Abstract
Background
Chronic pancreatitis (CP) is currently diagnosed using invasive endoscopic as well as radiation and non-radiation-based imaging techniques. However, urine can be safely and non-invasively collected and as such may offer a superior alternative to current techniques of CP diagnosis. We use mass spectrometry-based methods to discover proteins which are exclusive to or differentially abundant in urine of chronic pancreatitis patients.
Methods
We have performed a comparative quantitative proteomic analysis of urine collected from 5 healthy controls and 5 severe CP patients. Proteins from urine were fractionated briefly on SDS-PAGE and subsequently digested in-gel with trypsin. The resulting peptides were fractionated for 3 hours by reversed-phase liquid chromatography in-line with a mass spectrometer. ProteinPilot software and the QSPEC algorithm identified proteins and determined statistically significant differences between cohorts. In addition, we used a third cohort of non-CP disease patients to filter out those proteins which may be indicative of an ailment other than CP.
Results
We identified over 600 proteins from urine, of which several hundred were either exclusive to or differ quantitatively between healthy controls and severe CP patients. Members of the cathepsin protein family were of significantly higher abundance in the severe CP cohort. In addition, we have identified a core set of 50 proteins in all 15 samples, 25 of which showed no significant difference among the cohorts.
Conclusions
Proteomic analysis identified differentially abundant proteins in healthy controls and severe CP patients. Such proteins represent an initial set of targets for directed proteomics experiments for further validation studies. However, larger cohorts will be required to determine if these differences have statistically significant diagnostic potential.
Keywords: pancreas, cathepsin, urine, biomarkers, chronic pancreatitis
Background
Chronic pancreatitis (CP) is characterized by chronic inflammation and progressive fibrosis, clinically manifested as intense pain, and pancreatic exocrine and endocrine insufficiency. In the United States, exocrine pancreatic disorders affect over one million persons and cost approximately $3 billion annually. During the past decade, diseases of the exocrine pancreas have resulted in 277,000 hospitalizations and 475,000 ambulatory care visits per year [1]. Clinical diagnosis of chronic pancreatitis is primarily based on morphological and functional findings. Complications, such as bleeding and fistulae formation, preclude pancreatic biopsy for histologic diagnosis. The non-histological “surrogate” gold standard is pancreas function testing [2], which can diagnose only moderate to late stage chronic pancreatitis in which tissue damage and fibrosis are irreversible [3]. Radiologic imaging is also limited in diagnosing early disease as objective morphologic changes are only associated with moderate to advanced disease. Identification of biomarkers of early chronic pancreatitis would revolutionize diagnosis and potentially lead to novel therapies designed to retard, or modify disease progression, before irreversible organ damage and dysfunction become apparent.
We have previously investigated biomarker candidates of chronic pancreatitis in pancreatic fluid. Using the secretin-stimulated endoscopic pancreatic function test (ePFT), mass spectrometry analysis, and bioinformatic data processing, we showed that pathologic changes in the pancreas may be reflected in the pancreatic fluid proteome [4–9]. The secretin-stimulated ePFT can safely collect pancreatic fluid from the duodenum without cannulation of the pancreatic duct [10–12]. Pancreatic fluid is the proximal body fluid of the pancreas and is a reservoir of locally secreted biomolecules that are likely to include specific markers of disease. ePFT, however, is a relatively invasive procedure requiring endoscopy, sedation and/or anesthesia, and is performed at limited specialized centers. A chronic pancreatitis diagnostic biomarker panel, based on a non-invasively collected fluid, such as urine, would thus be preferred.
The analyses of proximal and systemic fluids have inherent advantages in medical testing. A proximal fluid directly represents a particular organ or region and is concentrated with secretions from surrounding cells. In contrast, systemic fluids, such as urine, represent the entire body and thus provide a snapshot of the whole organism under a given set of systemic conditions. The majority of the proteins detected in a systemic fluid are most likely not directly related to the disease or organ of interest [13]; analysis of systemic fluids may be confounded by proteins representing the normal or abnormal physiology of other organs and/or their pathophysiologic response to the disease of interest. These characteristics limit the utility of systemic fluids in investigating pathogenic and pathophysiologic mechanisms. However, systemic fluid analysis would be appropriate, clinically, for targeted biomarker assays for diagnosis and staging. Compared to other body fluids, urine, in particular, is readily available in large volumes, non-invasively collected, and the same individual can be sampled repeatedly and safely.
Mass spectrometry-based proteomic techniques can identify diagnostic urinary biomarkers to complement established diagnostic methods and improve early diagnosis of chronic pancreatitis [7]. Traditionally, pre-fractionation of proteins and/or peptides is performed prior to LC-MS/MS analysis. Greater sample complexity requires a greater degree of fractionation to achieve maximum proteome coverage. However, compared to whole cell lysates, the proteome of urine is suspected to be less complex [14–17]. As such, we have used a modified strategy to minimize sample processing by altering two parameters: 1) time of SDS-PAGE fractionation and 2) length of column and gradient during liquid chromatography. First, during a typical mini-gel SDS-PAGE fractionation, proteins migrate 7–8 cm and the gel lane is divided into 10–24 fractions, each to be processed separately via GeLC-MS/MS. However, we performed SDS-PAGE fractionation on the TCA-extracted protein sample until the proteins migrated only 1 cm into the gel, after which a single 1 cm gel slice is processed per lane. Second, following peptide extraction from gel slices, traditional LC-MS/MS methods then analyze the eluent from a 12–15 cm long reversed phase capillary column over a 1 hr period, however, we used a 45 cm-long column and eluted over a 3 hr period. This strategy simplifies sample processing while allowing efficient peptide fractionation upstream of mass spectrometric analysis.
We present the first comparative mass spectrometry-based proteomic analysis of urine in the study of chronic pancreatitis. The aim of this study is to determine the feasibility of identifying diagnostic protein biomarker candidates of chronic pancreatitis in urine using a new variation of well-established GeLC-MS/MS strategy employing minimal pre-fractionation.
Results
Several hundred proteins were identified in each specimen
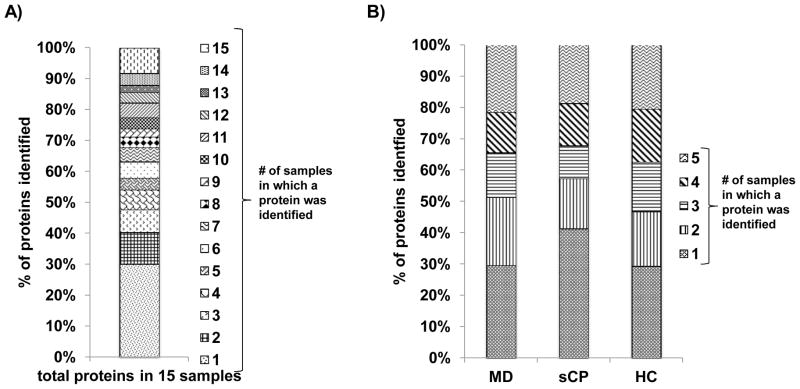
SDS-PAGE analysis (Figure 2) showed that, as expected, simple visualization of gels in not sufficient to determine proteomic differences between cohorts. Our subsequent mass spectrometry-based analysis of the gel lanes identified a total of 609 unique proteins in 15 samples (Supplemental Table 1). Figure 3A depicts the redundancy of each protein over all samples, i.e. proteins were grouped according to the number of samples in which they were identified. Approximately 25% of the proteins were identified in 10 or more samples and greater than 45% of the proteins were identified in 5 or more samples.
