Abstract
The pathophysiology of numerous human disorders, such as atherosclerosis, diabetes, obesity and Alzheimer's disease, is accompanied by increased production of reactive oxygen species (ROS). ROS can oxidatively damage nearly all biomolecules, including lipids, proteins and nucleic acids. In particular, (poly)unsaturated fatty acids within the phospholipid (PL) structure are easily oxidized by ROS to lipid peroxidation products (LPP) carrying reactive carbonyl groups. Carbonylated LPP are characterized by high in vivo toxicity due to their reactivity with nucleophilic substrates (Lys-, Cys-and His-residues in proteins or amino groups of phosphatidylethanolamines [PE]). Adducts of unsaturated LPP with PE amino groups have been reported before, whereas less is known about the reactivity of saturated alkanals – which are significantly increased in vivo under oxidative stress conditions – towards nucleophilic groups of PLs.
Here, we present a study of new alkanal-dipalmitoyl-phosphatidylethanolamine (DPPE) adducts by MS-based approaches, using consecutive fragmentation (MSn) and multiple reaction monitoring techniques. At least eight different DPPE–hexanal adducts were identified, including Schiff base and amide adducts, six of which have not been reported before. The structures of these new compounds were determined by their fragmentation patterns using MSn experiments. The new PE-hexanal adducts contained dimeric and trimeric hexanal conjugates, including cyclic adducts. A new pyridine ring containing adduct of DPPE and hexanal was purified by HPLC, and its biological effects were investigated. Incubation of peripheral blood mononuclear cells and monocytes with modified DPPE did not result in increased production of TNF-α as one selected inflammation marker. However, incorporation of modified DPPE into 1,2-dipalmitoleoyl-sn-phosphatidylethanolamine multilamellar vesicles resulted in a negative shift of the transition temperature, indicating a possible role of alkanal-derived modifications in changes of membrane structure. © 2014 The Authors. Journal of Mass Spectrometry published by John Wiley & Sons, Ltd.
Keywords: lipid oxidation, MSn, high resolution MS, multiple reaction monitoring, alkanals
Introduction
Human organism is continuously exposed to many different sources of ‘oxidative stress’: sun light, oxidants in food, air pollution, ionizing radiations, smoke and the metabolism itself may all lead to harmful effects.1–7 When those oxidant species cannot be balanced by antioxidants (e.g. vitamin C or tocopherol) or enzymatic systems such as superoxide dismutase and catalase present in the cells, a new (and potentially harmful) status called ‘oxidative stress’ is established. At this condition, free radicals such as O2•−or HO• and other reactive oxygen species (ROS) such as H2O2 or HOCl are in high concentrations8and can induce severe damage to the major (macro)molecules present in the cells, i.e. DNA,9 lipids10 and proteins.11,12 ROS-induced damages can accumulate over time and contribute to cell injury and development of human diseases. ROS have been implicated in the development of several diseases, such as Parkinson's disease, Alzheimer's disease, atherosclerosis, arthritis, diabetes and heart failure, as well as in the process of aging.13–18
Specifically, peroxidation of membrane (phospho)lipids (PLs) can impair membrane functions, decrease fluidity, lead to inactivation of membrane-bound receptors and increase permeability to ions and even cause membrane rupture.19 High levels of oxidative stress can even induce cell death directly by necrosis, while mild oxidative stress can trigger the process of apoptosis.20 Lipid peroxidation occurs when ROS react with the lipid-esterified fatty acyl residues, particularly with polyunsaturated ones.21 During this process, a variety of products can be formed, including reactive aldehydes that are generated upon the cleavage of the olefinic residues. In particular, malondialdehyde and 4-hydroxynonenal are known to covalently modify proteins through the formation of Schiff bases or Michael-type addition products.22 These modifications can lead to a reduced functionality23 or even a complete loss of the protein function.24
Reactive carbonyls can also be generated during various cooking or frying processes.25 For instance, during the pasteurization of milk, hexanal is one of the major aldehydes detectable.26 During deterioration of meat, propanal and hexanal were also detected and quantified in turkey breast.27 Finally, during the heat treatment of various kinds of fish, elevated amounts of aldehydes were also present.28 This is one reason why the consumption of fried food is often considered to possess a negative impact on the consumer's health.
Both endogenous and exogenous aldehydes are considered toxic because their carbonyl group can react with nucleophilic groups present in many macromolecules within the cell. Some of the major targets are represented by the amino group of proteins, in particular the amino residue of the side chain of lysine.29 Nevertheless, it should be noted that the amino group of the head group of phosphatidylethanolamine (PE) is also reactive with aldehydes.30 The reactivity of the amino groups is not always identical; it is known for instance that γ-ketoaldehyde is characterized by a higher reactivity towards PE head group than the amino groups of proteins.31 Several groups tried to study the biological relevance of modified PE or modified proteins by aldehydes.32,33 Although most of the adducts with unsaturated aldehydes were successfully identified,32 no particular attention was paid to adducts generated by saturated aldehydes such as hexanal and propanal. Those lipid peroxidation products are derived from oxidation of ω-6 and ω-3 fatty acids (or the related fatty acyl residues within a lipid),34 and it was demonstrated that those aldehydes can generate in vivo Schiff base adducts of PE and proteins.31
In a tissue, PE adducts can trigger inflammation and cell damage. Recent evidence indicates that they can promote platelets prothrombinase activity35 and induce auto-oxidation of low-density lipoproteins. Treatment of human umbilical vein endothelial cells (HUVEC) with modified PE resulted in an increase of the expression of PERK (protein kinase RNA-like endoplasmic reticulum kinase), IRE1 (serine/threonine-protein kinase/endoribonuclease) and ATF6 (Activating transcription factor 6) – all proteins involved in the unfolded proteins response.31 Modified PE is also capable of triggering inflammatory pathways: it was demonstrated in a cell culture model of inflammation (THP-1 monocyte adhesion to HUVEC) that secretion of markers of inflammation such as MCP1 and IL-8 increased rapidly after modified PE addition.31 Additionally, the pyrrole modified PE, a particular product of the reaction between γ-ketoaldehyde and PE, can induce negative curvature of lipid monolayers disrupting the membrane shape.31 Finally, the reaction between the reactive aldehydes such as hexanal and propanal and the amino groups of proteins resulted in amide type and Schiff base adducts, which were detected in atherosclerotic plaques of rats34and humans.31 Hexanoyl-modified PE species detected in urine36 had a positive correlation with other biomarkers of inflammation.
In this study, dipalmitoyl-phosphatidylethanolamine (DPPE) was incubated in vitro with hexanal, and adducts formed upon this reaction were detected using high resolution Orbitrap MS and structurally characterized by sensitive MSn in linear iontrap. After structural characterization of the known and the new compounds, the complex reaction mixture was separated by HPLC, and selected compounds were isolated for further characterization of possible structural or functional effects.
Material and methods
Chemicals
1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1,2-dipalmitoleoyl-sn-glycero-3-phosphoethanolamine (DiPoPE), 1-palmitoyl-2-linoleoyl-sn-phosphatidylcholine (PLPC) and stearylamine were purchased from Avanti Polar Lipids, Inc. (Alabaster, Alabama). Ammonium formate, propanal, butanal, pentanal, hexanal, octanal, nonanal and 7-(diethylamino)-coumarin-3-carbohydrazide (CHH) were supplied by Sigma-Aldrich GmbH, and sodium cyanoborohydride (NaCNBH4) was purchased from FlukaChemie GmbH (Buchs, CH). Ultra liquid chromatography grade acetonitrile and methanol were from Biosolve BV (Valkenswaard, Netherlands). Ethanol and acetic acid were from Carl Roth GmbH & Co. KG and chloroform was from Merck KGaA (Darmstadt, Germany). Peptide GGQEHFAHKLILR (N-terminus acetylated) was synthesized in-house by solid phase peptide synthesis on a multiple peptide synthesizer (SYRO 2000, MultiSynTech GmbH, Witten, Germany) using Fmoc-chemistry. l-glutamine, penicillin, streptomycin, lipopolysaccharide, peptidoglycan from Staphylococcus aureus, phorbol 12-myristate 13-acetate and ionomycin were purchased from Sigma-Aldrich GmbH.
Incubation of phospholipids with saturated aldehydes
DPPE in chloroform was evaporated to dryness and re-solubilized (0.1 mmol/l) in 100 µl of water or PBS (8 mmol/l Na2HPO4, 1 mmol/l KH2PO4, pH 7.4) by vortexing and sonicating for 10 min to form multilamellar vesicles (vesicles size 0.1–1 µm determined by dynamic light scattering; data not shown). Saturated aldehyde (propanal, butanal, pentanal, hexanal, octanal and nonanal; each aldehyde separately; 400 mmol/l) was added, and the sample was incubated for 1 h at 37 °C. Lipid extraction was performed by adding a mixture of chloroform : methanol (1 : 1; v/v). The lower organic phase was collected, diluted 1 : 5 (v/v) in ESI solution 1 (methanol : chloroform (2 : 1; v/v) containing 5 mmol/l ammonium formate) and directly analyzed by ESI-LTQ-Orbitrap MS.
Incubation of peptide with saturated aldehydes
Peptide Ac-GGQEHFAHKLILR (in water; 0.1 mmol/l) was incubated with propanal, butanal, pentanal, hexanal, octanal and nonanal (400 mmol/l; each aldehyde separately) for 24 h at 37 °C. Samples were diluted 1 : 5 (v/v) with ESI solution 2 (50% acetonitrile, 25% methanol, 1% acetic acid) and analyzed by ESI-LTQ-Orbitrap MS.
Incubation of stearylamine with hexanal
Stearylamine (in water, 0.1 mmol/l) was incubated with hexanal (400 mmol/l) for 1 h at 37 °C. Lipid extraction was performed by adding a mixture of chloroform : methanol (1 : 1; v/v). The lower organic phase was collected, diluted 1 : 5 (v/v) in ESI solution 1 and directly analyzed by ESI-LTQ-Orbitrap MS.
