Abstract
AIM
To assess the association between age-related macular degeneration (AMD) and three single nucleotide polymorphisms (SNPs) related to the vascular endothelial growth factor (VEGF) gene.
METHODS
The patients who were diagnosed with AMD were included in this prospective study. Three SNPs (rs1413711, rs2146323, and rs3025033) of the VEGF gene were genotyped by real-time polymerase chain reaction in the genomic DNA isolated from peripheral blood samples of the 82 patients and 80 controls.
RESULTS
The genotype frequencies of rs1413711 and rs2146323 were not significantly different between the study group and the control group (P=0.072 and P=0.058). However, there was a significant difference in the genotype frequencies of these SNPs between the wet type AMD and dry type AMD (P=0.005 and P=0.010, respectively). One of the SNPs (rs1413711) was also found to be associated with the severity of AMD (P=0.001) with significant genotype distribution between early, intermediate, and advanced stages of the disease. The ancestral alleles were protective for both SNPs while the polymorphic alleles increased the risk for dry AMD.
CONCLUSION
VEGF SNPs rs1413711 and rs2146323 polymorphisms are significantly associated with AMD subtypes in our population.
Keywords: age-related macular degeneration, vascular endothelial growth factor, single nucleotide polymorphism
INTRODUCTION
Age-related macular degeneration (AMD) is a multifactorial disease that represents the most common cause of irreversible visual impairment in the elderly population[1]. Patients with this disease eventually develop either neovascularization (wet type) or geographic atrophy. Wet type AMD is characterized by choroidal neovascularization (CNV), which causes exudative and hemorrhagic changes in the retina and eventually significant visual loss in a majority of the cases[2]–[4].
Risk factors of AMD are heterogeneous, including mainly increasing age and different genetic predispositions, together with several environmental/epigenetic factors[1]. There is extensive support for a strong genetic component in AMD pathogenesis[2].
VEGF is an important factor during angiogenesis, and elevated VEGF concentration in CNV suggested that VEGF is also important in CNV formation. Several studies found increased levels of VEGF in the vitreous samples of patients with wet AMD. The VEGF protein level was associated with single nucleotide polymorphisms (SNPs) in the promoter and untranslated region (UTRs) of the gene. So, a number of studies have been focused on the relation between the SNPs of the gene and AMD. But there is no consensus between the results, and it is thought that many factors such as ethnical differences, environmental factors, etc., might be responsible for this difference[5]. Recently, it has been reported that genetic variations might be associated with treatment response in wet type AMD patients[6]–[10]. Therefore, a better understanding of the relation between genetic factors and AMD may affect the therapeutic decisions in patients with AMD in the future.
Here, we report the association of VEGF gene rs1413711, rs2146323, and rs3025033 polymorphisms in a Turkish population with AMD, and to our knowledge, this is the first report from our country.
SUBJECTS AND METHODS
Patients and Controls
The patients who were diagnosed with AMD at the Department of Ophthalmology, Pamukkale University, were included in this prospective study. The control group consisted of the patients without visual impairment, drusen, or retinal pigment epithelium (RPE) changes under dilated fundus examination. A total of 162 subjects were included in the study. Of the 162 subjects, 82 (50.6%) had AMD and 80 (49.4%) comprised the control group.
The patients who were younger than 55y and had diabetes mellitus, glaucoma, high myopia (>6 D), retinal pathology, or any disease that can lead to CNV (angioid streak, inflammation, trauma etc.) were excluded from the study. Institutional review board approval was obtained prior to the beginning of the study, and all participants provided written informed consent. The methods used in this study conformed to the tenets of the Declaration of Helsinki (2008) of the World Medical Association.