Figure 2. SDS-PAGE 1 cm separation gel image.
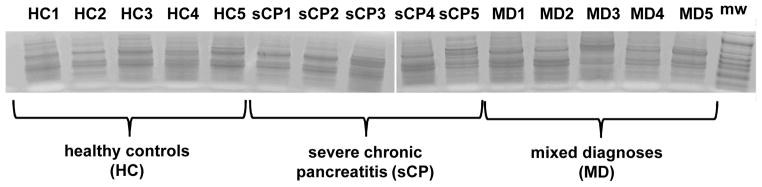
Fifteen samples - 5 each for healthy controls (HC), severe chronic pancreatitis (sCP), and mixed diagnoses (MD) - were analyzed via SDS-PAGE.
Figure 3. Bar graphs examining redundancy among identified proteins.
A) Redundancy of proteins identified in all 15 samples indicating the proportion of the total proteins identified in a specified number (1 through 15) of samples. B) Redundancy of proteins categorized by cohort (n=5), indicating the proportion of proteins identified in a specified number (1 through 5) of samples.
When examining each cohort individually (Figure 3B), the protein redundancy distribution within each cohort was similar to that of the whole sample set; approximately 30 – 45% of the identified proteins were identified in 5 or more samples and the largest proportion of proteins (25 – 40%) were identified in only one sample.
Differentially-expressed proteins were identified in severe chronic pancreatitis (sCP) versus healthy controls (HC)
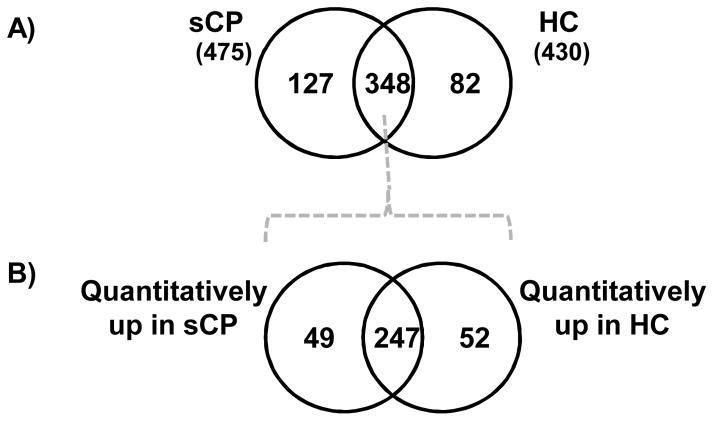
We identified a total of 430 unique proteins in healthy control subjects and 474 unique proteins in subjects with sCP. Comparing proteins identified in both cohorts, 127 proteins (Supplemental Table 2) were exclusive to the sCP cohort and 82 (Supplemental Table 3) were exclusive to the HC cohort (Figure 4A). The majority of the proteins exclusive to a specific cohort were identified in only 1 or 2 individual samples, making it difficult to assess the significance of these proteins as markers of pancreatic disease. However, we did identify several proteins in 3 or more of the 5 samples. In the sCP cohort, Ig lambda-7 chain C region, carbonic anhydrase 1 and neutrophil gelatinase-associated lipocalin were identified in 3 of the 5 samples. In the HC cohort, 6-phosphogluconolactonase appeared in all 5 sCP samples; ephrin type-B receptor, phosphatidylcholine -sterol acyltransferase, and N(G),N(G)-dimethylarginine dimethylaminohydrolase appeared in 4 samples; and Poliovirus receptor-related protein 4 and CD276 appeared in 3 of the 5 samples. These exclusive proteins, particularly those appearing in 4 or 5 samples, represent promising biomarker candidates and targets for further investigation.
Figure 4. Venn diagrams comparing identified proteins for the sCP and HC cohorts.
A) Proteins identified in sCP and HC cohorts were compared qualitatively to identify proteins exclusively to a particular cohort. B) The 348 common proteins were analyzed further using the QSPEC algorithm to identify statistically significant differences between the two cohorts.
Additionally, a total of 348 proteins were found to be common to both cohorts. Using the QSPEC algorithm, we compared the spectral counts of these common proteins to identify proteins present in statistically significant different abundance between the two cohorts. We identified 49 proteins (Supplemental Table 4) significantly more abundant in the sCP cohort and 52 proteins (Supplemental Table 5) significantly more abundant in the HC cohort (Figure 4B). Combining these two sets of proteins (those exclusive to and those statistically more abundant in a particular cohort) we determined that 176 (i.e., 127+49) and 134 (i.e., 82+52) proteins were differentially abundant in the sCP and HC cohorts, respectively, which we refer to as sCP′ and HC′.
A third (mixed diagnosis, MD) cohort was analyzed to control for pathologic changes in the urine proteome that may not be directly due to chronic pancreatitis
A cohort composed of patients with chronic abdominal pain, acute pancreatitis, and a participant with “borderline” chronic pancreatitis was included in our analysis as these individuals represent the intended use population, i.e., clinical patients commonly presenting to a tertiary care pancreas center for evaluation of abdominal pain.
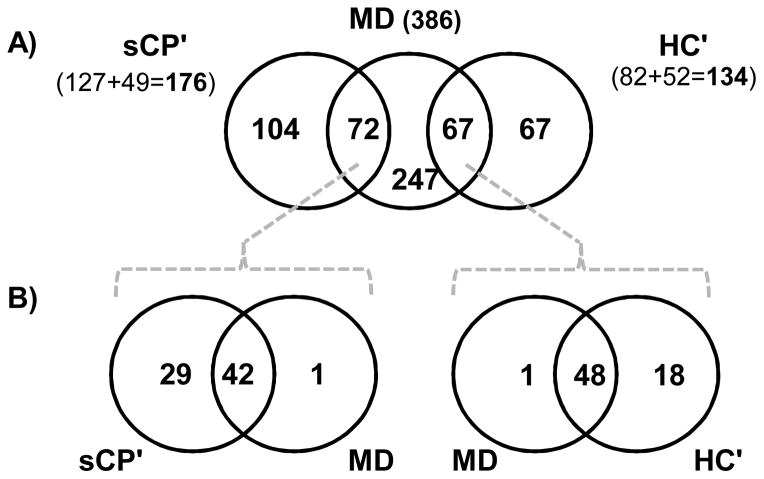
We identified a total of 386 unique proteins in this mixed diagnosis (MD) cohort. We compared the sCP′ protein subset to the proteins identified from MD cohort, as well as the HC′ protein subset to the proteins identified from MD cohort. In each of these comparisons, overlapping proteins (not exclusive to either the sCP′ or the HC′ cohort) will be eliminated from our list of biomarker candidates. Compared to the MD cohort, 104 (Supplemental Table 6) of the 176 proteins in the sCP′ subset remained exclusive to the sCP cohort, while 67 (Supplemental Table 7) of the 135 proteins in the HC′ subset continued to be exclusive to the HC cohort (Figure 5A). As such, 72 proteins were identified in both the sCP′ subset of proteins and the MD proteome, while 67 proteins were identified in both the HC′ subset and the MD proteome.