Reduction of Schiff base
Modified lipid extract (10 μl) was dried and dissolved in water. NaCNBH4 was added in the final concentration of 100 mmol/l and incubated 1 h on ice. Lipid extraction was performed by adding a mixture of chloroform : methanol (1 : 1; v/v). The lower organic phase was collected, diluted 1 : 5 (v/v) in ESI solution 1 and directly analyzed by ESI-LTQ-Orbitrap MS.
Derivatization of lipid extracts with 7-(diethylamino)-coumarin-3-carbohydrazide
Lipid extract (5 μl) was added to 50 μl of 3 mmol/l CHH in methanol. Sample was incubated 1 h (37 °C, 550 rpm), diluted 1 : 5 (v/v) in ESI solution (methanol : chloroform (2 : 1; v/v) containing 5 mmol/l ammonium formate) and directly analyzed by ESI-LTQ-Orbitrap MS.
Mass spectrometry
Aldehyde adducts of PE and peptide were analyzed by direct injection using robotic nanoflow ion source TriVersa NanoMate (AdvionBio Sciences, Ithaca NY) equipped with nanoelectrospray chips (1.5 kV ionization voltage, 0.4 psi back pressure) coupled to an LTQ-Orbitrap XL ETD mass spectrometer (Thermo Fischer Scientific GmbH, Bremen, Germany). The temperature of the transfer capillary was set to 200 °C, and the tube lens voltage to 115 V. Mass spectra were recorded from m/z 400–2000 in the Orbitrap mass analyzer at a mass resolution of 100,000 at m/z 400. Tandem mass spectra were acquired by performing CID (isolation width 1–1.5 u, normalized collision energy 25%, activation time 30 ms, activation Q 0.25) in the linear ion trap. Acquired data were analyzed by using Xcalibur software (version 2.0.7).
Concentration-dependent experiment
The DPPE (0.1 mmol/l) in chloroform was dried and re-suspended in 100 µl water. Saturated aldehyde was diluted with 10% aqueous ethanol. Diluted aldehyde was incubated with DPPE for 1 h at 37 °C. Lipid extraction was performed by adding chloroform : methanol (1 : 1; v/v). The lower part was diluted 1 : 5 in ESI solution 1 and analyzed by ESI-Orbitrap.
Multiple reaction monitoring for relative quantification of modified PE
Quantification of compounds was performed on an ESI-QTRAP 4000 (ABSciex, Darmstadt, Germany). All experiments were carried out in positive ion mode. The optimum multiple reaction monitoring (MRM) parameters for each compound (declustering, entrance and collision cell exit potential) were obtained using large scale reaction mixture (50 µg of DPPE and 20 mg of hexanal; incubated for 24 h). Sample was diluted and directly injected using a syringe infusion pump.
To relatively quantify the amount of each modification over different incubation time points, DPPE (0.1 mmol/l) was dried and incubated with 5% (v/v) of hexanal in water. After 0, 10, 20 and 30 min and 1, 2, 4, 6, 24 h, lipids were extracted as described (vide supra). Samples were diluted in ESI solution 1, and 30 µl was used (in triplicates) for direct injection followed by MRM analysis on the ESI-QTRAP. ESI parameters were ESI source curtain gas 30, CAD medium, ion spray voltage 5500 V, temperature 650 °C, source gas 1 50 and source gas 2 50. PLPC (1 pg/µl) was spiked in each sample as internal standard. Relative intensity was obtained by normalizing the intensity of each signal relative to the PLPC standard. Data were acquired and quantified with Analyst software version 1.6.
Separation of lipid-aldehyde products
Hexanal-modified DPPE was purified by HPLC (Beckmann System Gold, with 168NM Diode array detector, Beckmann Coulter GmbH, Brea, California). The mobile phase consisted of eluent A (methanol/acetonitrile/aq. 1 mmol/l ammonium acetate 60 : 20 : 20 (v/v/v)) and eluent B (1 mmol/l ammonium acetate in ethanol). DPPE was incubated with hexanal as described previously. Lipid extracts were dissolved in 20% B and separated on a Jupiter C5 column (5 µm; 150 × 4.60 mm, 300 Å; Phenomenex) with a flow rate of 1 ml/min. Gradient elution was conducted as follows: isocratic step at 20% B for 2 min, gradient increase up to 100% B in 10 min and isocratic step at 100% B for 8 min. Eluted lipid adducts were monitored at 256 and 206 nm using PDA detector. The fractions containing trimeric adducts were manually collected and identified by ESI-Orbitrap MS.
Determination of the phospholipid concentration
The concentration of the product of interest was determined by 1H NMR spectroscopy. As the product was planned to be subsequently used for the cellular experiments, no internal standard could be used. Therefore, the sample was dissolved in CDCl3 (99.6% 2H), and the intensity of the residual CHCl3 was used for the quantitative determination. All NMR measurements were performed on a Bruker DRX-300 spectrometer operating at 300.13 MHz for 1H. All spectra were recorded at 310 K using a 13C/1H ‘dual’ probe. One thousand and twenty-four transients were acquired to obtain a sufficiently high signal-to-noise (S/N) ratio. A repetition time of 10 s was used to account for T1 relaxation time effects. No window functions were used prior to Fourier transformation.
Differential scanning calorimetry
Lipid vesicles were obtained by dissolving DiPoPE (5.5 mg/ml) with or without aldehyde-modified PE (0.07 mg/ml) in chloroform and samples were dried for 2 h under nitrogen. Dry lipids were re-suspended in 20 mmol/l piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 1 mmol/l EDTA and 150 mmol/l NaCl (pH 7.4) by vortexing to form multilamellar vesicles. Differential scanning calorimetric measurements were performed on a Nano DSC Differential scanning calorimeter (TA Instruments, Eschborn, Germany) using a cell volume of 340 µl. The scan rate was 1 °C/min from 10 °C to 70 °C with a delay of 5 min between sequential scans in order to allow thermal equilibration. Four heating and cooling scans were acquired per sample. Calorimetry curves were analyzed with NanoAnalyze Data Analysis version 2.1.
Incubation of human monocytes and peripheral blood mononuclear cells with modified PEs
Human subjects were recruited among healthy blood donors. All experiments with human materials were approved by the local ethics committee, and informed consent was obtained from all subjects. Human peripheral blood mononuclear cells (PBMCs) were isolated from the peripheral blood using Ficoll® density gradient centrifugation. Human monocytes were isolated by negative magnetic separation (Miltenyi Biotech) as described previously.37 Human PBMCs and primary monocytes (3 × 105 cells per 200 µl) were incubated at 37 °C and 5% CO2 in RPMI1640 supplemented with 10% heat-inactivated fetal calf serum, 2 mmol/l l-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin. Human PBMCs and primary monocytes were treated with aldehyde-modified PE, dissolved in medium (10, 100 nmol/l or 1 µmol/l) for 4 h (primary monocytes and PBMCs), 16 h or 24 h (PBMCs). As positive controls, monocytes were incubated with lipopolysaccharides (100 ng/ml), and human PBMCs with peptidoglycan from Staphylococcus aureus (PNG-SA; 5 µg/ml), phorbol 12-myristate 13-acetate (PMA; 20 ng/ml) and ionomycin (0.5 µg/ml) for the same periods of time. Supernatants were collected and stored at −80 °C. Human tumor necrosis factor alpha (TNF-α) was measured by commercially available enzyme immunoassay following the manufacturer's protocol (OptEIA; BD Bioscienes). To assess statistical significance, students t-test was used. Prior to all comparisons, a normality test (Kolmogorov–Smirnov test) was performed.
Results
Despite multiple indications of high abundance of alkanals during lipid peroxidation, their reactivity towards nucleophilic substrates is much less studied than that of hydroxyl-alkenals or low molecular weight (di)aldehydes, such as acrolein and malondialdehyde. Nevertheless, multiple reports, especially in the field of food chemistry, indicate high concentrations of alkanals in biological samples. To investigate in more detail the reactivity of alkanals towards nucleophilic groups in PLs, DPPE was exposed to hexanal for 1 h at 37 °C. Reaction was performed in PBS (pH 7.4) or water. No differences were observed, and therefore, all subsequent reactions were performed in water to minimize the effects of salts during subsequent ESI-MS. A more detailed investigation of the influence of the supramolecular structure of PE (that forms hexagonal phases in contrast to the majority of other PLs) on the product yield was beyond the scope of this investigation.
Mass spectrometric analysis of DPPE-hexanal incubation mixtures
Incubation of DPPE (protonated ion at m/z 692.52) with hexanal for 1 h resulted in the appearance of eight signals (Fig. 1, A) with m/z 774.60, 790.60, 856.68, 874.69, 906.72, 936.74, 938.76 and 954.75. A Survey scan was acquired using high resolution MS (100 000 at m/z 400) on Orbitrap and allowed to obtain elemental composition of DPPE-hexanal adducts using 3 ppm mass accuracy. These products were characterized by the mass shifts to DPPE of 82, 98, 164, 182, 214, 244, 246 and 262 m/z units corresponding to addition of C6H10, C6H10O, C12H20, C12H22O, C13H26O2, C18H28, C18H30 and C18H30O groups to DPPE (Table 1). Taking into account the molecular weight of hexanal of 100.087 g/mol (monoisotopic mass) and the elemental composition calculated from high resolution MS scans, we propose formation of monomeric, dimeric and trimeric Schiff base adducts, and their hydrated forms on PE head group (Scheme 1) and the ion at m/z 790.60 was assigned as amide-linked type adduct between hexanal and the amino group of PE. Additionally, the ion at m/z 906.72 was proposed to represent a methanol adduct [M + CH3OH]+38–40 of the ion 874.69. In order to confirm the proposed Schiff base adducts, reduction with NaCNBH4 was performed. Although it was not possible to obtain a quantitative reduction, the ESI mass spectrum clearly indicated characteristic mass shifts of 2 amu in the case of monomeric Schiff base adducts (m/z 776.62), and 4 amu in the case of the dimer and trimer (m/z 860.71 and 940.77) (Fig. 1 B). Corresponding hydrated forms of the dimer and trimer were reduced with mass shift of 2 amu. The proposed amide-linkage adduct (m/z 790.60) was not reduced as expected.
