Abstract
Most biological rhythms are generated by a population of cellular oscillators coupled through intercellular signaling. Recent experimental evidence shows that the collective period may differ significantly from the autonomous period in the presence of intercellular delays. The phenomenon has been investigated using delay-coupled phase oscillators, but the proposed phase model contains no direct biological mechanism, which may weaken the model's reliability in unraveling biophysical principles. Based on a published gene regulatory oscillator model, we analyze the collective period of delay-coupled biological oscillators using the multivariable harmonic balance technique. We prove that, in contradiction to the common intuition that the collective period increases linearly with the coupling delay, the collective period turns out to be a periodic function of the intercellular delay. More surprisingly, the collective period may even decrease with the intercellular delay when the delay resides in certain regions. The collective period is given in a closed-form in terms of biochemical reaction constants and thus provides biological insights as well as guidance in synthetic-biological-oscillator design. Simulation results are given based on a segmentation clock model to confirm the theoretical predictions.
I. Introduction
Rhythms are fundamental to biological activities. With periods ranging from seconds in glycolytic oscillations to years in reproduction, these rhythms are among the most conspicuous properties of living systems [1], [2]. Underlying biological rhythms are networks of interacting cellular oscillators. These cellular oscillators can synchronize rhythms with a certain collective period, yet it remains an exciting challenge to understand the mechanism of how collective oscillation period arises from autonomous cellular oscillations.
Although it has been generally believed that the collective period is determined by the average of cell-autonomous periods, it is recently reported that in the presence of intercellular delays the collective period will be greatly altered from the averaged autonomous periods. This has been experimentally substantiated for coupled semiconductor lasers [3] and in the zebrafish segmentation clock [4]. This phenomenon is of fundamental importance in the study of biological rhythms, given the prevalence of time delays in biological interactions. One source of delay is a threshold effect, i.e., the concentration of an effector must exceed or fall below a certain value before the affected value is altered [5]. Both nonlinear chemical reaction kinetics and hysteresis can act like thresholds, even when the threshold values are not sharp. Secondly, there are known delays in transcription and translation associated with mRNA and protein processing in the nucleus and cytoplasm, respectively [6]. Finally, there are delays in the transport of intermediates between cellular compartments [6].
Based on phase oscillators, the authors in [3], [4], [7], [8] studied the collective period of delay-coupled oscillators. However, the phase oscillator model may be too phenomenological to reveal underlying principles, because it contains no direct biological mechanism for the cellular clock. In this paper, we analyze the influence of intercellular delay on the collective period using a gene regulatory biophysical clock model. To our knowledge, no analytical results have been reported on the collective period of delay-coupled mechanism-based gene regulatory oscillators to date. In our study, we use an oscillator model in which oscillations are induced by the direct autorepression of a gene by its own protein product. Biological rhythms as diverse as cell-cycles in bacterial [9], segmentation clocks in vertebrates [10], and circadian rhythms in mammals [11] depend heavily on autorepression. Following the same principles, we model the intercellular interaction as co-repression based on the fact that mutual repression between a pair of oscillators comprises a positive feedback loop between the oscillators, which has been reported to synchronize various biological oscillators [12]. This model is also inspired by the fact that intercellular repression is widespread in sensory pyramidal neurons [13], visual thalamus [14], and insulin secretion [15], to name a few examples.
We use the multivariable harmonic balance (MHB) technique to analyze the collective period of delay-coupled gene regulatory oscillator networks. The MHB technique, although approximate, has been shown to be reliable for the analysis of biochemical oscillating systems [16], [17], and can provide an effective way to characterize the frequency, amplitude, and phase of coupled oscillators [16], [18], [19], [20]. Due to multi-cellular structure, the solution to harmonic balance equations is very difficult to obtain. Here we circumvent the problem by restricting our attention to solutions corresponding to synchronized oscillations since we are interested in the collective period. The main contributions are as follows: 1) the collective period of delay-coupled gene regulatory oscillators is derived in terms of biochemical parameters, which gives insights into the basic mechanism of biological oscillations; 2) it is proven that the collective period is a periodic function of the intercellular delay in contrast to the linear function assumed in the existing literature (e.g., [10]); and 3) the region in which the collective period is larger/smaller than the autonomous period (oscillation period of isolated oscillators) and, the region in which the collective period increases/decreases with the intercellular delay are given explicitly.
