Version Changes
Updated. Changes from Version 1
In this revised version of the article, we have extended our software to include two new features: automatic partitioning of sets and layout algorithms adapted to be setaware. To incorporate these changes, we updated screenshots and descriptions of the UI, included a new paragraph in the Results as well as sentences in existing paragraphs, and added two new authors.
Abstract
setsApp ( http://apps.cytoscape.org/apps/setsapp) is a relatively simple Cytoscape 3 app for users to handle groups of nodes and/or edges. It supports several important biological workflows and enables various set operations. setsApp provides basic tools to create sets of nodes or edges, import or export sets, and perform standard set operations (union, difference, intersection) on those sets. Automatic set partitioning and layout functions are also provided. The sets functionality is also exposed to users and app developers in the form of a set of commands that can be used for scripting purposes or integrated in other Cytoscape apps.
Keywords: cytoscape, app, sets functionality
Introduction
Cytoscape 1, 2 provides an environment for the visualization and analysis of networks and associated annotations. The primary audience for Cytoscape is the biological community and Cytoscape supports a number of standard use cases for analyzing and visualizing biological data. Many of these use cases involve the selection of a number of nodes or edges based on some analysis or annotation and either performing an action on that selection or comparing those nodes or edges to a different set of nodes or edges that resulted from alternative analyses or analyses based on alternative annotations. The core capabilities for Cytoscape provide some tools to facilitate these types of comparisons but they can be counterintuitive or complicated to use. setsApp is a Cytoscape 3 application that provides a general set of tools for users and developers to define and maintain sets of nodes or edges and compare those sets using the standard set operations of union, intersection, and difference. Partition and layout features are also provided to assist in generating sets and performing set-aware layouts.
In this paper, we present the implementation of setsApp, in particular, how setsApp integrates with the Cytoscape command system, and then present a sample biological workflow using setsApp.
Implementation
Cytoscape provides two approaches to implementing apps: a simple app and a bundle app. Simple apps are implemented using the same general approach as in Cytoscape 2.8, but at the cost of significant flexibility. Bundle apps utilize Open Service Gateway Initiative (OSGi) 3 interfaces through APIs provided by Cytoscape to interact with the Cytoscape core functionality. setsApp is implemented as a bundle app and utilizes the Cytoscape 3.1.0 API. There are three main components to the setsApp implementation: the user interface, the command interface, and the underlying data model for maintaining sets of nodes and edges.
User interface
The setsApp user interface consists of menu items in the main Apps menu, node and edge context menus, and a panel added to the Control Panel (left or west) section of the Cytoscape user interface. The main feature of the Sets panel is the list of currently defined sets. Each set can be expanded to see all of the nodes or edges within that set, and context menus provide the ability to select, deselect, rename, or remove sets. As long as you have one set defined, Partition and Layout buttons will be active at the bottom of the panel. The partition feature will create new sets based on shared and excluded sets present in all currently defined sets. The layout feature will perform the selected layout algorithm taking into account set memberships, i.e., attempting to keep sets close together. When multiple sets are selected, the Set Operations buttons are enabled. This allows users to create new sets based on the union, intersection, or difference of other sets. Note that the results of a union or intersection are well-defined for multiple sets, but the difference operation is order dependent. If only two sets are selected, the order of selection is preserved. If more than two sets are selected, the order is the order of selection, so care must be taken when attempting to create a difference set of more than two sets. Sets can be created from the currently selected nodes or edges, or based on a particular node or edge attribute. When creating sets from attributes, the user will need to supply a prefix for the sets to be created and choose the attribute (currently only String attributes are supported) from a list. Sets can also be created by importing them from a simple text file. Each set can be individually exported to a text file.
setsApp provides a context menu for sets and set members in the control pane. In addition to having menu items to manage sets, the user may select all set members in the network, or if the set is expanded, individual members. This functionality presents an easy way for users to visualize the results of set operations and to perform interactive exploratory analysis.
The menus provided through the top-level Apps menu offer the same functionality as the Create set from menu in the setsApp control panel with the addition of a menu to import a set from a file. Node and edge context menus provide the user with the ability to add or remove the corresponding node or edge from sets.