Figure 1.
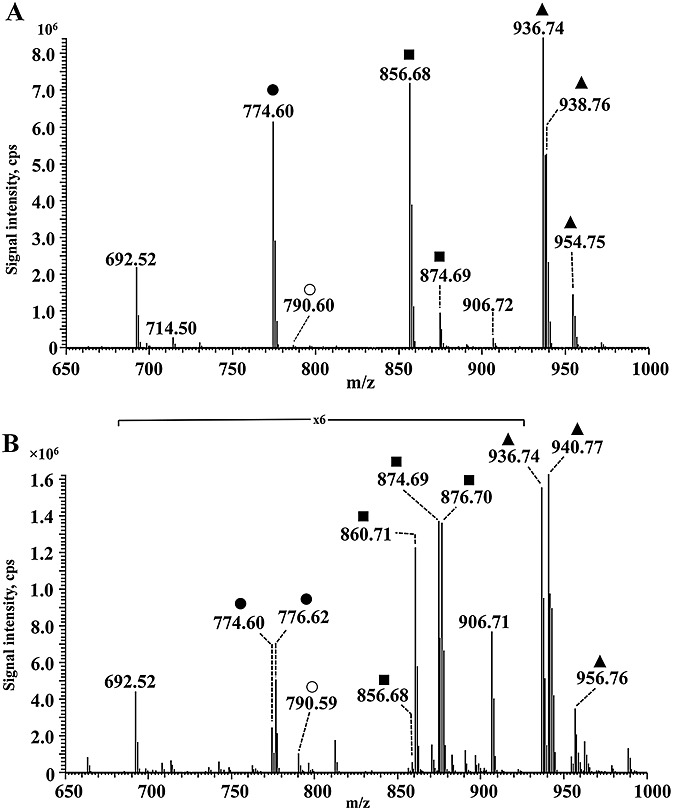
ESI-Orbitrap MS spectra of dipalmitoyl-phosphatidylethanolamine (DPPE)-hexanal incubation mixtures before (A) and after (B) reduction withNaCNBH4. Unmodified DPPE, amide-linked adducts, monomeric, dimeric, trimeric adducts with hexanal and its hydrated forms were detected. Single Schiff base adduct was detected (•) as well as amide adduct (○), dimeric hexanal adducts (▪) and trimeric hexanal adduct(▲).
Table 1.
Overview of detected m/z values for DPPE-hexanal adducts, their elemental composition, proposed chemical structure and optimized MRM transitions
| Detected m/z | Elemental composition | Mass accuracy (ppm) | Mass shift to unmodified DPPE | Chemical structure | m/z detected after reduction | MRM transition |
|---|---|---|---|---|---|---|
| 692.5222 | C37H75O8NP | −0.38 | — | DPPE | — | 692 → 551 |
| 774.6005 | C43H85O8NP | −0.27 | (C6H10) 82.07 | Schiff base | 776.6158 | 774 → 224 |
| 790.5952 | C43H85O9NP | −0.55 | (C6H10O) 98.07 | Hydrated Schiff base | — | 790 → 240 |
| 856.6788 | C49H95O8NP | −0.17 | (C12H20) 164.15 | Dimer | 860.7102 | 856 → 306 |
| 874.6893 | C49H97O9NP | −0.18 | (C12H22O) 182.17 | Hydrated Dimer | 876.7048 | 874 → 324 |
| 936.7416 | C55H103O8NP | 0.06 | (C18H28) 244.21 | Pyridinium ring adduct | — | 936 → 386 |
| 906.7154 | C50H101O10NP | −0.38 | (C13H26O2) 214.2 | MeOH adduct of 874.6 | — | — |
| 938.7530 | C55H105O8NP | −4.45 | (C18H30) 246.23 | Trimer | 940.7722 | — |
| 954.7516 | C55H105O9NP | −0.52 | (C18H30O) 262.22 | Hydrated trimer | 956.7667 | 954 → 404 |
DPPE, dipalmitoyl-phosphatidylethanolamine; MRM, multiple reaction monitoring.
Scheme 1.
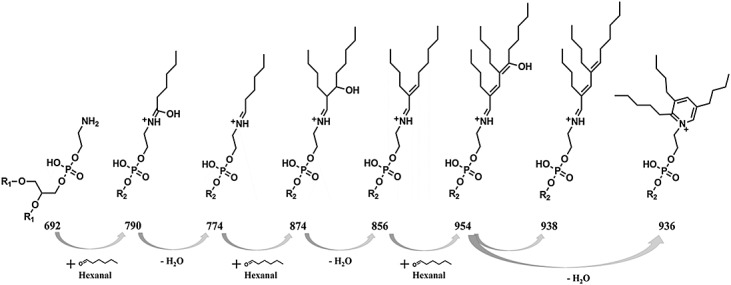
Proposed mechanism of consecutive formation of dimeric and trimeric dipalmitoyl-phosphatidylethanolamin-hexanal adducts.
MSn based identification of DPPE-hexanal adducts
In order to understand the chemical nature of new products obtained after DPPE incubation with hexanal, sequential fragmentation up to MS7 was performed using linear ion trap. DPPE was incubated with hexanal in water overnight to obtain higher yields of adducts, and reaction mixture was electro sprayed into LTQ-Orbitrap mass spectrometer using robotic ESI source.
Under CID conditions, DPPE decomposed into two major fragments at m/z 551.5 and 142.5 corresponding to the diacylglycerol (DAG) and head group, respectively. The same fragmentation pattern was also observed in all other modified PEs, and ions of the unmodified DAG (m/z 551.5) and the modified head group being detected. The product ion spectra of 774.60 m/z (Fig. S1), proposed as Schiff base adduct, showed the characteristic product ions at m/z 551.51 (DAG) and 224.15 (modified head group ion). Ion at m/z 224.15 was selected for MS3 CID analysis, and several product ions were obtained, including ion at m/z 126.10, which corresponds to the loss of the phosphoric acid (−98 amu) from the precursor. Furthermore, ions derived from the MS4 fragmentation using m/z 126.10 as a precursor (m/z 112.34, 98.15, 83.97, 69.98, 55.99) exhibited the mass differences of 14 amu suggesting the presence of a hexanal alkyl chain, thus confirming the structure of the Schiff base adduct. MSn CID spectra of reduced Schiff based adduct (m/z 776.62; Fig. S2) further confirmed the modification structure by the presence of all head group specific ions with a mass shift of 2 amu.
The structure of amide-like adduct at m/z 790.60 was investigated (Fig. S3). Head group ion at m/z 240.06 present in MS2 CID spectrum was selected for further fragmentation. In MS3 spectrum, the signal at m/z 222.17 was attributed to the loss of water from the head group, and this supports the presence of a hydroxyl group in the molecule. It was further supported by the loss of water from the ion at m/z 141.13 formed by the loss of 98 amu (loss of the phosphoric acid) resulting in m/z 123.95.
The structure of a dimeric hexanal adduct (m/z 856.68) was elucidated by several consecutive tandem MS experiments (Fig. S4). Product ion at m/z 306.25 observed in MS2 spectra was attributed to the modified head group. Fragmentation of ion at m/z 306.25 led to the formation of multiple product ions from which the ion at m/z 208.22 (loss of the phosphoric acid) was selected because of the high intensity and the interesting product ion pattern. Most of the observed product ions differed by the mass shift of 14 amu (m/z 180.20, 166.18, 152.13, 138.09 and 124.09), confirming the presence of the alkyl chain. Signal at m/z 152.13 corresponded to the loss of 56 amu (C4H8), suggesting the presence of a second aldehyde linked at position Cβ on the first alkyl chain (Scheme 1). Further fragmentation of the ion at m/z 152.13 (MS5) led to the formation of product ions (m/z 124.08, 109.98, 95.95, 81.94 and 67.98), which are characteristic of alkyl chain fragmentations. In order to further confirm the structure, fragmentation behavior of the corresponding reduced species (m/z at 860.71) was investigated (Fig. S5). MS2 fragmentation of the ion at m/z 860.71 resulted in four product ions (DAG fragment at m/z 551.39, modified head group fragment at m/z 310.18, loss of water and loss of 46 amu from precursor). Product spectrum of m/z 310.18 confirmed the assignment of the previous structure by the presence of the ions with a difference of 4 amu compared to unreduced dimer head group.
Ion 874.69 was assigned as a hydrated form of the m/z 856.68 adduct (Fig. S6). CID product ions of m/z 874.69 were formed by the loss of water from the precursor (m/z 856.59) and the second hexanal moiety (m/z 774.40). Additionally, unmodified DAG (m/z 551.54) and modified head group (m/z 324.12) fragments were detected as expected. Head group product ion was selected for further fragmentation, which resulted in a loss of water (m/z 306.28) and the second hexanal moiety (m/z 224.17). Analysis of head group ion confirmed the presence of a hydroxyl group in the dimer. Upon fragmenting the hydrated dimer head group, two main product ions were observed. The first signal at m/z 306.28 was formed after loss of water from the precursor. Sequential fragmentation of product ions at m/z 306.28 and 224.17 (MS4 and MS5, Fig. S6,C–F) resulted in the identical product ions previously observed and assigned to dimeric and monomeric Schiff base head groups (S4 and S1, respectively). Fragmentation of the reduced hydrated dimer (876.70 ion, Fig. S7) confirmed the proposed structure for the m/z 874.69 ion. Observed ion at m/z 326.15 in MS2 mass spectrum showed a difference of 2 amu for the head group fragment due to the reduction of the double bond.
