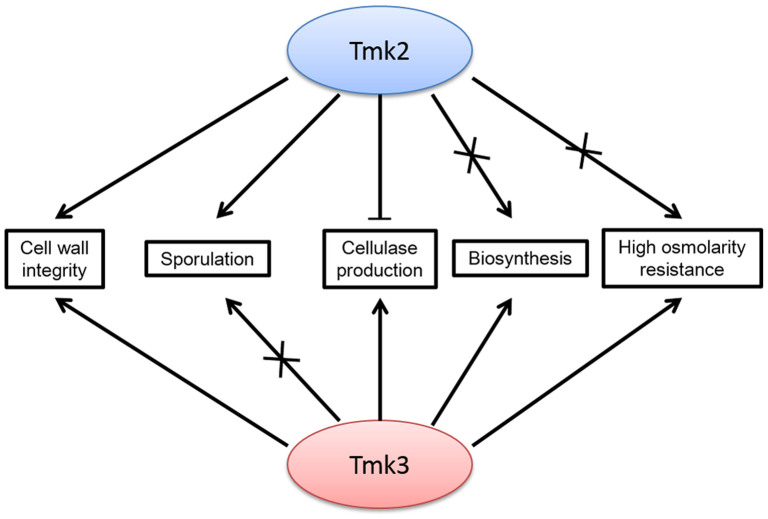
By summarizing the role of Tmk2 identified in this work and comparison with Tmk3, we can clearly see overlapping and distinct functions of Tmk2 and Tmk3 in H. jecorina (Fig. 6). They share similar roles in cell wall integrity maintenance, but have distinct functions in high osmolarity resistance, biosynthesis and sporulation. Our most interesting finding is that the deletion of tmk2 and tmk3 have drastically different consequences on cellulase production: downregulation for deletion of tmk3 and upregulation for deletion of tmk2. These observations also provide the answers to the questions raised in the beginning of this manuscript: Tmk2 also functions in cell wall integrity maintenance just like the other Slt2-like MAPKs; Tmk2 is far from simply a redundancy to Tmk3 in H. jecorina; Tmk2 also participates in cellulase production regulation, but in a different manner. A full scene of the MAPK functions in H. jecorina is still not revealed before further work could be carried out on the role of Tmk1, more detailed mechanisms of function for MAPKs, and the relationship between the three MAPKs, which hopefully will help us to more clearly understand fungal physiology in H. jecorina, in particular cellulase production and regulation mechanisms.
Figure 6. The overlapping and distinct roles of Tmk2 and Tmk3 in H. jecorina.
Methods
Strain and chemicals
H. jecorina Δku70 strain that is a derivative of the H. jecorina QM9414 uridine auxotrophic defective TU-6 strain (ATCC MYA-256) was used as the high transformation efficiency parent strain54. Wheat bran was a generous gift by Longlive Bio-Technology Co., Ltd., Yucheng, Shandong, China. Uridine was purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Calcofluor white (CFW), pNPG, pNPC and CMC were purchased from Sigma-Aldrich Corporation (St. Louis, MO, US). Congo red was purchased from Tianjin Damao Chemical Reagent Factory (Tianjin, China). All other chemicals were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
Sequence comparison
Sequence alignment between S. cerevisiae Slt2, H. jecorina Tmk2 and H. jecorina Tmk3 was carried out using the Clustal X2 software55.
Construction of H. jecorina Δtmk2
To construct H. jecorina Δtmk2, a knockout cassette was first constructed which contain the 1.5 kb upstream region of tmk2, pyr4 gene from H. jecorina, and 1.5 kb downstream region of tmk2. Transformation of the knockout cassette was carried out according to previously published protocols54. The transformants were grown on uridine-lacking minimal media plates for selection.