Command interface
In addition to the standard user interface described above, setsApp provides a number of “commands”. These commands may be used for scripting purposes or by other Cytoscape apps that wish to take advantage of the setsApp functionality. Table 1 provides a list of commands and the arguments.
Table 1. setsApp Commands.
Arguments with asterisks are required.
| Command | Arguments | Description |
|---|---|---|
| setsApp addTo |
edgeList=
list of edges*
nodeList= list of nodes* network= network to use name= name of set* |
Adds the listed nodes or edges to the named set. An
error will occur if the types (node or edge) do not match or if the set does not exist. The set name and one of edgeList or nodeList are required. |
| setsApp createSet |
edgeList=
list of edges*
nodeList= list of nodes* network= network to use name= name of set* |
Creates the set from the edge or node list. It throws an
error if both edge and node lists are provided. |
| setsApp difference |
name=
name of new set*
set1= name of the first set* set2= name of the second set* |
Performs a difference of two sets and puts the result into
a new set. |
| setsApp export |
Column=
column containing the
id key name= name of set* setFile= path to the file to import* |
Exports a set to the specified file using the designated
column to identify the node or edge. |
| setsApp import |
Column=
column containing the
id key Type= [Node|Edge]* name= name of set* setFile= path to the file to import* |
Exports a set to the specified file using the designated
column to match the node or edge identifier from the file. |
| setsApp intersect |
name=
name of new set*
set1= name of the first set* set2= name of the second set* |
Performs an intersection of two sets and puts the result
into a new set. |
| setsApp remove | name= name of set* | Removes (deletes) the set. |
|
setsApp
removeFrom |
edgeList=
list of edges*
nodeList= list of nodes* network= network to use name= name of set* |
Removes the listed nodes or edges from the named set.
An error will occur if the types (node or edge) do not match or if the set does not exist. The set name and one of edgeList or nodeList are required. |
| setsApp rename |
newName=
new name for the set
oldName= old (current) name for the set |
Renames an existing set. |
| setsApp union |
name=
name of new set*
set1= name of the first set* set2= name of the second set* |
Performs a union of two sets and puts the result into a
new set. |
A command is made available to Cytoscape by creating a standard Cytoscape TaskFactory with two new properties in the org.cytoscape.work package: ServiceProperties.COMMAND_NAMESPACE, which is always set to “ setsApp”; and ServiceProperties.COMMAND, which is the command name (e.g. “addTo”). The command arguments are implemented as Tunables within the Task called by the designated TaskFactory. Because there is no guarantee that the Task will be executed within the context of a GUI, care should be taken to make sure that the appropriate Tunable types are used. For example, the NodeList Tunable allows the command-line user to enter a list of nodes rather than assuming that the user will have selected nodes interactively.
For another Cytoscape app to use any of these commands, it would need to call one of the Cytoscape TaskManagers and provide it org.cytoscape.command.CommandExecutorTaskFactory’s createTaskIterator method with the appropriate argument map, command, and command namespace. The TaskObserver method may be used if the command returns any values. For example:
Listing 1. Example Command Usage.
SynchronousTaskManager tm =
serviceRegistrar. getService
(SynchronousTaskManager.
class
);
CommandExecutorTaskFactory cetf =
serviceRegistrar. getService
(CommandExecutorTaskFactory.
class
);
Map<String, Object> argMap =
new
HashMap<String, Object>();
argMap.put(
"name"
,
"NewSet"
);
// selected is a special keyword for the
NodeList tunable
argMap.put(
"nodeList"
,
"selected"
);
// the current network
argMap.put(
"network"
,
"current"
);
// Assumes that this implements TaskObserver
tm.execute(cetf.createTaskIterator(
"setsApp"
,
"createSet"
,
argMap,
this
),
this
);
Data model
The main model object for a node or edge set is the Set object, which stores a map of all of the nodes or edges in this set. A SetsManager provides the methods to create and destroy sets. The SetsManager also serves the critical function of serializing the information about Sets to the default hidden table (see CyNetwork.HIDDEN_ATTRS) for nodes or edges (depending on the type of the Set). Each Set is created as a boolean column in the hidden table which is set to true if the corresponding node or edge is in that set. By storing values in the default hidden tables, the information about sets is automatically saved in Cytoscape sessions and restored when sessions are reloaded. SetsManager implements SessionReloadedListener and recreates the Sets from the information stored in the hidden table columns.