All participants underwent a detailed ophthalmologic examination, including measurement of best corrected visual acuity, intraocular pressure, anterior segment examination with slit-lamp, and posterior segment examination with slit-lamp and fundoscopy lens. A colour fundus picture was obtained for all participants and a fluorescein angiography for the study group. The AMD stage assigned was based on the most severe eye at the time of recruitment. The severity of AMD was classified as early, intermediate, and advanced as previously defined in age related eye disease study (AREDS) report No:6 (early for grade 1 and 2, intermediate for grade 3, and advanced for grade 4)[11]. The size of drusen was classified as small (<63 µm), medium (63-125 µm), and large (>125 µm), whereas the type of drusen was classified as soft, hard, and confluent drusen.
Genotyping
Peripheral blood samples were collected from the subjects. Genomic DNA was extracted from all samples of 200 µL whole blood using a commercial kit (High Pure PCR Template Preparation Kit, Roche Diagnostics, Germany). Three SNPs, including rs1413711, rs2146323, and rs3025033 in the VEGF gene, were studied. Primers and probes that amplify 158 bp, 200 bp, and 124 bp target sequences (for rs1413711, rs2146323, and rs3025033, respectively) are shown in Table 1. Real-time PCR analyses were performed on the LightCycler 480 Real-Time PCR System (Roche Diagnostics). The cycling conditions for all SNPs were: 10min at 95°C for Taq activation, followed by 35 cycles of 95°C for 10s, 56°C (for rs1413711 and rs2146323) and 54°C (for rs3025033) for 10s, and 72°C for 10s. For the amplicon identification, the following conditions were used: 30s at 95°C, 45s at 40°C, and 0s at 85°C for one cycle.
Table 1. Primer and probe sequences for VEGF genotyping.
| SNP | Sequence (5′→3′) |
| rs1413711 | |
| Primer sense | 5′-TGACAATATTCTCCCGGGACC-3′ |
| Primer antisense | 5′-AGTGTGACCTTCAGAGGCCC-3′ |
| Probe-1 | 5′-CTTCCAAGGCCAGGGGGCA-3′-FLa |
| Probe-2 | b5′L640-AGGAGGGGCGGTTCTAGGCAGGCA-3′ |
| rs2146323 | |
| Primer sense | 5′-AAGCTTAGGGAAGTGCTTCAA-3′ |
| Primer antisense | 5′-CTGCGCTGATAGACATCCAT-3′ |
| Probe-1 | 5′-TGTAATGCCACTCTTTGGAGCTT-3′-FLa |
| Probe-2 | b 5′L640-GAATCAGGCAAGTCCTTCC-3′ |
| rs3025033 | |
| Primer sense | 5′-AAGACTTTGTGGGGATTTCCTA-3′ |
| Primer antisense | 5′-TTGGTTTCACATAGGGCCAA-3′ |
| Probe-1 | 5′-AGGGAAGTCCTTGGAGTGTCTCCCC-3′-FLa |
| Probe-2 | b 5′L640-CCCCAGCAATGTTCTTGTGGC-3′ |
aFL: Fluorescein; b640: LightCycler-Red 640.
Statistical Analysis
Statistical analysis was carried out by SPSS (version 11; SPSS Inc., Chicago, IL) software. The Chi-square analysis was used to test the significance of the differences of observed alleles and genotypes between groups. A logistic regression model was used to calculate the odds ratios and 95% confidence intervals (CI).
RESULTS
Patients' Characteristics
A total of 162 subjects were included in the study. Of 162 subjects, 82 (50.6%) had AMD and 80 (49.4%) comprised the control group. Table 2 shows the demographics of the study and the control groups. Because of the significant difference in age between the study and the control groups, the statistical analysis was adjusted by age. The type and the severity of AMD in the study group are shown in Table 3.