Growth of H. jecorina parent and Δtmk2 strains on plates
The spores of H. jecorina parent and Δtmk2 strains were first prepared by growing both strains on PDA plates in an incubator (Model DNP-9162, Shanghai Jing Hong Laboratory Instrument Co., Ltd., Shanghai, China) at 30°C and harvesting spores after 7 days. Spores were counted using a hemacytometer. Minimal media solution containing 0.5% NH4SO4, 0.06% MgSO4, 1.5% KH2PO4, 0.08% CaCl2, 0.00005% FeSO4·7H2O, 0.00016% MnSO4·H2O, 0.00014% ZnSO4·7H2O and 0.00002% CoCl2 was prepared. Minimal media plates containing 20 ml of minimal media solution, 0.4 g agar, 0.2 g carbon source and 20 mg uridine (parent strain only) were prepared. Avicel double layer plates were prepared as previously described5. Approximately 105 spores of H. jecorina parent and Δtmk2 strains were inoculated on PDA plates (4 replicates), Avicel double layer plates (3 replicates) and minimal media plates containing glucose (19 replicates for parent strain, 17 replicates for knockout strain), lactose (3 replicates) or glycerol (3 replicates). These plates except for Avicel double layer plates were incubated in an incubator (Model MJX-250, Ningbo Jiangnan Instrument Factory, Ningbo, China) at 30°C for 66 hours before checking for phenotypic features. The Avicel double layer plates were incubated in the same incubator at 30°C for 5 days prior to examination.
Microscopic observations
PDA agar plates were first prepared, from which a thin slice (~1 mm thick) of the media was cut off and laid carefully on top of a glass slide. A cover slide was then put on top of the media, creating a ‘sandwich'. A drop of water containing approximately 105 spores of H. jecorina parent or Δtmk2 strains was subsequently inoculated on the side of the cover slide. The inoculated sandwich was then transferred to a glass plate in which a piece of cotton rinsed with 30% glycerol was present to maintain moisture. The plate was then incubated at room temperature for 42 hours. The glass slides were then removed from the glass plates for microscopic observation with a bright field microscope (Nikon eclipse E100, 400 fold magnification).
Sensitivity to CR, CFW, NaCl and H2O2
Approximately 105 spores of H. jecorina parent and Δtmk2 strains were inoculated on PDA plates containing 25, 50, 75, 100, 150, 200 μg/ml CR, 20, 40, 60, 80 μg/ml CFW, 0.2, 0.4, 0.6, 0.8, 1.0 M NaCl, or 1, 2, 3, 4 mM H2O2. These plates were grown at 30°C in an incubator (Model MJX-250, Ningbo Jiangnan Instrument Factory, Ningbo, China) for 66 hours prior to examination. The diameters of colonies on each plate were measure for comparison. Three replicates of each experiment were performed.
Assay for sporulation
Approximately 105 spores of H. jecorina parent and Δtmk2 strains were inoculated on PDA plates and grown at 30°C in an incubator (Model MJX-250, Ningbo Jiangnan Instrument Factory, Ningbo, China) for 6 days until maximum sporulation was achieved. The spores were subsequently washed with washing solution (0.9% NaCl plus 0.05% Tween-80) and counted with a hemacytometer. Three biological replicates and three technical replicates for each biological replicate (a total of 9 replicates) were carried out for each strain.
Biochemical assays
H. jecorina parent and Δtmk2 strains were grown in liquid media containing 100 ml of minimal media solution, 2 g of wheat bran, 2 g of Avicel and 100 mg uridine (parent strain only). Approximately 106 spores were inoculated in liquid media to initiate growth. The cultures were shaken in a shaker (Model SKY-1112B, Shanghai Sukun Ltd., Shanghai, China) at 30°C and 200 rpm. Aliquots were drawn each day for biochemical assays. Tests for FPA, pNPCase, pNPGase, CMCase activities, extracellular protein levels and total ATP content were carried out essentially as previously described5. Three biological replicates for each activity were carried out.