A pyridine ring containing structure was suggested for the ion at m/z 936.74 (Fig. S8). MS2 spectrum showed the modified head group ion at m/z 386.23. By selecting it for MS3 fragmentation, it was possible to detect an ion at m/z 288.24 formed by the loss of 98 amu. A series of the signals with a mass shift of 2 amu was observed for the majority of the product ions, indicating the presence of a double bond along the alkyl chains. The proposed structure in Fig. S8, panels B and C, indicates one of the possible positions of the double bound. MS3 product ions at m/z 260.18, 246.17, 232.19 and 218.14, with a mass difference of 14 amu, provide information about the related alkyl chain. Ion at m/z 232.19 was selected for further fragmentation (MS4), and product ions at m/z 216.20, 202.04, 190.16, 174.06 and 160.11 were observed, indicating alkyl chain fragmentation (Fig. S8, C). Additionally, product ions at m/z 147.09 and 133.02 corresponded to the fragmentation after the loss of one of the alkyl chains from the ring.
Linear hexanal trimer was also detected (m/z 938.76); however, because of the instability of this compound, it was not possible to efficiently isolate this specific ion. The possible structure of this molecule was determined by MSn fragmentation of the reduced trimer ion at m/z 940.77 (Fig. S9). Trimer adduct formed an intense head group modified product ion at m/z 390.18. After MS3 fragmentation, product ion observed at m/z 292.25 indicated the loss of the phosphate from the head group. Ions at m/z 249.22, 236.05 and 222.19 suggested the fragmentation of the alkyl chain. Ion at m/z 224.05, with a mass difference of 2 amu from 222.19, may indicate the presence of a double bond in the molecule. Similarly, the MS4 spectrum (Fig. S9, C) of m/z 292.25 indicates the fragmentation of an alkyl chain, and the ion at m/z 236.23 may suggest the loss of C4H8 confirming the branching along the alkyl chain. Furthermore, from the MS3 spectrum (Fig. S9, B), three ions (m/z 236.23, 222.16, 208.15) were selected and fragmented in order to confirm the proposed structure. Product ions at m/z 236.23 (Fig. S9 panel D) indicated alkyl chain fragmentation (m/z 208.17, 194.32, 180.14, 166.11 and 152.06 with mass differences of 14 amu). Additionally, ion at m/z 180.14 can also be formed after loss of 56 amu (C4H8). Fragmentation of ion at m/z 222.24 (Fig. 9, E) resulted in ions at m/z 180.12, 166.08, 152.09 and 138.07 indicating the fragmentation along the alkyl chain. Similarly, the most intense product ion at m/z 180.12 supported the presence and the position of the second or third hexanal moiety. Similar results were obtained after fragmentation of the ion at m/z 138.13 (Fig. S9, F and G). Finally, fragmentation of the ion at m/z 208.15 (Fig. S9, H), further confirmed the presence of three hexanal moieties on the aldehdye modified head group.
Figure 9.
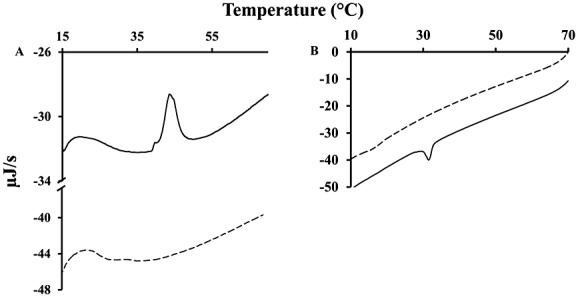
Heating (A) and cooling (B) differential scanning calorimetry plots of DiPoPE vesicles (black line) and DiPoPE vesicles containing pyridine ring modified dipalmitoyl-phosphatidylethanolamine (0.09 molar ratio; dotted line).
Particular attention was also given to ion at m/z 954.75. As shown in Fig. S10, fragmentation occurs by the initial loss of diacylglycerol. MS2 spectra reveal the presence of two ions, one at m/z 404.26 that corresponds to the head group of the hydrated trimer, followed by the ion at m/z 386.22 formed by loss of water. Fragmentation of the ion at m/z 404.26 resulted in ions at m/z 386.25, 324.30 and 306.29, which were attributed to the loss of water, 80 and 98 amu (attribute to the losses of phosphoric acid), respectively. The presence of the ion at m/z 263.18 was explained by the loss of ethanolamine (43 amu). Head group specific ion at m/z 306.29 was isolated for further fragmentation, and product ion at m/z 278.22 indicated the loss of 28 amu (–CH2CH2). Ion at m/z 152.11 indicated the loss of the terminal hexanal moiety (Fig. S10 panel C). Additional product ions at m/z 250.20, 222.29 and 178.13 were attributed to the fragmentation along the alkyl chain and confirmed structural assignment. Corresponding reduced ion was fragmented to confirm the elucidation of the structure (Fig. S11).
Evaluation of the reaction mechanism between DPPE and hexanal
Two different hypotheses can explain the formation of dimeric and trimeric PE-aldehyde adducts. First, the aldehyde present in a solution at higher concentrations can form dimeric and trimeric structures via an aldol-condensation mechanism. Carrying a free carbonyl group, these dimers and trimers can modify the primary amino group of the DPPE head group. Alternatively, formation of dimeric and trimeric DPPE–aldehyde adducts can occur via consecutive addition of aldehyde to the previously formed monomeric Schiff base via aldol-condensation mechanism. To verify the mechanism, several experiments were performed including evaluation of hexanal concentration influence, kinetic of adducts formation and identification of reactive carbonylated compounds in the incubation mixtures.
Thus, the dependence of adduct formation on the hexanal concentration was monitored by incubating 0.1 mmol/l DPPE with increasing concentrations of aldehyde for 1 h, and the resulting mixtures were analyzed by direct infusion on an ESI-Orbitrap MS. Relative intensity for each Schiff base product (Fig. 2A) and their hydrated analog (Fig. 2B) was normalized for each incubation. In the samples incubated with 0.1, 0.5 or 1 mmol/l hexanal, no products were detected. At 2.5 mmol/l hexanal, the first Schiff base adduct with m/z 774.5 was observed, and the intensity of the signal increased further, peaking at 25 and 400 mmol/l hexanal concentrations. All other adducts including amide-linked monomer, Schiff base dimer and its hydrated form, non- and hydrated trimer and cyclic pyridine ring compound followed the same behavior, but were detected only after incubation of DPPE with 25 mmol/l or higher concentrations of the hexanal. Based on the obtained data, it is likely that the monomeric Schiff base DPPE-hexanal adducts represent the first reaction products, which are necessary for the formation of dimeric and trimeric hexanal-modified PE species.
Figure 2.
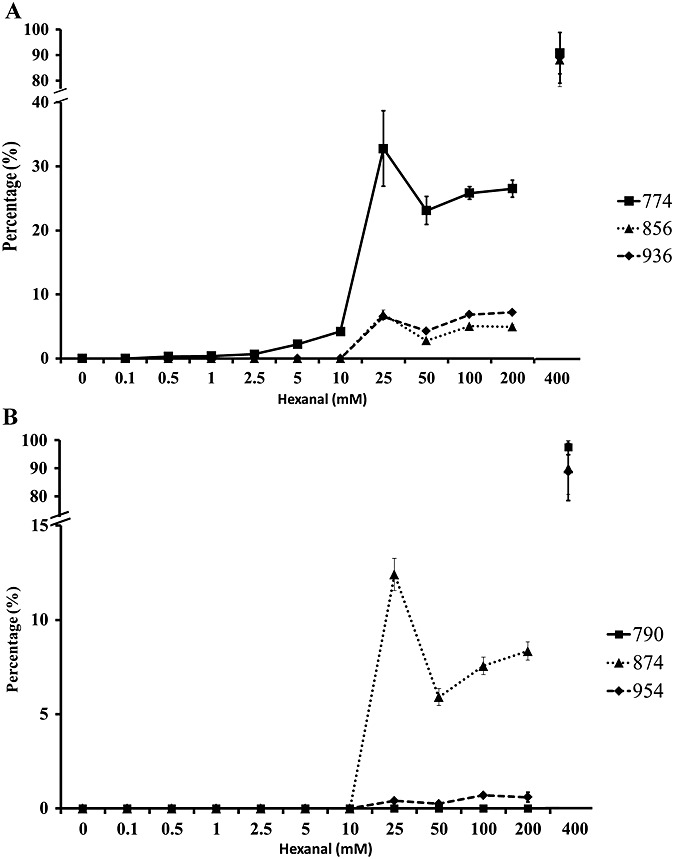
Concentration-dependent experiment. Dipalmitoyl-phosphatidylethanolamine (0.1 mM) was incubated with several concentration of hexanal in water for 1 h at 37 °C. Lipid was extracted and analyzed by ESI-Orbitrap. Absolute intensities recorded for each compound were normalized, and the highest value was defined as 100%. (A) Schiff base (774) dimer (856) and trimer adduct (936), (B) Amide adduct (790), and hydrated dimer (874) and trimer (954).
To monitor the kinetic of DPPE-hexanal adducts formation, an MRM-based method was designed. Specific product ions characteristic of modified head groups of PE-hexanal adducts previously determined in MS/MS experiments were used as specific transitions during direct infusion MRM experiments. Unmodified DPPE was monitored by transition 692 → 551, and each modified PE by the specific head group transitions (Table 1). Intensity of unmodified DPPE decreased gradually over the incubation time. Maximum formation of monomeric and dimeric Schiff bases was observed after 2 h of incubation (Fig. 3A). Importantly, significantly elevated levels of monomeric Schiff base adduct were detected already after 20 min of incubation. The maximum intensities after 2 h of incubation were characteristic for dimeric and trimeric hydrated forms as well (Fig. 3B), but the abundance of the trimer was continuously increasing over 6 h of incubations. However, after 58 h of incubation, the abundance of all adducts was decreased. Kinetic evaluations demonstrate maximum formation of monomeric and dimeric adducts after 2 h of incubation and consecutive formation of trimeric hexanal-DPPE adduct gradually increasing over the incubation time. The result of this time-dependent experiment is a clear indication that the generation of these adducts actually occurred in the reaction mixture – not during sample preparation, extraction using methanol and chloroform mixture or during the ESI process.
