Abstract
Exploring the molecular difference among breast cancer subtypes is of crucial importance in understanding its heterogeneity and seeking its effective clinical treatment. For this, several layers of information including immunohistochemical markers and a variety of high-throughput genomics approaches have been intensively used. Here we have explored the intrinsic differences among breast cancer subgroups defined by immunohistochemical expression (IHC) of hormone receptors ER and PR as well as human epidermal growth factor receptor 2 (HER2) using the mRNA and miRNA expression profiles of 115 tumors. A core basal group was further defined by epidermal growth factor receptor and cytokeratin 5/6 IHC expression and compared to triple negative group. A set of differentially expressed genes including 1015 mRNAs and 69 miRNAs was found to distinguish tumor subtypes whose generality was demonstrated using two independent data sets. The network was explored for each subtype and biomass synthesis signaling was found to play an important role in the core basal subgroup. This study contributes to elucidating the intrinsic relations among breast cancer subgroups defined by ER, PR and HER2 expression via integrating mRNA and miRNA expression. The results can avail functional studies of breast cancer with translational potential for clinical use.
Breast cancer is a heterogeneous disease where classically, immunohistochemistry (IHC) markers, together with clinicopathologic variables are used for prognosis prediction and treatment selection1,2. The commonly used IHC markers include estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2). ER-positive tumors account for >70% of breast cancer3, respond to anti-estrogen or aromatase inhibitors and have a more indolent clinical course2. ER-negative tumors are hormone-independent and characterized by a more aggressive behavior4. PR is induced by estrogen and is a favorable prognostic marker1. HER2 is the most well-known prognostic member of the epidermal growth factor receptor family, whose amplification or over-expression is predictive of poor survival1. HER2 status can be used to sub-classify also [ER−/PR−] tumors into distinct subgroups of [ER−/PR−]HER2+ (breast tumors with negative ER and PR status, and positive HER2 status) and [ER−/PR−]HER2− (breast tumors with negative ER, PR and HER2 status), where [ER−/PR−]HER2−, called triple negative phenotypic tumors (TNP) given their negative status of all the three receptors, forms a clinically challenging group with poor prognosis and difficult to treat5. A core basal subgroup (CoreBasal) with distinct clinical and immunophenotypic differences has been more recently distinguished within the triple negative group using the epidermal growth factor receptor (EGFR, also known as HER1) which is associated with a lack of ER and poor prognosis1 and cytokeratins such as KRT5, KRT6, KRT14, KRT17 which are over-expressed in core basal tumors6. Particularly, triple negative tumors with positive EGFR or KRT5/6 staining are referred to as core basal tumors7. The 2012 IMPAKT task force investigated the medical usefulness of current methods for breast cancer subgroup classification and recommended the use of ER and HER2 for the identification of clinically relevant breast cancer subgroups8.
Breast cancer subgroup classification based on IHC markers is widely used in both clinical and research settings due to its reliability and reproducibility9. Several so called intrinsic breast cancer subgroups have been identified by large-scale gene expression profiling and shown to be associated with differential prognosis of the patients10. In year 2000, Perou et al. proposed that breast tumors could be classified into at least four intrinsic subgroups, i.e., luminal-like, HER2 positive, basal-like and normal-like tumors11. These subgroups are similar but not identical to the classification according to receptor classification using IHC. Particularly, luminal A and luminal B intrinsic subgroups are often considered to represent the [ER+/PR+]HER2− (breast tumors with positive ER and PR status, and negative HER2 status) and [ER+/PR+]HER2+ (breast tumors with positive ER, PR and HER2 status) subgroups, respectively12, and the intrinsic HER2 subgroup refers to [ER−/PR−]HER2+ tumors12. Recently, it was reported that only part of luminal B tumors are HER2 positive and additional luminal B tumors can be distinguished from luminal A tumors by the proliferation marker Ki6713. MiRNAs, a category of small (approximately 22-nucleotide) non-coding RNAs with regulatory activity, have attracted much attention in tumor subgroup characterization due to their regulatory role in mRNA expression14. A number of differentially expressed miRNAs among breast cancer subgroups have been identified, and the miRNA patterns distinguish between the basal-like and luminal tumors (luminal A and luminal B)14.
A number of studies have been conducted to further define tumor subgroups, indicating also finer-grained subgroups within each major group15,16. Curtis et al. recently identified 10 intrinsic breast cancer subgroups by jointly clustering the copy number and gene expression data of 2000 breast tumors15. The Cancer Genome Atlas (TCGA) network investigated breast cancer subgroups by incorporating information from multiple platforms, i.e., genomic DNA copy number arrays, DNA methylation, exome sequencing, mRNA arrays, miRNA sequencing and reverse-phase protein arrays. They conclude that diverse genetic and epigenetic alterations converge phenotypically into four major expression-only breast tumor subgroups, i.e., luminal A, luminal B, HER2 positive and triple negative16.
With the aim of uncovering the intrinsic differences and heterogeneity of breast cancer at multiple levels, we integrated mRNA and miRNA expression and network analysis in characterizing breast cancer subgroups defined by expression of the major hormonal and growth factor receptors, i.e., ER, PR and HER2, and additionally identified the differences between the core basal and triple negative subgroups at the gene and network levels.
Results
Differentially expressed genes in each subgroup
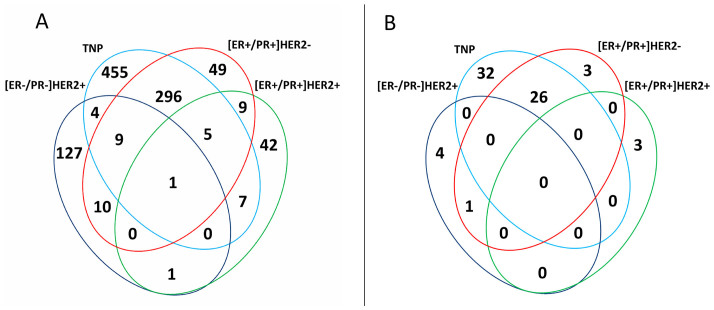
Among the 183 samples in the Helsinki Breast Cancer Study (HEBCS) data set, 115 invasive tumors could be determined by our available IHC marker information (Supplementary Tables 1 and 2). Four subgroups, i.e., [ER+/PR+]HER2−, [ER+/PR+]HER2+, [ER−/PR−]HER2+ and triple negative phenotype (TNP) tumors, were defined, with core basal (CoreBasal) tumors distinguished from the TNP subgroup as a separate group. As seen from table 1, we identified the mRNA and miRNA differentially expressed genes (diff-genes) for each of the five IHC defined subgroups (diff-gene sets are listed in Supplementary Tables 3 and 4), and the unified diff-gene set is the union of all the differentially expressed genes containing altogether 1015 mRNAs (the unified mRNA diff-genes) and 69 miRNAs (the unified miRNA diff-genes). The majority of the genes in the unified diff-gene set come from TNP (777 mRNAs, 58 miRNAs) and CoreBasal (535 mRNAs, 64 miRNAs) tumors, which were analysed using the least number of patients (18 TNP including 12 CoreBasal tumors), suggesting that the different number of genes among subgroups within the unified diff-gene set is not driven by the sample size but the intrinsic nature of the subgroups. The Venn diagrams of the diff-genes of the subgroups are presented in Figures 1A (mRNAs) and 1B (miRNAs), respectively. The TNP and [ER+/PR+]HER2− subgroups show the biggest overlap both for mRNAs and miRNAs. Specifically, 78% of [ER+/PR+]HER2− mRNA diff-genes overlap with TNP mRNA diff-genes and 87% of [ER+/PR+]HER2− miRNA diff-genes overlap with TNP miRNA diff-genes. Only one gene, KRT5, is common among the four sets of mRNA diff-genes, and is down-regulated in [ER+/PR+] tumors and up-regulated in [ER−/PR−] tumors. On the other hand, 84% of [ER−/PR−]HER2+ mRNA diff-genes are subgroup specific with only 16% overlap with the other subgroups, and [ER+/PR+]HER2+ miRNA diff-genes are exclusively subgroup specific.
