SUMMARY
Skeletal muscles are formed in numerous shapes and sizes, and this diversity impacts function and disease susceptibility. To understand how muscle diversity is generated, we performed gene expression profiling of two muscle subsets from Drosophila embryos. By comparing the transcriptional profiles of these subsets, we identified a core group of founder cell-enriched genes. We screened mutants for muscle defects and identified novel functions for Sin3A and 10 other transcription and chromatin regulators in the Drosophila embryonic somatic musculature. Sin3A is required for the morphogenesis of a musclesubset, and Sin3A mutants display muscle loss and misattachment. Additionally, misexpression of identity gene transcription factors in Sin3A heterozygous embryos leads to direct transformations of one muscle into another, while overexpression of Sin3A results in the reverse transformation. Our data implicate Sin3A as a keybuffer controlling muscle responsiveness to transcription factors in the formation of muscle identity, thereby generating tissue diversity.
Keywords: muscle, myogenesis, founder cells, Sin3A, chromatin, transcriptional regulation, Drosophila
INTRODUCTION
Skeletal muscles exist in a range of shapes and sizes specific to their functions. This diversity manifests itself not only in distinct morphologies, but in differing susceptibilities to diseases like muscular dystrophy (Cardamone et al., 2008). The Drosophila somatic musculature provides an ideal system to address the mechanisms underlying muscle diversity. In the Drosophila embryo a repeated pattern of 30 distinct muscle fibers is present in each abdominal hemisegment (Figure 1A). Despite similarities, such as shared expression of contractile proteins, each muscle fiber can be distinguished by properties like its size, shape orientation, number of nuclei, innervation and tendon attachment sites (Baylies et al., 1998). Muscle fibers arise by the iterative fusion of two types of myoblasts, called founder cells (FCs) and fusion competent myoblasts (FCMs), to form a syncytium (Rochlin et al., 2010). In fusion mutants, single FCs develop and extend processes towards their attachment sites, indicating that FCs contain the information required to direct morphogenesis of each specific muscle (Rushton et al., 1995). This information is encoded by DNA-binding transcription factors expressed in incompletely overlapping subsets of FCs, known as the FC identity genes (Beckett and Baylies, 2006; Tixier et al., 2010). Over a dozen identity genes have been identified, including the zinc-finger transcription factor Krüppel (Kr) (Ruiz-Gómez et al., 1997), the homeodomain proteins slouch (slou) (Dohrmann et al., 1990), apterous (ap) (Bourgouin et al., 1992) and araucan/caupolican (ara/caup) (Carrasco-Rando et al., 2011).
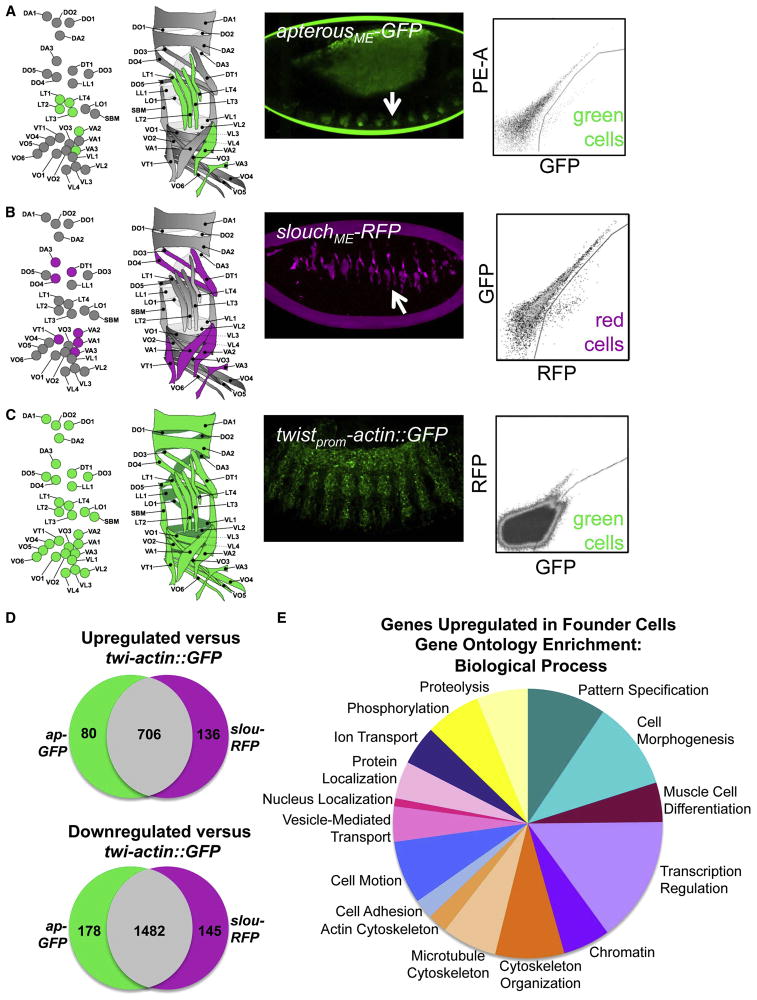
Figure 1. FACS and Transcriptional Profiling of Drosophila Muscle Founder Cells.
(A–C) Fluorescent transgenes marked subsets of muscle cells for FACS. Diagram of FCs labeled by each transgene at stage 13 (left) and into which muscles those FCs develop at stage 16 (right). Confocal projections show transgene expression at stage 13 and representative sort plots show positive cells. “PE-A” designates the fluorophorephycoeryhtrin. (D) Overlap between the gene lists derived from the microarray results. (E) GO Biological Process categories found to be enriched in FCs. See also Tables S1–S3.
The process by which identity genes determine final muscle characteristics is not fully understood. The combinatorial code hypothesis predicts that each FC has a distinct gene expression profile leading to the adoption of unique muscle characteristics (Tixier et al., 2010). While previous studies examined the differences in gene expression between FCs and FCMs, no study has compared distinct groups of FCs (Artero et al., 2003; Busser et al., 2012; Estrada et al., 2006). It is therefore unknown how different each set of FCs is from another set, and what genes contribute to the establishment of muscle diversity.
The chromatin regulator Sin3A is conserved from yeast to humans, serving as a scaffold protein for the Sin3/histone deacetylase (HDAC) complex (Grzenda et al., 2009; Silverstein and Ekwall, 2005). Though initially characterized as a repressor, Sin3/HDAC has been implicated in both positive and negative regulation of transcription via its role in chromatin modification. Sin3/HDAC has many roles in development, particularly in muscle (Cunliffe, 2008; Sharma et al., 2008; Sheeba et al., 2007; Shi and Garry, 2012; van Oevelen et al., 2010; 2008). In chick, Sin3A has been shown to be important in the development of rostral pre-somitic mesoderm and fully formed somites (Sheeba et al., 2007; Tomancak et al., 2002). Mice with reduced levels of Sin3A in muscle exhibit disruption of the sarcomere, and depletion of Sin3A from C2C12 myoblast cells results in downregulation of a number of genes important for somitogenesis and sarcomere formation (van Oevelen et al., 2010). Interestingly, no prior role has been discovered in these systems for Sin3A in the formation of specific muscle identities.
An intriguing aspect of muscle development in Drosophila has been that direct transformations of one muscle into another have been very rarely observed, though they might have been expected when identity genes are misregulated. These observations have led us to hypothesize that only certain muscles in the pattern are competent to respond to individual identity genes. One explanation for the differential competency of muscles is alteration in chromatin structure, but no chromatin regulators had been shown to be important for this process in Drosophila. Here, we use a combination of fluorescence-activated cell sorting (FACS), microarray and mutant analysis to identify 11 genes not previously characterized in Drosophila embryonic muscle development. We report a novel role for the chromatin regulatorSin3A in muscle morphogenesis, as well as in the adoption of muscle identity. Our work indicates that one important role for Sin3A is to guarantee that cells are competent to respond to information from identity genes, thus ensuring muscle identity and ultimately, tissue diversity.