Table 2. Demographics of the study and the control groups x±s.
| Parameters | AMD | Control | P |
| Age (a) | 71.7± 5.3 | 62.8± 5.2 | <0.001 |
| Gender n(%) | 0.534 | ||
| F | 37 (45.1) | 40 (50) | |
| M | 45 (54.9) | 40 (50) |
Table 3. The type and the severity of AMD in the study group.
| Parameters | n (%) |
| AMD type | |
| Wet | 43 (52.4) |
| Dry | 39 (47.6) |
| Severity of AMD | |
| Early | 11 (13.4) |
| Intermediate | 21 (25.6) |
| Advanced | 50 (62) |
| Size of Drusen | |
| Small (<63µm) | 44 (53.7) |
| Medium (63-125µm) | 27 (32.9) |
| Large (>125µm) | 11 (13.4) |
| Type of Drusen | |
| Hard | 45 (54.9) |
| Soft | 33 (40.2) |
| Confluent | 4 (4.9) |
Vascular Endothelial Growth Factor Genotype Frequencies and Association of Patient Characteristics with the Genotype
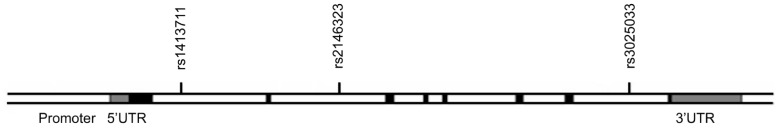
Genotype distribution of the all SNPs in the VEGF gene in patients with AMD and healthy controls are listed in Tables 4 and 5. The positions of the three SNPs analyzed in this study are shown in Figure 1. We could not find a significant difference in rs1413711 genotypes between the patients and controls (P=0.72), but there was a significant difference between wet and dry AMD with regard to rs1413711 genotypes (P=0.005). We also found that the presence of an A allele was significantly associated with the disease (OR=2.25; 95%CI 1.15-4.43).
Table 4. Genotype and allele frequencies of VEGF gene polymorphisms in AMD patients in comparison to controls.
| SNP | Genotype/allele | Patients n (%) | Controls n (%) | OR (95% CI) |
| rs1413711 | GG | 21 (25.6) | 34 (42.5) | 0.47 (0.24-0.91)a |
| GA | 2 (2.4) | 1 (1.3) | 1.98 (0.18-22.22) | |
| AA | 59 (72.0) | 45 (56.2) | 2.00 (1.04-3.84)a | |
| GG vs (GA+AA) | 2.25 (1.15-4.43)a | |||
| G | 44 (26.8) | 69 (43.1) | 0.48 (0.30-0.77)a | |
| A | 120 (73.2) | 91 (56.9) | 2.07 (1.30-3.30)a | |
| rs2146323 | CC | 16 (19.5) | 29 (36.3) | 0.43 (0.21-0.87)a |
| CA | 17 (20.7) | 20 (25.0) | 0.79 (0.38-1.64) | |
| AA | 49 (59.8) | 31 (38.7) | 2.35 (1.25-4.41)a | |
| CC vs (CA+AA) | 2.02 (0.99-4.16)a | |||
| C | 49 (29.9) | 78 (48.8) | 0.45 (0.28-0.71)a | |
| A | 115 (70.1) | 82 (51.2) | 2.23 (1.42-3.52)a | |
| rs3025033 | AA | 80 (97.6) | 79 (98.8) | |
| AG | 1 (1.2) | 1 (1.2) | ||
| GG | 1 (1.2) | 0 (0) | ||
| A | 161 (98.2) | 159 (99.4) | ||
| G | 3 (1.8) | 1 (0.6) |
VEGF: Vascular Endothelial Growth Factor; OR: Odds Ratio; CI: Confidence Interval; aThe significant P values.