Table 1. Gene number in each diff-gene set. The unified diff-genes includes the unique genes in the diff-gene sets of [ER+/PR+]HER2−, [ER+/PR+]HER2+, [ER−/PR−]HER2+ and TNP subgroups.
| [ER+/PR+]HER2− | [ER+/PR+]HER2+ | [ER−/PR−]HER2+ | TNP | CoreBasal | Union | |
|---|---|---|---|---|---|---|
| mRNA | 379 | 65 | 152 | 777 | 535 | 1015 |
| miRNA | 30 | 3 | 5 | 58 | 64 | 69 |
Figure 1. Venn diagram of subgroup specific diff-genes, indicating numbers of genes in the different or overlapping groups.
(A) mRNA, (B) miRNA.
Generality of the differentially expressed genes
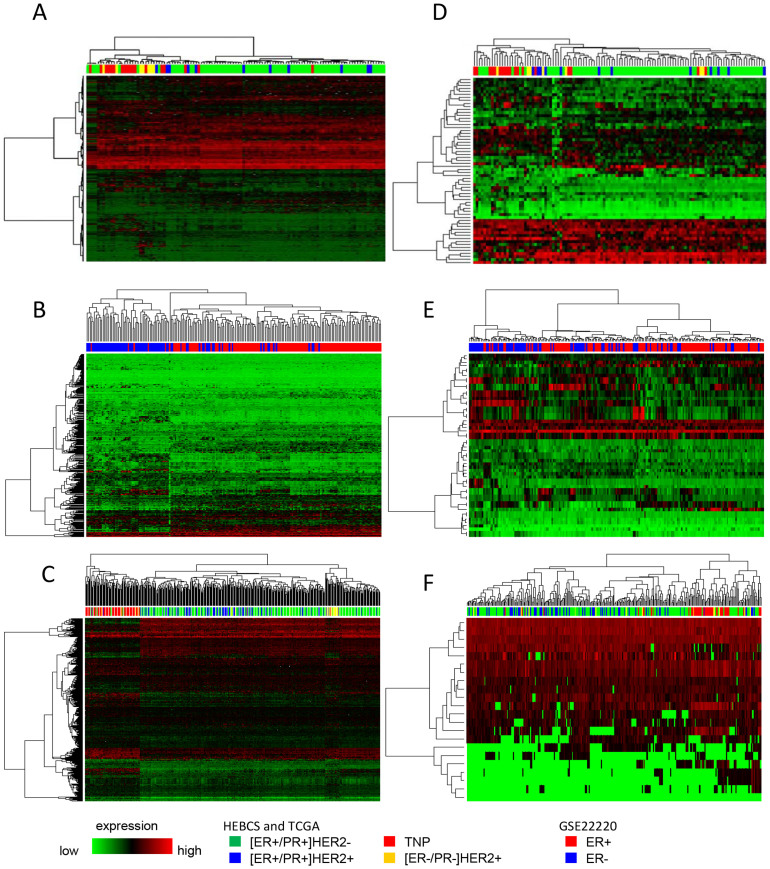
The subgroups could be well differentiated with our diff-genes in the public data sets, especially for mRNA (Figure 2). MiRNA diff-genes performed less accurately as compared with mRNA diff-genes. As miRNA and mRNA data share the same sample set, this difference is probably due to the complexity of the conversion from miRNA expression to the phenotypic differences among different breast tumor subgroups, which is mediated and influenced by many factors such as post-transcriptional regulations and generally large number of varieties of miRNA gene targets. Nevertheless, we could not rule out the possible effects of a small sample size. Numerically, the computational classifier comprised of our unified diff-genes was able to correctly predict the subgroup in 75% of the tumors in the public data sets (table 2), implying that our diff-genes, although identified from relatively small datasets, have a good generality.
Figure 2. Tumors from HEBCS and TCGA clustered by four breast cancer subgroups and GSE22220 by two subgroups on the basis of mRNA and miRNA unified diff-genes.
(A) Applying the mRNA unified diff-genes to the HEBCS data set. (B) Applying the mRNA unified diff-genes to the GSE22220 data set. (C) Applying the mRNA unified diff-genes to the TCGA data set. (D) Applying the miRNA unified diff-genes to the HEBCS data set. (E) Applying the miRNA unified diff-genes to the GSE22220 data set. (F) Applying the miRNA unified diff-genes to the TCGA data set. The mRNA and miRNA unified diff-genes are listed in supplementary Tables 3 and 4, respectively.
Table 2. Cross-validation of the diff-gene sets. ‘SVM’ and ‘KNN’ represent support vector machine and k-nearest neighbor classifiers, respectively, where k = 10 in KNN.