RESULTS
FACS Purification and Microarray Analysis of Muscle Founder Cells
To identify FC-specific genes and other factors contributing to muscle diversity, we performed microarray analysis on FAC-sorted populations of FCs from dissociated embryos at late stage 12/early stage 13 (8–10 hours after egg laying, AEL). These embryos expressed one of two fluorescent transgenes in a subset of muscle FCs under the control of an identity gene promoter: apME-GFP (Figure 1A) or slouME-RFP (Figure 1B) (Richardson et al., 2007; Schnorrer et al., 2007). twistprom-actin::GFP labels both FCs and FCMs and was used as a control (Figure 1C) (Richardson et al., 2007). Sorting of FC subsets has not been done previously. These cells represented a very small percentage of the overall cells in the embryo; for apME-GFP, the sorted cells comprised only 0.02% of total dissociated embryonic cells. We, however, purified sufficient material for sample amplification to be unnecessary. RNA from these samples was used for microarray analysis (see Materials and Methods). We analyzed these microarray data, generating lists of genes that are differentially regulated in FCs (Table S1).
We used several approaches to validate our array data. First, we confirmed the presence of expected muscle genes, like the mesodermal transcription factors twist, DMef2, and pox meso in our list of upregulated genes (Baylies and Bate, 1996; Duan et al., 2007; Lilly et al., 1994; Nguyen et al., 1994; Taylor et al., 1995). Our lists shared a large number of genes with an RNAi screen in fly adult muscle (Table S2) (Schnorrer et al., 2010). Next, we cross-referenced over 100 up-regulated genes with a mesodermal whole genome chromatin immunoprecipitation (ChIP) study and discovered that 94% of these genes had Twist or DMef2 binding sites within 30 kilobases, suggesting somatic mesoderm expression (Zinzen et al., 2009) (Table 1 and data not shown).
Table 1. Genes that were identified as upregulated in the somatic mesoderm by microarray analysis.
Genes were scored as having proximal Twist or DMef2 binding sites if a prior whole genome ChIP on Chip experiment (Zinzen et al., 2009) identified a binding site within 30 kb of the gene. Whether these sites are located 5′ or 3′ to the coding region, or within intronic sequences, is noted. See Also Figure S1.
| Gene | Flybase Gene ID | Mammalian Homolog | Protein Type | Mesodermal-Specific Transcription Factor Binding Sites |
|---|---|---|---|---|
| chn | FBgn0015371 | - | zinc-finger containing | Twi and DMef2 5′ and intron |
| Lid | FBgn0031759 | Kdm5 | histone demethylase | Twi and DMef2 3′ |
| Med13 | FBgn0003415 | Med13 | Mediator complex member | Twi and DMef2 intron, 3′ |
| crp | FBgn0001994 | Tcfap4 | bHLH | Twi and DMef2 5′ and intron |
| Gug | FBgn0010825 | Atrophin1 and Rere | chromatin regulator | Twi and DMef2 5′ and intron |
| nom | FBgn0037617 | - | zinc-finger containing | Twi 5′ |
| Elo-B | FBgn0023212 | Elongin-B | elongation factor | Twi and DMef2 5′ and 3′ |
| lola | FBgn0005630 | ZBTB20 | zinc-finger containing | Twi and DMef2 5′, intron and 3′ |
| Alh | FBgn0261238 | AF10/AF17 | zinc-finger containing | Twi and DMef2 5′, intron and 3′ |
| Kdm2 | FBgn0037659 | Kdm2 | histone demethylase | Twi and DMef2 5′, intron and 3′ |
| sin3A | FBgn0022764 | Sin3A | chromatin regulator | Twi and DMef2 5′ and intron |
Upon comparison of our lists of upregulated genes, we observed significant overlap between the two muscle subsets. This result suggested that a core gene expression profile defines FCs, with few genes controlling later diversification of muscle types (Figure 1D). In situ hybridization analysis of over 150 array-identified genes revealed that 80% were expressed within the somatic mesoderm (Table S3, Figure S1). A comparison of our upregulated lists of genes with ImaGO terms describing embryonic gene expression revealed an enrichment for the terms “muscle cell primordium” and “embryonic and larval somatic muscle” (Table S2) (Tomancak et al., 2002). Using the Database for Annotation, Visualization and Integrated Discovery (DAVID), we also identified a number of Biological Function Gene Ontology (GO) categories that were enriched in our upregulated gene lists (Figure 1E) (Huang et al., 2009a; 2009b). Significantly enriched GO categories represent many functions important for myogenesis, including muscle cell differentiation pattern specification, cell morphogenesis, actin cytoskeleton regulation, and microtubule-based processes.
Many of the genes common to both FC subsets encode general regulators of transcription, including TAFs, elongation factors and components of transcriptional activator and repressor complexes. Such genes might be expected to be essential for a regulatory cascade, altering expression of downstream signaling and cell morphogenesis molecules responsible for differentiating FCs from FCMs. We focused on transcription factors and chromatin regulators to identify new regulators of muscle morphogenesis.
Identification of Regulators of Drosophila Embryonic Muscle Morphogenesis
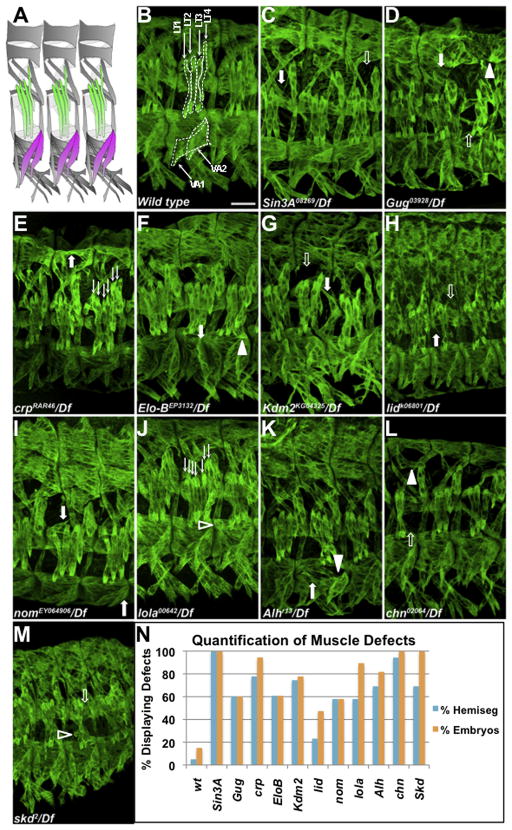
We conducted further analysis on FC-upregulated genes with confirmed mesodermal expression by in situ hybridization (Figure S1). We assessed the muscle pattern in available mutants and identified muscle defects in embryos homozygous for mutations in 11 genes not previously implicated in somatic muscle morphogenesis (Figure 2A–N; Table 1, Figure S2). These encode the previously uncharacterized zinc-finger protein CG8145, which we have named Numerous disordered muscles (Nom); zinc-finger proteins Longitudinals lacking (Lola), Alhambra (Alh) and Charlatan (Chn); bHLH protein Cropped/dAP-4 (Crp); elongation factor and E3 ubiquitin ligase complex member Elongin-B (Elo-B); Mediator complex member Med13/Skuld/Blind spot (Med13); and chromatin modifiers Little imaginal discs (Lid), Lysine-specific demethylase 2 (Kdm2), Grunge/Atrophin (Gug) and Sin3A (Figure 2B–M) (Aso and Conrad, 1997; Boube et al., 2000; Erkner et al., 2002; Escudero, 2005; Gildea et al., 2000; King-Jones et al., 1999; Lagarou et al., 2008; Linder et al., 2001; Pennetta and Pauli, 1998; Seeger et al., 1993). With the exception of the previously uncharacterized gene nom, all other genes are conserved in vertebrates (Table 1). Mutants displayed a range of defects, including muscle loss, missing or incorrect attachments, shape changes or extra muscles. Gene-specificity for each mutant allele was confirmed by examining embryos bearing the mutant allele in trans to a deficiency (Figure 2), and, when available, multiple mutant alleles for each gene were screened (data not shown). Homozygous mutants were also found to have muscle defects (Figure S2). No two genes displayed the same pattern of embryonic muscle defects, suggesting at least some independent functions for each factor.