II. Model description
We adopt the well-established Hill-type auto-repression model of cellular oscillators [10], [21]:
| (1) |
where m(t) ≥ 0 is the instantaneous concentration of a mRNA which codes for a protein with instantaneous concentration p(t) ≥ 0. The protein acts as a repressor. The constants a > 0 and b, c > 0 denote production and decay rates, respectively. Tm > 0 denotes the time delay between the initiation of transcription and the arrival of the mature mRNA molecule in the cytoplasm. Tp > 0 denotes the time delay between the initiation of translation and the emergence of a complete functional protein molecule. f(p) is the rate of production of new mRNA molecules, and the constants k and p0 represent the action of an inhibitory protein that acts as a dimer. ν is the Hill coefficient, which describes the cooperativity of end product repression.
We use co-repression to establish the intercellular signaling [12]. This is based on the facts that 1) mutual repression between a pair of biological oscillators constitutes a positive feedback loop coupling between the oscillators, which is regarded as the most prevalent induction scheme of synchronization in homogeneous biological oscillators [12]; and 2) intercellular repression is widespread in gene regulatory networks [22]. The ith oscillator's dynamics is described by:
| (2) |
where and denotes index set {j = 1, 2, . . . , N, j ≠ i} and ; m0 is a constant; τ is the time delay in intercellular interaction. Since is a decreasing function of mj(j ≠ i), the intercellular coupling between oscillator i and oscillator j is repressive.
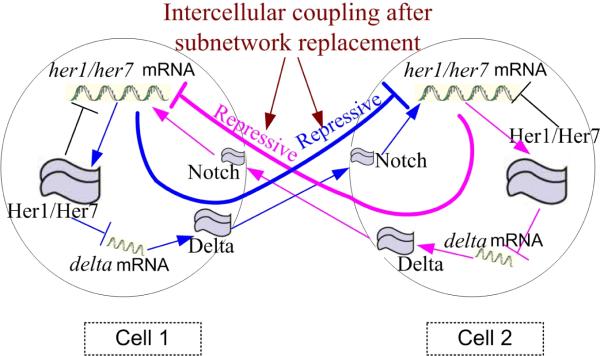
Remark 1: It is worth noting that the intercellular signaling can be a complex cascade composed of many intermediate molecules. For example, in the zebrafish segmentation clock, the intercellular signaling is a cascade [10]: her1/her7 mRNA in cell 1 → Her1/Her7 in cell 1 ⊣ delta mRNA in cell 1 → Delta in cell 1 → Notch in cell 2 → her1/her7 mRNA in cell 2, where “→” denotes activation and “⊣” denotes repression (cf. Fig. 1). In this case, by using the subnetwork replacement technique [23], the interaction cascade can be replaced with “her1/her7 mRNA in cell 1 ⊣ her1/her7 mRNA in cell 2” without changing the qualitative characteristics of intercellular interaction [23]. Given this, (2) can be used to model many biochemical oscillator networks such as the insulin secreting pancreatic islets [12], ovulation regulation networks [12], and neural stem cell maintenance networks [24], to name a few.
Fig. 1.
Modeling the intercellular coupling in the segmentation clock [10]. Utilizing the subnetwork replacement technique [23], the cascade (the thin blue/pink arrows/bars) can be replaced with the repressive interaction (the thick blue/pink bar ⊣.
III. The collective period
To facilitate analysis, we recast (2) in a matrix form:
| (3) |
where M(t) = [m1(t) m2(t) . . . mN(t) ]T, P(t) = [p1(t) p2(t) . . . pN(t) ]T, and .