| Diff-gene | HEBCS | TCGA | ||
|---|---|---|---|---|
| SVM | KNN | SVM | KNN | |
| mRNA | 0.757 | 0.748 | 0.735 | 0.723 |
| miRNA | 0.765 | 0.757 | 0.549 | 0.594 |
Intrinsic relationship among subtypes revealed by differentially expressed genes
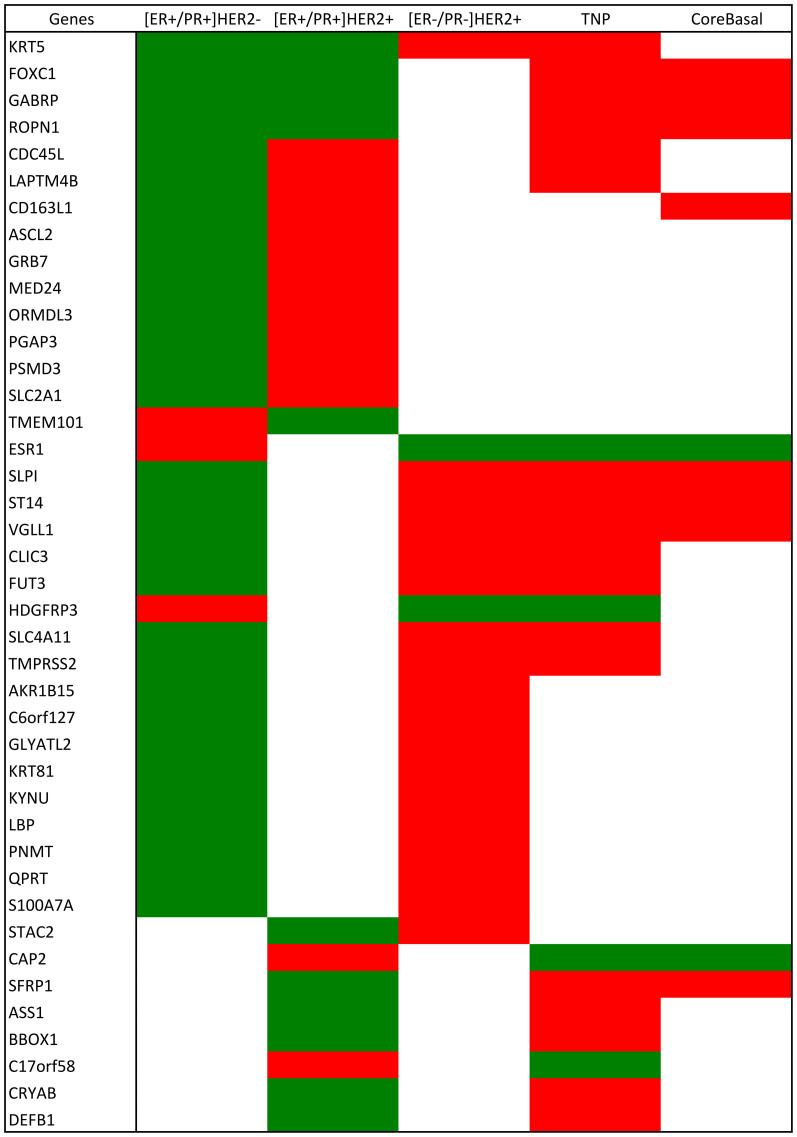
Most of the genes present in multiple tumor subgroups are oppositely regulated between [ER+/PR+] and [ER−/PR−] tumors (Figure 3). We compared mRNA diff-genes between the two [ER+/PR+] subgroups defined by HER2. Four genes are down-regulated in all ER positive tumors and eleven genes are oppositely regulated between [ER+/PR+]HER2− and [ER+/PR+]HER2+ tumors. The [ER−/PR−]HER2+ subgroup presented 20 genes oppositely regulated compared with ER positive tumors. Among the 20 genes, 19 are differentially expressed between [ER−/PR−]HER2+ and [ER+/PR+]HER2−, and two are differentially expressed between [ER−/PR−]HER2+ and [ER+/PR+]HER2+. One gene, AACS, is up-regulated both in [ER−/PR−]HER2+ and [ER+/PR+]HER2− tumors. We found 10 genes altered in the same direction between [ER−/PR−]HER2+ and TNP tumors. The [ER−/PR−]HER2+ subgroup has 10 genes coherently regulated with the TNP subgroup, and 10 genes over-expressed only in [ER−/PR−]HER2+ but not the TNP tumors. The regulatory direction of these genes was also checked in CoreBasal tumors, where 9 genes were disrupted in the same direction in TNP and CoreBasal tumors, including 7 elevated genes and 2 suppressed ones (ESR1, CAP2). The gene CD163L1 was found up-regulated only in CoreBasal tumors but no differential expression was observed among TNP tumors.
Figure 3. Genes shared in the mRNA diff-gene sets among breast cancer subgroups.
Genes shared in the mRNA diff-gene sets among [ER+/PR+]HER2−, [ER+/PR+]HER2+, [ER−/PR−]HER2+ and TNP subgroups. The regulation direction of the listed genes are also checked in the CoreBasal subgroup. The over- and under- expression are colored in red and green, respectively, based on the log2-transformed fold change.
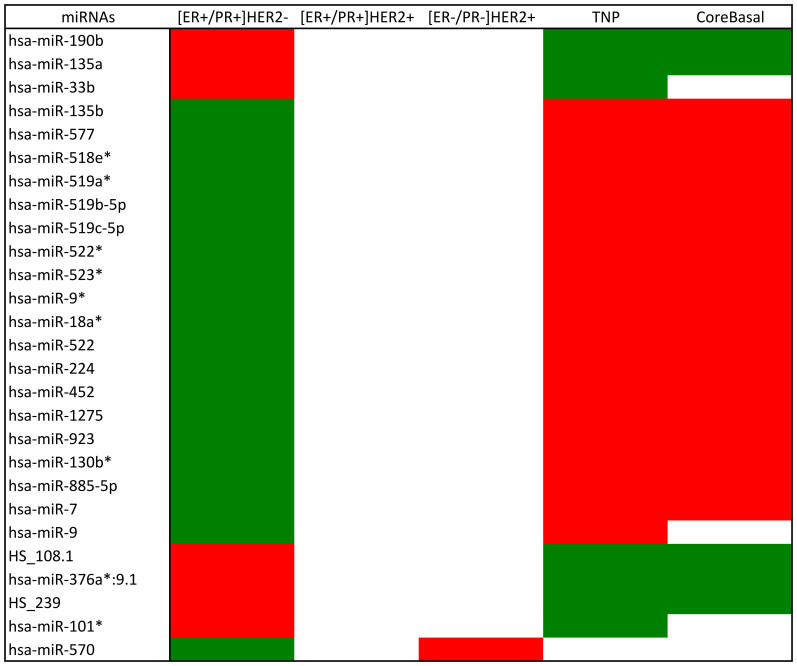
Except for hsa-miR-570 that is oppositely regulated between [ER+/PR+]HER2− and [ER−/PR−]HER2+ tumors, all miRNAs differentially expressed in multiple tumor subgroups are reversely deregulated in the [ER+/PR+]HER2− and TNP subgroups (Figure 4). More precisely, there are 7 miRNAs induced in [ER+/PR+]HER2− tumors and repressed in the TNP subgroup and 19 miRNAs deregulated reversely. According to the miRWalk prediction, two miRNA pairs (miR-135a and miR-135b, miR-519b-5p and miR-519c-5p) in Figure 4 share the same targets (Supplementary Tables 5 and 6) and four miRNAs bind to ER related genes (i.e., hsa-miR-135a, hsa-miR-135b and hsa-miR-365 target ESRR1, hsa-miR-7 targets ESRRG and ESR2).
Figure 4. MicroRNAs shared in the miRNA diff-gene sets among breast cancer subgroups.
MiRNAs shared in the miRNA diff-gene sets among [ER+/PR+]HER2−, [ER+/PR+]HER2+, [ER−/PR−]HER2+ and TNP subgroups. The regulation direction of the listed miRNAs are also checked in the CoreBasal subgroup. The over- and under- expression are colored in red and green, respectively, based on the log2-transformed fold change.
There are 353 and 121 genes uniquely present in TNP and CoreBasal mRNA diff-genes, respectively (Supplementary Table 7), and 9 and 16 miRNAs specifically shown in these two subgroups correspondingly (Supplementary Table 8).
