Abstract
Assays based on Bioluminescence Resonance Energy Transfer (BRET) provide a sensitive and reliable means to monitor protein-protein interactions in live cells. BRET is the non-radiative transfer of energy from a 'donor' luciferase enzyme to an 'acceptor' fluorescent protein. In the most common configuration of this assay, the donor is Renilla reniformis luciferase and the acceptor is Yellow Fluorescent Protein (YFP). Because the efficiency of energy transfer is strongly distance-dependent, observation of the BRET phenomenon requires that the donor and acceptor be in close proximity. To test for an interaction between two proteins of interest in cultured mammalian cells, one protein is expressed as a fusion with luciferase and the second as a fusion with YFP. An interaction between the two proteins of interest may bring the donor and acceptor sufficiently close for energy transfer to occur. Compared to other techniques for investigating protein-protein interactions, the BRET assay is sensitive, requires little hands-on time and few reagents, and is able to detect interactions which are weak, transient, or dependent on the biochemical environment found within a live cell. It is therefore an ideal approach for confirming putative interactions suggested by yeast two-hybrid or mass spectrometry proteomics studies, and in addition it is well-suited for mapping interacting regions, assessing the effect of post-translational modifications on protein-protein interactions, and evaluating the impact of mutations identified in patient DNA.
Keywords: Cellular Biology, Issue 87, Protein-protein interactions, Bioluminescence Resonance Energy Transfer, Live cell, Transfection, Luciferase, Yellow Fluorescent Protein, Mutations
Introduction
Both classical linkage and next-generation sequencing analyses of human disorders are revealing the clinical relevance of proteins involved in a range of biological pathways. It is often the case that, prior to their identification in such studies, there has been little or no investigation of the biological role of these proteins. One fruitful avenue to begin exploring the biological function of a protein of interest is to identify which other proteins it interacts with in its physiological context. Characterizing molecular networks in this fashion provides insights into the biological pathways underlying the human phenotype.
The most frequently used large-scale screening approaches for identifying candidate interaction partners for proteins of interest are yeast two-hybrid screening1 and mass spectrometry-based proteomics2. These methods can be very successful in suggesting potential interacting proteins, but are vulnerable to false positive results. Therefore, confirmation of an interaction identified by yeast two-hybrid or mass spectrometry screening requires validation of the interaction using a second technique. Typically a co-immunoprecipitation or pull-down assay is used for this purpose3. One disadvantage of using such techniques for validation is the requirement for cell lysis, which destroys the intracellular conditions that may be essential for maintaining certain protein interactions. A second disadvantage is that weak or transient protein interactions may be disrupted during washing steps. Furthermore, these assays demand significant hands-on time, are limited in the number of samples that can be processed simultaneously, and often require time-consuming optimization of reagents and protocols.
To overcome some of the problems associated with co-immunoprecipitation experiments, several assays have been developed based on fluorescent and bioluminescent proteins that can be used in live cells. The first such assays were based on Fluorescence (or Förster) Resonance Energy Transfer (FRET), the non-radiative transfer of energy between two fluorescent proteins with overlapping emission and excitation spectra4. The efficiency of energy transfer is strongly distance-dependent, therefore observation of the FRET phenomenon requires that the donor and acceptor fluorophores be in close proximity. To test for an interaction between two proteins of interest, one protein is expressed as a fusion with the donor fluorophore (commonly cyan fluorescent protein; CFP) and the second as a fusion with the acceptor fluorophore (commonly yellow fluorescent protein; YFP). An interaction between the two proteins of interest may bring the donor and acceptor fluorophores sufficiently close for energy transfer to occur, which will result in a measurable increase in the emission of light from the YFP acceptor relative to the CFP donor. FRET has been successful in detecting protein-protein interactions in live cells4. The main drawback of using FRET for detecting protein-protein interactions is the requirement for external illumination for excitation of the donor fluorophore. External illumination results in high background in the emission signal, unwanted excitation of the acceptor, and photobleaching of both donor and acceptor fluorophores. These effects reduce the sensitivity of the assay for detecting protein-protein interactions.
A modification of the FRET assay which overcomes the problem of high background from external illumination is the Bioluminescence Resonance Energy Transfer (BRET) assay5,6. In the BRET system the donor fluorophore is replaced by a luciferase enzyme. Thus the energy for the excitation of the acceptor fluorophore is generated within the system by the oxidation of a luciferase substrate, rendering external illumination unnecessary. In the most common configuration of this assay, the donor is Renilla reniformis luciferase and the acceptor is YFP (for a discussion of alternative donor and acceptor proteins see Pfleger et al.5). Accordingly, in this system, a protein of interest is fused to luciferase and a potentially-interacting protein to YFP, or vice versa. The BRET assay requires the addition of coelenterazine as a substrate for luciferase. Because coelenterazine is cell-permeable, it is possible to perform BRET assays in live cells. However, native coelenterazine is unstable in aqueous solution, and the enzyme-independent breakdown of coelenterazine both reduces the concentration of substrate available for the assay and generates autoluminescence, which reduces the sensitivity of measurements of luciferase activity. The use of BRET in live cells has been facilitated by the development of protected coelenterazines, which are stable in aqueous solution but are cleaved by cytosolic esterases after diffusion across the cell membrane to generate active coelenterazine inside the cell7.
Following addition of substrate to cells expressing luciferase- and YFP-fusion proteins, energy transfer resulting from protein-protein interactions is quantified by monitoring emission from luciferase and YFP. Because protein interactions can be monitored directly in live cells in multi-well plates, the BRET assay constitutes a simple, scalable method for validating putative interactions that is cost- and time-efficient.
In addition to validating putative interactors identified in proteomic screening studies, the BRET system can also be used to test candidate interactors arising from prior biochemical and structural studies on the protein of interest. Once the existence of a protein-protein interaction has been established (either by using the BRET assay or by other techniques), there is potential for the BRET assay to be employed further to characterize the interaction. For example, the interacting regions can be mapped by generating truncated versions of the proteins, and the involvement of specific residues in the interaction can be demonstrated by creating point mutations. Furthermore, the modulatory effect of posttranslational modifications or small molecules (such as drugs or ligands) on protein-protein interactions can be investigated8-10.
The BRET assay also has great potential for investigating mutations identified in patient DNA. In cases where a causative role for a mutation has been established, studying the effect of the mutation on protein-protein interactions using BRET may reveal more about the molecular etiology of the phenotype11. Since the advent of next-generation sequencing methodologies, it is increasingly common for several potentially-damaging mutations to be identified within an individual, in which case it is unclear which are relevant to the phenotype12. In this situation the BRET assay may be valuable in evaluating the impact of mutations on protein function and hence their relevance to the disorder.
Protocol
1. Creation of Plasmids
Subclone the cDNAs for each protein of interest into both the pLuc and pYFP vectors, using standard molecular biology techniques (Figure 1). For detailed protocols see Green et al13.
Sequence all constructs to verify that the proteins of interest are in frame with the Luc/YFP sequence with no intervening stop codons.