Figure 5. Venn diagrams comparing differentially expressed proteins for the sCP and HC cohorts with those identified in the MD cohort.
A) Exclusive and statistically different proteins were combined appropriately into 2 subsets, sCP′ and HC′, which were compared qualitatively with the MD cohort to eliminate proteins that are not specific to the chronic pancreatitis disease state. B) Proteins that were shared between MD and the other cohorts were subjected to QSPEC analysis and proteins with no significant difference between sCP′ and MD, as well as those with no significant difference between HC′ and MD, were removed from the set of potential chronic pancreatitis targets.
We also investigated if differences existed quantitatively in the proteins determined to be in common between the sCP′ and MD cohorts, as well as the HC′ and MD cohorts. Using spectral counting and QSPEC, we compared the MD cohort to the HC′ and sCP′ for significant differences in the abundance of common proteins (Figure 5B). Those proteins found with similar abundance likely represent confounding pathophysiological processes, and not chronic pancreatitis. Consequently these proteins were eliminated as potential diagnostic biomarkers of chronic pancreatitis. First, we examined the comparison of the HC′ and MD cohorts (Figure 5B left). This analysis showed that of the 72 proteins common to both sCP′ and MD, 1 was of greater abundance in the MD cohort, 42 were of similar abundance. More importantly, 29 proteins (Supplemental Table 8) were of significantly greater abundance in the sCP′ subset, representing the diagnostic biomarker candidates of chronic pancreatitis. Second, we examined the comparison of the HC′ and MD cohorts (Figure 5B right). Of the 67 proteins common to both HC′ and MD, 18 proteins (Supplemental Table 9) were of significantly higher abundance in HC′ subset and thus remained potentially useful targets for diagnostic testing. Additionally, 1 protein was more abundant in the MD cohort and 48 did not differ in abundance. Thus, a total of 49 proteins were eliminated as potential biomarker candidates of a normal pancreas. Therefore, our filtering strategy reduced potential confounding by eliminated many proteomic changes representing a disease state, but not necessarily chronic pancreatitis.
We identified a core set of proteins common to all 15 samples
Of the 609 proteins identified, 50 were present in all 15 samples (Table 2). These 50 proteins are readily identified using our minimal sample processing GeLC-MS/MS workflow. An ideal biomarker would be detected without the need for excessive enrichment or sample processing. Differentially expressed proteins readily identified with mass spectrometry are advantageous over those requiring large-scale preparation, as long as sensitivity and specificity are not compromised. Paired comparisons of the cohorts using QSPEC analysis identified 25 proteins common to all cohorts and present in statistically similar abundance. These proteins include beta-2-glycoprotein, fibronectin, insulin-like growth factor-binding protein 7, osteopontin, prothrombin, and serotransferrin. If validated in larger cohorts, several of these proteins may serve as candidates for normalization controls, which may be integrated into future, directed mass spectrometry workflows.
Table 2.
Core set of proteins identified in all 15 samples.
| Protein names | UniProt entry | spectral counts
|
significant difference (p<0.05) | ||
|---|---|---|---|---|---|
| ΣHC | ΣsCP | ΣMD | |||
| Alpha-1-antichymotrypsin | P01011 | 556 | 392 | 598 | no |
| Alpha-1-antitrypsin | P01009 | 1527 | 606 | 1264 | |
| Alpha-2-HS-glycoprotein | P02765 | 485 | 383 | 397 | no |
| Apolipoprotein D | P05090 | 2000 | 1512 | 1375 | |
| Basement membrane-specific heparan sulfate protein | P98160 | 2261 | 2535 | 1981 | |
| Beta-2-glycoprotein 1 | P02749 | 330 | 322 | 364 | no |
| Cathepsin D | P07339 | 355 | 760 | 270 | |
| CD44 antigen | P16070 | 261 | 298 | 352 | no |
| CD59 glycoprotein | P13987 | 148 | 283 | 211 | |
| Clusterin | P10909 | 612 | 550 | 435 | no |
| Collagen alpha-1 | P12109 | 1215 | 658 | 693 | |
| Cubilin | O60494 | 1348 | 1075 | 1122 | no |
| EGF-containing fibulin-like extracellular matrix protein 1 | Q12805 | 318 | 479 | 295 | no |
| Fibronectin | P02751 | 587 | 673 | 775 | no |
| Galectin-3-binding protein | Q08380 | 513 | 504 | 461 | no |
| Gelsolin | P06396 | 868 | 1287 | 664 | no |
| Hemopexin | P02790 | 496 | 353 | 534 | no |
| Ig alpha-1 chain C region | P01876 | 821 | 982 | 886 | no |
| Ig gamma-1 chain C region | P01857 | 1949 | 1350 | 1794 | |
| Ig gamma-2 chain C region | P01859 | 2268 | 1483 | 1942 | no |
| Ig heavy chain V-III region BRO | P01766 | 225 | 264 | 292 | no |
| Ig kappa chain C region | P01834 | 3456 | 8024 | 3137 | |
| Ig kappa chain V-III region VG | P04433 | 141 | 246 | 117 | |
| Insulin-like growth factor-binding protein 7 | Q16270 | 437 | 407 | 386 | no |
| Inter-alpha-trypsin inhibitor heavy chain H4 | Q14624 | 1455 | 1376 | 1036 | |
| Keratin, type I cytoskeletal 10 | P13645 | 937 | 1014 | 1399 | no |
| Keratin, type I cytoskeletal 9 | P35527 | 861 | 787 | 1378 | |
| Keratin, type II cytoskeletal 1 | P04264 | 1409 | 1543 | 2209 | |
| Kininogen-1 | P01042 | 2920 | 1983 | 2549 | |
| Lysosomal alpha-glucosidase | P10253 | 885 | 762 | 1046 | no |
| Monocyte differentiation antigen CD14 | P08571 | 590 | 568 | 452 | no |
| Non-secretory ribonuclease | P10153 | 374 | 607 | 591 | |
| Osteopontin | P10451 | 790 | 812 | 895 | no |
| Pancreatic alpha-amylase | P04746 | 2035 | 868 | 4547 | |
| Peptidoglycan recognition protein 1 | O75594 | 343 | 244 | 264 | no |
| Plasma serine protease inhibitor | P05154 | 737 | 370 | 492 | no |
| Polymeric immunoglobulin receptor | P01833 | 1393 | 1569 | 1300 | no |
| Pro-epidermal growth factor | P01133 | 1730 | 587 | 892 | |
| Prostaglandin-H2 D-isomerase | P41222 | 1395 | 3798 | 1617 | |
| Protein AMBP | P02760 | 2019 | 6600 | 3094 | |
| Prothrombin | P00734 | 406 | 483 | 332 | no |
| Retinol-binding protein 4 | P02753 | 337 | 483 | 315 | |
| Secreted and transmembrane protein 1 | Q8WVN6 | 242 | 311 | 133 | no |
| Serotransferrin | P02787 | 1933 | 1449 | 2179 | no |
| Serum albumin | P02768 | 16657 | 13767 | 20708 | |
| Uromodulin | P07911 | 8539 | 2502 | 5943 | |
| Vasorin | Q6EMK4 | 689 | 442 | 495 | |
| Vesicular integral-membrane protein VIP36 | Q12907 | 847 | 460 | 503 | |
| Vitamin D-binding protein | P02774 | 407 | 208 | 425 | |
| Zinc-alpha-2-glycoprotein | P25311 | 872 | 1885 | 1107 | |
HC, healthy controls; sCP, severe chronic pancreatitis; MD, mixed diagnosis samples.