Real-time PCR reactions
Real-time PCR (qPCR) reactions were carried out under identical conditions as in previous work on Tmk35. The reactions start with a preincubation step at 95°C for 300 seconds, followed by 40 cycles of three-step amplification (10 seconds at 95°C, 10 seconds at 59°C, 10 seconds at 72°C) and a melting step (10 seconds at 95°C, 60 seconds at 65°C, 1 second at 97°C). Data analysis was carried out using Microsoft Excel. Primers and amplicon sizes are indicated in Supplementary Information 4 online.
For evaluation of transcriptional abundance of chitin synthase- and β-1,3-glucan synthase-coding genes, approximately 106 spores of H. jecorina parent and Δtmk2 strains were inoculated in liquid media containing 100 ml minimal media solution, 2 g glucose and 100 mg uridine (parent strain only). The cultures were shaken in a shaker (Model SKY-1112B, Shanghai Sukun Ltd., Shanghai, China) at 30°C and 200 rpm for 3 days. The cultures were subsequently harvested for total RNA extraction. The cDNA was synthesized using PrimeScriptTM RT reagent kit with gDNA eraser (Perfect Real Time) from Takara Bio Inc. (Shiga, Japan). qPCR reactions were carried out on a LightCycler 96 Real-Time PCR system (Roche Applied Science, Mannheim, Germany) using FastStart Essential DNA Green Master (Roche Applied Science, Mannheim, Germany) as the dye. Three individual biological replicates and three individual technical replicates for each biological sample (a total of 9 replicates for each reaction) were carried out.
For evaluation of transcriptional levels of cellulase-coding genes and transcription factor-coding genes, H. jecorina parent and Δtmk2 strains were grown essentially as described above except for the inclusion of 2 g wheat bran and 2 g Avicel instead of glucose in the media. The synthesis of cDNA and performance of qPCR reactions were carried out similarly as described above. Nine biological replicates and three technical replicates for each biological replicate were carried out for cellulase-coding genes and transcription factor-coding genes (a total of 27 replicates).
Statistical analysis
For tests of significant difference between two sets of data, two-tailed Student's t-tests were carried out. P<0.05 was considered significant different. A minimum of three replicates were present in each set of data.
Author Contributions
M.W., Y.D., Q.Z., K.L. and B.J. constructed the knockout strain and carried out phenotypic analyses. Y.D., Q.Z. and F.W. carried out qPCR analyses. M.W. and X.F. conceived of the study, interpreted data, prepared figures and wrote the manuscript. All authors reviewed the manuscript. X.F. oversaw and coordinated the project.
Supplementary Material
Supplementary information
Acknowledgments
We thank Ms. Shaoli Hou for technical assistance. This work was supported by The National High-tech R&D program of China (863 program, No. 2011AA022302), National Natural Science Foundation of China (No. 31200051), National Energy Applied Technology Research and Demonstration Project (No. NY20130402), Shandong Province Natural Science Foundation (No.ZR2012CQ022), and Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry.