Perform functional assays as desired to confirm that the fusion proteins retain biological activity. In the case presented here the subcellular localization of YFP fusion proteins was ascertained by fluorescence microscopy.
Make expression plasmids for appropriate Luc and YFP control proteins by engineering the relevant targeting signals into the pLuc and pYFP expression vectors. In the case presented here, generate nuclear-targeted Luc and YFP constructs by inserting a nuclear localization signal into the pLuc and pYFP vectors.
Make a positive control construct in which Luc and YFP are fused into a single polypeptide. In the case presented here, generate a positive control in which the YFP coding sequence is inserted into the pLuc vector.
Select a neutral filler plasmid to be used to equalize the mass of DNA in transfection mixes. Use a plasmid that has no eukaryotic promoter and will therefore be transcriptionally inactive in mammalian cells, such as a bacterial cloning vector.
2. Preparation of DNA Mixes
Estimate the concentration of all the plasmids described in Section 1 based on absorbance at 260 nm (1 absorbance unit is equivalent to 50 µg/ml of DNA). Determine the molecular mass of each plasmid by multiplying the number of base pairs by 650 Da. Use the concentration and molecular mass to calculate the molar concentration of each plasmid preparation. Dilute the plasmid DNA preparations to a concentration of 36 nM. These will be the working stocks that will be used to prepare the DNA mixes for transfection.
Prepare a control DNA mix containing 1,800 ng of filler plasmid in a final volume of 20 µl of water.
Prepare a control DNA mix containing 5 µl of the pLuc-control construct. Add filler plasmid to bring the total DNA mass to 1,800 ng. Add water to bring the final volume to 20 µl.
Prepare a control DNA mix containing 5 µl of pLuc-control construct and 5 µl of pYFP-control construct. Add filler plasmid to bring the total DNA mass to 1,800 ng. Add water to bring the final volume to 20 µl.
Prepare a control DNA mix containing 5 µl of positive control construct. Add filler plasmid to bring the total DNA mass to 1,800 ng. Add water to bring the final volume to 20 µl.
Prepare the following DNA mixes to test for homodimerization of a protein of interest, X. For each DNA mix combine 5 µl of the relevant pLuc construct and 5 µl of the pYFP construct. Add filler plasmid to bring the total DNA mass to 1,800 ng. Add water to bring the final volume to 20 µl. A) pLuc-control and pYFP-X B) pLuc-X and pYFP-control C) pLuc-X and pYFP-X
Prepare the following DNA mixes to test for an interaction between a pair of proteins of interest, X and Y. For each DNA mix combine 5 µl of the relevant pLuc construct and 5 µl of the pYFP construct. Add filler plasmid to bring the total DNA mass to 1,800 ng. Add water to bring the final volume to 20 µl. A) pLuc-control and pYFP-X B) pLuc-X and pYFP-control C) pLuc-control and pYFP-Y D) pLuc-Y and pYFP-control E) pLuc-X and pYFP-Y F) pLuc-Y and pYFP-X
3. Transfection
Harvest subconfluent HEK293 cells from a 75 cm2 flask. Dilute 10% of the total cells into 13 ml of culture medium. Dispense 130 µl of cell suspension into each well of a white clear bottomed 96-well tissue culture plate. Culture cells for 24 hr.
Calculate the number of wells to be transfected by multiplying the number of DNA mixes by 3. Bring serum-free culture medium to room temperature. Prepare a master mix containing 6.3 µl of serum-free medium and 0.18 µl transfection reagent per well. Mix by vortexing and incubate at room temperature for 5 min.
Prepare transfection mixes by adding 2 µl of DNA mix to 20 µl of serum-free medium/transfection reagent master mix. Do not vortex. Incubate at room temperature for 10 min.
Transfect three wells with each transfection mix dispensing 6.5 µl of transfection mix per well. Culture the cells for a further 36-48 hr.
4. Measurement of BRET Signal
Dissolve live-cell luciferase substrate at 34 mg/ml in DMSO by vortexing.
Dilute reconstituted live-cell luciferase substrate at 1:1,000 in substrate dilution medium pre-warmed to 37 °C. Allow 50 µl of substrate dilution medium per well. Vortex to mix. A precipitate may form, but will not interfere with the assay.
Aspirate the culture medium from the 96-well plate. Dispense 50 µl of diluted live-cell luciferase substrate into each well. Culture cells for at least 2 hr (up to 24 hr).
Remove the lid from the 96-well plate and incubate the plate for 10 min at room temperature inside the luminometer.
Measure emission from Luc and YFP one well at a time. Measure emission from Luc using a filter blocking wavelengths longer than 470 nm. Measure emission from YFP using a 500-600 nm band-pass filter. Integrate emission signals over 10 sec.
5. Data Analysis
Average the Luc emission readings from the 3 wells that received only the filler plasmid. Subtract this value from all other Luc emission readings to produce the background-subtracted Luc emission values.
Average the YFP emission readings from the 3 wells that received only the filler plasmid. Subtract this value from all other YFP emission readings to produce the background-subtracted YFP emission values.
For each well, divide the background-subtracted YFP emission value by the background-subtracted Luc emission value to give the uncorrected BRET ratio.
Average the uncorrected BRET ratios from the three wells that were transfected with only the pLuc-control plasmid. Subtract this value from all the other uncorrected BRET ratios to give the corrected BRET ratios.
For each of the remaining sets of corrected BRET ratios, average the values from the three transfected wells to obtain a final BRET ratio.
Representative Results
The principle of the BRET assay is illustrated in Figure 2. The assay setup used throughout the experiments presented here is depicted in Figure 3. The detection of a strong BRET signal from cells transfected with a luciferase-YFP fusion protein confirmed that energy transfer was observable in this experimental setup (Figure 4).
Our research focuses on the role of the FOXP family of transcriptional repressors in brain development. Heterozygous mutations affecting FOXP2 lead to a rare form of speech and language disorder, of which the most prominent feature is developmental verbal dyspraxia -difficulty with producing the complex sequences of orofocial muscle movements required for speech14-16. FOXP1 mutations have been reported in patients with autism spectrum disorders and intellectual disability accompanied by severe speech and language problems17-20. The interaction of FOXPs with other proteins is being investigated to gain insights into the molecular mechanisms relevant to the role of these proteins in brain development.
FOXP2 is known to form homo- or heterodimers with other FOXP family members21, therefore FOXP2 homodimerization was used to validate the BRET assay (Figure 4). To exclude the possibility that interaction of FOXP2 fusion proteins in this assay was due simply to protein overexpression, the specificity of the interaction was demonstrated by introducing a mutation into the leucine zipper dimerization domain of FOXP2 (in-frame deletion of residue E400), which is known to disrupt homo- and heterodimerization21 (Figure 5A). When Luc-FOXP2.DE400 and YFP-FOXP2 fusion proteins were co-expressed, a reduction in the BRET signal was observed compared to when Luc-FOXP2 and YFP-FOXP2 were co-expressed (Figure 5B). This signal reduction was not due to an alteration in the subcellular localization of the mutant protein (Figure 5C) or to a difference in expression levels as assessed by western blotting (data not shown). The signal reduction was also observed when Luc-FOXP2 was expressed with YFP-FOXP2.DE400 (data not shown). Thus BRET is effective in detecting protein homodimerization, and the interaction of proteins remains specific when these proteins are overexpressed as luciferase- and YFP-fusions. Furthermore, the use of BRET in combination with targeted mutation of proteins has the potential to identify individual residues involved in protein-protein interactions.