Figure 3.
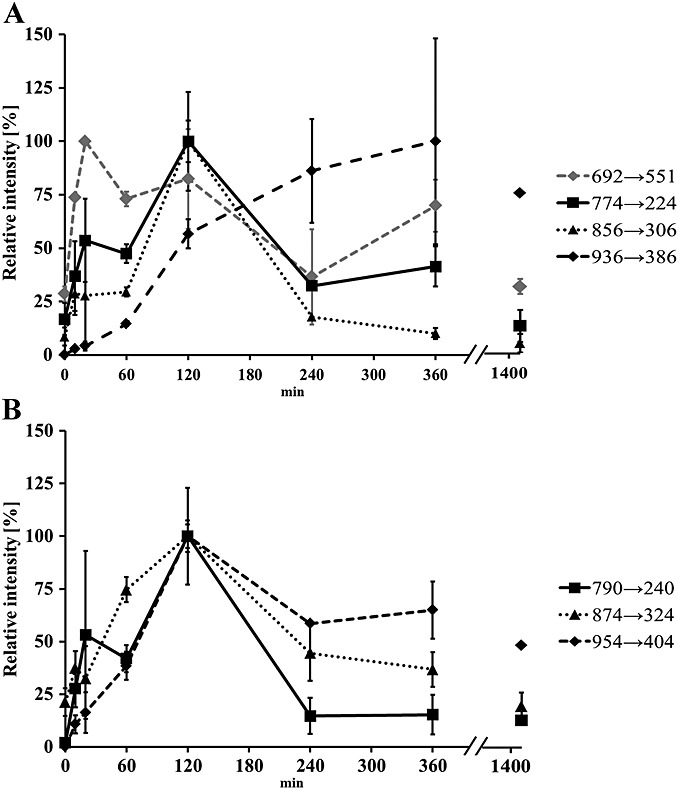
Time dependence of dipalmitoyl-phosphatidylethanolamine (DPPE), DPPE-hexanal Schiff base adducts (A), amide-linked and hydrated Schiff base adducts (B) formation monitored in MRM experiment. DPPE (0.1 mM) was mixed with hexanal 5% (v/v) in water, and the intensity of each compound was quantified (y-axis) after 10, 20, 30, 60, 120, 240 and 360 min and 24 h after the start of the incubation (x-axis). (A) unmodified dipalmitoyl-phosphatidylethanolamine (692), Schiff base (774), dimer (856) and trimer (936), (B) hydrated form of monomer (790) dimer (874) and trimer (954).
Finally, in order to verify the presence of reactive free dimeric and trimeric aldehyde adducts in the reaction mixture, hexanal with and without DPPE was incubated with CHH.41 CHH specifically reacts with free carbonyl groups forming a stable hydrazone. It was revealed by MS that only the single molecule of hexanal reacted with CHH forming a product ion at m/z 358.21. On the contrary, no adducts with the dimer and trimer of hexanal were observed (Fig. 4). DPPE after incubation with hexanal was also derivatized with CHH. Similarly, the MS spectrum showed only the hexanal-CHH derivative but no additional products.
Figure 4.
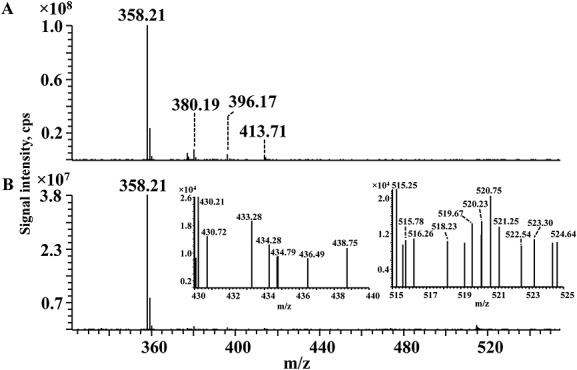
ESI-Orbitrap mass spectra of hexanal (A) and dipalmitoyl-phosphatidylethanolamine-hexanal mixture (B) derivatized with CHH for detection of reactive carbonylated compounds. Ion at m/z 358.21 corresponds to the CHH-single hexanal derivative. Inserts represent the m/z ranges of expected CHH-derivatives of dimeric and trimeric hexanal products.
All the data presented previously indicate that PE-hexanal dimeric and trimeric adducts are formed by the consecutive addition of monomeric hexanal molecules on the head group of DPPE starting from a monomeric Schiff base via a β-aldol-condensation mechanism.
Reactivity of different alkanals towards DPPE primary amino group
To check the reactivity of other alkanals towards the amino group of PEs, DPPE was incubated with a set of aldehydes, including propanal, butanal, pentanal, hexanal, octanal and nonanal (Fig. 5A). The expected monomers, dimers and trimers could be observed for all incubations. However, amide-linkage adducts were detected only in the case of the propanal, butanal, pentanal and hexanal adducts. Additionally, hydrated forms of the dimer and trimer were present only for hexanal, octanal (only hydrated dimer) and nonanal. Interestingly, formation of cyclic trimers as in the case of the hexanal pyridine ring adduct was detected only for nonanal (m/z 1062.91).
Figure 5.
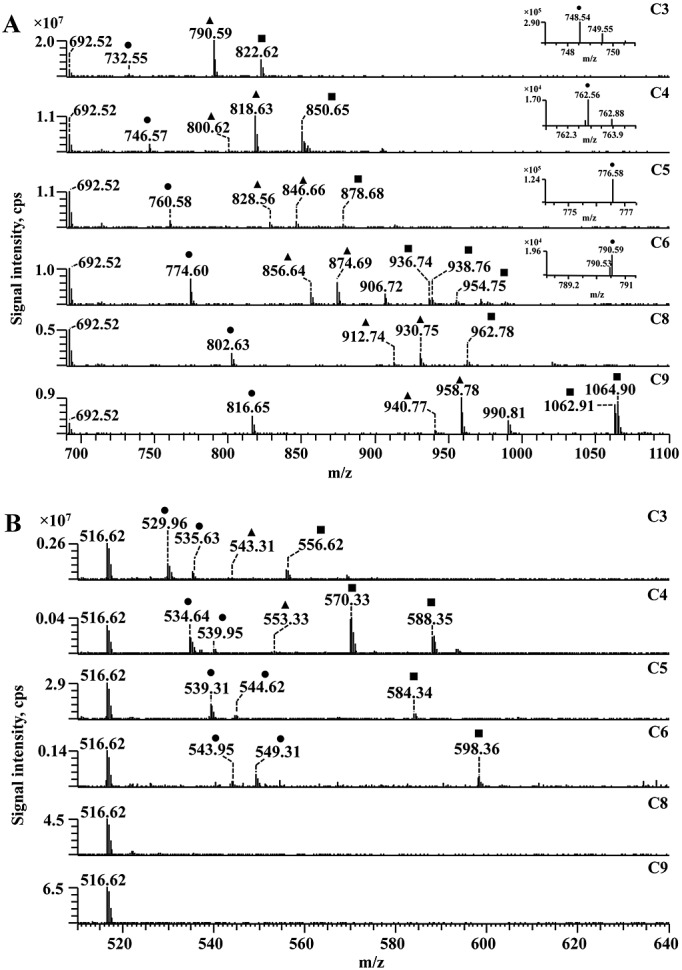
ESI-Orbitrap MS spectra of dipalmitoyl-phosphatidylethanolamine (A) and model lysine containing peptide (B) incubated with different saturated aldehydes. For peptide-alkanal incubations, all adducts correspond to the triply charge state. Different adducts are indicated by the following symbols: • monomers (Schiff base adduct, hydrated Schiff base); ▪ dimeric adducts and hydrated forms; and ▲trimer adducts and hydrated forms. Saturated aldehydes were propanal (C3), butanal (C4), pentanal (C5), hexanal (C6), octanal (C8) and nonanal (C9).
Furthermore, we investigated the potential generation of mixed dimeric and trimeric adducts formation. DPPE was incubated with an equimolar mixture of pentanal and hexanal overnight, and resulting products were analyzed by high resolution MS (Fig. 6). Multiple products were observed, including monomeric, dimeric and trimeric adducts of pentanal (m/z 760.58, 828.65 and 894.69) and hexanal (m/z 774.60, 856.68 and 936.74) to DPPE. In addition, there were pentanal–hexanal dimers (m/z 842.66) and pentanal–pentanal–hexanal (m/z 908.71) and pentanal–hexanal–hexanal (m/z 922.72) trimers.
Figure 6.
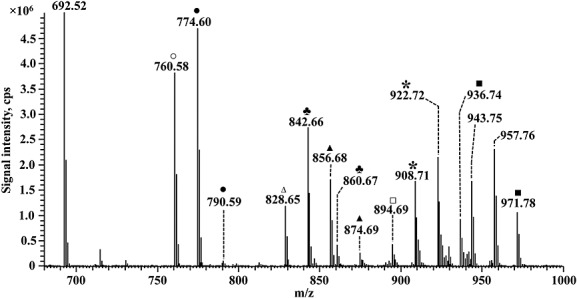
ESI-Orbitrap MS spectrum of dipalmitoyl-phosphatidylethanolamine (DPPE) incubated with hexanal and pentanal. Hexanal and pentanal 5% (v/v) were incubated in water with DPPE (0.1 mM) for 1 h 37 °C. The following Hexanal-DPPE adducts were detected: • monomer; ▪ dimer; and ▲trimer. Additionally, the following Pentanal-DPPE adducts were detected: ○ monomer; □ dimer; Δ trimer;♣ dimeric adduct; and ∗ trimeric adduct containing a combination of hexanal and pentanal with DPPE.