The most biologically significant property of (1) is that it can have oscillating solutions, which is the focus of this paper. (Note that the HMB technique does not give a necessary and sufficient condition for such solutions [19], [20].) It is noteworthy that although the period of a single cellular oscillator has been studied [16], [17], no analytical results exist addressing the collective period of repressively coupled gene regulatory oscillators. Building upon our recent study on delay-free coupled Goodwin oscillators [25], we propose to study delay-coupled oscillators using the MHB technique [18], [26]. Since multi-cellular structure leads to high-dimensional harmonic balance equations, it is very difficult to derive the solution. Here we are interested in the collective period, so we restrict our attention to solutions corresponding to synchronized oscillations, which gives a way to solve the problem. The definition of synchronized oscillation is provided below. We assume that the parameters are chosen such that synchronized solution exists.
Definition 1: (2) is synchronized if and for all 1 ≤ i, j ≤ N.
Theorem 1: Using harmonic balance technique, mi(t) and pi(t) can be approximated by (14) and (15) respectively, and the collective period of (2) (when synchronized) can be obtained as , where w is the minimum positive solution to
| (4) |
and
| (5) |
In (5), κ denotes the ratio between the Fourier coefficients of with respect to pi and , respectively (Due to the structure of , κ is independent of i and j). The value of κ (which is not needed in the analysis in this paper) can be calculated numerically [27].
The proof is given in the appendix.
Remark 2: Although the collective period of coupled oscillators has been studied based on the simple phase model in [3], [4], [7], [8], [28], to our knowledge, no analytical result exists addressing the collective period of delay-coupled gene regulatory oscillators.
From the proof of Theorem 1, one gets that the influence of intercellular coupling is represented by ξ. So by setting ξ = 0, one gets the autonomous period of a single gene regulatory oscillator, which is the same as the existing result in [17]:
Corollary 1: The autonomous period of gene regulatory oscillator (1) can be obtained as
| (6) |
where wA is the minimum positive solution to
| (7) |
Proof: The corollary can be easily obtained by setting ξ in (4) to 0.
IV. The influence of intercellular delay on the collective period
Although the system of equations in (4) and (5) cannot be solved analytically, it can be used to analyze the influence of intercellular delay τ on the collective period T:
Theorem 2: Define , then when the network in (2) is synchronized, the relationship between collective period T and intercellular delay τ is as follows:
- when τ satisfies
T is identical to the autonomous period TA in (6);(8) - when τ satisfies
for some k = 2n–1, (n = 0, 1, 2, . . .), then T is smaller than the autonomous period TA;(9) - when τ satisfies
for some k = 2n, (n = 0, 1, 2, . . .), then T is larger than the autonomous period TA;(10)
Moreover,
- when τ resides in the following region,
for some k = 0, 1, 2, . . ., then T increases with τ;(11) - when τ resides in the following region,
for some k = 0, 1, 2, . . ., then T decreases with τ;(12)
The proof is given in the appendix.
Remark 3: Theorem 2 is consistent with the simulation results in [29], which show that depending on its magnitude, intercellular delay can both increase and decrease the collective oscillation period of stellate cell networks. It is also reminiscent of the simulation results in [30], which show that the intercellular delay periodically regulates the collective period of inter-coupled Hodgkin-Huxley neurons. From a mathematical point of view, periodic regulation is also reasonable since any intercellular delay τ has the same effect as τ + nT (n = 1, 2, . . .) due to the periodic oscillation (repetition) of mi(t). Therefore, periodic regulation of the collective period by intercellular delay may be a general rule of delay coupled biochemical oscillator networks, and the results obtained in this paper can provide insights into biological oscillator network behavior understanding as well as guidance in synthetic biological oscillator network design.