To assess the effectiveness of the BRET assay in detecting heterotypic interactions, the interaction of FOXP2 with FOXP1 was tested (Figure 6). A BRET signal was observed in both possible configurations of the assay (i.e. with FOXP2 as the donor or as the acceptor fusion protein). Therefore the BRET assay is able to detect both homo- and heterotypic interactions. In addition, the suitability of the BRET assay for detecting interaction of FOXP2 with non-FOXP proteins was tested by examining the interaction with CtBP1, a transcriptional co-repressor and known FOXP2 interaction partner21. The interaction between CtBP1 and FOXP2 was originally suggested by a yeast two-hybrid screen and was subsequently validated by co-immunoprecipitation experiments21. The FOXP2-CtBP1 interaction was detected by the BRET assay when FOXP2 was the donor fusion protein and CtBP1 was the acceptor, but not in the reverse configuration (Figure 7A). Such outcomes are to be expected with the BRET system (see Discussion section). The interaction between FOXP2 and CtBP1 was observed even though only a minor proportion of CtBP1 was localized to the nucleus (Figure 7B), highlighting the sensitivity of this assay. Thus the BRET assay is useful for the validation of putative interactions identified through yeast two-hybrid screening.
Finally, the BRET assay was employed to examine the effects of pathogenic mutations in FOXP2 on protein-protein interactions. Two point mutations in FOXP2 have been reported to cause a rare autosomal dominant disorder affecting speech and language (Figure 8A). The R553H missense mutation was discovered through a linkage study in a large family in which half the members were affected by the disorder14. The R328X nonsense mutation was discovered through targeted sequencing of the FOXP2 locus in probands with a diagnosis of developmental verbal dyspraxia22. The ability of the mutant proteins to dimerize with wild-type FOXP2 was assessed using the BRET assay. The results using wild-type FOXP2 as a donor and mutant FOXP2 as the acceptor are shown in Figure 8B. The same results were observed using mutant FOXP2 as the donor and wild-type as the acceptor (data not shown). The R553H mutation, which is within the DNA-binding domain of FOXP2, did not affect the ability of the mutant protein to dimerize with the wild-type. The pathogenic mechanism of this mutation may therefore include a dominant negative effect resulting from dimerization of the mutant with the normal protein23. The R328X mutation introduces a premature stop codon, with the result that the encoded protein lacks the leucine zipper dimerization domain and the DNA-binding domain, and shows some cytoplasmic mislocalization (Figure 8C). Consistent with the absence of the dimerization domain and mislocalization to the cytoplasm, the truncated protein shows greatly reduced interaction with wild-type FOXP2. This mutation is therefore likely to be pathogenic due to haploinsufficiency. Thus the BRET assay can provide insight into the biological effects of mutations discovered in patients.
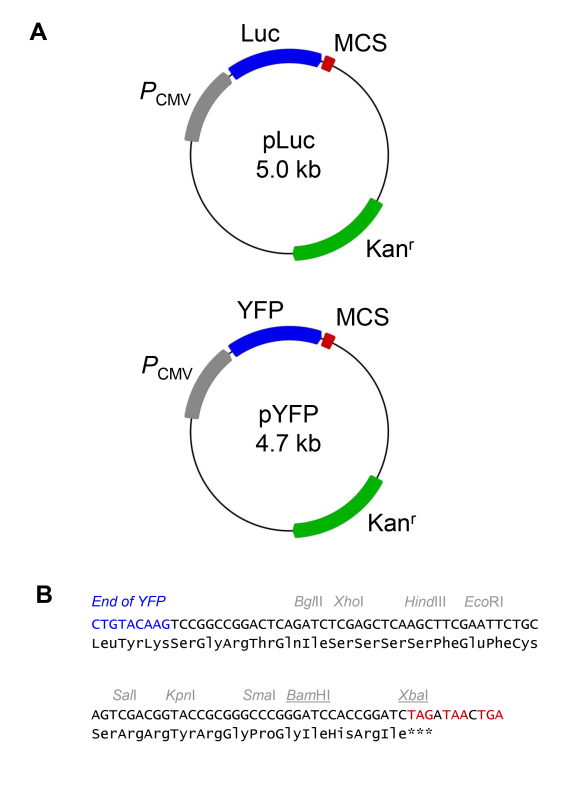 Figure 1. Vectors for expression of YFP- and luciferase-fusion proteins. A) Circle maps of the pYFP and pLuc vectors. The pLuc vector is used for expression of luciferase fusion proteins as BRET donors. The pYFP vector is used for expression of YFP fusion proteins as BRET acceptors. Vectors have a CMV promoter (PCMV) for high-level expression in mammalian cells, a multiple cloning site (MCS) for insertion of cDNAs for proteins of interest, and a kanamycin resistance gene (Kanr) and bacterial replication origin for propagation of the plasmid in bacteria. B) Multiple cloning site of the pLuc and pYFP vectors. The end of the YFP coding sequence is shown in blue. Selected restriction sites are annotated. Note that the XbaI site is blocked by overlapping dam methylation in standard strains of E. coli. cDNAs coding for proteins of interest should be cloned in-frame with the amino acid sequence shown. In the experiments described here, inserts were cloned into the BamHI and XbaI restriction sites (underlined). Stop codons have been removed from the end of the luciferase and YFP coding sequences to provide an open reading frame encompassing the luciferase/YFP and subcloned cDNA of interest. Stop codons are present at the end of the multiple cloning site in all three reading frames (shown in red), therefore it is not essential to include a stop codon in the inserted cDNA.
Figure 1. Vectors for expression of YFP- and luciferase-fusion proteins. A) Circle maps of the pYFP and pLuc vectors. The pLuc vector is used for expression of luciferase fusion proteins as BRET donors. The pYFP vector is used for expression of YFP fusion proteins as BRET acceptors. Vectors have a CMV promoter (PCMV) for high-level expression in mammalian cells, a multiple cloning site (MCS) for insertion of cDNAs for proteins of interest, and a kanamycin resistance gene (Kanr) and bacterial replication origin for propagation of the plasmid in bacteria. B) Multiple cloning site of the pLuc and pYFP vectors. The end of the YFP coding sequence is shown in blue. Selected restriction sites are annotated. Note that the XbaI site is blocked by overlapping dam methylation in standard strains of E. coli. cDNAs coding for proteins of interest should be cloned in-frame with the amino acid sequence shown. In the experiments described here, inserts were cloned into the BamHI and XbaI restriction sites (underlined). Stop codons have been removed from the end of the luciferase and YFP coding sequences to provide an open reading frame encompassing the luciferase/YFP and subcloned cDNA of interest. Stop codons are present at the end of the multiple cloning site in all three reading frames (shown in red), therefore it is not essential to include a stop codon in the inserted cDNA.