Gene ontology analysis revealed different classes of proteins which were identified exclusively or in higher abundance in the sCP and HC cohorts
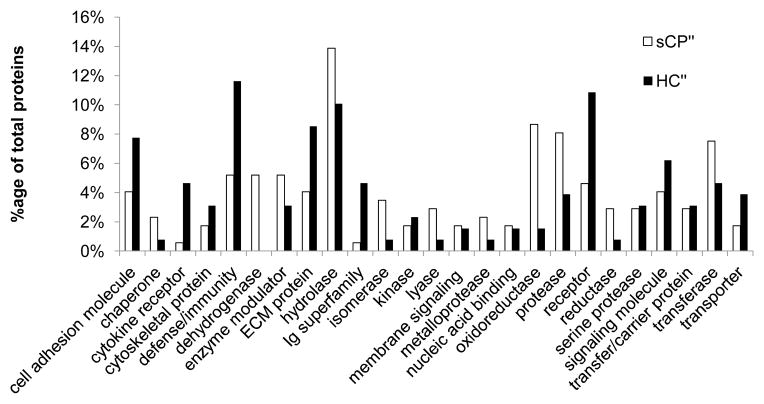
Using the DAVID interface, we categorized the potential biomarker candidate proteins according to their functional class (Figure 6). We compared the proteins in the sCP′ subset (which includes the 104 proteins that are exclusive to the sCP cohort, and the 29 proteins that were of higher abundance after filtering out those which overlap with the mixed diagnosis (MD) cohort) and those in the HC′ subset (which includes the 67 proteins that are exclusive to the HC cohort and the 18 proteins that were of higher abundance, after filtering out those which overlap with the mixed diagnosis (MD) cohort). The sCP cohort proteins correspond to those listed in Supplementary Tables 6 and 8 and the HC cohort proteins correspond to those listed in Supplementary Tables 7 and 9. A wide range of protein classes were identified; of note, proteases and other enzyme classes (hydrolase, oxidoreductase, and transferase) were more commonly identified in the sCP cohort. Proteins in this class included both secreted and lysosomal proteins: lysosomal protective protein, dipeptidyl peptidase 4, plasma glutamate carboxypeptidase, dipeptidyl-peptidase 1, pepsin, and members of the cathepsin family (B, D, and Z). The HC cohort urine was rich in defense/immunity-related proteins, cell adhesion molecules, receptors, and extracellular matrix (ECM) proteins. Of note are the defense/immunity-related proteins which included nectin 3, neogenin, and several immunogloblins which appear to be more highly abundant in the HC cohort. The investigation of these protein classes will allow us to target, not only single proteins, but classes, with protein microarrays ELISA, western blotting and/or targeted mass spectrometry experiments.
Figure 6. Gene ontology classification of proteins differentially abundant in severe chronic pancreatitis (sCP) and healthy controls (HC).
Functional class categorization of the aforementioned protein subsets according to Panther classification using the DAVID interface.
Discussion
We used a modified GeLC-MS/MS strategy for a chronic pancreatitis biomarker discovery assay to identify potential protein targets in urine for subsequent validation. Using this strategy, we identified a total of 609 proteins in the urine of 15 individuals. We qualitatively and quantitatively compared the proteins identified in severe chronic pancreatitis (sCP) and healthy control (HC) urine. We established subsets of differentially abundant proteins (i.e., exclusive to and of statistically greater abundance): 176 proteins in the sCP cohort and 134 proteins in the HC cohort. To isolate chronic pancreatitis as the cause for the enrichment of certain proteins, we eliminated proteins which were also present in a mixed diagnosis (MD) cohort. The MD cohort represents the typical clinical presentation of referred patients: individuals with chronic abdominal pain, acute pancreatic, and probable, but not severe, chronic pancreatitis. This strategy removed approximately one third of the proteins in each subset. Furthermore, gene ontology analysis identified certain classifications of proteins, such as proteases (particularly serine proteases) and defense/immune proteins as potential targets for directed analysis. In addition, of the 609 proteins identified, 50 were identified in all 15 samples, and as such may be useful for normalization in downstream assays.
Of particular interest for further investigation may be the cathepsin family of proteins. Three cathepsin subtypes (cathepsin B, D, and Z) were found to be more abundant in sCP compared to the HC and MD cohorts. This family of proteins is involved in mammalian cellular turnover, including bone absorption [18]. In a prior study, low-trauma bone fracture appeared to have a higher prevalence in chronic pancreatitis [19]. However, cathepsins have a wide range of cellular functions, including amyloid plaque breakdown in strokes [20], cancer [21], and arthritis [22] but have not been thoroughly studied in pancreatic disease. The role of these particular members of the cathepsin family in bone turnover and its relationship to chronic pancreatitis merits further investigation.
Relative to recent studies, in which thousands of proteins have been identified in urine, 609 proteins is a small number to be identified via mass spectrometry-based proteomics [14–17]. However, our goal was not to obtain a comprehensive proteomic profile of urine, but rather to use minimal sample processing to discover potential protein targets which could be utilized in efficient directed mass spectrometry experiments on larger cohorts. By limiting fractionation, we will likely identify the most abundant proteins. As such, for translational studies seeking an ELISA or dip-stick test as a diagnostic, enrichment and fractionation is a deterrent to general acceptance and the characterization of abundant proteins is a more practical strategy.
Our previous biomarker-driven studies focused on the proteomic analysis of pancreatic fluid [4, 7–9]. While pancreatic fluid is the proximal exocrine secretion of the pancreas, its collection is invasive, requires sedation and is performed only at a limited number of specialized centers. Biomarkers found in systemic fluids may be less specific than those obtained from proximal organ fluids, as the origin of the suspected biomarkers may be difficult to attain. Moreover, the complexity of systemic fluids poses a considerable challenge to investigating the mechanisms of disease. However, our aim is to pursue a non-invasively-collected fluid (i.e., urine) as an alternative for use in biomarker discovery due to the relative ease of collection and analysis, a favored characteristic of a diagnostic clinical test.