V. Application to a segmentation clock model
We apply the obtained results to the segmentation clock model proposed in [10], [31]. The segmentation clock is a population of coupled cellular gene regulatory oscillators in the embryo that drive the sequential subdivision of the presomitic mesoderm into multicellular blocks termed somites. In [10], Lewis formulated each cellular oscillator of the zebrafish segmentation clock as a feedback loop in which Her1 or Her7 protein directly binds to the regulatory DNA of its own gene to inhibit transcription. Let mi(t) and pi(t) be the concentrations of her1/her7 mRNA and the corresponding protein in the ith (i = 1, 2, . . . , N) cell at time t, then the dynamics of pi(t) and mi(t) in an isolated cell are governed by (1) with parameters given as follows [10]: b = c = 0.231, a = 4.5 protein molecules per mRNA molecule per minute, k = 33 molecules per diploid cell per minute, ν = 2, and p0 = 40 molecules. Lewis modeled the intercellular coupling pathway from cell i to cell j as her1/her7 mRNA in cell i → Her1/Her7 in cell i ⊣ delta mRNA in cell i → Delta in cell i → Notch in cell j → her1/her7 mRNA in cell j. According to Remark 1, the intercellular signaling reduces to her1/her7 mRNA in cell i ⊣ her1/her7 in cell j. So the interaction between any two oscillators is mutual repressive and the dynamics of the oscillator network can be formulated by (2).
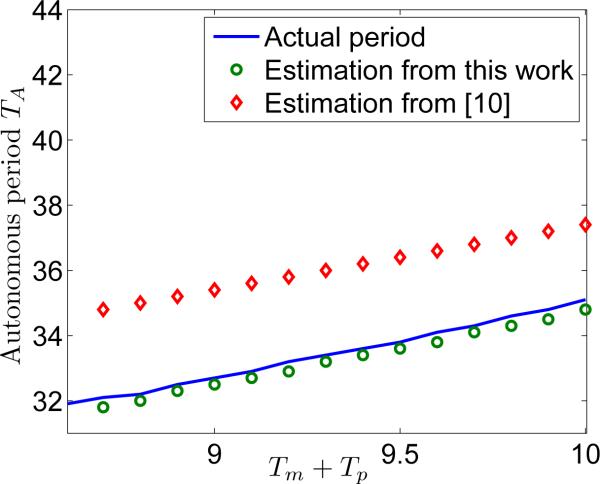
Reference [10] gives an analytic approximation of the autonomous oscillation period, i.e.,
| (13) |
thus, we can compare it with our results in (7). The comparison under different values of Tm + Tp is given in Fig. 2, where it can be seen that our result is more accurate.
Fig. 2.
Comparison of estimated autonomous period with [10]. The actual period is calculated from direct numerical simulation of the model in [10].
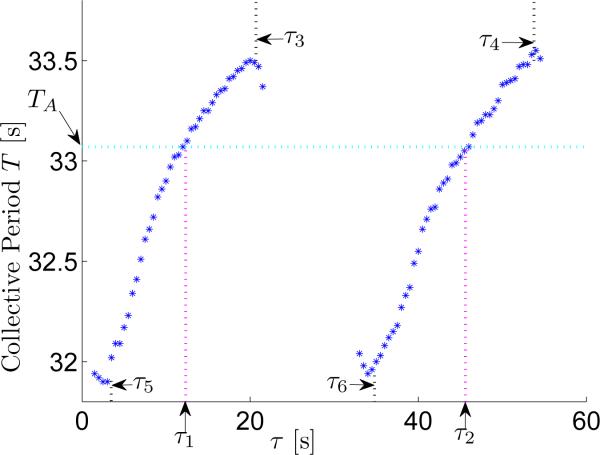
Next we show that our analytical prediction is consistent with numerical simulation of the network of gene regulatory oscillators. We set N = 9 and m0 identical to p0, i.e., 40 molecules. We fixed Tm and Tp to 7.1s and 1.7s, respectively, which gives an autonomous oscillation period TA = 33.07s. The results of simulating the network under different intercellular delays, τ, are given in Fig. 3. The collective period is represented by blue asterisks (when 0 < τ < 1.5 and 21.5 < τ < 33, the network did not synchronize, so there is no collective period under these delays). We can see that at and where , the collective period is identical to the autonomous period. This verifies Theorem 2, which predicts that if the network can be synchronized and the intercellular delay is expressed by for k = 0, 1, 2, . . ., then the collective period will be identical to TA. From the figure, we can also see that when or , the collective period is smaller than TA, and otherwise it is larger than the autonomous period TA, which is also consistent with the statement in Theorem 2. Furthermore, the results in Fig. 3 also roughly follow the monotonicity property prediction in Theorem 2, which states that when and , the collective period decreases with τ, otherwise the collective period increases with τ (Note that since the prediction is based on the assumption that all oscillators are synchronized, there is a small prediction error when delay τ gets close to the non-synchronizable regions).