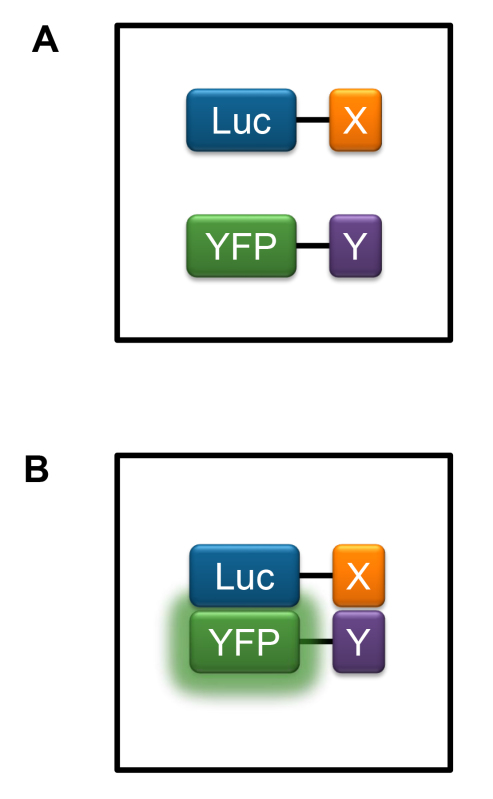 Figure 2. Principle of the BRET system. Proteins of interest, X and Y, are fused to a donor luciferase enzyme (Luc, peak emission 475 nm) and an acceptor fluorescent protein (YFP, peak emission 530 nm), respectively. Fusion proteins are expressed in cultured mammalian cells and following addition of a cell-permeable luciferase substrate, emission from Luc is measured using a filter blocking wavelengths longer than 470 nm and emission from YFP is measured using a 500-600 nm band-pass filter. A) No interaction between protein X and protein Y. Energy transfer from the Luc to YFP does not occur. B) Interaction between protein X and protein Y. Resonance energy transfer occurs from Luc to YFP causing an increase in the ratio of emission measured using the 500-600 nm band-pass filter compared to the filter blocking wavelengths longer than 470 nm.
Figure 2. Principle of the BRET system. Proteins of interest, X and Y, are fused to a donor luciferase enzyme (Luc, peak emission 475 nm) and an acceptor fluorescent protein (YFP, peak emission 530 nm), respectively. Fusion proteins are expressed in cultured mammalian cells and following addition of a cell-permeable luciferase substrate, emission from Luc is measured using a filter blocking wavelengths longer than 470 nm and emission from YFP is measured using a 500-600 nm band-pass filter. A) No interaction between protein X and protein Y. Energy transfer from the Luc to YFP does not occur. B) Interaction between protein X and protein Y. Resonance energy transfer occurs from Luc to YFP causing an increase in the ratio of emission measured using the 500-600 nm band-pass filter compared to the filter blocking wavelengths longer than 470 nm.
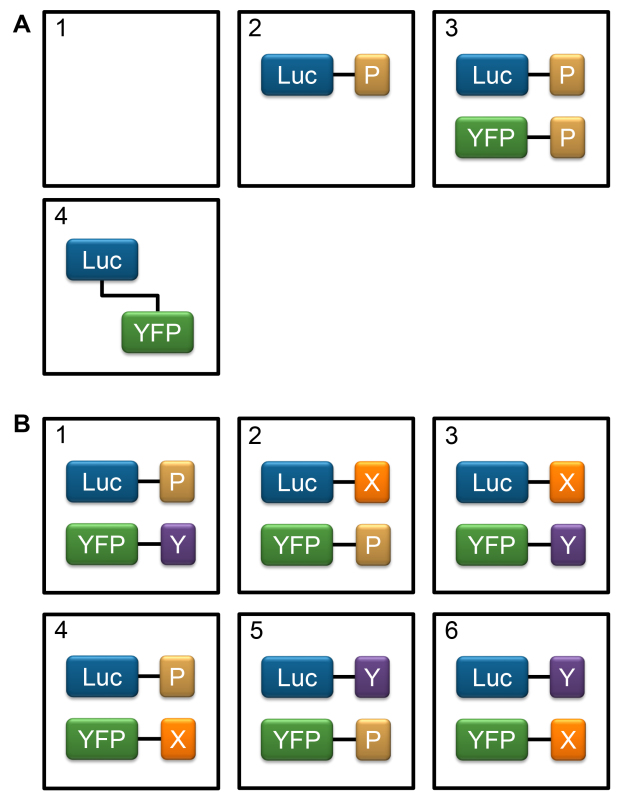 Figure 3. Assay setup. The assay setup to test for an interaction between two proteins of interest, X and Y. Proteins of interest are fused to luciferase (Luc) as the BRET donor and yellow fluorescent protein (YFP) as the BRET acceptor. In the control proteins, Luc and YFP are fused to a peptide containing a targeting signal for the relevant subcellular compartment (P), if appropriate. A) Controls to be included in each plate. 1) Mock-transfection control to establish background levels of luminescence and fluorescence resulting from luminometer noise and enzyme-independent breakdown of substrate; 2) pLuc-control only transfection to establish the proportion of luciferase emission which is detected within the emission range of YFP in the absence of a BRET acceptor; 3) Transfection of pLuc-control and pYFP-control to establish the baseline BRET signal resulting from Luc-YFP interaction; 4) Transfection of Luc-YFP fusion as a positive control to ensure that a BRET signal can be measured. B) Set of experimental conditions to test for an interaction between two proteins of interest, X and Y. 1, 2, 4, 5) Controls for non-specific interaction between the proteins of interest and Luc/YFP; 3) Test condition with X as the donor and Y as the acceptor; 6) Test condition with Y as the donor and X as the acceptor.
Figure 3. Assay setup. The assay setup to test for an interaction between two proteins of interest, X and Y. Proteins of interest are fused to luciferase (Luc) as the BRET donor and yellow fluorescent protein (YFP) as the BRET acceptor. In the control proteins, Luc and YFP are fused to a peptide containing a targeting signal for the relevant subcellular compartment (P), if appropriate. A) Controls to be included in each plate. 1) Mock-transfection control to establish background levels of luminescence and fluorescence resulting from luminometer noise and enzyme-independent breakdown of substrate; 2) pLuc-control only transfection to establish the proportion of luciferase emission which is detected within the emission range of YFP in the absence of a BRET acceptor; 3) Transfection of pLuc-control and pYFP-control to establish the baseline BRET signal resulting from Luc-YFP interaction; 4) Transfection of Luc-YFP fusion as a positive control to ensure that a BRET signal can be measured. B) Set of experimental conditions to test for an interaction between two proteins of interest, X and Y. 1, 2, 4, 5) Controls for non-specific interaction between the proteins of interest and Luc/YFP; 3) Test condition with X as the donor and Y as the acceptor; 6) Test condition with Y as the donor and X as the acceptor.