A major advantage of the methodology described herein is the minimal sample processing necessary for proteomic analysis. Apart from TCA precipitation, to concentrate and extract proteins from the non-protein constituents of urine, no sample enrichment is performed. Although SDS-PAGE was used to visualize proteins prior to tryptic digestion, gel-free in-solution digestion may be an equivalent alternative. We do caution that for protease-rich fluids, such as pancreatic fluid, in-solution digestion may promote undesired proteolysis by endogenous enzymes producing non-tryptic peptides, which will complicate database searching and protein identification. Regarding pancreatic fluid, we postulate that the denaturing conditions of SDS-PAGE and subsequent fixing of the protein in the polyacrylamide matrix prevents refolding of protease active sites, limiting non-tryptic cleavage of proteins. However, in urine such spurious protease activity may not be as prevalent, but caution may be warranted.
The use of lengthy reversed-phase columns and extended liquid chromatography gradients provide fractionation that is directly in-line with the mass spectrometer. The typical column length for a nanospray mass spectrometry experiment is 12–18 cm, however, we used 45 cm for the present study, along with an extension of the gradient from 1 to 3 hr. Coinciding with the advent of ultra-high pressure liquid chromatography (UHPLC) systems, longer columns, typically packed with reversed-phase C18 particles of smaller diameter, can be packed and show promise for the in-line fractionation of complex digests [23, 24]. Additional pre-fractionation, at the subcellular level and/or via full 24 fractions on SDS-PAGE, strong cation exchange chromatography, isoelectric focusing, or OFFGEL (Agilent), fractionation would increase the proteome coverage. However, such procedures would concomitantly increase sample processing times and may result potentially in sample loss due to the additional processing procedures. Future studies may further lengthen the column to provide greater protein loading capacity and elongate the gradient for greater fractionation.
To increase further the proteome coverage, depletion of highly abundant proteins may be justified, although caveats persist. The depletion of relatively high abundance proteins, such as albumin and immunoglobulins found in our core proteome subset, may be advantageous for biomarker discovery. Albumin can be depleted using dye-based columns, often with Cibachrome Blue dye [25], or monoclonal antibodies that improve specificity [26]. Additional abundant proteins are depleted often to improve identification of low abundance proteins. For example, multiple affinity removal columns capable of removing up to 14 of the most abundant proteins, simultaneously, are commercially available (MARS - Multiple Affinity Removal System, Agilent) [27] [26]. However, depletion of certain proteins, such as albumin (that functions as carrier proteins), may result in the loss or co-depletion of other significant proteins [28]. Furthermore, each additional step in the workflow may result in greater sample loss and variability.
Protein biomarkers of chronic pancreatitis in urine could potentially offer an innovative and non-invasive modality for diagnosis, staging and prognostication of the disease. In addition to urine, blood can also be collected non-invasively and used in protein-based chronic pancreatitis biomarker discovery. The human blood proteome is a reflection of the specific physiological state at a given point in time, rich in potential biomarkers from a variety of organ systems. Consistently detected alterations in the proteome can potentially be used for disease diagnosis, staging and prognosis [29]. Blood (both serum and plasma) has one of the highest dynamic ranges of any body fluid, with specific protein concentrations spanning over 10 orders of magnitude [30]. Although the dynamic range of the urine proteome still spans several orders of magnitude, its simplicity compared to serum makes it an attractive alternative in the search for diagnostic biomarkers of chronic pancreatitis [31]. For example, during urine production, renal blood flow undergoes glomerular ultrafiltration, greatly reducing urine protein complexity, thereby simplifying analysis and preparation methods [32–34]. In addition, up to 30% of proteins in urine are not of urinary tract origin, but are from distant sites [35]. Although urine was chosen over blood derivatives for analysis because of this decreased complexity and narrower dynamic range, the methods described herein are also relevant to proteomic biomarker discovery in blood, as well as other body fluids [36].
Conclusions
The diagnosis of early chronic pancreatitis remains elusive. Early detection is vital to initiate treatment within the therapeutic window before irreversible pancreatic scarring and dysfunction, and before symptoms become debilitating. Here we have presented a mass spectrometry-based strategy for chronic pancreatitis biomarker discovery in urine. Our data show both qualitative and quantitative differences between the urine proteomes of healthy subjects and that of subjects with severe chronic pancreatitis. To control for potential confounding diagnoses, we also compared a mixed disease cohort representing typical patients who are commonly referred to a pancreas center, whose protein profile may reflect a diseased state, but not necessarily that of chronic pancreatitis. Based on these investigations, we have assembled a subset of potential diagnostic biomarkers for chronic pancreatitis. Several of these proteins can be categorized as proteins having roles in defense and immunity and proteases. As such, future studies may develop assays to target these particular classes of proteins. The specific proteins may also serve as targets for further studies using directed mass spectrometry approaches or classical biochemistry techniques, such as ELISA or western blotting.
Methods
Study Population
This protocol was approved by the Institutional Review Board at Brigham and Women’s Hospital (BWH) (IRB # 2007-P-002480/1 for severe chronic pancreatitis (sCP) and mixed disease (MD) cohorts, and 2011P-001391 for healthy control volunteers). The experimental cohorts were assembled from adult patients referred to the Center for Pancreatic Disease (BWH) for evaluation of abdominal pain, and the healthy control (HC) adult volunteers were recruited from the general population. Patient characteristics are listed in Table 1.
Table 1.
Cohort characteristics
| Healthy controls (HC) | Severe chronic pancreatitis (sCP) | Mixed diagnoses (MD) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| HC1 | HC2 | HC3 | HC4 | HC5 | sCP1 | sCP2 | sCP3 | sCP4 | sCP5 | MD1 | MD2 | MD3 | MD4 | MD5 | |
| Gender | M | M | M | M | M | F | M | M | M | M | M | F | M | F | M |
| Race | W | B | W | W | W | W | W | W | W | W | W | H | W | W | W |
| Age (years) | n/a | n/a | n/a | n/a | n/a | 53 | 44 | 78 | 68 | 33 | 74 | 42 | 69 | 39 | 54 |
| MANNHEIM diagnosis | n/a | n/a | n/a | n/a | n/a | Def CP | Def CP | Def CP | Def CP | Def CP | AP | AP | Prob CP | CAP | CAP |
| Atlanta AP severity | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | Severe | Mild | n/a | n/a | n/a |
| [HCO3] | n/a | n/a | n/a | n/a | n/a | 38 | 50 | 36 | 39 | 60 | n/a | n/a | n/a | n/a | 100 |
| Smoking | n/a | n/a | n/a | n/a | n/a | 2 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 2 |
| Alcohol | n/a | n/a | n/a | n/a | n/a | 1 | 2 | 1 | 2 | 1 | 2 | 0 | 0 | 2 | 2 |
AP, acute pancreatitis; n/a, available; M, male; F, female; W, white; H, Hispanic; B, black; CP, chronic pancreatitis; Def CP, definite chronic pancreatitis; Prob CP, probable chronic pancreatitis; CAP, chronic abdominal pain. Smoking and Alcohol: 0, never smoking; 1, past; 2, current.