Figure 2. Mutants for Genes Identified by Microarray Display Muscle Phenotypes.
(A) Diagram of wild-type muscle pattern of three hemisegments at stage 16. The LT muscles are green and the VA muscles are magenta (B–M) Stage 16 embryos stained with anti-Myosin heavy chain (MHC). In this and all following figures, unless indicated, approximately 3 hemisegments are shown, Scale bar, 25 μm. In the wild-type embryo, the LT and VA muscles of one hemisegment are outlined with dashed lines. Mutant phenotypes are indicated by filled arrows (misshapen), open arrows (missing muscles), line arrows (extra muscles), filled arrowheads (misattached muscles) and open arrowheads (unattached myospheres). (N) Percentage of hemisegments (blue) and embryos (orange) displaying aberrant phenotypes in each mutant background. Five abdominal hemisegments from at least 20 embryos for each genotype were quantified. See also Figures S2–S4.
To show that the observed muscle defects in these mutants were the result of loss of gene function in the mesoderm, we examined the cuticle of mutant embryos to determine whether overall embryonic patterning was disrupted; we did not detect significant defects for most of the genes (Figure S3). Finally, where available, we stained embryos expressing the FC-specific transgene rplacZ with antibodies against these proteins to show that they were expressed in the muscle. We detected expression of Alh, Chn, Med13/Skd, Sin3A and Crp in FCs (Figure S4). While some of the genes were expressed in most or all FCs (eg., Sin3A, Med13/Skd), others were expressed only in a subset of FCs (eg., Chn). Additionally, we detected Elo-B with an antibody against the human protein in the musculature of stage 16 embryos (Figure S4).
We performed a comparative analysis to test whether other studies supported a role for these newly identified genes in muscle development. Global chromatin immunoprecipitation studies identified Twi or DMef2 binding sites within the introns of or in close proximity to all 11 genes (Table 1) (Zinzen et al., 2009). Additionally, RNA Polymerase II has been shown to bind to the promoters of Med13, crp, lola, chn, Elo-B, Alh, Kdm2, lid and Sin3A in purified muscle cells (Weake et al., 2011). This study also found that the transcriptional co-activator complex SAGA bound to the promoters of lola, chn, Elo-B, Alh, Kdm2, lid and Sin3A in muscle cells and showed that Chn, Alh and Sin3A interact directly with the SAGA complex (Weake et al., 2011). In aggregate, comparison of our data to these whole genome studies confirmed the roles of these factors in the somatic musculature, but underscored the importance of in vivo experiments to demonstrate functional significance and identify specific muscles and pathways where these factors play a role.
Sin3A, EloB, Gug/Atrophin, crp and lola have also been previously implicated in the development of adult muscle (Schnorrer et al., 2010). Sin3A has been well-characterized as a general chromatin regulator and has been shown to play an important role in vertebrate somitogenesis, myotube differentiation, and sarcomere formation (Cunliffe, 2008; Sheeba et al., 2007; Shi and Garry, 2012; van Oevelen et al., 2010). Since the role of chromatin regulators in the establishment of muscle identity has never been characterized, we investigated Sin3A in the generation of muscle diversity.
Sin3A Mutants Display Muscle Defects
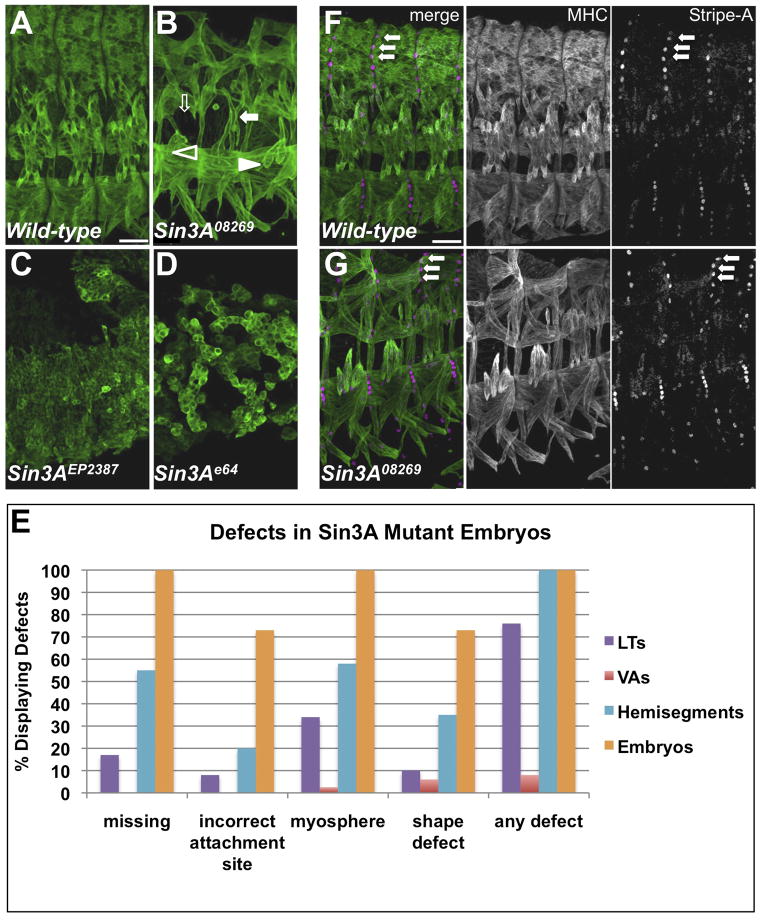
We examined several loss-of-function Sin3A alleles for aberrant muscle phenotypes (Figure 3A–D). Homozygous mutants for Sin3A08269, a partial loss-of-function allele caused by a transposable element insertion into the first intron, exhibited a range of muscle phenotypes (Figures 2C, 3B, S2C). Muscle defects were observed in 100% of embryos and 100% of abdominal hemisegments, and 100% of homozygous mutants died as embryos (Figures 2N, 3E, S2N, data not shown). The EMS/X-ray-generated allele Sin3Ae64 and the transposable element insertion allele Sin3AEP2387 showed strong patterning defects, including germband retraction failure (Figure 3C–D). In mutants carrying these two alleles, we were unable to distinguish a segmentally repeated muscle pattern and could detect unfused myoblasts. Together, these phenotypes suggest that the Sin3Ae64 and Sin3AEP2387 alleles have early defects in embryonic patterning and mesodermal specification, which is consistent with the abnormal cuticle pattern in these mutants (Figure S3 and data not shown). In contrast, the Sin3A08269 allele generated a recognizable muscle pattern and did not display early defects. For these reasons, we focused subsequent studies on the Sin3A08269 allele.
Figure 3. Mutant Phenotypes of and Identity Gene Expression in Sin3A Mutant Embryos.