Fig. 3.
Verification using a segmentation clock model described in [10].
VI. Discussion
The collective period of coupled biological oscillators are attracting increased attention as more experimental evidence shows that it can differ significantly from the autonomous period. Existing analytical results are based on sinusoidally delay-coupled phase oscillators, which may be too phenomenological to reveal underlying mechanisms. In fact, from the numerical results in Fig. 3, it can be seen that the variation of the collective period with the intercellular delay is quite different from a sinusoidal curve, which further substantiates the necessity of non-phase model based studies. However, to our knowledge, there are no existing analytical results addressing the collective period of delay-coupled oscillators based on biophysical regulatory models. Using a gene regulator oscillator model of the cellular oscillation in the segmentation clock, we study the collective period of delay-coupled cellular oscillators using the multivariable harmonic balance technique. Due to the multiple oscillator structure, the harmonic balance equations are very difficult to solve. We circumvent the problem by ignoring solutions corresponding to unsynchronized oscillations, which are not relevant to our study, but will greatly facilitate the analysis.
When there is an intercellular delay, we prove that the collective period is a periodic function of the intercellular delay. This argues against the existing assumption that the collective period increases linearly with the intercellular delay. More surprisingly, we prove that the collective period may even decrease with the intercellular delay when the delay resides in certain regions. The results are confirmed by numerical simulations on a published gene regulatory oscillator network model of the segmentation clock. The results are given in a closed-form expression in terms of the biochemical reaction constants and thus provide biological insight as well as guidance in synthetic-biological-oscillator design.
Acknowledgments
The work was supported in part by U.S. ARO through Grant W911NF-07-1-0279, NIH through Grant GM096873, the Institute for Collaborative Biotechnologies through grants W911NF-09-0001 and W911NF-09-D-0001-0027 from the U.S. ARO, JSPS through grant No. 23-9203, and Grant-in-Aid for Scientific Research (A) through grant No. 21246067.
Appendix: Harmonic balance investigation
The dynamics of the coupled gene regulatory oscillators in (3) can be transformed into the frequency domain as shown in Fig. 4.
Fig. 4.
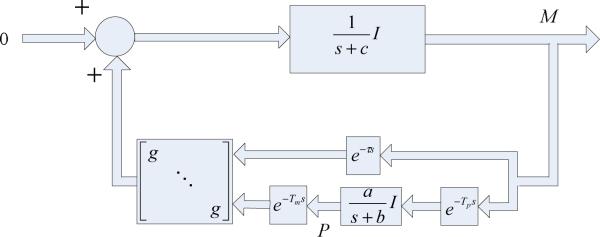
Schematic diagram of the frequency domain formulation of (3). P(s) and M(s) are the respective element-wise Laplace transform of P(t) and M(t) in (3).
According to the harmonic balance technique [26], since and in Fig. 4 are low pass filters, the higher order harmonics of oscillations in the closed-loop system in Fig. 4 can be safely neglected. Thus the wave forms of mi(t) and pi(t) can be approximated by their zero-order and first-order harmonic components [26]:
| (14) |
| (15) |
where αi, xi and βi, yi denote the amplitudes of the zero-order and first-order harmonics, respectively, and w and ϕi, φi denote the oscillation frequency and the phases, respectively.