The subgroup clustering pattern generated according to the identified diff-genes are the same when applied to the HEBCS and TCGA data sets, illustrating the comparability of the data sets (Supplementary Figure 1). The grouping of [ER−/PR−]HER2+ samples differs when miRNA and mRNA diff-genes are used, suggesting that the mRNA expression pattern of [ER−/PR−]HER2+ is more similar to TNP tumors as compared with its miRNA expression profile which is more closely grouped with the ER positive tumors.
Statistically, the mRNA data is highly correlated between HEBCS and TCGA data sets for all the four subgroups, while the miRNA data is less well correlated between different data sets except for [ER−/PR−]HER2+ tumors (table 3). However, when ER positive ([ER+/PR+]HER2− and [ER+/PR+]HER2+) and ER negative ([ER−/PR−]HER2+ and TNP) tumors are considered, the correlation across different data sets are significantly improved for both miRNAs and mRNAs. TNP tumors have the most varied mRNA expression pattern compared with the other subgroups.
Table 3. Correlation between HEBCS and TCGA data.
| Subtypes | Cora1 (mRNA) | Pb1 (mRNA) | LMc1 (mRNA) | Pd1 (mRNA) | Cora2 (miRNA) | Pb2 (miRNA) | LMc2 (miRNA) | Pd2 (miRNA) |
|---|---|---|---|---|---|---|---|---|
| [ER+/PR+]HER2− | 0.378 | <0.001 | 0.34 | <0.001 | 0.367 | 0.112 | 0.094 | 0.181 |
| [ER+/PR+]HER2+ | 0.358 | <0.001 | 0.323 | <0.001 | 0.266 | 0.256 | 0.069 | 0.325 |
| [ER−/PR−]HER2+ | 0.171 | 0.012 | 0.123 | 0.05 | 0.469 | 0.038 | 0.11 | 0.085 |
| TNP | 0.133 | 0.03 | 0.137 | 0.035 | 0.391 | 0.089 | 0.056 | 0.37 |
| ER+ | 0.37 | <0.001 | 0.332 | <0.001 | 0.315 | 0.048 | 0.081 | 0.093 |
| ER− | 0.15 | 0.002 | 0.133 | 0.003 | 0.439 | 0.005 | 0.084 | 0.053 |
a1Spearman correlation score between HEBCS and TCGA data sets for each mRNA diff-gene set.
b1p-value of a1.
c1Coefficient of the linear model between HEBCS and TCGA data sets for each mRNA diff-gene set.
d1p-value of c1.
a2Spearman correlation score between HEBCS and TCGA data sets for each miRNA diff-gene set.
b2p-value of a2.
c2Coefficient of the linear model between HEBCS and TCGA data sets for each miRNA diff-gene set.
d2p-value of c2.
Network and pathway analysis using differentially expressed genes among subtypes
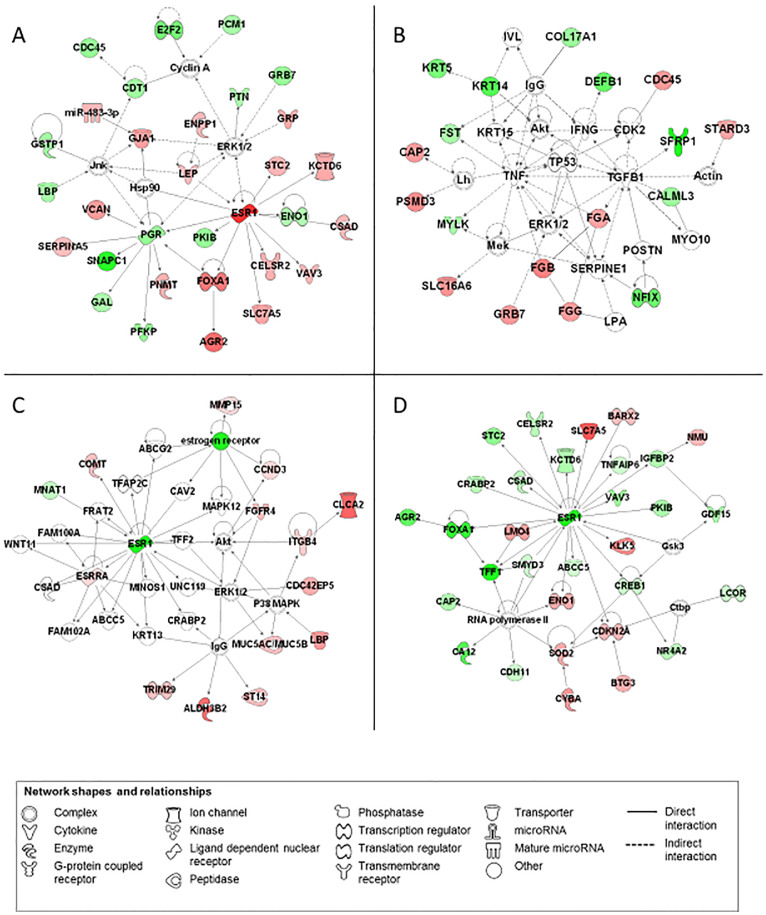
The most significant networks of each subtype are shown in Figure 5 (the diff-genes of each subtype including genes shared among subtypes) and Supplementary Figure 2 (the diff-genes of each subtype that are mutually exclusive among subtypes), with information for the top five networks listed in Supplementary Tables 9 and 10, accordingly.
Figure 5. Schematic representations of the most significant networks generated by IPA.
(A) [ER+/PR+]HER2−, (B) Triple negative, (C) [ER+/PR+]HER2+ and (D) [ER−/PR−]HER2+. Molecules shown in red (increased expression) and green (decreased expression) identified in our set of genes. Molecules with no coloring represent genes not present in our data set but relevant to biological pathways and incorporated to generate networks (description and fold change associated with differentially expressed genes are described in Supplementary Tables 3 and 4).
The most significant network covers 46% ([ER−/PR−]HER2+) to 91% (TNP) of the diff-genes for each subgroup. Each of the networks is assigned with functional annotations associated with genes in a specific network. Genes in the top networks of the [ER+/PR+]HER2− and TNP subgroups associate with cancer, system disorder and reproductive system disease (Figures 5A and 5D, Supplementary Table 9), and genes in the top network of the HER2+ subgroups are related to cellular development (Figures 5B and 5C, Supplementary Table 9).
The most significant network generated using the subgroup specific diff-genes includes 26% ([ER+/PR+]HER2+) to 80% (TNP) diff-genes per subgroup. The genes in [ER+/PR+]HER2− and [ER+/PR+]HER2+ associate with cell death and survival and, in addition, genes in [ER+/PR+]HER2+ are also involved in cell cycle, DNA replication, recombination and repair (Supplementary Figures 2A and 2B, Supplementary Table 10). The genes in the most significant networks for HER2+ subgroups show also associations with dermatological diseases and conditions, with [ER−/PR−]HER2+ specifically related to tissue and cell morphology (Supplementary Figure 2C). The diff-genes specific for the TNP subgroup are associated with hematological system development and function, tissue morphology and cancer (Supplementary Figure 2D).