(A–D) Wild-type (OreR) and Sin3A homozygous mutant embryos stained for MHC. Scale bar, 25 μm. In panel (B), mutant phenotypes are indicated by filled arrows (misshapen), open arrows (missing muscles), filled arrowheads (misattached muscles) and open arrowheads (unattached myospheres). (E) Quantification of mutant phenotypes in Sin3A08269 homozygous embryos. Five abdominal hemisegments from at least 10 embryos for each genotype were quantified. (F–G) Wild-type and Sin3A08269 embryos stained for MHC (white in single channel, green in merge) and StripeA (white in single channel, magenta in merge). Arrows point to SrA-expressing tendon cells. See also Figure S5.
The segmentally-repeated muscle pattern is generated in part in response to signals from the overlying ectoderm to the mesoderm during development (Baylies and Michelson, 2001). To ensure that the muscle defects in Sin3A08269 embryos were not due to a disruption of ectodermal patterning, we stained embryos for StripeA (SrA), a marker of mature tendon cells (Figure 3F–G) (Volk, 1999). SrA-expressing tendon cells are visible along the segment border in Sin3A08269 mutants (Figure 3G, arrows), indicating that these cells are properly specified and positioned. These results are consistent with the wild-type cuticle pattern in Sin3A08269 mutants (Figure S3). The Sin3A08269 allele presents a unique genetic tool to examine the role of Sin3A in the generation of muscle diversity.
To confirm the muscle defects in Sin3A08269 mutants, we analyzed two sets of muscles in the muscle pattern: the four lateral transverse muscles (LTs 1–4), and two ventral acute muscles (VA1 and VA2). The centrally located LTs have simple, elongated shapes, dorsal-ventral orientations and single attachment sites at the dorsal and ventral ends of the muscles. LT4 has a characteristic positional shift dorsally. The ventrally located VA1 and VA2 muscles are flag shaped, extending diagonally from an attachment to the posterior segment border in each hemisegment. VA2 has a distinctive downward curve, while VA1 extends across the hemisegment. In Sin3A08269 mutants, the LTs were often misshapen and frequently attached incorrectly (Figure 3B,E). We also frequently observed missing or unattached LTs; unattached muscles did not adopt an elongated morphology (Figure 3B,E). We observed VA1 and VA2 defects much less frequently, but did detect muscle misattachments and mild changes in muscle shape (Figure 3B,E).
To test whether the LT and VA FCs have been specified correctly in Sin3A08269 mutants, we examined expression of the identity gene transcription factors Kr and Slou. LT2, LT4, VA1 and VA2 are specified correctly in Sin3A08269 mutants (Figure 4A–F). Additionally, in Sin3A08269 mutants, all four LT muscles expressed an apME-NLS::dsRed transgene that is detectable in the nuclei (Figure 4E–J) (Richardson et al., 2007; Metzger et al., 2012). It was apparent from the expression of apME-NLS::dsRed transgene that fusion occurs in the Sin3A08269 mutant, but due to morphological defects, the nuclei are not well spread out, making determination of final nuclei number difficult (Figure 4E–J). The expression of Kr, Slou and the apME-NLS::dsRed transgene confirmed that the LT and VA muscles retained their essential identities, despite the morphological changes undergone by these muscles in Sin3A08269 mutants.
Figure 4. Identity Gene Expression in Sin3A Mutant Embryos.
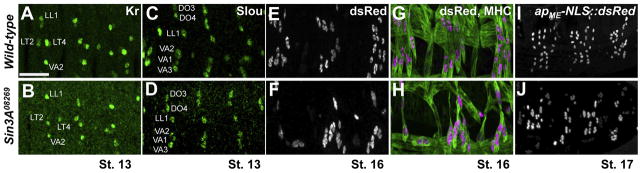
(A–J) Wild-type and Sin3A08269 mutants stained with antibodies against FC markers. Scale bar, 25 μm.(A–B) Stage 13 embryos stained with anti-Kr (green). (C–D) Stage 13 embryos stained with anti-Slou (green). (E–F) Wild-type and Sin3A08269 embryos expressing the apME-NLS::dsRed transgene (G–H) Stage 16 embryos stained with anti-MHC (green) and anti-dsRed (white in single channel, magenta in merge) to shown nuclear pattern. (I–J) Live images of stage 17 embryos expressing the apME-NLS::dsRed transgene (white).
The misattachment phenotypes observed in Sin3A08269 mutants were reminiscent of mutations in the integrin-encoding genes multi edematous wings (mew, αPS1 integrin) and myospheroid (mys, βPS integrin). These integrins are critical for the formation of muscle and tendon attachments (Volk, 1999). Additionally, Sin3A positively regulates the vertebrate homologs Itga7 and Itgb1 in C2C12 myoblasts (van Oevelen et al., 2010). We examined Mew and Mys expression in Sin3A08269 embryos by staining and quantitative PCR analysis and found reduced expression in Sin3A08269 mutants (Figure S5). Interestingly, greater differences in Integrin expression were seen in the LTs. While Mew and Mys expression at the segment border was disrupted, expression at the ends of the LT muscles was completely absent. This reduction in Integrin expression most likely contributes to the attachment defects observed in Sin3A08269 mutants.
Genetic Interaction between Sin3A and Identity Gene Transcription Factors
Studies in vertebrate muscle have pointed to a role for chromatin structure during many steps of skeletal myogenesis (Sartorelli and Juan, 2011; Fong et al., 2012). Recent work in neurons has also demonstrated a role for chromatin regulators in modulating the response to transcription factors in the determination of neuronal cell fate (Poole et al., 2011; Tursun et al., 2011; Weinberg et al., 2013). We hypothesized that Sin3A performs an analogous function in the Drosophila musculature, thereby altering identity gene transcription factor response. We therefore genetically reduced the dosage of Sin3A by half and tested whether overexpression of identity genes in this background led to changes in the muscle pattern.
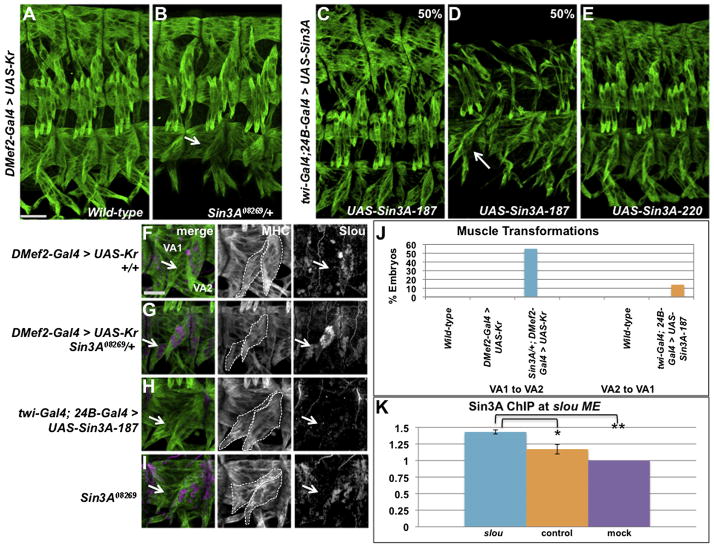
The VA muscles provide one of the few examples of direct muscle transformation (Ruiz-Gómez et al., 1997). Previous experiments found that driving UAS-Kr expression with the strong mesodermal driver twi-Gal4;24B-Gal4 results in the transformation of muscle VA1 to VA2 at a low penetrance (Ruiz-Gómez et al., 1997). In contrast, we observed no muscle defects when the DMef2-Gal4 driver was used to express UAS-Kr (Figure 5A). The lack of muscle defects in these embryos provided a sensitive assay to examine the effects of Sin3A loss on muscle morphogenesis. We tested whether overexpression of Kr by DMef2-Gal4 would lead to more severe muscle defects in a Sin3A08269 heterozygous background, and found, remarkably, that 55% of embryos reproducibly displayed the muscle transformation of VA1 into VA2 (Figure 5B, J).