Materials
SeeBluePlus2 Pre-Stained standard (LC5925), LDS (lithium dodecyl sulfate) sample buffer (NP0008), NuPAGE 4–12% Bis-Tris polyacrylamide gels (NP0335), SimplyBlue Coomassie stain (LC0665), and MES-SDS (2-(N-morpholino)ethanesulfonic acid-sodium dodecyl sulfate) electrophoresis buffer (NP002) were from Invitrogen (Carlsbad, CA). Sequencing-grade modified trypsin (V5111) was obtained from Promega (Madison, WI). Other reagents and solvents were from Sigma-Aldrich and Burdick & Jackson, respectively.
Experimental workflow
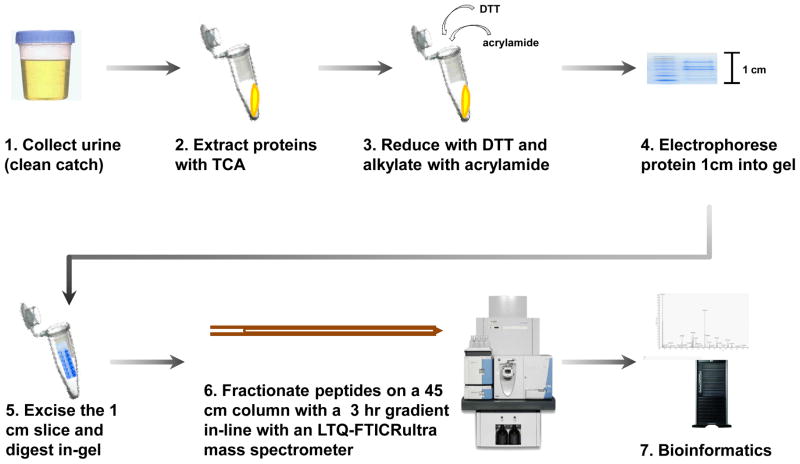
The general experimental workflow (Figure 1) was as follows: 1) Urine samples were collected (clean-catch); 2) proteins were extracted from the sample by trichloroacetic acid (TCA) precipitation; 3) proteins were reduced with dithiotreitol (DTT) and alkylated with acrylamide; 4) proteins were electrophoresed into an SDS-PAGE gel for a length of 1 cm; 5) proteins from each sample were in-gel tryptically digested in a single gel slice; 6) peptides were fractionated using a 45 cm capillary reversed-phase (C18) column with a 3 hr linear gradient, in-line with an LTQ-FT Ultra mass spectrometer; 7) bioinformatics processing using ProteinPilot for database searching, QSPEC [37] to determine statistically significant differences in spectral counts between cohorts, and DAVID [38, 39] to perform gene ontology analysis.
Figure 1. Experimental workflow.
The method was as follows: 1) Urine was collected via clean-catch. 2) Proteins were extracted with trichloroacetic acid (TCA) precipitation. 3) Proteins were reduced with dithiotreitol (DTT) and alkylated with acrylamide. 4) Proteins were separated via SDS-PAGE for a length of 1 cm. 5) The single band was excised and in-gel tryptically digested. 6) Peptides were fractionated on a 45 cm column with a 3 hr gradient in-line with an LTQ-FTICR Ultra mass spectrometer. And 7) Bioinformatics tools, such as ProteinPilot, QSPEC, and DAVID were used to identify, quantify, and characterize the collected data.
Sample preparation
Urine collection and storage
Urine from the subjects’ second void of the day was collected using the clean-catch method. The urine was promptly aliquoted and stored at −80°C until analysis. The urinary protein concentration was determined using the bicinchoninic acid (BCA) colorimetric assay.
TCA precipitation of urine
A total of 125 μL of ice-cold 100% TCA was added to 1 mL of each urine sample. The samples were vortexed for 5 seconds and incubated at 4°C for 2 hours. The samples were then centrifuged at 20,000×g at 4°C for 30 minutes and the supernatant carefully aspirated and discarded. One milliliter of 100% ice-cold acetone was added to the pellets, which were then briefly vortexed and incubated at −20°C for 1 hour. The samples were centrifuged again at 20,000×g at 4°C for 30 minutes, and the pellets washed twice with 100% ice-cold acetone. The final pellets were allowed to air dry at room temperature.
SDS-PAGE analysis
LDS sample buffer, with 50mM DTT, was added to each sample to achieve a 1X concentration. The samples were incubated at 56°C for 30 min and allowed to cool. The samples were then alkylated with 1% acrylamide and incubated for 30 min at 23°C. Proteins (approximately 100 μg) were fractionated by SDS-PAGE at 150 volts in MES buffer for a total migration length of 1 cm. Gels were rinsed in deionized water for 10 min, fixed in 45% methanol/45% water/10% acetic acid for 30 min, stained with SimplyBlue Coomassie for 1 hour, and destained overnight in deionized water.
GeLC-MS/MS analysis
Proteins in the entire 1 cm gel section underwent standard in-gel tryptic digestion using established techniques [40, 41]. Peptides were extracted from each gel section and fractionated by a nanoflow reversed-phase ultra-high pressure liquid chromatography system (nanoLC, Eksigent) in-line with a linear trap quadrupole-Fourier transform ion cyclotron mass spectrometer (LTQ-FT Ultra, Thermo Scientific). The reversed-phase liquid chromatography columns (45 cm long × 150 μm ID, New Objective, Woburn, MA) were packed in-house (Magic C18, 5 μm, 100 Å, Michrom BioResources). Samples were analyzed with a 3-hr linear gradient (5–29% acetonitrile with 0.2% formic acid), and data were acquired in a data-dependent manner using 6 MS/MS scans for every full scan spectrum.
Bioinformatics and Data Analysis
Mascot generic files (“mgf”) were generated using MSconvert software [42]. All MS data generated from the gel sections were searched against the UniProt database (downloaded November 11, 2011) using the Paragon Algorithm [43] integrated into the ProteinPilot search engine (v. 4; ABSciex). Search parameters were set as follows: sample type, identification; Cys alkylation, acrylamide; Instrument, Orbitrap/FT (1–3 ppm), LTQ MS/MS; special factors, gel-based ID; ID focus, Biological Modifications; database, UniProt database; detection protein threshold, 95.0%; and search effort, thorough ID. Species specificity was Homo sapiens. We used Proteomics System Performance Evaluation Pipeline (PSPEP) to determine the cutoff that would be result in a 1% global false discover rate at the protein level.
Venn diagrams
The VENNY on-line Venn diagram plotter was used to obtain lists of unique and common proteins among the cohorts investigated [44].
QSPEC spectral counting analysis
Relative protein quantification was performed using a label-free technique, spectral counting, which compared the number of identified peptide-spectra matches for the same protein across multiple data sets. To search for differences in the protein profile among data sets, spectral counts were normalized based on the total spectral counts, as outlined previously [45]. Briefly, spectral counts of each protein were first divided by the total spectral counts of all proteins from the same sample, and then scaled by multiplying the total spectral counts of each sample by the number of spectral counts of the sample with the highest number of spectral counts. Significance analysis of our normalized spectral count data was performed using QSPEC, a recently published algorithm for determining the statistical significance of differences in spectral count data from two cohorts [37]. This algorithm used the Bayes Factor, in lieu of the p-value, as a measure of evidential strength [46, 47]. By convention, a Bayes factor greater than 10 suggested strong evidence that a particular protein was differentially expressed between the two cohorts; thus a value of 10 was used as our significance threshold [48].