References
- Schmoll M. The information highways of a biotechnological workhorse-signal transduction in Hypocrea jecorina. BMC Genomics 9, 430 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R. & Nevalainen H. Trichoderma reesei RUT-C30-thirty years of strain improvement. Microbiology 158, 58–68 (2012). [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin M. C., Albertyn J., Alexander M. & Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. et al. A mitogen-activated protein kinase Tmk3 participates in high osmolarity resistance, cell wall integrity maintenance and cellulase production regulation in Trichoderma reesei. Plos One 8, e72189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L. P., Nicole M. C., Duplessis S. & Ellis B. E. Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant cell 24, 1327–1351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. & Prade R. A. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 43, 1065–1078 (2002). [DOI] [PubMed] [Google Scholar]
- Dixon K. P., Xu J. R., Smirnoff N. & Talbot N. J. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant cell 11, 2045–2058 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. M. et al. Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 51, 1267–1277 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lamm R., Pillonel C., Lam S. & Xu J. R. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68, 532–538 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Jarana J., Sousa S., González F., Rey M. & Llobell A. ThHog1 controls the hyperosmotic stress response in Trichoderma harzianum. Microbiology 152, 1687–1700 (2006). [DOI] [PubMed] [Google Scholar]
- Xue T., Nguyen C. K., Romans A. & May G. S. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot. Cell 3, 557–560 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbeck J., Pahlman A. K., Akhtar N., Blomberg A. & Adler L. Purification and characterization of two isoenzymes of DL-glycerol-3-phosphatase from Saccharomyces cerevisiae: Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 271, 13875–13881 (1996). [DOI] [PubMed] [Google Scholar]
- Albertyn J., Hohmann S., Thevelein J. M. & Prior B. A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14, 4135–4144 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y. et al. HOG-MAPK signaling regulates the adaptive responses of Aspergillus fumigatus to thermal stress and other related stress. Mycopathologia 174, 273–282 (2012). [DOI] [PubMed] [Google Scholar]
- Luo X. et al. The MAP kinase Bbslt2 controls growth, conidiation, cell wall integrity, and virulence in the insect pathogenic fungus Beauveria bassiana. Fungal Genet. Biol. 49, 544–555 (2012). [DOI] [PubMed] [Google Scholar]
- Hou Z. et al. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe. In. 15, 1119–1127 (2002). [DOI] [PubMed] [Google Scholar]
- Park G., Pan S. & Borkovich K. A. Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa. Eukaryot. Cell 7, 2113–2122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiante V., Heinekamp T., Jain R., Hartl A. & Brakhage A. A. The mitogen-activated protein kinase MpkA of Aspergillus fumigatus regulates cell wall signaling and oxidative stress response. Fungal Genet. Biol. 45, 618–627 (2008). [DOI] [PubMed] [Google Scholar]
- Xu J. R., Staiger C. J. & Hamer J. E. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. U. S. A. 95, 12713–12718 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey G., Held K., Scheffer J., Tenberge K. B. & Tudzynski P. CPMK2, an SLT2-homologous mitogen-activated protein (MAP) kinase, is essential for pathogenesis of Claviceps purpurea on rye: evidence for a second conserved pathogenesis-related MAP kinase cascade in phytopathogenic fungi. Mol. Microbiol. 46, 305–318 (2002). [DOI] [PubMed] [Google Scholar]
- Kumar A. et al. Overlapping and distinct functions of two Trichoderma virens MAP kinases in cell-wall integrity, antagonistic properties and repression of conidiation. Biochem. Biophys. Res. Commun. 398, 765–770 (2010). [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Van der Lee T., Waalwijk C. & Gert H. J. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant Microbe. In. 19, 389–398 (2006). [DOI] [PubMed] [Google Scholar]
- Fujioka T. et al. MpkA-Dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6, 1497–1510 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F. et al. A fungal cell wall integrity-associated MAP kinase cascade in Coniothyrium minitans is required for conidiation and mycoparasitism. Fungal Genet. Biol. 49, 347–357 (2012). [DOI] [PubMed] [Google Scholar]
- Sanz A. B. et al. Chromatin remodeling by the SWI/SNF complex is essential for transcription mediated by the yeast cell wall integrity MAPK pathway. Mol. Biol. Cell 23, 2805–2817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui O. & Hahn M. The Slt2-type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Mol. Plant Pathol. 8, 173–184 (2007). [DOI] [PubMed] [Google Scholar]
- Kojima K., Kikuchi T., Takano Y., Oshiro E. & Okuno T. The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium. Mol. Plant Microbe. In. 15, 1268–1276 (2002). [DOI] [PubMed] [Google Scholar]