Table 5. The genotype and allele frequencies of VEGF SNPs according to the type of AMD.
| Parameters | Patients n (%) | Controls n (%) | OR (95% CI) |
| rs1413711 | |||
| Wet AMD | |||
| GG | 17 (39.5) | 34 (42.5) | 0.89 (0.42-1.88) |
| GA | 0 (0) | 1 (1.3) | - |
| AA | 26 (60.5) | 45 (56.2) | 1.19 (0.56-2.50) |
| GG vs (GA+AA) | 1.13 (0.53-2.41) | ||
| G | 34 (39.5) | 69 (43.1) | 0.86 (0.51-1.47) |
| A | 52 (60.5) | 91 (56.9) | 1.16 (0.68-1.98) |
| Dry AMD | |||
| GG | 4 (10.3) | 34 (42.5) | 0.16 (0.05-0.48)a |
| GA | 2 (5.1) | 1 (1.3) | 4.27 (0.38-48.60) |
| AA | 33 (84.6) | 45 (56.2) | 4.28 (1.61-11.35)a |
| GG vs (GA+AA) | 6.47 (2.10-19.93)a | ||
| G | 10 (12.8) | 69 (43.1) | 0.19 (0.09-0.40)a |
| A | 68 (87.2) | 91 (56.9) | 5.16 (2.48-10.74)a |
| rs2146323 | |||
| Wet AMD | |||
| CC | 12 (27.9) | 29 (36.3) | 0.68 (0.30-1.53) |
| CA | 12 (27.9) | 20 (25.0) | 1.16 (0.50-2.68) |
| AA | 19 (44.2) | 31 (38.7) | 1.25 (0.59-2.65) |
| CC vs (CA+AA) | 1.27 (0.56-2.86) | ||
| C | 36 (41.9) | 78 (48.8) | 0.76 (0.45-1.28) |
| A | 50 (58.1) | 82 (51.2) | 1.32 (0.78-2.24) |
| Dry AMD | |||
| CC | 4 (10.3) | 29 (36.3) | 0.20 (0.07-0.62)a |
| CA | 5 (12.8) | 20 (25.0) | 0.44 (0.15-1.28) |
| AA | 30 (76.9) | 31 (38.7) | 5.27 (2.21-12.58)a |
| CC vs (CA+AA) | 4.29 (1.38-13.37)a | ||
| C | 13 (16.7) | 78 (48.8) | 0.21 (0.11-0.41)a |
| A | 65 (83.3) | 82 (51.2) | 4.76 (2.43-9.31)a |
VEGF: Vascular Endothelial Growth Factor; OR: Odds Ratio; CI: Confidence Interval; aThe significant P values.
Figure 1. The approximate positions of the three SNPs of VEGF gene analyzed in our study. Black, gray, and white boxes indicate the exons, untranslated regions (UTRs) and the introns, respectively.
For rs2146323, there was no significant difference between patients and controls (P=0.058). We have also observed that the frequency of the genotypes is significantly higher in patients with dry AMD compared to patients with wet AMD (P=0.010). The presence of at least one A allele was significantly associated with the disease (OR= 2.02; 95% CI 0.99-4.16). In the case of dry AMD, this association was more prominent (OR=4.29; 95% CI 1.38-13.37) compared to wet AMD (OR=1.27; 95% CI 0.56-2.86).
The AA genotype at rs3025033 was the most prevalent genotype in both patients and controls (97.6% and 98.8%, respectively). There was no difference in genotype distribution between dry and wet AMD groups (P=0.616).
We found that the presence of ancestral allele (G) in rs1413711 was protective for all AMD patients (OR 0.47 95% CI 0.24-0.91); however, the risk for AMD was increased in patients with AA genotype (OR 2.00 95% CI 1.04-3.84), and AA genotype comprised a highly increased risk for dry AMD (OR 4.28 95% CI 1.61-11.35). The association between the disease and the presence of at least one A allele at the rs1413711 was particularly observed in the case of dry AMD, where the risk was almost 7 times higher than the control group (OR 6.47, 95% CI 2.10-19.93).
As for rs2146323, the ancestral C allele was highly protective (OR 0.45 95% CI 0.28-0.71), whereas homozygous presence of the A allele was found to pose a highly increased risk for dry AMD (OR 5.27 95% CI 2.21-12.28). The presence of at least one A allele was associated with dry AMD (OR 4.29, 95% CI 1.38-13.37).