Figure 5. Alteration of Muscle Identity in Sin3A Gain and Loss of Expression Embryos.
(A–B) Stage 16 embryos stained with anti-MHC. Arrow points to the VA1 to VA2 muscle transformation in Sin3A08269/+; DMef2-Gal4 > UAS-Kr embryos. 55% of embryos in (B) display the muscle transformation, n=20. Scale bar, 25 μm.(C–E) Stage 16 embryos stained with anti-MHC expressing either the 187 or 220 kDa isoforms of UAS-Sin3A under the control of twi-Gal4; 24B-Gal4. Arrow points to VA2 to VA1 muscle transformation, 14% of embryos, n=56. (F–I) Slou expression (white in single channel, magenta in merge) in the VA muscles of stage 16 Sin3A08269 mutant embryos, Scale bar, 15μm. Embryos are also stained with anti-MHC (green) to show muscle pattern, and VA1 and VA2 are outlined with dashed lines. One hemisegment is shown. Arrows point to muscle VA2. (J) Quantification of ventral muscle transformations in Sin3A gain and loss of function embryos. Five abdominal hemisegments from at least 20 embryos for each genotype were quantified. (K) ChIP was performed on extracts from wild-type embryos at stage 13 using anti-Sin3A antibody. The presence of the slou mesodermal enhancer or a control region was assayed in precipitated samples by quantitative PCR. Sin3A binding was significantly enriched at the slou mesodermal enhancer (blue) compared to the control region (orange, p=0.045), as well as relative to mock (purple, p=0.003). Error bars indicate standard deviation. See also Figure S6.
Since reduction of Sin3A expression affects VA muscle identity, we next examined the effect of Sin3A overexpression on the muscle pattern. In Drosophila, multiple Sin3A protein isoforms are expressed in the embryo that differ in their C-terminal domain (Spain et al., 2010). We drove expression of the 187 and 220 kDa isoforms of Sin3A with DMef2-Gal4 or twi-Gal4; 24B-Gal4. Similar to what we and others have observed with overexpression of Kr, we observed much stronger defects in embryos where twi-Gal4;24B-Gal4 was used to drive transgene expression rather than DMef2-Gal4 (data not shown) (Ruiz-Gómez et al., 1997). Embryos in which UAS-Sin3A-187 was misexpressed in the mesoderm displayed a range of defects in the somatic musculature (Figure 5C–D). While half of the embryos displayed a wild-type pattern (Figure 5C), the other 50% of embryos show missing and misshapen muscles (Figure 5D). Strikingly, we detect the opposite VA2 to VA1 muscle transformation in a subset (14%) of embyros (Figure 5D, J). Misexpression of UAS-Sin3A-220 in the mesoderm did not result in severe muscle defects or VA2 to VA1 muscle transformations (Figure 5E), suggesting that the Sin3A-187 isoform is the more active isoform in the mesoderm. This result is consistent with a role for Sin3A-187 in differentiating cells (Spain et al., 2010).
To confirm the VA muscle transformations, we stained embryos for Slou expression. Slou protein is expressed in VA2 in wild-type embryos and is activated by Kr (Figure 5F) (Ruiz-Gómez et al., 1997). In Sin3A08269 heterozygous embryos in which UAS-Kr is being driven by DMef2-Gal4, Slou was visible in two VA muscles, confirming the VA1 to VA2 transformation (Figure 5G). We did not observe ectopic expression of Slou elsewhere in the muscle pattern (data not shown). In twi-Gal4; 24B-Gal4 > UAS-Sin3A-187 embryos, we no longer detect Slou staining in either of the two ventral muscles, consistent with a VA2 to VA1 transformation (Figure 5H) (Ruiz-Gómez et al., 1997).
We next examined expression of slou transcript in wild-type and Sin3A08269 homozygous embryos by quantitative RT-PCR. slou expression was significantly increased in Sin3A08269 homozygous mutants (Figure S6A), demonstrating that, in the whole embryo, reduction of Sin3A affected the levels of slou transcript. Despite this overall increase in expression, we do not see changes to the pattern of Slou expression in Sin3A08269 mutants without misexpression of Kr (Figure 5I). Loss of Sin3A appears to permit increased Slou expression only in muscles where Slou is normally expressed. This result suggests that Slou is depressed by loss of Sin3A, but requires positive activation of Slou by Kr and other Slou activators. Finally, ChIP experiments in wild-type embryos revealed an enrichment of Sin3A at the slou mesodermal enhancer, suggesting that the repression of slou by Sin3A was direct (Figure 5K).
To test whether the muscle transformations were the result of a specific genetic interaction between Sin3A and Kr, or if Sin3A had a more general effect on muscle responsiveness to identity gene expression, we misexpressed UAS-ara with DMef2-Gal4. As with Kr, we observed an enhancement of aberrant muscle phenotypes by misexpressing ara in Sin3A08269 heterozygous embryos when compared with misexpression in a wild-type background, including an increased number of LT muscles (Figure S6B–C). We concluded that muscle changes are not confined to a specific genetic interaction between Sin3A and Kr, but are likely an effect of global changes to chromatin structure that could affect responsiveness of target genes to all identity genes.
To explore the genetic interactions between Sin3A, Kr and ara further, we examined embryos transheterozygous for Sin3A08269 and either the Kr mesodermal loss-of-function allele P[ry+ KrCD]bw Kr1, or the ara and caup deficiency Df IroDFM3 (Carrasco-Rando et al., 2011; Ruiz-Gómez et al., 1997). We failed to observe mesodermal defects in these transheterozygous embryos (Figure S6D–F). This result is consistent with a mechanism in which the activity of Sin3A in wild-type embryos opposes the activities of Kr and Ara.
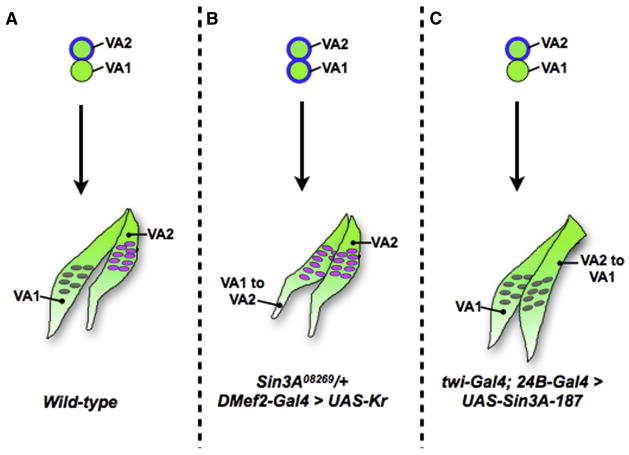
Kr is a positive regulator of slou in the ventral muscles (Ruiz-Gómez et al., 1997). Taken together, our work demonstrates a role for Sin3A in this process: titrating the ability of muscles to respond to the Kr signal (Figure 6). In wild-type embryos, the VA1 and VA2 FCs are derived from the same Kr-expressing muscle progenitor. Kr expression is turned off in VA1, but remains on to activate slou in VA2. Our data suggest thatSin3A maintains slou in a repressed state in VA1, potentially helping to mediate the transition of this FC from a Kr-positive to a Kr-negative state. Loss of Sin3A alleviates this repression, allowing slou to be ectopically activated by Kr in VA1 and transforming it to VA2. This interpretation explains the differences in phenotypes seen when different Gal4 drivers are used: while moderate Kr levels (such as those driven by DMef2-Gal4) are insufficient to drive an increase in slou expression if that locus has not been derepressed by reduction of Sin3A activity, embryos expressing higher Kr levels (such as those driven by twi-Gal4; 24B-Gal4) are able to activate expression of slou. Similarly, high levels of Sin3A overexpression, such as those driven by twi-Gal4; 24B-Gal4, are required to prevent activation of slou by Kr in VA2. It is possible these differences are also a result of the slightly earlier expression of twi-Gal4; 24B-Gal4. Consistent with a role for Sin3A as a factor that modulates muscle response to identity genes, we do not observe the VA2 to VA1 muscle transformation in 100% of muscles or hemisegments in embryos where Sin3A is overexpressed, suggesting that the transformation depends on the endogenous state of gene expression in individual embryos. We conclude that Sin3A activity could sensitize certain FCs and muscles to identity gene transcription factors, refining the combinatorial code and ensuring muscle diversity.