DAVID Functional Annotation Bioinformatics Microarray Analysis
We used the DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Database (http://david.abcc.ncifcrf.gov/) interface [38, 39] to classify proteins according to the Panther Protein Class [49].
Supplementary Material
Summary of all proteins (n=609) identified.
Proteins (n=127) exclusive to sCP cohort (not appearing in HC).
Proteins (n=82) exclusive to HC cohort (not appearing in sCP).
Proteins (n=49) of higher abundance in sCP compared to HC cohorts, as determined via QSPEC.
Proteins (n=52) of higher abundance in HC compared to sCP cohorts, as determined via QSPEC.
Proteins (n=104) exclusive to sCP′ cohort (i.e., exclusive to or of higher abundance in sCP cohortversus HC cohort) compared to MD cohort.
Proteins (n=67) exclusive to HC′ cohort (i.e., exclusive to or of higher abundance in HC cohort versus CP cohort) compared to MD cohort.
Proteins (n=29) of higher abundance in sCP′ cohort (i.e., exclusive to or of higher abundance in sCP cohort versus HC cohort) compared to MD cohort.
Proteins (n=18) of higher abundance in HC′ cohort (i.e., exclusive to or of higher abundance in HC cohort versus CP cohort) compared to MD cohort.
Acknowledgments
Funds were provided by the following NIH grants: 1 F32 DK085835-01A1 (JP), 1 R21 DK081703-01A2 (DC) and 5 P30 DK034854-24 (Harvard Digestive Diseases Center; DC). We would like to thank the Burrill family for their generous support through the Burrill Research Grant. We would also like to thank members of the Steen Laboratory at Children’s Hospital Boston, particularly John FK Sauld and Ali Ghoulidi for their technical assistance and critical reading of the manuscript. In addition, we thank members of the Center for Pancreatic Disease at Brigham and Women’s Hospital, particularly Shadeah Suleiman for her technical assistance.
Abbreviations
- CP
chronic pancreatitis
- GeLC-MS/MS
in-gel tryptic digestion followed by liquid chromatography-tandem mass spectrometry
- HC
healthy controls
- LTQ-FTICR
linear trap quadrupole- Fourier transform ion cyclotron resonance mass spectrometry
- MD
mixed diagnosis
- sCP
severe chronic pancreatitis
- TCA
trichloroacetic acid
Footnotes
Conflicts of interests
The authors declare no competing interests.
Author contributions
JP carried out the experiments. SB and VK provided technical assistance. JP and VK drafted the original manuscript. JP, HS, and DC conceived of the study, and participated in its design and coordination. All authors helped to draft the manuscript and approved the final manuscript.
Contributor Information
Joao A. Paulo, Department of Pathology, Children’s Hospital Boston, Boston, MA. Proteomics Center at Children’s Hospital Boston, Boston, MA. Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Vivek Kadiyala, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Scott Brizard, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Peter A. Banks, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Darwin L. Conwell, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Hanno Steen, Departments of Pathology, Children’s Hospital Boston and Harvard Medical School, Proteomics Center at Children’s Hospital Boston, Boston, MA.
References
- 1.James S. Opportunities and challenges at NIDDK in digestive diseases research. Gastroenterology. 2007;132:1219–1220. doi: 10.1053/j.gastro.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 2.DiMagno EP, Go VL, Summerskill WH. Relations between pancreatic enzyme ouputs and malabsorption in severe pancreatic insufficiency. N Engl J Med. 1973;288:813–815. doi: 10.1056/NEJM197304192881603. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury R, Bhutani MS, Mishra G, Toskes PP, Forsmark CE. Comparative analysis of direct pancreatic function testing versus morphological assessment by endoscopic ultrasonography for the evaluation of chronic unexplained abdominal pain of presumed pancreatic origin. Pancreas. 2005;31:63–68. doi: 10.1097/01.mpa.0000164451.69265.80. [DOI] [PubMed] [Google Scholar]
- 4.Paulo JA, Kadiyala V, Lee LS, Banks PA, Conwell DL, Steen H. Proteomic Analysis (GeLC-MS/MS) of ePFT-Collected Pancreatic Fluid in Chronic Pancreatitis. J Proteome Res. 2012 doi: 10.1021/pr2011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulo JA, Lee LS, Banks PA, Steen H, Conwell DL. Difference gel electrophoresis identifies differentially expressed proteins in endoscopically collected pancreatic fluid. Electrophoresis. 2011;32:1939–1951. doi: 10.1002/elps.201100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulo JA, Lee LS, Wu B, Banks PA, Steen H, Conwell DL. Cytokine profiling of pancreatic fluid using the ePFT collection method in tandem with a multiplexed microarray assay. J Immunol Methods. 2011;369:98–107. doi: 10.1016/j.jim.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulo JA, Lee LS, Wu B, Banks PA, Steen H, Conwell DL. Mass spectrometry-based proteomics of endoscopically collected pancreatic fluid in chronic pancreatitis research. Proteomics Clin Appl. 2011;5:109–120. doi: 10.1002/prca.201000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulo JA, Lee LS, Wu B, Repas K, Banks PA, Conwell DL, Steen H. Optimized sample preparation of endoscopic collected pancreatic fluid for SDS-PAGE analysis. Electrophoresis. 2010;31:2377–2387. doi: 10.1002/elps.200900762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulo JA, Lee LS, Wu B, Repas K, Mortele KJ, Banks PA, Steen H, Conwell DL. Identification of pancreas-specific proteins in endoscopically (endoscopic pancreatic function test) collected pancreatic fluid with liquid chromatography--tandem mass spectrometry. Pancreas. 2010;39:889–896. doi: 10.1097/MPA.0b013e3181cf16f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conwell DL, Zuccaro G, Jr, Vargo JJ, Morrow JB, Obuchowski N, Dumot JA, Trolli PA, Burton A, O’Laughlin C, Van Lente F. An endoscopic pancreatic function test with cholecystokinin-octapeptide for the diagnosis of chronic pancreatitis. Clin Gastroenterol Hepatol. 2003;1:189–194. doi: 10.1053/cgh.2003.50028. [DOI] [PubMed] [Google Scholar]
- 11.Conwell DL, Zuccaro G, Jr, Vargo JJ, Trolli PA, Vanlente F, Obuchowski N, Dumot JA, O’Laughlin C. An endoscopic pancreatic function test with synthetic porcine secretin for the evaluation of chronic abdominal pain and suspected chronic pancreatitis. Gastrointest Endosc. 2003;57:37–40. doi: 10.1067/mge.2003.14. [DOI] [PubMed] [Google Scholar]
- 12.Wu B, Conwell DL. The endoscopic pancreatic function test. Am J Gastroenterol. 2009;104:2381–2383. doi: 10.1038/ajg.2008.181. [DOI] [PubMed] [Google Scholar]