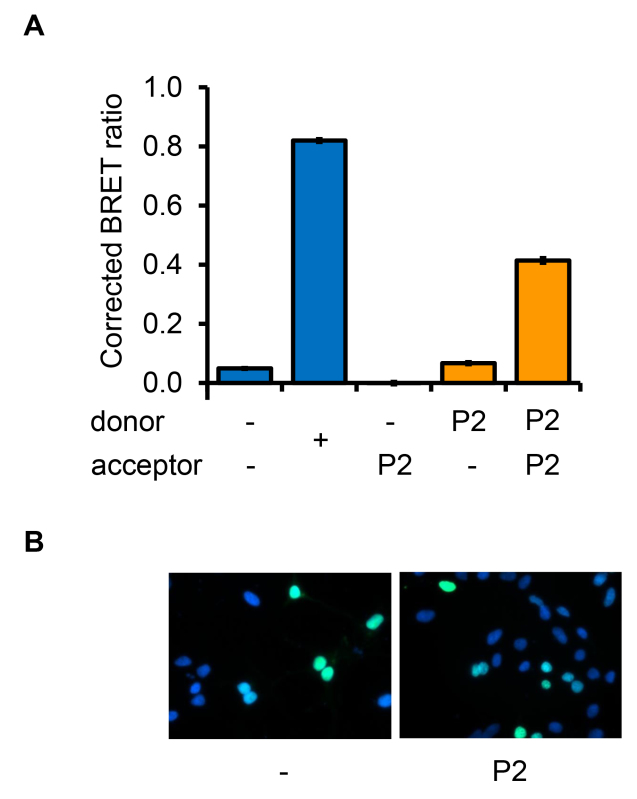 Figure 4. Detection of FOXP2 homodimerization. A) To validate the BRET assay for the detection of FOXP2 homodimers, HEK293 cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either FOXP2 (P2) or a nuclear localization signal (-). The nuclear-targeted luciferase and YFP proteins serve as negative controls. As a positive control for detection of a BRET signal, cells were also transfected with a construct for expression of a luciferase-YFP fusion protein (+). B) Fluorescence microscopy images of cells transfected with YFP fused to a nuclear localization signal (-) or with YFP fused to FOXP2 (P2).
Figure 4. Detection of FOXP2 homodimerization. A) To validate the BRET assay for the detection of FOXP2 homodimers, HEK293 cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either FOXP2 (P2) or a nuclear localization signal (-). The nuclear-targeted luciferase and YFP proteins serve as negative controls. As a positive control for detection of a BRET signal, cells were also transfected with a construct for expression of a luciferase-YFP fusion protein (+). B) Fluorescence microscopy images of cells transfected with YFP fused to a nuclear localization signal (-) or with YFP fused to FOXP2 (P2).
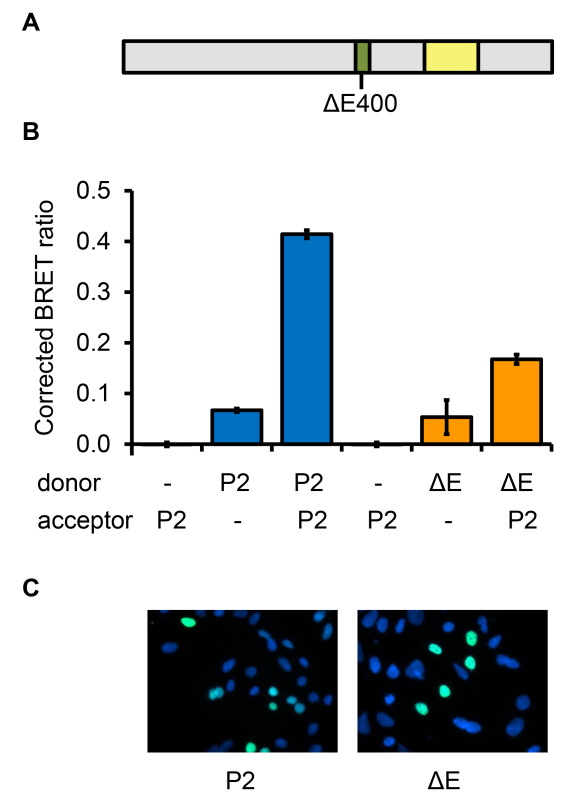 Figure 5. Use of dimerization-deficient FOXP2 to demonstrate assay specificity. A) Schematic diagram of the FOXP2 protein showing the position of the DE400 mutation, which disrupts dimerization. The leucine zipper dimerization domain is shown in green and the DNA-binding domain in yellow. B) To test whether overexpression of proteins may lead to false-positive results in the BRET assay, the specificity of the assay was assessed using the FOXP2.DE400 variant. Cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to FOXP2 (P2), FOXP2.DE400 (DE) or a nuclear localization signal (-). C) Fluorescence microscopy images of cells transfected with YFP fused to FOXP2 (P2) or to FOXP2.DE400 (DE).
Figure 5. Use of dimerization-deficient FOXP2 to demonstrate assay specificity. A) Schematic diagram of the FOXP2 protein showing the position of the DE400 mutation, which disrupts dimerization. The leucine zipper dimerization domain is shown in green and the DNA-binding domain in yellow. B) To test whether overexpression of proteins may lead to false-positive results in the BRET assay, the specificity of the assay was assessed using the FOXP2.DE400 variant. Cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to FOXP2 (P2), FOXP2.DE400 (DE) or a nuclear localization signal (-). C) Fluorescence microscopy images of cells transfected with YFP fused to FOXP2 (P2) or to FOXP2.DE400 (DE).
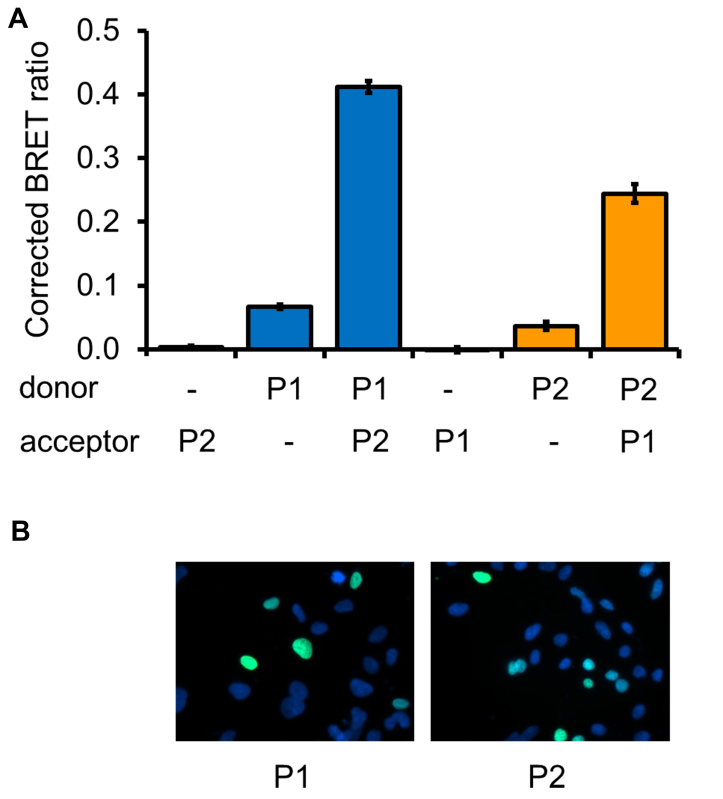 Figure 6. Detection of FOXP1-FOXP2 heterodimerization. A) To validate the BRET assay for the detection of heterodimerization between the homologous proteins FOXP2 and FOXP1, cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either FOXP2 (P2), FOXP1 (P1) or a nuclear localization signal (-). B) Fluorescence microscopy images of cells transfected with YFP fused to FOXP1 (P1) or to FOXP2 (P2).