Figure 6. A Model for VA1 and VA2 Identity.
Diagram showing the development of the VA1 and VA2 muscle FCs (top, green circles) into muscles (green, bottom). Kr expression is depicted by a blue line around the FC. The fate of the ventral muscles is shown in (A) wild-type, (B) Sin3A08269/+; DMef2-Gal4 > UAS-Kr and (C) twi-Gal4; 24B-Gal4 > UAS-Sin3A-187.
DISCUSSION
Prior FAC-sorting and transcriptional profiling of Drosophila muscle cells has focused on identifying differences between FCs and FCMs and examining global effects of identity gene misexpression (Artero et al. 2001; Busser et al., 2012; Estrada et al., 2006). Two of these studies used genetic manipulation to enrich for FC and FCM populations (Artero et al., 2001; Estrada et al., 2006). Our unique sorting strategy purified very small populations of FC subsets from wild-type embryos for analysis. By this approach, we observed significant overlap in the transcriptional profiles of these two muscle subsets, defining a group of factors common to FCs and suggesting that a relatively small number of genes encode factors responsible for muscle diversity in each population, while the rest comprise a “molecular signature” for all FCs. We then screened homozygous mutant embryos for muscle defects. The strong combination of FACS, microarray and mutant analysis has so far identified 11 factors with novel roles in Drosophila embryonic muscle development, 10 of which have already been shown to have mammalian homologs. While this work has focused on a number of factors that were found to be enriched in both FC subsets, examination of FC subset-specific factors is ongoing and promises to yield additional genes regulating morphogenesis of muscle subsets.
Our screen identified a number of ubiquitously-expressed essential genes, which can be difficult to identify using forward genetic screening due to maternal loading and early functions during embryogenesis. Factors like Med13, Elo-B and Sin3A are required for the regulation of transcription in most cells and tissues. Our approach enabled the discovery of muscle-specific roles for these general transcriptional regulators, broadening our understanding of the genetic network that controls muscle development and setting the stage for future studies into the molecular mechanisms that regulate the establishment of muscle diversity.
Our array and screen identified novel roles for 11 transcription factors and chromatin modifiers, effectively doubling the list of transcriptional regulators known to control muscle development in the Drosophila embryo. The conservation of these factors opens up the possibility that roles for these factors will be found in vertebrate myogenesis, as well. The various mutant phenotypes we observed suggest that the newly identified genes may have defined functions in specific aspects of muscle development: FC specification or cell viability for muscle loss, muscle guidance and myotendinous junction formation for muscle misattachment and FC specification and cytoskeleton regulation for shape changes. Future work will focus on identification of specific regulatory targets to determine the distinct roles played by these new factors at particular steps in development.
Interestingly, although we detect expression of Sin3A in all FCs in the muscle pattern, we detect phenotypic changes in Sin3A08269 mutants primarily within the LTs. It is clear from analysis of the stronger Sin3Ae64 and Sin3AEP2387 alleles that Sin3A is required throughout the musculature for earlier myogenesis steps like myoblast fusion. We attribute the lack of defects in all 30 muscles in Sin3A08269 mutants to two causes: i) the weaker nature of the Sin3A08269 allele; and ii)the integration of Sin3A into the complicated genetic network of previously characterized identity genes. Our subsequent experiments in which misexpression of Kr in Sin3A08269 heterozygotes leads to VA muscle transformations supports the theory that Sin3A still plays a role in muscles even if we do not see perturbations to those muscles in Sin3A08269 homozygotes. Additionally, we found that overexpression of the Sin3A-187 isoform in the mesoderm led to muscle transformations, while overexpression of the Sin3A-220 isoform did not. This result is consistent with previous studies showing that Sin3A-187 is the dominant isoform in differentiated tissues and has greater histone deacetylase activity than Sin3A-220. The data suggest that Sin3A-187 isoform is more active in the muscle tissue at genes required for the specification of muscle fate.
How is muscle identity regulated by identity genes? The idea of a combinatorial code specifying muscle fate predicts that the DNA-binding transcription factors work cooperatively and/or antagonistically to generate a muscle-specific transcriptional profile. The incomplete overlap of identity gene expression generates some specificity, as does the timing of identity gene expression (Carrasco-Rando et al., 2011; Tixier et al., 2010). But how do identity genes work once they have bound to the DNA? What factors ensure that the correct FCs respond to signals from identity genes? Our results suggest Sin3A is a factor that titrates the responsiveness of muscles to identity genes. Genetic reduction of Sin3A levels leads VA1 to be susceptible to overexpression of the identity gene Kr, ectopically activating Slou expression in this muscle and resulting in VA1 to VA2 transformation. In contrast, overexpression of Sin3A prevents activation of Slou in VA2, and drives VA2 to VA1 transformation.
One hypothesis to explain our results is that the ability of Kr to activate Slou expression in a given muscle depends on the histone modification landscape over the slou promoter, which is in turn regulated by Sin3A histone deacetylase activity. This model suggests that one critical factor contributing to muscle competence is chromatin structure. Our model is built upon observations of specific single muscle transformations in the embryonic somatic musculature. The challenge going forward will be to design methods to monitor transcription factor binding, chromatin structure and gene expression changes at the molecular level in a relatively small percentage of cells in the embryo. The muscle transformation results we observe are similar to studies from the C. elegans nervous system, in which chromatin regulators have been shown to play important roles in the terminal differentiation of neurons (Doitsidou et al., 2013; Poole et al., 2011; Tursun et al., 2011; Weinberg et al., 2013). Our work extends these findings to the Drosophila musculature, suggesting a more global role for chromatin regulation in tissue diversity and differentiation. It is possible that the other chromatin modifiers found in our screen, Lid and Kdm2, will be found to play roles in this process as well.
Our results draw a distinction between Sin3A and the well-characterized identity genes. While Sin3A is required for the correct formation of muscles, its absence does not change FC specification. This lack of phenotype likely indicates that other positive regulators of transcription are required to drive fate changes in these muscles; alternatively, it could also be that a stronger depletion of Sin3A would cause muscle transformations, but that this phenotype is obscured by the earlier patterning defects we observe with stronger alleles. It is important to note that Sin3A08269 homozygous mutant embryos do not display VA1 to VA2 transformations, despite overall derepression of slou transcript. While loss of Sin3A disrupts the muscle pattern, it is only in concert with identity gene misexpression that we detect its role in muscle diversity, underscoring the importance for the identity genes in prescribing final muscle characteristics.
Additionally, though overexpression of Sin3A in the mesoderm can lead to VA2 to VA1 transformations, this change does not happen in every hemisegment. Sin3A is just one factor contributing to muscle identity in the context of each muscle’s specific network of gene expression. Our experiments reveal the strong commitment to a specific cell fate each muscle makes during development. The somatic musculature is particularly resistant to perturbations; we show that overexpression of Kr can only cause muscle transformations when a strong Gal4 driver is used. In this way, formation of muscle identity is analogous to a buffered solution. In this system, reduction of Sin3A is like removing the buffer, allowing more frequent muscle identity changes. Sin3A, then, positively contributes to the establishment of muscle diversity by fortifying the muscles against shifts in gene expression.