Results
A simple example use case might be the exploration of the data set from Ideker et al., 2001 4, which measured the change in expression for 331 genes after a systematic deletion of genes known to be involved in the Saccharomyces cerevisiae switch to galactose metabolism. This data was combined with known protein-protein and protein-DNA interactions to explore the biological response to deletions in the presence (the G in the column names) or absence of galactose in the medium. This data set is now included as a sample with Cytoscape downloads ( galFiltered.cys). In our workflow we use Cytoscape’s Select panel to select all proteins that are underexpressed (gal1RGexp < -0.5 fold change) in the deletion of GAL1 ( Figure 1). That selection is saved as a set named GAL1- ( Apps→ SetsApp→Create node set). We then select all of the proteins that are overexpressed (gal1RGexp > 0.5 fold change) in the deletion of GAL1 and save that selection as a set named GAL1+. Repeating this for GAL4 (column gal4RGexp) and GAL80 (column gal80Rexp) results in 6 sets altogether: GAL1+, GAL1-, GAL4+, GAL4-, GAL80+, GAL80- ( Figure 2). Note that the data for GAL1 and GAL4 is in the presence of galactose, but the data for GAL80 is in the absence of galactose since GAL80 is a known repressor of GAL4.
Figure 1. Screenshot of Cytoscape’s Select panel with underexpressed genes in the gal1RGexp condition being selected.
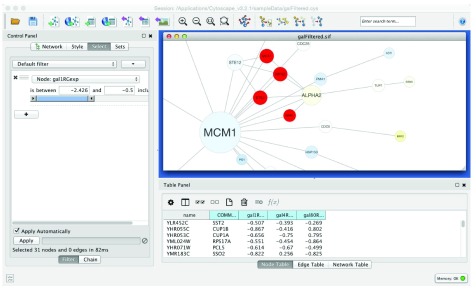
Nodes that match the gal1RGexp condition are highlighted in red.
Figure 2. Screenshot of Cytoscape showing the Sets panel with all condition sets created.
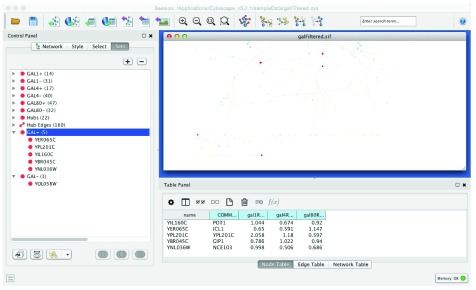
Nodes in the GAL+ set are highlighted in red.
Given those six sets of genes, we can explore the data sets by looking at combinations of the sets. For example, we could look at the intersection of all of the underexpressed proteins by selecting each of GAL1-, GAL4-, and GAL80- in the Sets panel and pressing the Intersection button in the Set Operations box near the bottom of the panel. If we name the resulting set GAL- we see that it contains a single gene: YOL058W (ARG1). In this data set of 331 genes, only this one gene is repressed for all three of the deleted GAL genes. In the absence of galactose when GAL80 is deleted, ARG1 is underexpressed, and in the presence of galactose when either GAL1 or GAL4 are deleted the gene is also underexpressed. Looking at the expression significance values in the Node Table Panel of Cytoscape (gal1RGsig, gal4RGsig, and gal80Rsig) this is a highly significant result, although there is no direct correlation between the galactose switch and arginine biosynthesis regulation that we were able to find in the literature. On the other hand, ARG1 is regulated by the GCN4 activator which is known to repress protein synthesis during periods of stress or starvation 5, which explains the significant down-regulation of ARG1. We can perform a similar analysis to understand the consistent up-regulation of the five genes in the GAL+ set corresponding to gene symbols: GIP1, NCE103, YIG1, POT1, and ICL1. Figure 2 shows the results of the intersection operations.