The mRNA diff-genes of the CoreBasal subgroup are largely involved in pathways controlling biomass synthesis, such as glycolysis and steroid biosynthesis (Supplementary Table 11). [ER+/PR+]HER2+ and TNP tumors are enriched with genes communicating with NF-κB (Supplementary Figure 3).
Discussion
We compared our mRNA diff-genes with the gene signature published in17, which is the first gene signature reported using mRNA profiling and has been implemented for tumor subgroup prediction. There were 81 genes in common (as listed in Supplementary Table 12), among which some are the IHC markers used for tumor subgroup discrimination including ESR1 and KRT5, some are known prognostic markers such as GATA3 whose expression level is strongly associated with the differentiation level of breast cancer18, and some are shared among any of the four tumor subgroups in our mRNA diff-genes (as listed in Figure 3). The shared genes are KRT5, FOXC1, GABRP, LAPTM4B, GRB7, MED24, SLPI and VGLL1, most of which are generally differentially expressed between ER positive and ER negative tumors, demonstrating the fundamental differences between the two tumor classes and the critical role of ER signaling in driving the phenotypic differences of breast cancer.
Several miRNAs are also consistent with the previous studies. Blenkiron et al. carried out a study in 2007, where the miRNA features were determined for subgroups classified using mRNA data14. Due to the different subgroup categorization methods used, finding the direct correspondence between the two studies is difficult. However, hsa-miR-135b is up-regulated in TNP tumors and down-regulated in [ER+/PR+]HER2− tumors in this study and is elevated in basal-like tumors and suppressed in luminal tumors in [14].
ESR1, the discriminative factor between ER positive and ER negative tumors that mediates the biological effects of estrogens through direct binding to the estrogen response elements (EREs) of the target genes or via protein-protein interactions with other DNA-binding transcription factors in the nucleus19, is oppositely regulated in the presented diff-gene sets (Figure 3). There are four miRNAs targeting ESR1-related proteins from our miRNA diff-genes, i.e., hsa-miR-135a, hsa-miR-135b, hsa-miR-365 and hsa-miR-7. Specifically, the first three miRNAs target ESRRA and hsa-miR-7 is predicted to bind both ESRRG and ESR2 by miRWalk20. ESRR1 and ESRRG belong to the ERR subfamily, which is highly similar to the ESR subfamily regarding its amino acid composition and function21. It has been reported that ESRRA, ESR2 and ESR1 share 70% amino acid identity21 and ERRs share common target genes and pathways with ERs22. Thus, it is reasonable to assume that these four miRNAs are crucial in distinguishing ER positive and ER negative breast cancer given their regulatory roles in ESR1-related proteins. Previously, a predictive signature containing 6 miRNAs, i.e., hsa-miR-135b, hsa-miR-190, hsa-miR-217, hsa-miR-218, hsa-miR-299, hsa-miR-342, was identified to be associated with estrogen signaling using artificial neural networks23. Also, high hsa-miR-135a expression is associated with good prognosis in ER positive tumors24 and hsa-miR-7 is one of the four miRNAs (hsa-miR-7, hsa-miR-128a, hsa-miR-210 and hsa-miR-516-3p) that are linked with ER positive and lymph node negative breast cancer25. Thus, this study confirmed three miRNAs from the previous reports and identified a new miRNA (hsa-miR-365) that plays critical roles in differentiating tumors by ER status. In addition, hsa-miR-365 has been reported to be cooperatively regulated by SP1 and NF-κB26, where NF-κB is a known mechanism or target in ER- or basal like breast cancer27, further confirming its differentiating roles. The miRNAs diff-genes targeting non-ESR1 related mRNA diff-genes and differentially expressed by the ER status are also found. For instance, hsa-miR-224 is suppressed in the [ER+/PR+]HER2− subgroup and elevated in TNP tumors (including the CoreBasal subgroup) according to our results. By checking its targets using miRWalk20 and the identified diff-genes, the DNA repair gene, MBD428, was found to be up-regulated in [ER+/PR+]HER2− mRNA diff-genes, and the tumor suppressor whose loss is characteristic of basal-like tumors, INPP4B29, was found down-regulated in TNP mRNA diff-genes. Also reversely regulated between [ER+/PR+]HER2− and TNP (including CoreBasal) tumors are hsa-miR-135a and hsa-miR-135b, which share the same targets, indicating that these miRNAs, although closely related and targeting the same genes, function in different tumor subgroups and have different or even opposite regulatory roles in tumor progression. However, this reasoning highly depends on the accuracy of the computational prediction, and needs to be tested in vivo.
HER2 is a critical gene in ligand-activated signalling pathways that regulate cell proliferation and cell death30. Several genes are oppositely regulated by HER2 status (Figure 3), and have been indicated for potential roles in controlling cell growth. For example, CDC45L is known to be associated with proliferation31, and ASCL2 is involved in Wnt signalling and is a putative regulator of cell growth32. Several genes up-regulated in [ER−/PR−]HER2+ tumors are closely located with the HER2 gene at chromosome region 17q12 whose amplification is a common mechanism for HER2 activation33, e.g. PNMT is mapped onto the same chromosome with 17439 bp away from and co-expressed with HER234. Genes whose deregulation is associated with HER2 status and cell growth are also found from the network analysis. For instance, AGTR1 is a central node in the [ER+/PR+]HER2− subgroup (Supplementary Figure 2A) and specifically over-expressed in such tumors as well as involved in cell growth35. No obvious difference is observed between genes differentially expressed in [ER−/PR−]HER2+ and TNP tumors, indicating the more critical roles of ER compared with HER2 in tumor subgroup classification.
Overall, proliferation is an important character in differentiating breast cancer subgroups, with poor prognosis associated with high tumor cell proliferation36. Several proliferation markers were found significant the diff-gene sets. For example, topoisomerase IIα (TOP2A), a proliferation maker whose over-expression is reported to be associated with shorter survival and HER2 amplification (or over-expression) in breast cancer37, is a diff-gene of [ER+/PR+]HER2+ tumors in this study. Since TOP2A is the target enzyme for top IIα inhibitors such as anthracycline, our results are consistent with the efficacy of anthracycline treatment in [ER+/PR+]HER2+ breast cancer. Indeed, cells co-amplifying TOP2A and HER2 are reported to be highly sensitive to anthracyclines38. Similarly, other proliferation markers such as thymidine kinase (TK)39 and mini-chromosome maintenance protein 2 (MCM2)40 were found to be characteristic of [ER+/PR+]HER2− and TNP tumors, respectively. Several genes associated with cellular proliferation were found oppositely regulated between ER positive and ER negative tumors in this study. For example, KRT5, which has a role in maintaining cell proliferation potential in the basal layer of stratified epithelia and modulating PI3K/Akt-mediated cell proliferation41, is down-regulated in ER positive and up-regulated in ER negative tumors. In addition, there are 8 diff-genes associated with cell division and 8 related to cyclin dependent kinases or their inhibitors, most of which are over-expressed in TNP tumors and down-regulated in [ER+/PR+]HER2− tumors (Supplementary Table 13), consistent with the positive association of proliferation with TNP tumor aggressiveness.