Regulators of chromatin structure have been increasingly shown to play important roles in muscle development and disease (Fong et al., 2012; Puri and Sartorelli, 2010; Sartorelli and Juan, 2011). Sin3A, in particular, has been shown to regulate the expression of several genes that have been implicated in muscular dystrophies (van Oevelen et al., 2010). While chromatin regulators have been implicated in muscle cell differentiation, Sin3A is the first to be shown to have a role in the formation of muscle identity. A key gap in our knowledge about the specification of muscle identity has been that misexpression of identity genes results in relatively few direct muscle transformations. Our results add a layer of complexity to the combinatorial code model for the generation of muscle diversity, suggesting that the epigenetic landscape of a particular muscle sets the stage for identity gene response, either by modulating transcription factor binding or fine tuning the activity of factors once bound to mesodermal enhancers. This knowledge will be critical to synthesize muscles of specific sizes and shapes for use in stem cell transfer therapies to treat muscle disease.
EXPERIMENTAL PROCEDURES
Fly Stocks
Drosophila stocks and crosses were grown on standard cornmeal medium at 25°C. The GAL4-UAS system (Brand and Perrimon, 1993) was used for expression studies. OreR was used as a control strain. Embryos were staged according to Campos-Ortega and Hartenstein (1985). Approximate ages are: stage 12 (7:20–9:20 h AEL), stage 13 (9:20–10:20 h AEL) and stage 16 (13:00–16:00 h AEL). For a list of fly stocks used in this study, please see Supplemental Experimental Procedures.
FACS, RNA Isolation and Microarray Analysis
Flies expressing GFP or RFP transgenes were maintained at 25° C on a 12-hour light/12-hour dark cycle. After a 2-hour pre-lay, flies were allowed to lay on plates for 2 hours; these plates were removed and aged for an additional 8 hours. Embryos were dissociated into a single cell suspension (Dobi et al., 2011). Cells were sorted using standard methods on a Beckton-Dickson FACS Aria Cell Sorter (Estrada and Michelson, 2008). Total cellular RNA from each sample was prepared from sorted cells using TRIzol LS Reagent without amplification (Invitrogen). Total cellular RNA was labeled and hybridized to Drosophila Affymetrix GeneChip 2.0 arrays according to the manufacturers instructions. Three independent sorts were made for each transgene, and each RNA sample was independently labeled and hybridized.
For apME-GFP, we sorted 1,000,000 GFP-positive cells from 4 × 108 starting cells, a positive-rate of 0.03%, which yielded 1 μg of RNA. The purity upon resort of the cells was 74%. The purity of S59ME-RFP and twiprom-actin-GFP was 87% and 95%, respectively, with positive-rates and yields also correspondingly higher.
Affymetrix microarray data was normalized and compared using Partek Genomics Suite Software. apterousME-GFP and slouchME-RFP-positive cells were compared to twiprom-actin-GFP-expressing cells to generate lists of differentially regulated genes, fold change ≥ 1.7, p-value < 0.05. The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 was used to assign Gene Ontology classifications to the genes identified by the array (Huang et al., 2009a; 2009b). Default parameters were used to perform Functional Annotation of gene lists, p-value < 0.05.
Accession Numbers
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE58738 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58738).
In situ hybridization
Probes for in situ hybridization were made using clones from the DGC collection (Stapleton et al., 2002). Hybridizations were performed as in Leatherbarrow and Halfon (2009). For additional detail, please see Supplemental Experimental Procedures.
Immunohistochemistry
Embryos were fixed in 4% paraformaldehyde/heptane for all immunohistochemistry. Primary antibodies were detected using Alexa-conjugated secondary antibodies (1:400; Invitrogen). Embryos were mounted in Vectashield (Vector Laboratories, CA) or ProLong Gold antifade reagent (Invitrogen). Double or triple staining using anti-β-galactosidase or anti-GFP was used to identify the presence of marked balancer chromosomes. For antibody sources and concentrations, please see Supplemental Experimental Procedures.
Fluorescent images were acquired on a Leica SP5 laser-scanning confocal microscope equipped with a 631.4 NA HCX PL Apochromat oil objective and LAS AF 2.2 software. Images were processed using Adobe Photoshop CS4. Maximum intensity projections of confocal z-stacks were rendered using Volocity Visualization software (Improvision).
Production of cDNA and Quantitative PCR
Wild-type (OreR) and Sin3A08269 homozygous embryos were collected, dechorionated and staged by embryo morphology. 10–20 St. 13 or St. 16 embryos/sample were lysed and transferred to TriReagent (Sigma). Total mRNA was prepared according to manufacturer’s instructions, and resuspended in RNAse-free water. 1 microgram of total mRNA was used to prepare first-strand cDNA using the Superscript III Reverse Strand Synthesis System (Invitrogen). For quantitative PCR, first-strand cDNA product was 1:5 in Blue qPCR SYBR Low Rox Master Mix (ThermoScientific). qPCR was conducted using an Applied Biosystems 7500 Real Time PCR System. qPCR results were analyzed and relative ratios of Slou transcripts (normalized to rp49 levels) in OreR and Sin3A08269 mutants were determined using described methods (Pfaffl, 2001). For primer sequences, please see Supplemental Experimental Procedures.
ChIP
We performed ChIP on st. 13 wild-type (OreR) embryos (Nien et al., 2011). ChIP lysates were immunoprecipitated with pre-immune serum or anti-Sin3A (Spain et al., 2010). Gene specific qPCR of purified DNA was performed as described above. Significance of binding enrichment was determined using a student’s t-test. For a list of primers, please see Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We would like to thank F. Schnorrer, S. Abmayr, J. Reinitz, C. Rushlow, L. Pile, J. Tresiman, M. Lehmann, E. Lai, S. Cohen, M. Ruiz-Gomez, A. Nose, T. Volk, the Developmental Studies Hybridoma Bank and the Bloomington Stock center for fly stocks and reagents; the MSKCC Flow Cytometry Core for cell sorting; the MSKCC Genomics Core for microarray hybridization and analysis; H. Huddleston, O. Stovicek, M. Cruz and J. Leatherbarrow for technical assistance; and the members of the Baylies Laboratory for help and discussion. This work was supported by a NIAMS National Research Service Award (1F32AR057290) to K.C.D., American Cancer Society award RSG-09-097-01-DDC to M.S.H., NIGMS R01-056989 to M. K. B. and NCI P30 CA008748 to MSKCC.
Footnotes
AUTHOR CONTRIBUTIONS
K.C.D. performed experiments and data analysis. M.S.H. performed in situ hybridization analysis. K.C.D. and M.K.B. conceived the project, and K.C.D., M.K.B. and M.S.H. wrote and edited the paper.