We can also explore data sets by using layout algorithms that are informed by the sets we created earlier. By default, Cytoscape provides over a dozen layout algorithms that spatially place nodes in order to help elucidate meaningful patterns of relationships in networks. Most layout algorithms take into account connectivity between nodes. SetsApp augments Cytoscape by supplying two additional layout algorithms that take set membership into account. The setsbased grid layout places nodes in the same set together into independent grids, i.e., a grid of grids. This is ideal for quickly separating nodes in different sets for manual manipulation later on. The setsbased force directed layout employs the Prefuse force directed layout provided by Cytoscape but also tries to put nodes in the same set closer together in the network. Users can adjust the force between nodes in a given set relative to connected nodes and thereby emphasize or diminish the grouping based on set membership by changing values in the layout settings panel ( Layout→ Settings... ).
Conclusions
There are many Cytoscape workflows that could take advantage of the features of setsApp. Any workflow that might want to look for groups of nodes or edges that share multiple traits, or that explicitly do not share those traits. While it is possible to duplicate many of the final results enabled by setsApp by using Cytoscape 3.1’s new Select panel, a user would need to know in advance exactly the combination of features that were biologically interesting. setsApp provides an alternative that allows users to explore various combinations of nodes and edges and to save such selections for later uses. We also provide layout algorithms that take set membership into consideration.
In the workflow we developed above, we combined the functionality of Cytoscape’s Select panel with setsApp to explore combinations of sets of genes based on shared properties. There are many more sophisticated apps available to Cytoscape users that could be used to do a more thorough analysis of this data set including jActiveModules 6, clusterMaker 7 and RINalyzer 8, however, the workflow above demonstrates the utility of a simple set-oriented approach to exploring networks.
Software availability
Software available from: http://apps.cytoscape.org/apps/setsApp
Latest source code: https://github.com/RBVI/setsApp
Source code as at the time of publication: https://github.com/F1000Research/setsApp
Archived code as at the time of publication: http://www.dx.doi.org/10.5281/zenodo.10424 9
License: Lesser GNU Public License 3.0: https://www.gnu.org/licenses/lgpl.html
Funding Statement
JHM, SL and AP were funded by NIGMS grants P41-GM103504 and P41-GM103311. AW and TF were funded by NIGMS grant P41-GM103311. NTD was partially funded by a Boehringer Ingelheim Fonds travel grant, and her research was conducted in the context of the DFG-funded Cluster of Excellence for Multimodal Computing and Interaction. MA was financially supported by the projects GANI_MED and BioTechMed-Graz.
v2; ref status: indexed
References
- 1. Shannon P, Markiel A, Ozier O, et al. : Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cline MS, Smoot M, Cerami E, et al. : Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–82. 10.1038/nprot.2007.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. OSGi Alliance. OSGi service platform: release 3, March 2003. IOS Press: Ohmsha, Amsterdam; Washington, DC,2003. Reference Source [Google Scholar]
- 4. Ideker T, Thorsson V, Ranish JA, et al. : Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–34. 10.1126/science.292.5518.929 [DOI] [PubMed] [Google Scholar]
- 5. Hinnebusch AG: Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272(35):21661–4. 10.1074/jbc.272.35.21661 [DOI] [PubMed] [Google Scholar]
- 6. Ideker T, Ozier O, Schwikowski B, et al. : Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–40. 10.1093/bioinformatics/18.suppl_1.S233 [DOI] [PubMed] [Google Scholar]
- 7. Morris JH, Apeltsin L, Newman AM, et al. : clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics. 2011;12:436. 10.1186/1471-2105-12-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doncheva NT, Assenov Y, Domingues FS, et al. : Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc. 2012;7(4):670–85. 10.1038/nprot.2012.004 [DOI] [PubMed] [Google Scholar]
- 9. Morris JH, Wu A, Doncheva NT, et al. : F1000Research/SettsApp. ZENODO. 2014. Data Source [Google Scholar]