Again, because the higher order harmonics of oscillations in the closed-loop system can be safely neglected, g(•) in (2) can be approximated legitimately by its describing functions based on multivariable Fourier analysis [27]:
| (16) |
where denotes the index set {j = 1, 2, . . . , N, j ≠ i} and
| (17) |
| (18) |
| (19) |
The describing function ηi and ξi,j are the gains of g(•) when the inputs are two sinusoids of amplitudes βi and yj, respectively, and the output is approximated by the first-order harmonic [26], [27]. Combining (2) and (16) one gets that ξi,j denotes oscillator j's influence on oscillator i, namely, the influence of intercellular coupling.
Based on the harmonic balance technique [26], pi(t) (i = 1, 2, . . . , N) in Fig. 4 must satisfy:
| (20) |
where
| (21) |
In its present form, analytical treatment of equation (20) is very difficult. However, considering that we are interested in the collective period, we can restrict our attention to solutions that describe synchronized oscillations of the oscillator network. This provides a clue to simplify the dynamics: synchrony indicates that the phases of each oscillator ϕi and φi (i = 1, 2, . . . , N) are identical, respectively.
A. Proof of Theorem 1
Given that the cellular oscillators are homogeneous and the coupling is symmetric, the amplitudes of all oscillators βi and yi (i = 1, 2, . . . , N) are respectively identical, too. Since ξi,j and ηi are determined by βi and yi, we further have ξ1,2 = . . . = ξ1,N = . . . = ξN–1,N = ξ and η1 = η2 = . . . = ηN = η. Using these properties, we can reduce (20) to:
| (22) |
where
| (23) |
It can be verified that H(jw) has two eigenvalues:
| (24) |
of multiplicity 1 and
| (25) |
of multiplicity N – 1, and only δ1 corresponds to eigenvectors with identical elements.
As analyzed above, when the network is synchronized, P and M are vectors of identical elements. Thus we have
| (26) |
According to (18), η is real, thus the item on the right hand side of (26) must be real, i.e., its imaginary part is zero.
Since can be rewritten as
it follows that the imaginary part of the right hand side of (26) is zero given that (4) is satisfied. Denote the ratio between the Fourier coefficients of with respect to pi and mj respectively as κ (Note that due to the structure of , κ is independent of i and j, and its exact value—which is not needed in the analysis in this paper—can be calculated numerically [27]), (5) is obtained from the equality of both sides of (26). Hence the theorem is proven.
B. Proof of Theorem 2
When the oscillator network is synchronized, the collective period is determined by (4), which can be rewritten as
| (27) |
From trigonometric addition formulas, (27) is equivalent to
| (28) |
Eqn. (28) can be further transformed into
| (29) |
Setting
| (30) |
and using the linear combination rule of sine and cosine, we can recast (29) into
| (31) |
where .
It can be easily verified that g(wA) = 0. So we have
| (32) |
Therefore, when (8) holds, the collective period T is the same as the autonomous period TA.
Since w is the minimal positive value satisfying (7) and (4) from the harmonic balance technique [26], one can verify that where . This is because when the respective two sides of (7) and (4) have different signs—excluding the possibility of solutions, whereas when , the respective two sides of (7) and (4) can be verified to have an intersection via trend analysis. (Note that: ① ξ < 0 holds since g(•) decreases with mi(t) and hence has a negative describing function [16], [27]; ② holds due to the fact that the half lives of molecules, and , need to be much smaller than Tm + Tp to guarantee sustained oscillations [32], [33].) Therefore, we have tan w(Tm + Tp) < 0 and bc–w2 > 0. Hence g(w) in (30) satisfies g(w) > 0 if and only if w > wA holds, and g(w) satisfies g(w) < 0 if and only if w < wA holds. Therefore, from (31), we have w > wA when sin(w(Tm+Tp–τ)+ϕ) > 0 is satisfied, and we have w < wA when sin(w(Tm + Tp – τ) + ϕ) < 0 is satisfied (notice that cos w(Tm+Tp) < 0 for and ξ < 0). Using the properties of sinusoidal functions, we have w > wA when τ satisfies (9) for k = 2n – 1 (n = 0, 1, 2, . . .), and w < wA when τ satisfies (10) for k = 2n (n = 0, 1, 2, . . .).