In the analysis of clinical variables and VEGF SNPs, there was no difference between patients and controls with regard to sex and type of drusen. However, the presence of AA genotype at rs1413711 was significantly related to the stage of disease (P=0.001) (Table 6), and the presence of one A allele was significantly associated with the size of drusen (P=0.042).
Table 6. Genotype distribution of VEGF SNPs by AMD stage.
| Genotype | Early n (%) | Stages of AMD |
||
| Intermediate n (%) | Advanced n (%) | P | ||
| rs1413711 | ||||
| GG | 1 (9.1) | 2 (9.5) | 18 (36.0) | 0.001 |
| GA | 2 (18.2) | 0 | 0 | |
| AA | 8 (72.7) | 19 (90.5) | 32 (64.0) | |
| Rs2146323 | ||||
| CC | 0 | 4 (19.0) | 12 (24.0) | 0.085 |
| CA | 3 (27.3) | 1 (4.8) | 13 (26.0) | |
| AA | 8 (72.7) | 16 (76.2) | 25 (50.0) | |
| rs3025033 | ||||
| AA | 11 (100.0) | 21 (100.0) | 48 (96.0) | 0.859 |
| AG | 0 | 0 | 1 (2.0) | |
| GG | 0 | 0 | 1 (2.0) | |
DISCUSSION
In this study, we analyzed three intronic SNPs along the VEGF gene and found that the ancestral alleles of both rs1413711 and rs2146323 were protective against AMD, whereas the presence of the polymorphic alleles increased the risk for the disease, particularly in the case of dry type, in a Turkish population. Additionally, rs1413711 was significantly associated with the stage of AMD.
Since introduction as a risk factor for AMD, the genetic variations have been widely studied in different populations. Studies from different populations have revealed the association of VEGF polymorphism and AMD types, and also the impact of VEGF polymorphisms to the outcomes of anti-VEGF treatment. However, there is inconsistency among the findings of those studies.
Association between rs1413711 and AMD was previously studied, but the results of these studies are inconsistent. In a study conducted in a Caucasian population from Northern Europe, Churchill et al[12] genotyped 45 patients with AMD and 94 healthy controls and reported that the GG genotype in rs1413711 SNP was significantly associated with AMD, whereas the presence of A allele was potentially protective. In a Taiwan Chinese cohort of 190 late AMD patients and 180 controls, Lin et al[13] genotyped five SNPs and did not found an association between rs1413711 and AMD. In another study with 159 neovascular AMD and 140 age- and sex-matched controls from China, Qu et al[14] found no association between the VEGF SNPs, including rs1413711 and AMD. Almeida et al[15] genotyped the SNP in 160 patients with AMD and 140 healthy controls in a Brazilian cohort, and they found an association between the mutant allele and the disease. In a recent meta-analysis study, Lu et al[16] reported that there was a significant relation between rs1413711 polymorphism and AMD risk among Europeans. We found that A is the risk allele for rs1413711 in our study group while G had a protective effect. G is the ancestral allele for this SNP in the dbSNP database (http://www.ncbi.nlm.nih.gov/snp). This SNP is highly variable in our study group with an allele frequency of A, 73.2% in cases and 56.9% in controls.
Association between rs1413711 and response to anti-VEGF treatment in AMD is also studied widely. In two recent papers Yuan et al[17] and Dos Reis Veloso et al[18] studied this association. Yuan et al[17] found TT and CT genotypes were associated with improved visual acuity in anti-VEGF treatment, while the others[18] found no association between the VEGF genotypes and the treatment outcome.
Haines et al[19] analyzed eight genes including VEGF in two independent datasets consisting of a family-based association dataset (162 families) and a case control dataset (399 cases and 159 controls) for genetic linkage and association with AMD. They studied five SNPs in the VEGF gene including rs2146323 and found no significant association of this SNP with AMD. Also, Churchill et al[12] tested rs2146323 in their study but could not find any association with AMD. Fang et al[20] tested nine VEGF SNPs including rs1413711, and rs2146323 in a cohort of 515 Caucasian wet AMD patients for association with AMD and found that none of the investigated SNPs were associated with the disease. In contrast to these studies, we found that ancestral C allele of rs2146323 was highly protective against dry AMD, and also homozygosity of the A allele was associated with the disease.