Figure 6. Detection of FOXP1-FOXP2 heterodimerization. A) To validate the BRET assay for the detection of heterodimerization between the homologous proteins FOXP2 and FOXP1, cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either FOXP2 (P2), FOXP1 (P1) or a nuclear localization signal (-). B) Fluorescence microscopy images of cells transfected with YFP fused to FOXP1 (P1) or to FOXP2 (P2).
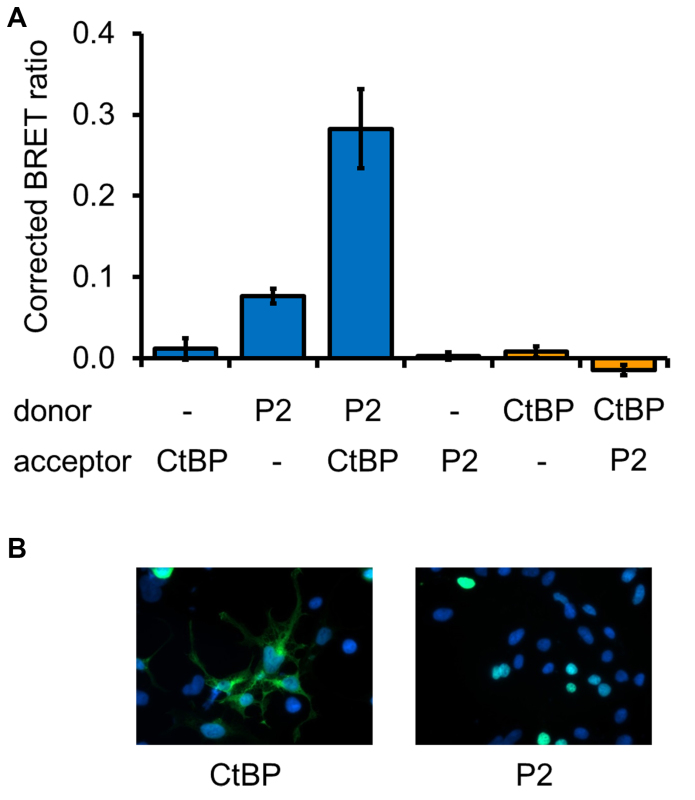 Figure 7. Validation of putative FOXP2 interactions from yeast two-hybrid screening. A) CtBP1 was identified as a candidate FOXP2-interacting protein by yeast two-hybrid screening. To test the suitability of the BRET assay for validation of putative interactions from such screens, cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either FOXP2 (P2), CtBP1 (CtBP) or a nuclear localization signal (-). B) Fluorescence microscopy images of cells transfected with YFP fused to CtBP1 (CtBP) or to FOXP2 (P2).
Figure 7. Validation of putative FOXP2 interactions from yeast two-hybrid screening. A) CtBP1 was identified as a candidate FOXP2-interacting protein by yeast two-hybrid screening. To test the suitability of the BRET assay for validation of putative interactions from such screens, cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either FOXP2 (P2), CtBP1 (CtBP) or a nuclear localization signal (-). B) Fluorescence microscopy images of cells transfected with YFP fused to CtBP1 (CtBP) or to FOXP2 (P2).
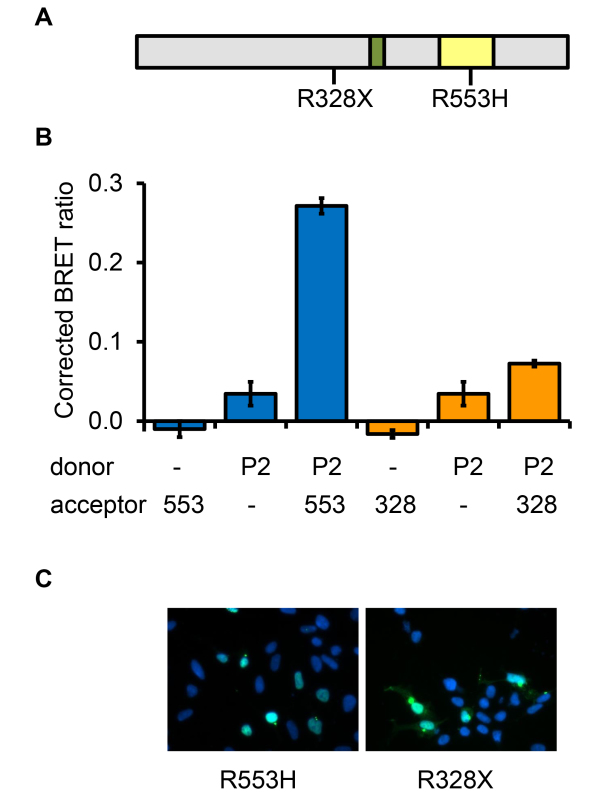 Figure 8. Evaluating the effects of pathogenic FOXP2 mutations on protein-protein interactions. A) Schematic diagram of the FOXP2 protein showing the positions of the R553H and R328X mutations which cause a rare form of speech and language disorder. The leucine zipper dimerization domain is shown in green and the DNA-binding domain in yellow. B) To assess the utility of the BRET assay for evaluating the impact of pathological mutations on protein-protein interactions, the effects of the R553H and R328X mutations on the ability of the mutant FOXP2 proteins to dimerize with normal FOXP2 were examined. Cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either normal FOXP2 (P2), FOXP2.R553H (553), FOXP2.R328X (328), or a nuclear localization signal (-). C) Fluorescence microscopy images of cells transfected with YFP fused to FOXP2.R553H (R553H) or to FOXP2.R328X (R328X). In this image FOXP2.R328X is predominantly nuclear, but in other cells within the population shows a more cytoplasmic distribution.