Reactivity of alkanals towards other nucleophilic substrates
In order to check if reactive alkanals can modify other biologically relevant nucleophilic substrates, stearylamine and synthetic peptides containing lysine residues were incubated with hexanal in water. MS analysis of stearylamine subsequent to incubation with hexanal showed the presence of unmodified stearylamine (m/z 270.31) and the corresponding trimer at m/z 514.53. No monomeric or dimeric adducts were detected in these experiments (Fig. 7).
Figure 7.
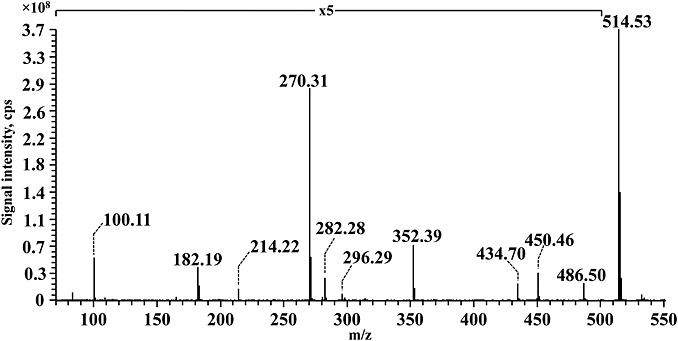
ESI-Orbitrap MS spectrum of hexanal incubated with stearylamine. Ions at m/z 270.31 and 514.53 correspond to unmodified dipalmitoyl-phosphatidylethanolamine and the pyridine trimer hexanal–stearylamine adduct, respectively.
Peptide Ac-GGQEHFAHKLILR was incubated with different alkanals, including propanal, butanal, pentanal, hexanal, octanal and nonanal. Modified peptides were characterized using direct infusion high resolution MS (Fig. 5B). All previously described adducts, including monomeric Schiff base, amide-linked adduct, dimer and trimer, were detected with a peptide in the case of propanal and butanal modifications. For pentanal and hexanal incubations, however, only monomeric Schiff base, amide-linked adduct and trimeric adducts were detected. In general, the signal of the amide type adduct was more intense in the case of the peptide when compared to DPPE incubations. Surprisingly, no adducts were observed after incubation of peptide with octanal and nonanal. Figure 8 exemplifies the MS spectrum obtained for the propanal-modified peptide. Modified peptide was detected as a triply charge ion (similarly, doubly charge ions were also investigated to confirm the structure of the identified adducts; data not shown). Ion at m/z 516.62 corresponded to the unmodified peptide, and ions at m/z 529.26 (+40 amu) and 535.96 (+56 amu) were assigned to the propanal Schiff base and amide-like type adducts, respectively. Ion at m/z 543.30 (+80 amu) corresponded to the presence of dimeric adduct on the lysine residue. To confirm Schiff base adducts, peptide-propanal adducts were reduced with NaCNBH4 (Fig. 8B). Surprisingly, intensities of the reduced adducts were much higher than unreduced ones, and the signal of unmodified peptide was present at a very low level. Ions at m/z 530.63 and 544.65 confirmed reduction of monomeric and dimeric Schiff base adducts. Additionally, the ion of the reduced trimer was clearly present (m/z 557.65). Tandem mass spectrometry was used to confirm the presence of propanal adduct on the lysine residue (data not shown).
Figure 8.
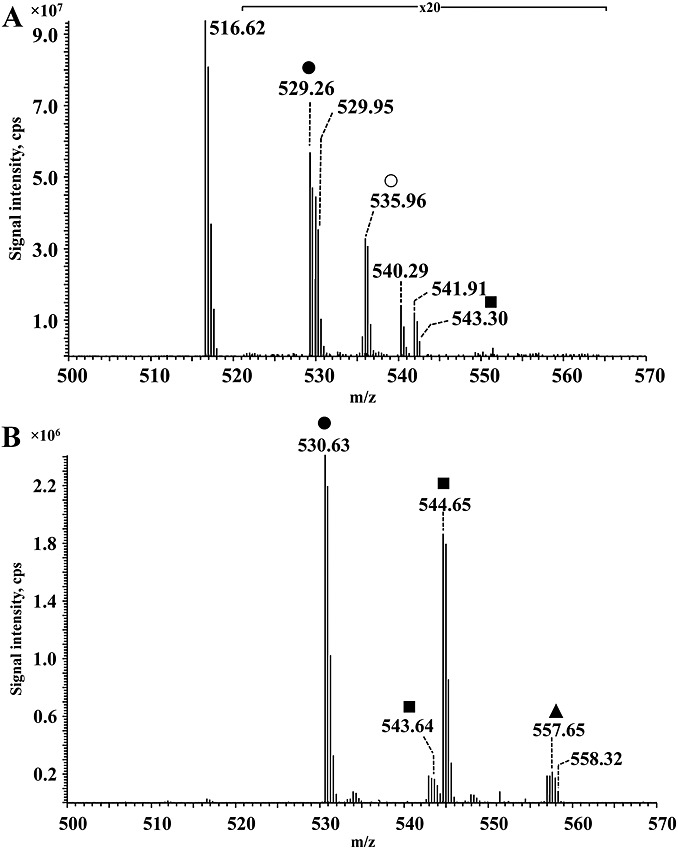
ESI-Orbitrap MS spectra of a peptide incubated with propanal before (A) and after (B) reduction withNaCNBH4. Peptide was incubated with 5% (v/v) propanal. Peptide and adducts are in triply charge state. (A) Unreduced, (B) reduced sample. Hexanal–peptide adducts were detected: • monomer;○ amide adduct; ▪ dimer; and ▲trimer.
Evaluation of potential membrane structure effects and possible biological effects of pyridinium-containing DPPE hexanal adduct
Because of the different structures of the herein discussed hexanal-DPPE adducts, the biological effects induced by these compounds may be different. However, such detailed investigations were beyond the scope of this paper: because the cyclic trimer adduct of hexanal and DPPE was the most abundant reaction product, it could be most easily purified by HPLC, which resulted in quantities sufficient for the cellular experiments. Therefore, only these products were investigated.
In order to investigate possible biological effects of alkanal adducts on the PE head group, a stable cyclic trimer adduct of hexanal and DPPE42 was purified by HPLC, and its concentration was determined using high resolution 1H NMR. The effect of the pyridine ring containing DPPE (m/z 936.5) on membrane curvature was determined using DiPoPE vesicles and differential scanning calorimetry (DSC) experiments. As shown in Fig. 9, the DSC profile of DiPoPE vesicles is characterized by a transition temperature (TH) at 43.9 °C in heating curves and 31.4 °C in cooling curves. Those peaks can be explained by the lipid phase shift from the lamellar (Lα) to the hexagonal (HII) phase. Addition of pyridine modified DPPE (Fig. 9, dotted line) altered the TH of DiPoPE vesicles. In both heating and cooling scans, the peak of DiPoPE TH disappeared. This effect is normally explained by the induction of a negative membrane curvature.43,44 Thus, the hydrophobic modification on the head group by the pyridine ring carrying long alkyl chains may be retained in the hydrophobic part leading to a macroscopic change of the lipid bilayer.31
Finally, in order to investigate possible pro-inflammatory effects of modified DPPE, which have previously been reported for some other PE head group modifications,45 isolated human primary monocytes and PBMCs were stimulated with pyridine ring containing PE, and induction of TNF-α expression and secretion was monitored. However, no increased induction of TNF-α expression was observed after cell stimulation by modified PE (Fig. S12, 13).
Discussion
Similar to proteins, lipid modifications also seem to play a major role in oxidative stress-related diseases.46 During lipid peroxidation, a large variety of products are formed, but only some of them were characterized so far.47 α,β-unsaturated aldehydes are commonly studied because of their high abundance and reactivity towards nucleophilic substrates.48 However, saturated aldehydes such as propanal, hexanal and nonanal are also produced during lipid peroxidation in large quantities.49 Reactivity of saturated aldehydes towards nucleophilic substrates in proteins were evaluated in several studies, and some possible modifications were described.29,34 However, less is known about the reactivity of these aldehdyes towards aminophospholipids, such as phosphatidylamines (PE) and phosphatidylserines.50 Thus, experiments were performed here in order to understand and elucidate the reactivity of saturated aldehydes with the primary amino group of DPPE. By combining high resolution Orbitrap mass spectrometry and MSn capabilities of a linear ion trap, it was possible to detect and identify several compounds arising from the reaction between DPPE and hexanal. High resolution and mass accuracy acquisition using Orbitrap mass analyzer allowed to identify the elemental composition of seven DPPE-hexanal adducts. In addition to the known Schiff base adducts and amide-linked DPPE derivatives, five new compounds were detected. Based on the elemental composition and ability to be reduced in the presence of NaCNBH4, new DPPE-hexanal adducts were assigned as dimeric and trimeric hexanal adducts and their hydrated forms. Further evaluation and confirmation of chemical structures was performed using sequential fragmentation of adducts in the linear ion trap. High sensitivity of LTQ mass analyzer allowed to perform up to MS7 tandem mass experiments, which resulted in the identification of linear hexanal-dimers and trimers as well as cyclic pyridine ring adducts on the DPPE head group.
Formation of dimeric and trimeric hexanal adducts on DPPE primary amino group can occur via two different mechanisms. Dimeric and trimeric hexanal derivatives can be formed already in solution at high hexanal concentrations. Alternatively, hexanal can form a Schiff base with DPPE primary amine followed by the addition of second hexanal via aldol condensation on the Cβ atom of the first molecule resulting in a branched dimer. Addition of the third hexanal moiety can occur via the same reaction, resulting in a linear branched trimer, which can be further cyclized to the stable pyridine ring adduct with three alkyl chains. Time- and concentration-dependent MS-based experiments were performed and indicated that consecutive addition of each hexanal moiety to the primary amine of DPPE head group via β-aldol-condensation occur if an excess of hexanal is used. Hydrated dimer and trimer identified in reaction mixtures can represent intermediates of adducts formation further confirming consecutive aldol-condensation mechanism. Additionally, using carbonyl group-specific derivatization, it was shown that only monomeric hexanal but not carbonyl-containing hexanal dimer or trimer were present both in pure hexanal solution as well as in the incubation mixtures with DPPE. Although the product yields are surely dependent on the reaction conditions, for instance, the solvent system, we studied exclusively the reactions in water because water is the only solvent with biological significance.