Immonen et al[7] analyzed the association of rs2146323 and rs3025033 with the risk of AMD, CNV size and configuration, and the anatomic response to photodynamic therapy in 162 patients with exudative AMD and 85 age-matched controls. They found no association between the SNPs and the presence of the disease, lesion size and configuration. The genotype frequencies of the rs2146323 were significantly different between the photodynamic therapy (PDT) responders and non-responders, but rs3025033 genotype distribution was even between the two groups. Our findings for rs3025033 was not suitable for a statistical analysis because homozygosity of the ancestral A allele was almost always present in cases and controls except for 2 individuals (1 wet AMD and 1 control) with AG and 1 individual (wet AMD) with GG genotype.
In conclusion, we found that the polymorphisms of VEGF are associated with the dry AMD in our population. Since the genetic variations were shown to be associated with treatment response in AMD patients receiving anti-VEGF treatment, further data from future studies would determine whether VEGF polymorphisms might play a role in treatment decisions in a perspective of personalized medicine.
Acknowledgments
Foundation: Supported by Pamukkale University Scientific Research Unit (No.2011TPF025)
Conflicts of Interest: Bulgu Y, None; Cetin GO, None; Caner V, None; Cetin EN, None; Yaylali V, None; Yildirim C, None.
REFERENCES
- 1.Parmeggiani F, Romano MR, Costagliola C, Semeraro F, Incorvaia C, D'Angelo S, Perri P, De Palma P, De Nadai K, Sebastiani A. Mechanism of inflammation in age-related macular degeneration. Mediators Inflamm. 2012;2012:546786. doi: 10.1155/2012/546786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorin MB. Genetic insights into age-related macular degeneration: controversies addressing risk, causality, and therapeutics. Mol Aspects Med. 2012;33(4):467–486. doi: 10.1016/j.mam.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green WR. Histopathology of age-related macular degeneration. Mol Vis. 1999;5:27. [PubMed] [Google Scholar]
- 4.Ferris FL, 3rd, Fine SL, Hyman L. Age-related macular degeneration and blindness due to neovascular maculopathy. Arch Ophthalmol. 1984;102(11):1640–1642. doi: 10.1001/archopht.1984.01040031330019. [DOI] [PubMed] [Google Scholar]
- 5.Hawkins BS, Bird A, Klein R, West SK. Epidemiology of age-related macular degeneration. Mol Vis. 1999;5:26. [PubMed] [Google Scholar]
- 6.Abedi F, Wickremasinghe S, Richardson AJ, Makalic E, Schmidt DF, Sandhu SS, Baird PN, Guymer RH. Variants in the VEGFA gene and treatment outcome after anti-VEGF treatment for neovascular age-related macular degeneration. Ophthalmology. 2013;120(1):115–121. doi: 10.1016/j.ophtha.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Immonen I, Seitsonen S, Tommila P, Kangas-Kontio T, Kakko S, Savolainen ER, Savolainen MJ, Liinamaa MJ. Vascular endothelial growth factor gene variation and the response to photodynamic therapy in age-related macular degeneration. Ophthalmology. 2010;117(1):103–108. doi: 10.1016/j.ophtha.2009.06.037. [DOI] [PubMed] [Google Scholar]
- 8.Francis PJ. The influence of genetics on response to treatment with ranibizumab (Lucentis) for age-related macular degeneration: the Lucentis Genotype Study (an American Ophthalmological Society thesis) Trans Am Ophthalmol Soc. 2011;109:115–156. [PMC free article] [PubMed] [Google Scholar]