Figure 8. Evaluating the effects of pathogenic FOXP2 mutations on protein-protein interactions. A) Schematic diagram of the FOXP2 protein showing the positions of the R553H and R328X mutations which cause a rare form of speech and language disorder. The leucine zipper dimerization domain is shown in green and the DNA-binding domain in yellow. B) To assess the utility of the BRET assay for evaluating the impact of pathological mutations on protein-protein interactions, the effects of the R553H and R328X mutations on the ability of the mutant FOXP2 proteins to dimerize with normal FOXP2 were examined. Cells were transfected with constructs for expression of luciferase (donor) or YFP (acceptor) fused to either normal FOXP2 (P2), FOXP2.R553H (553), FOXP2.R328X (328), or a nuclear localization signal (-). C) Fluorescence microscopy images of cells transfected with YFP fused to FOXP2.R553H (R553H) or to FOXP2.R328X (R328X). In this image FOXP2.R328X is predominantly nuclear, but in other cells within the population shows a more cytoplasmic distribution.
Discussion
The design of the fusion protein expression constructs is a critical step in setting up the BRET assay. In the experiments presented here, the proteins of interest were fused to the C-terminus of luciferase or YFP. It is also possible, and may be necessary, to fuse proteins to the N-terminus of luciferase/YFP. For some proteins, fusions may only be accepted at either the N- or C-terminus in order to avoid disruption of protein structure and function. Furthermore, for transmembrane proteins in which the N and C termini reside in different subcellular compartments, the luciferase/YFP protein must be fused to the terminus which is in the same compartment as the interaction partner. For example, in studies of the interactions between transmembrane G-protein coupled receptors and cytoplasmic β-arrestin, the luciferase was fused to the intracellular carboxyl tail of the transmembrane receptor8. In other cases the geometry of a protein-protein interaction may lead to energy transfer being more efficient using one of the two possible combinations of fusion proteins. Occasionally energy transfer may not be detected at all if using only one combination, leading to false negative results.
The sequence of the peptide linker between luciferase/YFP and the protein of interest may also affect the efficiency of resonance energy transfer. The linker should be long enough to allow luciferase/YFP and the protein of interest to fold without hindrance. A longer linker will impart greater flexibility to the polypeptide at the junction between luciferase/YFP and the protein of interest, which may increase BRET efficiency by facilitating alignment of the donor and acceptor dipoles. However, BRET efficiency may decrease if there is too much flexibility in the linker. In the experiments here, cloning of the proteins of interest into the BamHI and XbaI sites of the expression vectors resulted in a 23-amino acid linker, as shown in Figure 1. This linker has been successful for detecting interactions between several pairs of proteins.
Before proceeding to BRET experiments it should be ensured that the fusion proteins retain normal biological activity. The functioning of YFP can be assessed by direct visualization using a fluorescence microscope. The functioning of luciferase can be assayed using commercially available kits. The effect of fusion on the subcellular localization of proteins of interest can be assessed by immunostaining, or in the case of YFP-fusions, by direct visualization. The effect of fusion on the function of the protein of interest can be investigated by protein-specific assays - e.g., for a transcription factor it may be appropriate to assay DNA-binding activity. Note that luminescence measurements made during the experiment provide convenient in-assay confirmation of luciferase activity.
It is important to consider what the appropriate control constructs are for your protein of interest. YFP and luciferase are predominantly cytoplasmic and are therefore appropriate controls when the proteins of interest are cytoplasmic. However, when the proteins of interest are localized to other subcellular compartments it may be desirable to modify luciferase and YFP with peptide signals to direct their trafficking to the relevant compartment. Here the proteins of interest were transcription factors that are localized to the nucleus, therefore luciferase and YFP were modified by the addition of a nuclear localization signal. Specifically, a peptide including the nuclear localization signal of the SV40 large T antigen (CGYGPKKKRKVGGLDN) was appended to the C-terminus of luciferase and YFP by ligating synthetic double-stranded oligonucleotides between the BamHI and XbaI sites of the pLuc and pYFP vectors. Addition of this signal caused a significant redistribution of protein to the nucleus (Figure 4B).
Including the appropriate controls is crucial to the interpretation of BRET experiments. A mock-transfection control using an inert filler plasmid is essential to establish background levels of luminescence and fluorescence resulting from luminometer noise and enzyme-independent breakdown of substrate. With modern luminometers and luciferase substrates background luminescence levels are typically very low. Transfection with the appropriate control luciferase construct in the absence of a YFP construct is important to establish the proportion of luciferase emission which is detected within the emission range of YFP in the absence of a BRET acceptor. Transfection with the control luciferase and control YFP constructs provides the baseline BRET signal resulting from Luc-YFP interaction. For every pair of proteins that are tested for interaction, it is important to transfect each one alone with the appropriate control construct, as illustrated in Figure 3B, to control for any non-specific interaction between the proteins being tested and luciferase/YFP. Particularly when performing a BRET experiment for the first time, it is important to include a Luc-YFP fusion protein as positive control to ensure that a BRET signal is observable in your experimental setup. Here the coding sequence of YFP was cloned into pLuc at the BamHI and XbaI sites to generate a fusion protein in which YFP is fused to the C-terminus of luciferase.
In the protocol described here, the composition of the DNA mixes for transfection was calculated with the assumption that the average size of the Luc/YFP fusion protein expression constructs is not greater than 7.5 kb. If the expression constructs are significantly larger than this, then the number of moles of each expression construct added to the transfection mix may have to be reduced so that the total amount of DNA in the mix does not exceed the maximum permitted by the amount of transfection reagent. Alternatively the amount of transfection reagent and the amount of DNA in all the transfection mixes can be increased following the manufacturer’s guidelines. Experimental manipulation of 96-well plates is expedited by the use of multichannel pipettes and aspirators. It is desirable to minimize variation between samples in the length of transfection mix incubation time after addition of DNA to the serum-free medium/transfection reagent mix, therefore begin adding transfection mixes to cells as soon as the first mix has been incubating for 10 min. Because the BRET acceptor proteins are YFP fusions, it is possible to assess transfection efficiency by fluorescence microscopy or by measuring fluorescence in a microplate reader. Success of a BRET experiment depends on a good level of transfection so it is advisable to ensure that transfection is satisfactory before proceeding to addition of substrate and measurement of the BRET signal. The YFP tag also enables quantification of acceptor protein expression levels using a microplate reader. Quantification of acceptor protein levels may be useful for optimization and trouble-shooting of BRET experiments.
Cells should be incubated for at least 2 hr with luciferase substrate before measuring the BRET signal to ensure that a stable signal has been achieved. It is also acceptable to incubate cells with substrate for up to 24 hr. In this case the total luminescence counts will be lower, but the BRET ratios should be unchanged. Longer incubations with substrate may be more convenient (e.g., overnight) and also allow the measurement of BRET signals before and after an experimental manipulation such as addition of a pharmaceutical compound. Ideally, optimize cell seeding density so that cells reach confluency around the time of measurement. If cell density is above optimum then it may be advantageous to measure the BRET signal at an earlier time point to avoid cell overgrowth. If using a cell type other than HEK293, be sure to optimize cell culture and transfection conditions. BRET experiments have also been successfully performed in HeLa cells using the protocol described here.