Furthermore, it was demonstrated that other alkanals (from propanal till decanal) can react with the primary amino group of DPPE in a similar manner forming monomeric Schiff base, amide-linked, dimer, trimer and their hydrated forms, thereby illustrating the versatility of the related reaction mechanism. However, formation of cyclic trimeric adducts was demonstrated only for hexanal and nonanal. Moreover, using mixtures of pentanal and hexanal incubated with DPPE, ‘mixed’ dimeric and trimeric adducts could be identified, i.e. the complexity of possible PE-alkanal derivatives was increased. Reactivity of alkanals towards other nucleophilic substrates was evaluated using model peptides containing lysine residues. Co-incubation of the peptide with different alkanals resulted in the formation of all previously described adducts in the case of propanal and butanal incubations. For alkanals with longer carbon chains such as pentanal and hexanal only monomeric Schiff base, amide-linked adduct and trimeric adducts were detected. No adducts were observed after incubation of the peptide with octanal and nonanal probably because increased hydrophobicity of the long chain alkanals make it less assessable for hydrophobic peptides. Interestingly, signals of amide type adduct were more intense in the case of peptide when compared to DPPE incubations. The pKa values for lysine (10.5) and phosphatidyethanolamine (9.6)51–53 amino groups are not significantly different. Thus, differences in the reactivities of the peptide and DPPE towards alkanals with different lengths of the carbon chains are presumably primarily caused by the differences in the different hydrophobicities of the peptide and the lipid substrate.
The alkanal-modified PE species described are also of potential relevance in several biological or chemical processes. The high concentration of alkanals used in these experiments is most likely not comparable with any human pathologies. However, such adducts might be formed during the heating process of cooking, pasteurization of milk and treatment of fish flesh.26–28 Biological relevance of modified DPPE was studied using a purified pyridine adduct. It was previously demonstrated that hydrophobic modifications of PE head group might modify the lipid distribution in the membrane. Hydrophobic parts of the head groups could be retained in the lipid phase leading to a structural change of the surrounding lipids and the total membrane curvature. Using DSC, a shift of the TH of DiPoPE vesicle spiked with DPPE modified by a cyclic trimer was observed. The lowering of the TH could indicate that modified PE can induce a negative membrane curvature (i.e. concave shape) in PL bilayers. Such structural changes into the membrane could decrease the microfluidity with a hardening of the membrane.54 Severe curvature may also lead to a loss of stability and permeability,55 and in some cases, a loss of macroscopic structure was observed.56,57
The ability of the pyridine ring adduct of hexanal with DPPE to induce expression of TNF-α, one of the main cytokines of acute inflammatory response, was evaluated using human monocytes and PBMCs, but no increased release of TNF-α was observed. This result can be explained by the fact that modified PE may not directly be involved in the inflammation process but that a second mediator is required.58 It is also known that oxidized lipids can induce negative effects and even suppress the specific pro-inflammatory response in human monocytes.59,60 Evaluation of other possible biological effects will be performed in additional studies with the particular focus on the role of those PE modifications on the living organism.
Conclusions
Using high resolution Orbitrap MS and MSn on a linear ion trap, seven different adducts of DPPE and hexanal were identified and structurally characterized. Two previously described products, such as Schiff base and amide-linkage adducts, were detectable. Additionally, dimeric, linear and cyclic trimeric adducts and their hydrated forms were identified for alkanals with different lengths of the carbon chain. MS-based kinetics and concentration-dependent experiments as well as carbonyl-specific derivatization allowed to propose consecutive addition mechanisms of stable cyclic trimer formation via aldol condensation.
Acknowledgments
Financial support from the European Regional Development Fund (ERDF, European Union and Free State Saxony), the ‘Bundesministerium für Bildung and Forschung’ (BMBF) and the Leipzig Interdisciplinary Research Cluster of Genetic Factors, Clinical Phenotypes and Environment (LIFE Center, Universität Leipzig) is gratefully acknowledged. This publication is supported by LIFE – Leipzig Research Center for Civilization Diseases, Universität Leipzig. This project was funded by means of the European Social Fund and the Free State of Saxony.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
Supporting info item
References
- 1.Marionnet C, Pierrard C, Lejeune F, Sok J, Thomas M, Bernerd F. Different oxidative stress response in keratinocytes and fibroblasts of reconstructed skin exposed to non extreme daily-ultraviolet radiation. PloS one. 2010;5:e12059. doi: 10.1371/journal.pone.0012059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romieu I, Castro-Giner F, Kunzli N, Sunyer J. Air pollution, oxidative stress and dietary supplementation: a review. Eur. Respir. J. 2008;31:179–197. doi: 10.1183/09031936.00128106. [DOI] [PubMed] [Google Scholar]
- 3.Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int. J. Radiat. Biol. 1994;65:27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal R. Smoking, oxidative stress and inflammation: impact on resting energy expenditure in diabetic nephropathy. BMC Nephrol. 2005;6:13. doi: 10.1186/1471-2369-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gornati R, Colombo G, Clerici M, Rossi F, Gagliano N, Riva C, Colombo R, Dalle-Donne I, Bernardini G, Milzani A. Protein carbonylation in human endothelial cells exposed to cigarette smoke extract. Toxicol. Lett. 2013;218:118–128. doi: 10.1016/j.toxlet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Colombo G, Dalle-Donne I, Orioli M, Giustarini D, Rossi R, Clerici M, Regazzoni L, Aldini G, Milzani A, Butterfield DA, Gagliano N. Oxidative damage in human gingival fibroblasts exposed to cigarette smoke. Free Radic Biol Med. 2012;52:1584–1596. doi: 10.1016/j.freeradbiomed.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Alfadda AA, Sallam RM. Reactive oxygen species in health and disease. J. Biomed. Biotechnol. 2012:936486. doi: 10.1155/2012/936486. DOI: 10.1155/2012/936486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiller J, Fuchs B, Arnhold J, Arnold K. Contribution of reactive oxygen species to cartilage degradation in rheumatic diseases: molecular pathways, diagnosis and potential therapeutic strategies. Curr. Med. Chem. 2003;10:2123–2145. doi: 10.2174/0929867033456828. [DOI] [PubMed] [Google Scholar]
- 9.Dizdaroglu M. Oxidatively induced DNA damage: mechanisms, repair and disease. Cancer Lett. 2012;327:26–47. doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Massey KA, Nicolaou A. Lipidomics of polyunsaturated-fatty-acid-derived oxygenated metabolites. Biochem. Soc. Trans. 2011;39:1240–1246. doi: 10.1042/BST0391240. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Garcia A, Rodriguez-Rocha H, Madayiputhiya N, Pappa A, Panayiotidis MI, Franco R. Biomarkers of protein oxidation in human disease. Curr. Med. Chem. 2012;12:681–697. doi: 10.2174/156652412800792543. [DOI] [PubMed] [Google Scholar]
- 12.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 13.Varcin M, Bentea E, Michotte Y, Sarre S. Oxidative stress in genetic mouse models of Parkinson's disease. Oxid. Med. Cell. Longev. 2012:624925. doi: 10.1155/2012/624925. DOI: 10.1155/2012/624925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y, Wang X. Antioxidant therapies for Alzheimer's disease. Oxid. Med. Cell. Longev. 2012:472932. doi: 10.1155/2012/472932. DOI: 10.1155/2012/472932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong BW, Meredith A, Lin D, McManus BM. The biological role of inflammation in atherosclerosis. Can. J. Cardiol. 2012;28:631–641. doi: 10.1016/j.cjca.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 16.Selvaraju V, Joshi M, Suresh S, Sanchez JA, Maulik N, Maulik G. Diabetes, oxidative stress, molecular mechanism, and cardiovascular disease--an overview. Toxicol. Mech. Methods. 2012;22:330–335. doi: 10.3109/15376516.2012.666648. [DOI] [PubMed] [Google Scholar]
- 17.Selman C, Blount JD, Nussey DH, Speakman JR. Oxidative damage, ageing, and life-history evolution: where now? Trends Ecol. Evol. 2012;27:570–577. doi: 10.1016/j.tree.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 2006;10:389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziobro A, Duchnowicz P, Mulik A, Koter-Michalak M, Broncel M. Oxidative damages in erythrocytes of patients with metabolic syndrome. Mol. Cell. Biochem. 2013;378:267–273. doi: 10.1007/s11010-013-1617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura T, Cho DH, Lipton SA. Redox regulation of protein misfolding, mitochondrial dysfunction, synaptic damage, and cell death in neurodegenerative diseases. Exp. Neurol. 2012;238:12–21. doi: 10.1016/j.expneurol.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niki E, Yoshida Y, Saito Y, Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. BiochemBiophys. Res. Commun. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 22.Perluigi M, Coccia R, Butterfield DA. 4-Hydroxy-2-nonenal, a reactive product of lipid peroxidation, and neurodegenerative diseases: a toxic combination illuminated by redox proteomics studies. Antioxid. Redox. Signal. 2012;17:1590–1609. doi: 10.1089/ars.2011.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szweda LI, Uchida K, Tsai L, Stadtman ER. Inactivation of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal. Selective modification of an active-site lysine. J. Biol. Chem. 1993;268:3342–3347. [PubMed] [Google Scholar]