- 9.Nakata I, Yamashiro K, Nakanishi H, Tsujikawa A, Otani A, Yoshimura N. VEGF gene polymorphism and response to intravitreal bevacizumab and triple therapy in age-related macular degeneration. Jpn J Ophthalmol. 2011;55(5):435–443. doi: 10.1007/s10384-011-0061-z. [DOI] [PubMed] [Google Scholar]
- 10.Kloeckener-Gruissem B, Barthelmes D, Labs S, Schindler C, Kurz-Levin M, Michels S, Fleischhauer J, Berger W, Sutter F, Menghini M. Genetic association with response to intravitreal ranibizumab in patients with neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52(7):4694–4702. doi: 10.1167/iovs.10-6080. [DOI] [PubMed] [Google Scholar]
- 11.Age-Related Eye Disease Study Research Group The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study Report Number 6. Am J Ophthalmol. 2001;132(5):668–681. doi: 10.1016/s0002-9394(01)01218-1. [DOI] [PubMed] [Google Scholar]
- 12.Churchill AJ, Carter JG, Lovell HC, Ramsden C, Turner SJ, Yeung A, Escardo J, Atan D. VEGF polymorphisms are associated with neovascular age-related macular degeneration. Hum Mol Genet. 2006;15(19):2955–2961. doi: 10.1093/hmg/ddl238. [DOI] [PubMed] [Google Scholar]
- 13.Lin JM, Wan L, Tsai YY, Lin HJ, Tsai Y, Lee CC, Tsai CH, Tseng SH, Tsai FJ. Vascular endothelial growth factor gene polymorphisms in age-related macular degeneration. Am J Ophthalmol. 2008;145(6):1045–1051. doi: 10.1016/j.ajo.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 14.Qu Y, Dai H, Zhou F, Zhang X, Xu X, Zhang X, Bi H, Pan X, Wang H, Jiang H, Yin N, Dang G. Vascular endothelial growth factor gene polymorphisms and risk of neovascular age-related macular degeneration in a Chinese cohort. Ophthalmic Res. 2011;45(3):142–148. doi: 10.1159/000319543. [DOI] [PubMed] [Google Scholar]
- 15.Almeida LN, Melilo-Carolino R, Veloso CE, Pereira PA, Miranda DM, De Marco LA, Nehemy MB. Homozygosity for the +674C>T polymorphism on VEGF gene is associated with age-related macular degeneration in a Brazilian cohort. Graefes Arch Clin Exp Ophthalmol. 2012;250(2):185–189. doi: 10.1007/s00417-011-1807-5. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Shi Y, Xue C, Yin J, Huang Z. Pooled-analysis of the associations between three polymorphisms in the VEGF gene and age-related macular degeneration. Mol Biol Rep. 2012;39(6):6547–6553. doi: 10.1007/s11033-012-1483-5. [DOI] [PubMed] [Google Scholar]
- 17.Yuan D, Yuan D, Liu X, Yuan S, Xie P, Liu Q. Genetic association with response to intravitreal Ranibizumab for neovascular age-related macular degeneration in the Han Chinese population. Ophthalmologica. 2013;230(4):227–232. doi: 10.1159/000355068. [DOI] [PubMed] [Google Scholar]
- 18.Dos Reis Veloso CE, Frota de Almeida LN, Recchia FM, Pelayes D, Nehemy MB. VEGF gene polymorphism and response to intravitreal Ranibizumab in neovascular age-related macular degeneration. Ophthalmic Res. 2014;51(1):1–8. doi: 10.1159/000354328. [DOI] [PubMed] [Google Scholar]
- 19.Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, Olson L, Kenealy SJ, Hauser M, Gilbert JR, Pericak-Vance MA. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci. 2006;47(1):329–335. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 20.Fang AM, Lee AY, Kulkarni M, Osborn MP, Brantley MA., Jr Polymorphisms in the VEGFA and VEGFR-2 genes and neovascular age-related macular degeneration. Mol Vis. 2009;15:2710–2719. [PMC free article] [PubMed] [Google Scholar]