In the experiments described here, luminescence counts were measured at room temperature, which is the optimum temperature for Renilla luciferase activity. If performing a time-course experiment in the luminometer it is preferable to set the temperature at 37 °C. We have performed the same experiments at room temperature and at 37 °C and no significant difference in the results was observed. If performing experiments at room temperature, the plate should be allowed to equilibrate 10 min inside the luminometer prior to measurement to allow stabilization of Renilla luciferase activity. Here, luminescence signals were integrated over 10 sec, which typically provides high sensitivity, especially for proteins with low expression levels. However, signals may be integrated over shorter time periods if this provides sufficient sensitivity. Due to the stability of the luminescence signal produced by the live-cell luciferase substrate, it is acceptable to measure for up to 10 sec per well at each wavelength.
An interaction between two proteins of interest is confirmed by the BRET assay when the signal from the test condition significantly exceeds the signals from the relevant control conditions. The absence of a BRET signal is not necessarily confirmation that an interaction does not occur. In the case of an unexpected or negative result, the total luminescence and fluorescence counts should be examined to ensure that all wells were transfected correctly.
The BRET signal is also affected by the ratio of protein expression levels therefore it is useful to examine the relative expression levels of the two proteins of interest by comparing the fluorescence counts from wells transfected with YFP fusions of the two proteins (or the luminescence counts from wells transfected with the luciferase fusions). When one fusion protein expresses more strongly than the other, better results are likely to be obtained if the protein with the lower expression level is fused to luciferase, which was the case with FOXP2 and CtBP1 in the experiments presented here. It is also possible to manipulate protein levels by adjusting the molar ratio of expression plasmids in the transfection mix. Energy transfer between certain pairs of fusion proteins may be very inefficient due to the geometry of the protein complex. In such cases it may be worthwhile to try fusing the proteins of interest to the alternative termini of luciferase/YFP.
It is necessary to confirm that the BRET signal represents a physiological protein-protein interaction and is not simply due to protein overexpression. For this reason it is important to avoid gross overexpression of proteins. Specificity of a protein-protein interaction in the BRET system can further be confirmed by introducing mutations into the proteins that are known to reduce the affinity of the interaction, as demonstrated here using the DE400 variant of FOXP2. The mutant proteins can further be used in competition and saturation binding BRET assays. In a competition assay, the concentrations of donor and acceptor fusion proteins are kept constant while increasing the concentration of an untagged version of one of the interaction partners as a competitor. Untagged wild-type protein should compete with the tagged version for binding to its interaction partner, resulting in a decrease in the BRET signal, whereas the mutant protein should exhibit a reduced ability to compete out the interaction. In a saturation binding assay, the concentration of the donor fusion protein is kept constant, while increasing the concentration of the acceptor. For a specific interaction the BRET signal should eventually plateau as all donor molecules become saturated with the acceptor. For a non-specific interaction, the BRET signal will typically show a small linear increase due to an increase in random collisions between the donor and acceptor molecules. In practice, for interactions of moderate affinity between soluble proteins, it may be difficult to achieve a high enough acceptor:donor ratio to observe saturation while also maintaining sufficient levels of the donor protein for signal detection.
Caution should be exercised in the interpretation of differences between BRET ratios. The BRET assay does not provide a quantitative measure of the affinity of a protein-protein interaction because the efficiency of energy transfer is influenced by factors other than the strength of the protein-protein interaction. Such factors include the ratio of the levels of the fusion proteins within the cell, the geometry of the interaction, and the rates of association and dissociation of the complex. The ratios obtained with two different pairs of proteins therefore cannot necessarily be compared, but comparisons may be possible between different variants of the same protein, such as with the wild-type and ∆E400 variants of FOXP2 shown here. Occasionally, slightly negative BRET ratios may be observed. Negative ratios may result from the error in measurement of the baseline ratio in cells transfected with only the pLuc-control vector. Very high ratios are often indicative of protein aggregation, which can be confirmed by observation of transfected cells with a fluorescence microscope. It should be determined if aggregation is biologically relevant or an artifact of protein overexpression.
In conclusion, the BRET assay described here is a powerful approach to investigate protein-protein interactions in live cells. The application of BRET has been demonstrated for the purposes of validating candidate interactors from proteomic screening studies, identifying specific residues involved in protein-protein interactions, and evaluating the effects of mutations found in patient DNA. Because interactions are detected in real time without the requirement for cell lysis, the BRET signal can be measured before and after a treatment such as the addition of a pharmaceutical compound or ligand7. The BRET assay shows great promise for use in functional genomics research to assess the biological relevance of genetic variants discovered through next-generation sequencing.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported by the Max Planck Society.
References
- Suter B, Kittanakom S, Stagljar I. Two-hybrid technologies in proteomics research. Curr Opin Biotechnol. 2008;19:316–323. doi: 10.1016/j.copbio.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Berggard T, Linse S, James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- Ciruela F. Fluorescence-based methods in the study of protein-protein interactions in living cells. Curr Opin Biotechnol. 2008;19:338–343. doi: 10.1016/j.copbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Pfleger KD, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer. BRET). Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- Boute N, Jockers R, Issad T. The use of resonance energy transfer in high-throughput screening. BRET versus FRET. Trends Pharmacol Sci. 2002;23:351–354. doi: 10.1016/s0165-6147(02)02062-x. [DOI] [PubMed] [Google Scholar]
- Pfleger KD, et al. Extended bioluminescence resonance energy transfer (eBRET) for monitoring prolonged protein-protein interactions in live cells. Cell Signal. 2006;18:1664–1670. doi: 10.1016/j.cellsig.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Kocan M, Dalrymple M, Seeber R, Feldman B, Pfleger K. Enhanced BRET technology for the monitoring of agonist-induced and agonist-independent interactions between GPCRs and β-arrestins. Frontiers in Endocrinology. 2011;1 doi: 10.3389/fendo.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute N, Pernet K, Issad T. Monitoring the activation state of the insulin receptor using bioluminescence resonance energy transfer. Mol Pharmacol. 2001;60:640–645. [PubMed] [Google Scholar]
- Perroy J, Pontier S, Charest PG, Aubry M, Bouvier M. Real-time monitoring of ubiquitination in living cells by BRET. Nat Methods. 2004;1:203–208. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- Roduit R, Escher P, Schorderet DF. Mutations in the DNA-binding domain of NR2E3 affect in vivo dimerization and interaction with CRX. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriziotis P, Fisher SE. Neurogenomics of speech and language disorders: the road ahead. Genome Biol. 2013;14:204. doi: 10.1186/gb-2013-14-4-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. Fourth Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Fisher SE, Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25:166–177. doi: 10.1016/j.tig.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Graham SA, Fisher SE. Decoding the genetics of speech and language. Curr Opin Neurobiol. 2013;23:43–51. doi: 10.1016/j.conb.2012.11.006. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D, et al. Identification of FOXP1 deletions in three unrelated patients with mental retardation and significant speech and language deficits. Hum Mutat. 2010;31:1851–1860. doi: 10.1002/humu.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, et al. De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet. 2010;87:671–678. doi: 10.1016/j.ajhg.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDermot KD, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, et al. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15:3154–3167. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]