- Igbaria A. et al. Distinct and combined roles of the MAP kinases of Cochliobolus heterostrophus in virulence and stress responses. Mol. Plant Microbe. In. 21, 769–780 (2008). [DOI] [PubMed] [Google Scholar]
- Jain R. et al. The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol. Microbiol. 82, 39–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink H. J. & Osmani S. A. A mitogen-activated protein kinase (MPKA) is involved in polarized growth in the filamentous fungus, Aspergillus nidulans. FEMS Microbiol. Lett. 173, 117–125 (1999). [DOI] [PubMed] [Google Scholar]
- Bennett L. D., Beremand P., Thomas T. L. & Bell-Pedersen D. Circadian activation of the mitogen-activated protein kinase MAK-1 facilitates rhythms in clock-controlled genes in Neurospora crassa. Eukaryot. Cell 12, 59–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Mendoza A. et al. Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 100, 15965–15970 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K., Latha J., Hadar R. & Horwitz B. A. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2, 446–455 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reithner B. et al. Signaling via the Trichoderma atroviride mitogen-activated protein kinase Tmk 1 differentially affects mycoparasitism and plant protection. Fungal Genet. Biol. 44, 1123–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd L. R., Weimer P. J., van Zyl W. H. & Pretorius I. S. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliahu N., Igbaria A., Rose M. S., Horwitz B. A. & Lev S. Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryot. Cell 6, 421–429 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmén M., Saloheimo A., Onnela M. L. & Penttila M. E. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63, 1298–1306 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg D. & Mandels G. R. Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J. Bacteriol. 139, 761–769 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll M., Franchi L. & Kubicek C. P. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot. Cell 4, 1998–2007 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll M., Schuster A., Silva Rdo N. & Kubicek C. P. The G-alpha protein GNA3 of Hypocrea jecorina (Anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot. Cell 8, 410–420 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel C. et al. Light-dependent roles of the G-protein alpha subunit GNA1 of Hypocrea jecorina (anamorph Trichoderma reesei). BMC biology 7, 58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisch D., Kubicek C. P. & Schmoll M. New insights into the mechanism of light modulated signaling by heterotrimeric G-proteins: ENVOY acts on gna1 and gna3 and adjusts cAMP levels in Trichoderma reesei (Hypocrea jecorina). Fungal Genet. Biol. 48, 631–640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A., Tisch D., Seidl-Seiboth V., Kubicek C. P. & Schmoll M. Roles of protein kinase A and adenylate cyclase in light-modulated cellulase regulation in Trichoderma reesei. Appl. Environ. Microbiol. 78, 2168–2178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker A. R., Grosstessner-Hain K., Wurleitner E. & Mach R. L. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5, 2128–2137 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peij N. N. M., Gielkens M. M. C., de Vries R. P., Visser J. & de Graaff L. H. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64, 3615–3619 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro N., Saloheimo A., Ilmén M. & Penttilä M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276, 24309–24314 (2001). [DOI] [PubMed] [Google Scholar]
- Coradetti S. T. et al. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc. Natl. Acad. Sci. U. S. A. 109, 7397–7402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro N., Ilmén M., Saloheimo M. & Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69, 56–65 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S., Nakamura A., Hidaka M., Masaki H. & Uozumi T. Isolation of the creA gene from the cellulolytic fungus Humicola grisea and analysis of CreA binding sites upstream from the cellulase genes. Biosci. Biotechnol. Biochem. 62, 2364–2370 (1998). [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Y., Zhong Y., Qu Y. & Wang T. Ras GTPases modulate morphogenesis, sporulation and cellulase gene expression in the cellulolytic fungus Trichoderma reesei. PLoS One 7, e48786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. et al. Long-term strain improvements accumulate mutations in regulatory elements responsible for hyper-production of cellulolytic enzymes. Sci. Rep. 3, 1569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards W. C. An enzyme system to liberate spore and mycelial protoplasts from a dimorphic fungal plant pathogen Ophiostoma ulmi (Buism.) Nannf. Physiol. Mol. Plant Pathol. 44, 311–319 (1994). [Google Scholar]
- Zhang G. et al. Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J. Biotechnol. 139, 146–151 (2009). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information