The [ER+/PR+]HER2− and [ER+/PR+]HER2+ subgroups are always grouped together regardless of the data set (HEBCS, TCGA) used or the type of the genes (mRNA or miRNA) applied (Supplementary Figure 1), indicating their close relationships at both the mRNA and miRNA levels. However, the grouping of ER negative tumors does depend on the type of the genes, i.e., looser grouping between [ER−/PR−]HER2+ and TNP tumors is observed at the miRNA level than mRNA level. These results are consistent with the fact that ER positive and ER negative tumors are two major classes in breast cancer9, which could be well distinguished from each other at both the miRNA and mRNA levels. However, fine-grained classification within each major group may not be as clear as such especially at the miRNA level within the ER positive tumors. Similarly, [ER−/PR−]HER2+ tumors are shown to be more closely related to the TNP subgroup regarding their mRNA expression compared with the miRNA profiling. Overall, it is not surprising that the consistency scores within the major groups of ER positive or ER negative tumors are higher than those within each finer-grained subgroup, since more associations and less clear boundaries exist among subgroups than the two major branches.
Due to the more aggressive features and lack of therapeutic targets, triple negative and especially core basal tumors have become a key topic that has gained particular clinical and research interest7. We identified the core basal tumors within the TNP subgroup using EGFR and KRT5/6 to study the properties differentiating the core basal subgroup from TNP tumors with more aggressiveness. There are 111 mRNAs and 16 miRNAs whose expression is aberrant only in CoreBasal tumors (Supplementary Tables 7 and 8). Among these mRNAs, EIF4EBP1, LRRC6, RBM34, CKS1B, ADAM15, ZBP1, GGPS1, RCOR3, LOC149134 are amplified in 11%, 10%, 6%, 5%, 5%, 5%, 5%, 5%, 5% of the 825 breast invasive carcinomas in TCGA16, respectively, as queried using cBio Cancer Genomics Portal (cbioportal.org). Network analysis shows that CoreBasal specific mRNAs and miRNAs diff-genes are mostly related to infectious diseases, inflammation and cell signaling as compared to TNP tumors (Supplementary Table 10, Supplementary Figure 3), indicating the importance of tumor-promoting inflammation in facilitating neoplasias to acquire core cancer hallmarks42 in this subgroup.
Signalings controlling the biomass synthesis process such as glycolysis or gluconeogenesis and steroid biosynthesis are associated with the 111 mRNAs (Supplementary Table 7) differentially expressed in CoreBasal tumors (Supplementary Table 11), suggesting the crucial importance of another emerging cancer hallmark, i.e., reprogramming of energy metabolism, in this subgroup. NF-κB pathway was revealed as an important signaling in two clinically challenging subgroups27, i.e., [ER+/PR+]HER2+ and TNP, as represented by its central roles played in these networks (Supplementary Figures 2B and 2D). NF-κB aberration has been mostly reported in basal like breast cancer27. NF-κB is a critical transcription factor in regulating inflammation, whose crosstalk with glucocorticoid receptor has been proposed as the potential target for breast cancer treatment27.
Conclusion
We study the intrinsic differences of five breast cancer subgroups defined using the major receptor IHC markers, by analysing the differentially expressed genes among them and the pathways and networks within each subtype. Network analysis revealed high level relationships and interactions among the mRNA and miRNA diff-genes within the tumor subgroups. Further intrinsic relationships among the four tumor subgroups were revealed at the mRNA and miRNA levels. A novel miRNA differentiating breast tumors with different ER status, hsa-miR-365, was identified and worth testing in vivo. Energy metabolism reprogramming was found to play important roles in forming the aggressive nature of the CoreBasal subgroup.
Due to the limited sample size in our study, we did not aim to define fine-grained subgroups or detect subtle differences between breast tumors. Instead, we studied the differences among subtypes revealed by our dataset and explored their generality using two independent publically available datasets. On the other hand, we focused on the major characteristics of the four subgroups defined by the receptor immunohistochemistry markers as well as the differences between TNP and CoreBasal tumors here, where other markers such as those for proliferation could also be explored given data. Altogether, our results complement other relevant investigations, and avail functional studies of breast cancer with further translational potential for clinical use.
Methods
Tumor samples
The mRNA and miRNA HEBCS data were used for determining differentially expressed genes. There are 183 primary breast tumor samples, among which 151 were collected as a part of the unselected series at the department of Oncology of the Helsinki University Central Hospital (HUCH) in 1997, 1998 and 200043 and at the department of Surgery from 2001 to 200444. The remaining 32 patients belong to an ongoing collection of additional familial breast cancer series from the department of Clinical Genetics at HUCH. Out of the 183 samples, 115 tumors have unambiguous IHC marker information, which have been used for subgroup determination and further analysis.
Immunohistochemical marker status determination
The information on ER and PR status was collected from the pathology reports (positive when >10% of cells were stained)45. HER2 status was determined by gene amplification with chromogenic in situ hybridization (CISH) and immunohistochemical staining. In particular, samples with ≤5 replications from CISH or less than 10% of the cells stained were considered negative, those with ≥6 replications or over 90% cells stained were defined as positive, with CISH data preferred46. The EGFR and CK5/6 expression were analyzed using immunohistochemistry with more than 10% of the cells stained taken as positive and otherwise negative9,45.
Gene expression profiling
For mRNA data, total RNA was extracted from the 183 primary breast tumors, and the samples were processed and hybridized to Illumina HumanHT-12_V3 Expression BeadChips, containing 24660 Entrez Gene entities, according to the manufacturer recommendations (http://www.illumina.com). Similarly, for the miRNA profiling, the processed samples (183 patients) were hybridized onto the Illumina HumanMI_V2 BeadChips, containing 1104 known and predicted miRNAs. Both mRNA (GSE24450 at the Gene Expression Omnibus (GEO) database47) and miRNA (GSE43040) expression profilings were carried out at SCIBLU Genomics Centre, Lund University, Sweden.
Public data sets
To explore the generality of the differences identified among breast cancer subtypes, public data sets were used to distinguish subtypes by the identified differentially expressed genes. They are the breast cancer data set from TCGA (http://cancergenome.nih.gov) retrieved on 21th November 2011, and the GSE22220 data set24 from GEO database47. We took the primary solid tumor data from the TCGA data set (available from TCGA portal at http://tcga.cancer.gov/dataportal), which included mRNA data on 451 samples and 17814 genes and miRNA data on 315 patients and 1046 genes. Samples with unambiguous IHC marker subgroup classification were used in the analysis, including 433 and 311 samples from the mRNA and miRNA data sets, respectively. The mRNA data has been produced using Agilent 244 K Custom Gene Expression G4502A-07-3 platform, and the miRNA data has been generated by miRNA sequencing technique using IlluminaGA_miRNASeq. The level 3 data was used for both mRNA and miRNA data sets, i.e., the mRNA data was lowess normalized followed by log2-transformation of the ratio between two channels, and miRNA was normalized using appropriate method. To make the distribution of miRNA data comparable with the mRNA data, the logarithm 2 transformation was taken before usage.