Next we proceed to prove that when τ satisfies (11), the collective period T increases with τ, and when τ satisfies (12), the collective period decreases with τ.
It can be verified that g(w) is an increasing function of w for . Given that ξ < 0 and cos w(Tm+Tp) < 0 when , the right hand side of (31) increases with sin(w(Tm+Tp – τ)+ϕ), which is a decreasing function of τ when τ satisfies (11). So w is a decreasing function of τ when τ satisfies (11). Similarly, we can prove that the right hand side of (31) increases with sin(w(Tm + Tp – τ) + ϕ), which is an increasing function of τ when τ satisfies (12). So w is an increasing function of τ when τ satisfies (12). Recall that T is equal to and hence is a decreasing function of w, we have the second part of Theorem 2.
Contributor Information
Yongqiang Wang, Department of Chemical Engineering, University of California, Santa Barbara, California 93106-5080 USA. wyqthu@gmail.com..
Yutaka Hori, Department of Information Physics and Computing, The University of Tokyo, Tokyo 113-8656 Japan. Yutaka_hori@ipc.i.u-tokyo.ac.jp..
Shinji Hara, Department of Information Physics and Computing, The University of Tokyo, Tokyo 113-8656 Japan. Shinji_hara@ipc.i.u-tokyo.ac.jp..
Francis J. Doyle, Department of Chemical Engineering, University of California, Santa Barbara, California 93106-5080 USA. frank.doyle@icb.ucsb.edu..
References
- 1.Goldbeter A. Biochemical oscillations and cellular rhythms: the molecular bases of periodic and chaotic behaviour. Cambridge University Press; London: 1996. [Google Scholar]
- 2.Winfree AT. The geometry of biological time. Springer; New York: 2001. [Google Scholar]
- 3.Wünsche H, Bauer S, Kreissl J, Ushakov O, Korneyev N, Henneberger F, Wille E, Erzgrässer H, Peil M, Elsässer W, Fischer I. Synchronization of delay-coupled oscillators: a study of semiconductor lasers. Phys. Rev. Lett. 2005;94:163901. doi: 10.1103/PhysRevLett.94.163901. [DOI] [PubMed] [Google Scholar]
- 4.Herrgen L, Ares S, Morelli LG, Schröter C, Jülicher F, Oates AC. Intercellular coupling regulates the period of the segmentation clock. Curr. Biol. 2010;20:1244–1253. doi: 10.1016/j.cub.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 5.Friesen WO, Block GD. What is a biological oscillator? Am. J. Physiol. 1984;246:847–853. doi: 10.1152/ajpregu.1984.246.6.R847. [DOI] [PubMed] [Google Scholar]
- 6.Fall C, Marland E, Wagner J, Tyson JJ, editors. Computational Cell Biology. Springer; New York: 2005. [Google Scholar]
- 7.Yeung M, Strogatz S. Time delay in the Kuramoto model of coupled oscillators. Phys. Rev. Lett. 1999;82:648–651. [Google Scholar]
- 8.Earl MG, Strogatz S. Synchronization in oscillator networks with delayed coupling: a stability criterion. Phys. Rev. E. 2003;67:036204. doi: 10.1103/PhysRevE.67.036204. [DOI] [PubMed] [Google Scholar]
- 9.Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis J. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 12.Kim J, Shin D, Jung S, Heslop-Harrison P, Cho K. A design principle underlying the synchronization of oscillations in cellular systems. J. Cell Sci. 2010;123:1537–543. doi: 10.1242/jcs.060061. [DOI] [PubMed] [Google Scholar]
- 13.Doiron B, Chacron MJ, Maler L, Longtin A, Bastian J. Inhibitory feedback required for network oscillatory responses to communication but not prey stimuli. Nature. 2003;421:539–543. doi: 10.1038/nature01360. [DOI] [PubMed] [Google Scholar]