References
- Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. 2001 doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development. 2003;130:6257–6272. doi: 10.1242/dev.00843. [DOI] [PubMed] [Google Scholar]
- Aso T, Conrad MN. Molecular cloning of DNAs encoding the regulatory subunits of elongin from Saccharomyces cerevisiae and Drosophila melanogaster. Biochem Biophys Res Commun. 1997;241:334–340. doi: 10.1006/bbrc.1997.7819. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Michelson AM. Invertebrate myogenesis: looking back to the future of muscle development. Curr Opin Genet Dev. 2001;11:431–439. doi: 10.1016/s0959-437x(00)00214-8. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M, Ruiz-Gómez M. Myogenesis: a view from Drosophila. Cell. 1998;93:921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int Rev Neurobiol. 2006;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [DOI] [PubMed] [Google Scholar]
- Boube M, Faucher C, Joulia L, Cribbs DL, Bourbon HM. Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes & Development. 2000;14:2906–2917. doi: 10.1101/gad.17900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgouin C, Lundgren SE, Thomas JB. Apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9:549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Busser BW, Shokri L, Jaeger SA, Gisselbrecht SS, Singhania A, Berger MF, Zhou B, Bulyk ML, Michelson AM. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Development. 2012;139:1164–1174. doi: 10.1242/dev.077362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardamone M, Darras BT, Ryan MM. Inherited myopathies and muscular dystrophies. Semin Neurol. 2008;28:250–259. doi: 10.1055/s-2008-1062269. [DOI] [PubMed] [Google Scholar]
- Carrasco-Rando M, Tutor AS, Prieto-Sánchez S, González-Pérez E, Barrios N, Letizia A, Martín P, Campuzano S, Ruiz-Gómez M. Drosophila araucan and caupolican integrate intrinsic and signalling inputs for the acquisition by muscle progenitors of the lateral transverse fate. PLoS Genet. 2011;7:e1002186. doi: 10.1371/journal.pgen.1002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Curr Opin Genet Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobi KC, Metzger T, Baylies MK. Characterization of early steps in muscle morphogenesis in a Drosophila primary culture system. Fly (Austin) 2011;5:68–75. doi: 10.4161/fly.5.2.15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann C, Azpiazu N, Frasch M. A new Drosophila homeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes & Development. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- Doitsidou M, Flames N, Topalidou I, Abe N, Felton T, Remesal L, Popovitchenko T, Mann R, Chalfie M, Hobert O. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes & Development. 2013;27:1391–1405. doi: 10.1101/gad.217224.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Zhang C, Chen J, Sink H, Frei E, Noll M. A key role of Pox meso in somatic myogenesis of Drosophila. Development. 2007;134:3985–3997. doi: 10.1242/dev.008821. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkner A, Roure A, Charroux B, Delaage M, Holway N, Coré N, Vola C, Angelats C, Pagès F, Fasano L, et al. Grunge, related to human Atrophin-like proteins, has multiple functions in Drosophila development. Development. 2002;129:1119–1129. doi: 10.1242/dev.129.5.1119. [DOI] [PubMed] [Google Scholar]
- Escudero LM. Charlatan, a Zn-finger transcription factor, establishes a novel level of regulation of the proneural achaete/scute genes of Drosophila. Development. 2005;132:1211–1222. doi: 10.1242/dev.01691. [DOI] [PubMed] [Google Scholar]
- Estrada B, Michelson AM. A genomic approach to myoblast fusion in Drosophila. Methods Mol Biol. 2008;475:299–314. doi: 10.1007/978-1-59745-250-2_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Choe SE, Gisselbrecht SS, Michaud S, Raj L, Busser BW, Halfon MS, Church GM, Michelson AM. An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2006;2:e16. doi: 10.1371/journal.pgen.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. Genetic and Epigenetic Determinants of Neurogenesis and Myogenesis. Developmental Cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim Biophys Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Korge G, Lehmann M. The helix-loop-helix proteins dAP-4 and daughterless bind both in vitro and in vivo to SEBP3 sites required for transcriptional activation of the Drosophila gene Sgs-4. J Mol Biol. 1999;291:71–82. doi: 10.1006/jmbi.1999.2963. [DOI] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JAA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes & Development. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Gerlach N, Jäckle H. The Drosophila homolog of the human AF10 is an HP1-interacting suppressor of position effect variegation. EMBO Rep. 2001;2:211–216. doi: 10.1093/embo-reports/kve039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genetics. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA. D-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci USa. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennetta G, Pauli D. The Drosophila Sin3 gene encodes a widely distributed transcription factor essential for embryonic viability. Dev Genes Evol. 1998;208:531–536. doi: 10.1007/s004270050212. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole RJ, Bashllari E, Cochella L, Flowers EB, Hobert O. A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002109. doi: 10.1371/journal.pgen.1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V. HDACs and sirtuins: targets for new pharmacological interventions in human diseases. Pharmacol Res. 2010;62:1–2. doi: 10.1016/j.phrs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: when it takes more to make one. Dev Biol. 2010;341:66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gómez M, Romani S, Hartmann C, Jäckle H, Bate M. Specific muscle identities are regulated by Krüppel during Drosophila embryogenesis. Development. 1997;124:3407–3414. doi: 10.1242/dev.124.17.3407. [DOI] [PubMed] [Google Scholar]
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Juan AH. Sculpting chromatin beyond the double helix: epigenetic control of skeletal myogenesis. Curr Top Dev Biol. 2011;96:57–83. doi: 10.1016/B978-0-12-385940-2.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F, Kalchhauser I, Dickson BJ. The Transmembrane Protein Kon-tiki Couples to Dgrip to Mediate Myotube Targeting in Drosophila. Developmental Cell. 2007;12:751–766. doi: 10.1016/j.devcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Schnorrer F, Schönbauer C, Langer CCH, Dietzl G, Novatchkova M, Schernhuber K, Fellner M, Azaryan A, Radolf M, Stark A, et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- Seeger M, Tear G, Ferres-Marco D, Goodman CS. Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron. 1993;10:409–426. doi: 10.1016/0896-6273(93)90330-t. [DOI] [PubMed] [Google Scholar]
- Sharma V, Swaminathan A, Bao R, Pile LA. Drosophila SIN3 is required at multiple stages of development. Dev Dyn. 2008;237:3040–3050. doi: 10.1002/dvdy.21706. [DOI] [PubMed] [Google Scholar]
- Sheeba CJ, Palmeirim I, Andrade RP. Chick Hairy1 protein interacts with Sap18, a component of the Sin3/HDAC transcriptional repressor complex. BMC Dev Biol. 2007;7:83. doi: 10.1186/1471-213X-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Garry DJ. Sin3 interacts with Foxk1 and regulates myogenic progenitors. Mol Cell Biochem. 2012;366:251–258. doi: 10.1007/s11010-012-1302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, Yu C, Carlson J, George R, Celniker S, Rubin GM. The Drosophila Gene Collection: Identification of Putative cDNAs for 70% of D. Melanogaster Genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain MM, Caruso JA, Swaminathan A, Pile LA. Drosophila SIN3 isoforms interact with distinct proteins and have unique biological functions. J Biol Chem. 2010;285:27457–27467. doi: 10.1074/jbc.M110.130245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MV, Beatty KE, Hunter HK, Baylies MK. Drosophila MEF2 is regulated by twist and is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech Dev. 1995;51:139–41. doi: 10.1016/0925-4773(94)00323-f. [DOI] [PubMed] [Google Scholar]
- Tixier V, Bataillé L, Jagla K. Diversification of muscle types: recent insights from Drosophila. Exp Cell Res. 2010;316:3019–3027. doi: 10.1016/j.yexcr.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:research 0088.1. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C, Bowman C, Pellegrino J, Asp P, Cheng J, Parisi F, Micsinai M, Kluger Y, Chu A, Blais A, et al. The mammalian Sin3 proteins are required for muscle development and sarcomere specification. Mol Cell Biol. 2010;30:5686–5697. doi: 10.1128/MCB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C, Wang J, Asp P, Yan Q, Kaelin WG, Kluger Y, Dynlacht BD. A role for mammalian Sin3 in permanent gene silencing. Molecular Cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T. Singling out Drosophila tendon cells: a dialogue between two distinct cell types. Trends Genet. 1999;15:448–453. doi: 10.1016/s0168-9525(99)01862-4. [DOI] [PubMed] [Google Scholar]
- Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, Florens L, Washburn MP, Abmayr SM, Workman JL. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes & Development. 2011;25:1499–1509. doi: 10.1101/gad.2046211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg P, Flames N, Sawa H, Garriga G, Hobert O. The SWI/SNF Chromatin Remodeling Complex Selectively Affects Multiple Aspects of Serotonergic Neuron Differentiation. Genetics. 2013;194:189–198. doi: 10.1534/genetics.112.148742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.