The GSE22220 data set is composed of the mRNA data (GSE22219) where the samples from 216 patients were hybridized onto Illumina HumanRef-8_V1 expression BeadChips containing 24332 probes, and miRNA data (GSE22216) which was generated using Illumina HumanMI_V1 BeadChips containing 734 probes for 207 patients. Both GSE22219 and GSE22216 are quantile normalized.
Identification of subgroups and their differentially expressed genes
Defining tumor subgroups using immunohistochemical markers
Three IHC markers, i.e., ER, PR and HER2, were used to define the breast tumors into four subgroups, i.e., [ER+/PR+]HER2−, [ER+/PR+]HER2+, [ER−/PR−]HER2+ and TNP tumors. Within the TNP subgroup, we also identified the CoreBasal tumors using information on EGFR and KRT5/6. In particular, [ER+/PR+]HER2− tumors are defined as ER positive or PR positive tumors with negative HER2 status, [ER+/PR+]HER2+ tumors are ER positive or PR positive tumors with positive HER2 status, [ER−/PR−]HER2+ tumors are those that have negative ER and PR statuses but are HER2 positive, tumors with negative ER, PR and HER2 status are defined as TNP subgroup, among which those that are EGFR or KRT5/6 positive are defined as Core Basal tumors. Collectively, [ER+/PR+]HER2− and [ER+/PR+]HER2+ tumors are similar to the luminal, and [ER−/PR−]HER2+ and TNP are close to the non-luminal tumors as categorized using the expression profiles.
Gene expression data preprocessing
HEBCS data was used to identify the differentially expressed genes for each breast tumor subgroup. Microarray raw data was imported into R48 and processed by the methods included in the BioConductor facilities. Briefly, after quality control, the data was normalized using the quantile method49 and the gene expression matrix was obtained by averaging the probes mapping to the same Entrez Gene IDs. For the miRNA data, the gene expression matrix was obtained by averaging the probes mapped to the same miRNA ID.
Identification of the differentially expressed genes
Differentially expressed miRNAs and mRNAs between the breast cancer subgroups, as defined by IHC markers, were retrieved using linear models followed by moderated t-test. Genes with p-value < 0.05 after Benjamini and Hochberg post hoc correction were considered differentially expressed. The miRNAs differentially expressed in [ER+/PR+]HER2+ subgroup were selected to have nominal p-value < 0.05. Using the average expression of the other subtypes as the reference cohort, the genes (including both mRNA and miRNA) with base-2 logarithmic fold change larger than 0.58 or smaller than −0.58 were defined as differentially expressed genes (named ‘diff-genes’) for each tumor subgroup.
The union of the differentially expressed genes for each subgroup was considered the unified diff-genes.
Generality exploration using public datasets
The subtypes were grouped using the unified diff-genes by two classes of machine learning methods, i.e., unsupervised clustering and supervised classification, using two public data sets, i.e., TCGA (http://cancergenome.nih.gov) and GSE2222024 from GEO database47. Both methods assign objects into proper groups, which differ in that classification requires prior information on the ground-truth categories and involves a training process for the classifier while clustering does not.
The expression patterns of the unified diff-genes were used to cluster the samples in the two public data sets (TCGA and GSE22220), respectively. The TCGA samples are composed of four groups, i.e., [ER+/PR+]HER2−, [ER+/PR+]HER2+, [ER−/PR−]HER2+ and TNP subgroups, where the sample categories were determined based on the four IHC markers. The GSE22220 samples can only be classified into ER+ and ER− tumors based on ER status given its limited IHC marker information. Hierarchical clustering method was used here, where the distance matrix and agglomeration method used were optimized to produce the optimal heatmaps. In particular, the ward agglomeration method was used to generate the HEBCS mRNA and GSE22220 miRNA clusterings, and the complete agglomeration method was used to generate HEBCS miRNA, GSE22220 mRNA, TCGA mRNA, and miRNA clusterings. The Euclidean distance matrix was used in all clusterings, except for the TCGA miRNA clustering where the manhattan distance matrix was used.
To assess the generality of our diff-gene sets numerically, the predictive power of the miRNA and mRNA unified diff-genes was evaluated by two classification methods, i.e., support vector machine (SVM) and k-nearest neighbor (KNN). Both SVM and KNN are supervised machine learning methods widely applied in classification. SVM constructs a set of hyperplanes in a high-dimensional space, and the classification is achieved by the hyperplane that has the largest distance to the nearest training data point of any class. KNN classifies an object by taking a vote of its ‘k’ nearest neighbors, and the object is assigned to the class that is voted by the majority of the ‘k’ neighbor. In this study, k = 10. Both methods are coupled with leave-one-out cross validation. We use HEBCS and TCGA data sets in the classification given that they have all the four subgroup grouping information available.
Network and pathway analysis
In order to understand the biological interactions among the subtypes as defined, we performed network analyses of the diff-gene sets using the Ingenuity Pathway Analysis (IPA) tool, (Ingenuity Systems, www.ingenuity.com). We used two sets of input for IPA, i.e., the combined mRNA and miRNA diff-genes for each subgroup and, in a separate analysis, the subgroup specific diff-genes (including both mRNAs and miRNAs). The number of molecules shown in the network was set to a default limit of 35, i.e., only the most important genes with the maximum connectivity were included. The resulting networks were scored through Fisher's exact test, with the most significant (highest IPA scoring) networks selected.
In addition, we used DAVID50 to predict the enrichment of biological themes by the mRNA diff-genes, which performs an enrichment analysis for a given gene list and compares them to the KEGG pathways.
Author Contributions
X.D. designed, implemented the study, and prepared the draft. A.C. helped in formatting. A.C. and Z.B. are involved in finalizing the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Supplementary Information
Dataset 1
Dataset 2
Dataset 3
Dataset 4
Dataset 5
Dataset 6
Dataset 7
Dataset 8
Dataset 9
Dataset 10
Dataset 12
Dataset 11
Dataset 13
Acknowledgments
We thank Tuomas Heikkinen, Päivi Heikkilä, Kristiina Aittomäki, Carl Blomgvist, Dario Greco and Heli Nevanlinna for providing HEBCS data and their comments. This work was supported by the National Natural Science Foundation of China (Grant number 31471251), Academy of Finland (132473), Helsinki University Central Hospital Research Fund, the Sigrid Juselius Foundation, and the Finnish Cancer Society and the European Community's Seventh Framework Programme under grant agreement 223175 (HEALTH-F2-2009-223175).