- 13.Issaq HJ, Xiao Z, Veenstra TD. Serum and plasma proteomics. Chem Rev. 2007;107:3601–3620. doi: 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- 14.Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 15.Kentsis A, Monigatti F, Dorff K, Campagne F, Bachur R, Steen H. Urine proteomics for profiling of human disease using high accuracy mass spectrometry. Proteomics Clin Appl. 2009;3:1052–1061. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieper R, Gatlin CL, McGrath AM, Makusky AJ, Mondal M, Seonarain M, Field E, Schatz CR, Estock MA, Ahmed N, et al. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4:1159–1174. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 18.Authier F, Posner BI, Bergeron JJ. Endosomal proteolysis of internalized proteins. FEBS Lett. 1996;389:55–60. doi: 10.1016/0014-5793(96)00368-7. [DOI] [PubMed] [Google Scholar]
- 19.Tignor AS, Wu BU, Whitlock TL, Lopez R, Repas K, Banks PA, Conwell D. High prevalence of low-trauma fracture in chronic pancreatitis. Am J Gastroenterol. 2010;105:2680–2686. doi: 10.1038/ajg.2010.325. [DOI] [PubMed] [Google Scholar]
- 20.Lipton P. Ischemic cell death in brain neurons. Physiol Rev. 1999;79:1431–1568. doi: 10.1152/physrev.1999.79.4.1431. [DOI] [PubMed] [Google Scholar]
- 21.Nomura T, Katunuma N. Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J Med Invest. 2005;52:1–9. doi: 10.2152/jmi.52.1. [DOI] [PubMed] [Google Scholar]
- 22.Raptis SZ, Shapiro SD, Simmons PM, Cheng AM, Pham CT. Serine protease cathepsin G regulates adhesion-dependent neutrophil effector functions by modulating integrin clustering. Immunity. 2005;22:679–691. doi: 10.1016/j.immuni.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Nagaraj N, Alexander Kulak N, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. System-wide Perturbation Analysis with Nearly Complete Coverage of the Yeast Proteome by Single-shot Ultra HPLC Runs on a Bench Top Orbitrap. Mol Cell Proteomics. 2012;11:M111 013722. doi: 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakur SS, Geiger T, Chatterjee B, Bandilla P, Frohlich F, Cox J, Mann M. Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol Cell Proteomics. 2011;10:M110 003699. doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thresher WC, Swaisgood HE. Characterization of specific interactions of coenzymes, regulatory nucleotides and cibacron blue with nucleotide binding domains of enzymes by analytical affinity chromatography. J Mol Recognit. 1990;3:220–228. doi: 10.1002/jmr.300030509. [DOI] [PubMed] [Google Scholar]
- 26.Bjorhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5:307–317. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- 27.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics. 2003;3:422–432. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, 3rd, Liotta LA, Veenstra TD, Conrads TP. An investigation into the human serum “interactome”. Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 29.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Fliser D, Novak J, Thongboonkerd V, Argiles A, Jankowski V, Girolami MA, Jankowski J, Mischak H. Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 32.Rosty C, Christa L, Kuzdzal S, Baldwin WM, Zahurak ML, Carnot F, Chan DW, Canto M, Lillemoe KD, Cameron JL, et al. Identification of hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein I as a biomarker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res. 2002;62:1868–1875. [PubMed] [Google Scholar]
- 33.Kishi T, Grass L, Soosaipillai A, Scorilas A, Harbeck N, Schmalfeldt B, Dorn J, Mysliwiec M, Schmitt M, Diamandis EP. Human kallikrein 8, a novel biomarker for ovarian carcinoma. Cancer Res. 2003;63:2771–2774. [PubMed] [Google Scholar]
- 34.Alatas F, Alatas O, Metintas M, Colak O, Harmanci E, Demir S. Diagnostic value of CEA, CA 15-3, CA 19-9, CYFRA 21-1, NSE and TSA assay in pleural effusions. Lung Cancer. 2001;31:9–16. doi: 10.1016/s0169-5002(00)00153-7. [DOI] [PubMed] [Google Scholar]
- 35.Decramer S, Gonzalez de Peredo A, Breuil B, Mischak H, Monsarrat B, Bascands JL, Schanstra JP. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Paulo J, Vaezzadeh A, Conwell D, Lee R, Steen H. Sample Handling of Body Fluids for Proteomics. In: Ivanov A, Lazarev A, editors. Sample Preparation in Biological Mass Spectrometry. New York, NY: Springer; 2011. [Google Scholar]
- 37.Choi H, Nesvizhskii AI. False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J Proteome Res. 2008;7:47–50. doi: 10.1021/pr700747q. [DOI] [PubMed] [Google Scholar]
- 38.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Neubauer G, Mann M. Mapping of phosphorylation sites of gel-isolated proteins by nanoelectrospray tandem mass spectrometry: potentials and limitations. Anal Chem. 1999;71:235–242. doi: 10.1021/ac9804902. [DOI] [PubMed] [Google Scholar]
- 41.Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Anal Chem. 2001;73:1440–1448. doi: 10.1021/ac001318c. [DOI] [PubMed] [Google Scholar]
- 42.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.VENNY. An interactive tool for comparing lists with Venn diagrams. [ http://bioinfogp.cnb.csic.es/tools/venny/index.html]
- 45.Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., 3rd Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 46.Goodman SN. Toward evidence-based medical statistics. 1: The P value fallacy. Ann Intern Med. 1999;130:995–1004. doi: 10.7326/0003-4819-130-12-199906150-00008. [DOI] [PubMed] [Google Scholar]
- 47.Goodman SN. Toward evidence-based medical statistics. 2: The Bayes factor. Ann Intern Med. 1999;130:1005–1013. doi: 10.7326/0003-4819-130-12-199906150-00019. [DOI] [PubMed] [Google Scholar]
- 48.Jeffreys H. Theory of probability. 3. Oxford: Clarendon Press; 1961. [Google Scholar]
- 49.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of all proteins (n=609) identified.
Proteins (n=127) exclusive to sCP cohort (not appearing in HC).
Proteins (n=82) exclusive to HC cohort (not appearing in sCP).
Proteins (n=49) of higher abundance in sCP compared to HC cohorts, as determined via QSPEC.
Proteins (n=52) of higher abundance in HC compared to sCP cohorts, as determined via QSPEC.
Proteins (n=104) exclusive to sCP′ cohort (i.e., exclusive to or of higher abundance in sCP cohortversus HC cohort) compared to MD cohort.
Proteins (n=67) exclusive to HC′ cohort (i.e., exclusive to or of higher abundance in HC cohort versus CP cohort) compared to MD cohort.
Proteins (n=29) of higher abundance in sCP′ cohort (i.e., exclusive to or of higher abundance in sCP cohort versus HC cohort) compared to MD cohort.
Proteins (n=18) of higher abundance in HC′ cohort (i.e., exclusive to or of higher abundance in HC cohort versus CP cohort) compared to MD cohort.