- 24.Pickering AM, Davies KJ. Degradation of damaged proteins: the main function of the 20S proteasome. Prog. Mol. Biol. Transl. Sci. 2012;109:227–248. doi: 10.1016/B978-0-12-397863-9.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ajuyah AO, Fenton TW, Hardin RT, Sim JS. Measuring lipid oxidation volatiles in meats. J. Food Sci. 1993;58:270–273. [Google Scholar]
- 26.Elisia I, Kitts DD. Quantification of hexanal as an index of lipid oxidation in human milk and association with antioxidant components. J. Clin. Biochem. Nutr. 2011;49:147–152. doi: 10.3164/jcbn.10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pignoli G, Bou R, Rodriguez-Estrada MT, Decker EA. Suitability of saturated aldehydes as lipid oxidation markers in washed turkey meat. Meat Sci. 2009;83:412–416. doi: 10.1016/j.meatsci.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 28.Yasuhara A, Shibamoto T. Quantitative analysis of volatile aldehydes formed from various kinds of fish flesh during heat treatment. J. Agric. Food Chem. 1995;43:94–97. [Google Scholar]
- 29.Kato Y, Mori Y, Makino Y, Morimitsu Y, Hiroi S, Ishikawa T, Osawa T. Formation of N ε-(hexanonyl)lysine in protein exposed to lipid hydroperoxide: a plausible marker for lipid hydroperoxide-derived protein modification. J. Biol. Chem. 1999;274:20406–20414. doi: 10.1074/jbc.274.29.20406. [DOI] [PubMed] [Google Scholar]
- 30.Guo L, Chen Z, Amarnath V, Davies SS. Identification of novel bioactive aldehyde-modified phosphatidylethanolamines formed by lipid peroxidation. Free Radic. Biol. Med. 2012;53:1226–1238. doi: 10.1016/j.freeradbiomed.2012.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L, Chen Z, Cox BE, Amarnath V, Epand RF, Epand RM, Davies SS. Phosphatidylethanolamines modified by γ-Ketoaldehyde (γKA) induce endoplasmic reticulum stress and endothelial activation. J. Biol. Chem. 2011;286:18170–18180. doi: 10.1074/jbc.M110.213470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L, Davies SS. Bioactive aldehyde-modified phosphatidylethanolamines. Biochimie. 2013;95:74–78. doi: 10.1016/j.biochi.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Laird JM, Lu L, Roychowdhury S, Nagy LE, Zhou R, Crabb JW, Salomon RG. Isolevuglandins covalently modify phosphatidylethanolamines in vivo: detection and quantitative analysis of hydroxylactam adducts. Free Radic. Biol. Med. 2009;47:1539–1552. doi: 10.1016/j.freeradbiomed.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hisaka S, Kato Y, Kitamoto N, Yoshida A, Kubushiro Y, Naito M, Osawa T. Chemical and immunochemical identification of propanoyllysine derived from oxidized n-3 polyunsaturated fatty acid. Free Radic. Biol. Med. 2009;46:1463–1471. doi: 10.1016/j.freeradbiomed.2009.02.030. [DOI] [PubMed] [Google Scholar]
- 35.Zieseniss S, Zahler S, Müller I, Hermetter A, Engelmann B. Modified phosphatidylethanolamine as the active component of oxidized low density lipoprotein promoting platelet prothrombinase activity. J. Biol. Chem. 2001;276:19828–19835. doi: 10.1074/jbc.M007506200. [DOI] [PubMed] [Google Scholar]
- 36.Kato Y, Yoshida A, Naito M, Kawai Y, Tsuji K, Kitamura M, Kitamoto N, Osawa T. Identification and quantification of Nɛ-(Hexanoyl)lysine in human urine by liquid chromatography/tandem mass spectrometry. Free Radic. Biol. Chem. 2004;37:1864–1874. doi: 10.1016/j.freeradbiomed.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Rossol M, Meusch U, Pierer M, Kaltenhäuser S, Häntzschel H, Hauschildt S, Wagner U. Interaction between transmembrane TNF and TNFR1/2 mediates the activation of monocytes by contact with T cells. J. Immunol. 2007;179:4239–4248. doi: 10.4049/jimmunol.179.6.4239. [DOI] [PubMed] [Google Scholar]
- 38.Grujic S, Vasiljevic T, Lausevic M, Ast T. Study on the formation of an amoxicillin adduct with methanol using electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 2008;22:67–74. doi: 10.1002/rcm.3333. [DOI] [PubMed] [Google Scholar]
- 39.Kite GC, Stoneham CA, Veitch NC, Stein BK, Whitwell KE. Application of liquid chromatography-mass spectrometry to the investigation of poisoning by Oenanthecrocata. J. Chromatogr. B. 2006;838:63–70. doi: 10.1016/j.jchromb.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 40.Jirásko R, Holčapek M, Kolářová L, Baul TSB. Electrospray ionization-multistage tandem mass spectrometry of complex multitin organometallic compounds. J. Mass Spectrom. 2007;42:918–928. doi: 10.1002/jms.1229. [DOI] [PubMed] [Google Scholar]
- 41.Milic I, Hoffmann R, Fedorova M. Simultaneous detection of low and high molecular weight carbonylated compounds derived from lipid peroxidation by electrospray ionization-tandem mass spectrometry. Anal. Chem. 2012;85:156–162. doi: 10.1021/ac302356z. [DOI] [PubMed] [Google Scholar]
- 42.Nakanishi T, Suyama K. Formation of phosphatidyl 1-(2-hydroxyethyl)-2,3,5-trialkyl pyridinium on heating phosphatidyl ethanolamine with n-alkanal. Agr. Biol. Chem. 1974;38:1141–1147. [Google Scholar]
- 43.Bach D, Epand RF, Epand RM, Miller IR, Wachtel E. The oxidized form of cholesterol 3β-hydroxy-5-oxo-5,6-secocholestan-6-al induces structural and thermotropic changes in phospholipid membranes. Chem. Phys. Lipids. 2009;161:95–102. doi: 10.1016/j.chemphyslip.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Alves ID, Goasdoué N, Correia I, Aubry S, Galanth C, Sagan S, Lavielle S, Chassaing G. Membrane interaction and perturbation mechanisms induced by two cationic cell penetrating peptides with distinct charge distribution. Biochim. Biophys. Acta. 2008;1780:948–959. doi: 10.1016/j.bbagen.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Simões C, Silva AC, Domingues P, Laranjeira P, Paiva A, Domingues MR. Modified phosphatidylethanolamines induce different levels of cytokine expression in monocytes and dendritic cells. Chem. Phys. Lipids. 2013;175-176:57–64. doi: 10.1016/j.chemphyslip.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Dessì M, Noce A, Bertucci P, Manca di Villahermosa S, Zenobi R, Castagnola V, Addessi E, Di Daniele N. Atherosclerosis, dyslipidemia, and inflammation: the significant role of polyunsaturated fatty acids. ISRN Inflamm. 2013;12:191823. doi: 10.1155/2013/191823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosário M, Domingues M, Reis A, Domingues P. Mass spectrometry analysis of oxidized phospholipids. Chem. Phys. Lipids. 2008;156:1–12. doi: 10.1016/j.chemphyslip.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Spickett CM. The lipid peroxidation product 4-hydroxy-2-nonenal: advances in chemistry and analysis. Redox Biol. 2013;1:145–152. doi: 10.1016/j.redox.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaar V, Ste-Marie L, Montgomery JA. Striatal metabolism of hexanal, a lipid peroxidation product, in the rat. Metab. Brain Dis. 1999;2:71–82. doi: 10.1023/a:1020701612639. [DOI] [PubMed] [Google Scholar]
- 50.Hisaka S, Yamada N, Naito K, Osawa T. The immunological and chemical detection of N-(hexanoyl)phosphatidylethanolamine and N-(hexanoyl)phosphatidylserine in an oxidative model induced by carbon tetrachloride. BiochemBiophys Res Commun. 2010;4:631–636. doi: 10.1016/j.bbrc.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 51.Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agmon N, Gutman M. Bioenergetics: proton fronts on membranes. Nat. Chem. 2011;3:840–842. doi: 10.1038/nchem.1184. [DOI] [PubMed] [Google Scholar]
- 53.Tsui FC, Ojcius DM, Hubbell WL. The intrinsic pKa values for phosphatidylserine and phosphatidylethanolamine in phosphatidylcholine host bilayers. Biophys. J. 1986;49:459–468. doi: 10.1016/S0006-3495(86)83655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith PES, Brender JR, Ramamoorthy A. Induction of negative curvature as a mechanism of cell toxicity by amyloidogenic peptides: the case of islet amyloid polypeptide. J. Am. Chem. Soc. 2009;131:4470–4478. doi: 10.1021/ja809002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borst W, Visser NV, Kouptsova O, Visser AJWG. Oxidation of unsaturated phospholipids in membrane bilayer mixtures is accompanied by membrane fluidity changes. Biochim. Biophys. Acta. 2000;1487:61–73. doi: 10.1016/s1388-1981(00)00084-6. [DOI] [PubMed] [Google Scholar]
- 56.Lucy JA. Functional and structural aspects of biological membranes: a suggested structural rolefor vitamin E in the control of membrane permeability and stability. Ann. N. Y. Acad. Sci. 1972;203:4–11. doi: 10.1111/j.1749-6632.1972.tb27849.x. [DOI] [PubMed] [Google Scholar]
- 57.Kiefer CR, Snyder LM. Oxidation and erythrocyte senescence. Curr. Opin. Hematol. 2000;7:113–116. doi: 10.1097/00062752-200003000-00007. [DOI] [PubMed] [Google Scholar]
- 58.Uchida K. Redox-derived damage-associated molecular patterns: ligand function of lipid peroxidation adducts. Redox Biol. 2013;1:94–96. doi: 10.1016/j.redox.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kannan Y, Sundaram K, Narasimhulu CA, Parthasarathy S, Wewers MD. Oxidatively modified low density lipoprotein (LDL) inhibits TLR2 and TLR4 cytokine responses in human monocytes but not in macrophages. J. Biol. Chem. 2012;287:23479–23488. doi: 10.1074/jbc.M111.320960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamilton TA, Ma GP, Chisolm GM. Oxidized low density lipoprotein suppresses the expression of tumor necrosis factor-alpha mRNA in stimulated murine peritoneal macrophages. J. Immunol. 1990;144:2343–2350. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item