- 14.Bal T, Debay D, Destexhe A. Cortical feedback controls the frequency and synchrony of oscillations in the visual thalamus. J. Neurosci. 2000;20:7478–7488. doi: 10.1523/JNEUROSCI.20-19-07478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertram R, Satin L, Zhang M, Smolen P, Sherman A. Calcium and glycolysis mediate multiple brsting modes in pancreatic islets. Biophys. J. 2004;87:3074–3087. doi: 10.1529/biophysj.104.049262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapp P. Mathematical techniques for the study of oscillations in biochemical control loops. Bull. Inst. Math. Appl. 1976;12:11–20. [Google Scholar]
- 17.Hori Y, Hara S. Proc. American Contr. Conf. San Francisco, USA: 2011. Time delay effects on oscillation profiles in cyclic gene regulatory networks: harmonic balance approach. pp. 2891–2896. [Google Scholar]
- 18.Iwasaki T. Multivariable harmonic balance for central pattern generators. Automatica. 2008;44:3061–3069. doi: 10.1016/j.automatica.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Zheng M, Friesen W, Iwasaki T. Multivariable harmonic balance analysis of neuronal oscillator for leech swimming. J. Comput. Neurosci. 2008;25:583–606. doi: 10.1007/s10827-008-0105-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Iwasaki T. Matrix perturbation analysis for weakly coupled oscillators. Syst. Control Lett. 2009;58:148–154. [Google Scholar]
- 21.Monk NAM. Oscillatory expression of Hes1, p53, and NF–κB driven by transcriptional time delays. Curr. Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- 22.Goodenough DA, Goliger JA, Paul DL. Connexins, connexons, and intercellular communication. Annu. Rev. Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Kim J, Kwon Y, Lee H, Heslop-Harrison P, Cho K. Reduction of complex singnaling networks to a representative kernel. Sci. Signal. 2011;4:ra35. doi: 10.1126/scisignal.2001390. [DOI] [PubMed] [Google Scholar]
- 24.Kageyama R, Niwa Y, Shimojo H, Kobayashi T, Ohtsuka T. Ultradian oscillations in notch signaling regulate dynamic biological events. Curr. Top. Dev. Biol. 2010;92:311–331. doi: 10.1016/S0070-2153(10)92010-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang YQ, Hori Y, Hara S, Doyle FJ., III The collective oscillation period of inter-coupled Goodwin oscillators.. Proc. 51th IEEE Conf. Decision Control; Maui, USA. 2012.pp. 1627–1632. [Google Scholar]
- 26.Khalil HK. Nonlinear systems. Prentice Hall; New Jersey: 2002. [Google Scholar]
- 27.Courant R, Hilbert D. Methods of Mathematical Physics, Volumn 1. Interscience Publishers; New York: 1953. [Google Scholar]
- 28.Schuster HG, Wagner P. Mutual entrainment of two limit cycle oscillators with time delayed coupling. Prog. Theor. Phys. 1989;81:939–945. [Google Scholar]
- 29.Wang S, Chandrasekaran L, Fernandez F, White J, Canavier C. Short conduction delays cause inhibition rather than excitation to favor synchrony in hybrid neuronal networks of the entorhinal cortex. PLoS Comput. Biol. 2012;8:e1002306. doi: 10.1371/journal.pcbi.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid G, Ao X, Haenggi P. In-phase and anti-phase synchronization in noisy Hodgkin-Huxley neurons. submitted. doi: 10.1016/j.mbs.2013.02.007. available at http://arxiv.org/abs/1206.4841. [DOI] [PubMed]
- 31.Giudicelli F, Özbudak EM, Wright GJ, Lewis J. Setting the tempo in development: an investigation of the zebrafish somite clock mechanism. PLoS Biol. 2007;5:1309–1323. doi: 10.1371/journal.pbio.0050150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hori Y, Kim T, Hara S. Existence criteria of periodic oscillations in cyclic gene regulatory networks. Automatica. 2011;47:1203–1209. [Google Scholar]
- 33.Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nature Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]