References
- Walker R. A. Immunohistochemical markers as predictive tools for breast cancer. J Clin Pathol 61, 689–696, DOI 10.1136/jcp.2006.041830 (2008). [DOI] [PubMed] [Google Scholar]
- Dunnwald L. K., Rossing M. A. & Li C. I. Hormone receptor status, tumor characteristics, and prognosis: A prospective cohort of breast cancer patients. Breast Cancer Res 9, R6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masood S. Estrogen and progesterone receptors in cytology: a comprehensive review. Diagn Cytopathol 8, 475–491 (1992). [DOI] [PubMed] [Google Scholar]
- Putti T. C. et al. Estrogen receptor-negative breast carcinomas: a review of morphology and immunophenotypical analysis. Mod Pathol 18, 26–35, 10.1038/modpathol.3800255 (2005). [DOI] [PubMed] [Google Scholar]
- Hudis C. A. & Gianni L. Triple-negative breast cancer: an unmet medical need. Oncologist 16, 1–11 (2011). [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E. et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 25, 2273–2284 (2006). [DOI] [PubMed] [Google Scholar]
- Rakha E. A. et al. Triple-Negative Breast Cancer: Distinguishing between Basal and Nonbasal Subtypes. Clin Cancer Res 15, 2302–2310, Doi 10.1158/1078-0432.Ccr-08-2132 (2009). [DOI] [PubMed] [Google Scholar]
- Guiu S. et al. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Ann Oncol 23, 2997–3006, 10.1093/annonc/mds586 (2012). [DOI] [PubMed] [Google Scholar]
- Blows F. M. et al. Subtyping of Breast Cancer by Immunohistochemistry to Investigate a Relationship between Subtype and Short and Long Term Survival: A Collaborative Analysis of Data for 10159 Cases from 12 Studies. PLoS Med 7, e1000279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørlie T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS 98, 10869–10874, DOI 10.1073/pnas.191367098 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M. et al. Molecular portraits of human breast tumors. Nature 406, 747–752 (2000). [DOI] [PubMed] [Google Scholar]
- Vallejos C. S. et al. Breast Cancer Classification According to Immunohistochemistry Markers: Subtypes and Association With Clinicopathologic Variables in a Peruvian Hospital Database. Clin Breast Cancer 10, 294–300 (2010). [DOI] [PubMed] [Google Scholar]
- Cheang M. C. et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101, 736–750, 10.1093/jnci/djp082 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C. et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 8, R214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352, 10.1038/nature10983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70, 10.1038/nature11412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørile T. et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. PNAS 100, 8418–8423 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R. et al. Identification of GATA3 as a Breast Cancer Prognostic Marker by Global Gene Expression Meta-analysis. Cancer Res 65, 11259–11264 (2005). [DOI] [PubMed] [Google Scholar]
- Björnström L. & Sjöberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol 19, 833–842, 10.1210/me.2004-0486 (2005). [DOI] [PubMed] [Google Scholar]
- Dweep H., Sticht C., Pandey P. & Gretz N. miRWalk - database: prediction of possible miRNA binding sites by ‘walking’ the genes of 3 genomes. J Biomed Inform 44, 839–847 (2011). [DOI] [PubMed] [Google Scholar]
- Chisamore M. J., Cunningham M. E., Flores O., Wilkinson H. A. & Chen J. D. Characterization of a Novel Small Molecule Subtype Specific Estrogen-Related Receptor α Antagonist in MCF-7 Breast Cancer Cells. PLoS One 4, e5624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Kiriyama Y., Lee K. Y. & Giguère V. Transcriptional regulation of the estrogen-inducible pS2 breast cancer marker gene by the ERR family of orphan nuclear receptors. Cancer Res 61, 6755–6761 (2001). [PubMed] [Google Scholar]
- Lowery A. J. et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 11, R27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffa F. M. et al. MicroRNA associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res 71, 5635–5645 (2011). [DOI] [PubMed] [Google Scholar]
- Foekens J. A. et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. PNAS 105, 13021–13026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng D. H. et al. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res 57, 5221–5225 (1997). [PubMed] [Google Scholar]
- Ling J. & Kumar R. Crosstalk between NFkB and glucocorticoid signaling: A potential target of breast cancer therapy. Cancer Lett 322, 119–126, DOI 10.1016/j.canlet.2012.02.033 (2012). [DOI] [PubMed] [Google Scholar]
- Petronzelli F. et al. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem 275, 32422–32429 (2000). [DOI] [PubMed] [Google Scholar]
- Fedele C. G. et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. PNAS 107, 22231–22236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye M. A. Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family members. Breast Cancer Res 3, 385–389 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollok S., Bauerschmidt C., Sänger J., Nasheuer H. P. & Grosse F. Human Cdc45 is a proliferation-associated antigen. FEBS J 274, 3669–3684 (2007). [DOI] [PubMed] [Google Scholar]
- Jubb A. M. et al. Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is upregulated in intestinal neoplasia. Oncogene 25, 3445–3457 (2006). [DOI] [PubMed] [Google Scholar]
- Kauraniemi P. & Kallioniemi A. Activation of multiple cancer-associated genes at the ERBB2 amplicon in breast cancer. Endocr Relat Cancer 13, 39–49 (2006). [DOI] [PubMed] [Google Scholar]
- Dressman M. A. et al. Gene expression profiling detects gene amplification and differentiates tumor types in breast cancer. Cancer Res 63, 2194–2199 (2003). [PubMed] [Google Scholar]
- Atkinson J. M. et al. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer cell 20, 384–399, 10.1016/j.ccr.2011.08.013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzabek K., Laudanski P., Dzieciol J., Dabrowska M. & Wolczynski S. Protein kinase C involvement in proliferation and survival of breast cancer cells. Folia Histochem Cytobiol 40, 193–194 (2002). [PubMed] [Google Scholar]
- Depowski P. L. et al. Topoisomerase II alpha expression in breast cancer: Correlation with outcome variables. Modern Pathol 13, 542–547, DOI 10.1038/modpathol.3880094 (2000). [DOI] [PubMed] [Google Scholar]
- Jarvinen T. A. et al. Amplification and deletion of topoisomerase IIalpha associate with ErbB-2 amplification and affect sensitivity to topoisomerase II inhibitor doxorubicin in breast cancer. Am J Pathol 156, 839–847 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colozza M. et al. Proliferative markers as prognostic and predictive tools in early breast cancer: where are we now? Ann Oncol 16, 1723–1739, 10.1093/annonc/mdi352 (2005). [DOI] [PubMed] [Google Scholar]
- Rosenwald A. et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer cell 3, 185–197 (2003). [DOI] [PubMed] [Google Scholar]
- Alam H., Sehgal L., Kundu S. T., Dalal S. N. & Vaidya M. M. Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol Biol Cell 22, 4068–4078 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. Hallmarks of Cancer: The Next Generation. Cell 144, 646–674, DOI 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- Syrjakoski K. et al. Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J Natl Cancer Inst 92, 1529–1531, DOI 10.1093/jnci/92.18.1529 (2000). [DOI] [PubMed] [Google Scholar]
- Fagerholm R. et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet 40, 844–853 (2008). [DOI] [PubMed] [Google Scholar]
- Eerola H. et al. Histopathological features of breast tumours in BRCA1, BRCA2 and mutation-negative breast cancer families. Breast Cancer Res: BCR 7, R93–100 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommiska J. et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene 27, 2501–2506, DOI 10.1038/sj.onc.1210885 (2008). [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M. & Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R. D. C. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2009). [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M. & Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193, DOI 10.1093/bioinformatics/19.2.185 (2003). [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13, 10.1093/nar/gkn923 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Dataset 1
Dataset 2
Dataset 3
Dataset 4
Dataset 5
Dataset 6
Dataset 7
Dataset 8
Dataset 9
Dataset 10
Dataset 12
Dataset 11
Dataset 13