Abstract
The purpose of this study is to demonstrate the ability of imaging Cathepsin E (Cath E) positive tumors in living animals through selective targeting of Cath E proteolytic activity using a sensitive molecular imaging agent. Methods: a peptide-based Cath E imaging probe and a control probe were synthesized for this study. Human Cath E-positive cancer cells (MPanc96-E) were implanted subcutaneously in nude mice. Tumor-bearing mice were examined in vivo with near-infrared fluorescence (NIRF) imaging at various time points after intravenous injection of the Cath E sensing imaging probe. Excised organs and tissues of interest were further imaged ex vivo. Results: upon specific Cath E proteolytic activation, the NIRF signal of the imaging probe a was converted from an optically quenched initial state to a highly fluorescent active state. Imaging probe a was able to highlight the Cath E-positive tumors as early as 24 h post injection. Fluorescent signal in tumor was 3-fold higher than background. The confined specificity of imaging probe a to tumor associated Cath E was verified by using control imaging probe b. Both in vivo and ex vivo imaging results confirmed the superior selectivity and sensitivity of imaging probe a in Cath E imaging. Conclusions: the small animal studies demonstrated the capability of probe a for imaging Cath E-positive tumors. The developed optical probe could be applied in early diagnostic imaging and guiding subsequent surgical procedure.
Introduction
While numerous molecular targets have been identified and several targeted molecular imaging agents have been reported for the detection of malignant tumors, only a handful of them showed limited clinical potential.1–6 This is in part due to the widespread expression of these molecular targets, in both cancerous and normal tissues.7–10 Cathepsin E (Cath E), a tumor associated protein which is an intracellular non-lysosomal aspartic proteolytic enzyme, was observed in several tumors including pancreatic ductal adenocarcinoma (PDAC), cervical, gastric and intestinal cancers.11–13 Due to its distinct expression in premature stages of tumors such as PDAC, Cath E has been suggested as a potential clinical tumor marker for early detection.2,3,14–20
Recent developments in molecular imaging provide technologies to study diseases noninvasively and allow the analysis of disease status from whole body and tissue levels down to cellular and molecular levels, offering comprehensive details of diseases and enhancing accuracy of diagnosis.2,3,17–20 Ideally, a specific molecular imaging probe targeting a specific cancer marker could facilitate early tumor detection, rather than relying on conventional invasive pathological analysis.4,18,19 PET imaging is a sensitive way of imaging and monitoring treatment response of many cancers. However, the usefulness of FDG-PET in some cancers is limited due to low glucose turnover at early stages.21,22 A number of studies have shown that FDG-PET is useful in defining regions of locally advanced pancreatic cancer; though false positive is possible in cystic tumors.23,24 To obtain a decisive diagnosis of an early cancer stage, patients are currently required to be subjected to invasive biopsies and histological confirmation.
Despite the promising advantages of utilizing Cath E upregulation for cancer detection, this idea has been hampered by the lack of Cath E specific reporters. Recently, we reported the ability of detecting proteolytic activity of Cath E in vitro with a fluorogenic peptide substrate, Ala-Gly-Phe-Ser-Leu-Pro-Ala-Lys-dArg-CONH2 25. Compared to other reported substrates,26–29 this sequence showed distinct Cath E selectivity that is suitable for enzyme sensing. Thus it was used to prepare the Cath E activatable optical imaging probe in this study. Previously we have developed several protease sensitive imaging probes using a polylysine-based template and applied them to image disease associated proteases in vivo.3,30,31 Conjugating the newly identified peptide sequence with the same polymeric template, the Cath E imaging probe a was prepared to be fluorescently silent in its intact state, but to emit bright fluorescence after proteolytic activation by its target protease (Fig. S1, ESI†). Here we report the whole body and longitudinal in vivo imaging of Cath E-positive tumor xenografts in the mouse model using the developed near infrared fluorescent (NIRF) Cath E imaging probe.
Results
Imaging probe preparation
The self-quenched imaging probes used in this study were designed based on our prior work on protease imaging.3,30,31 Multiple fluorochromes were linked onto a polymeric template through protease selective peptide substrates (Fig. S1B, ESI†). Near complete fluorescent quenching was achieved by the densely arranged fluorochromes which were held by the pegylated polylysine template, DPGC. Following intravenous injection, the circulating probes were activated/degraded by disease associated proteases, resulting in instant dequenching of fluorescence. Since the polymer template DPGC consisted of protease resistant d-polylysine, the selective proteolytic degradation could only occur within the inserted peptides and be controlled by the specific peptide sequences.
The Cath E probe and its control imaging probe were assembled following the previously developed protocol with minor modification. The NIR fluorochrome, Cy5.5, was attached to the peptide prior to the polymer conjugation. The peptides and Cy5.5 labeled peptides were analyzed by RP-HPLC and ESI-MS. The results revealed the high purity (>96%) and confirmed the identities of the precursor peptides (Table 1). The average loading of the peptide substrates ranged from 21–23 peptides per DPGC, resulting in a hefty fluorescent quenching. The fluorometric assessment showed that the assembled imaging probes a and b, compared with free Cy5.5 fluorochrome, emitted less than 5% of fluorescent signal (Table S2, ESI†). This significant fluorescence quenching granted low background signal, while the in vitro validation and in vivo imaging demand the least initial fluorescence setting to obtain a maximized signal to background ratio.
Table 1.
Peptide substrates used for imaging probes preparation and their characterization
| Imaging probe | Protease-selective peptide sequence | Calculated mol. wt. | Observed [M + H]1+ /1 |
|---|---|---|---|
| a | Cy5.5-Ala-Gly-Phe-Ser-Leu-Pro-Ala-Gly-Cys-CONH2. | 1720.27 | 1721.28 |
| b | Cy5.5-Gly-Ser-Pro-Ala-Phe-Leu-Ala-Gly-Cys-CONH2. | 1720.27 | 1721.00 |
In vitro validation of imaging probes with Cath E and Cath D
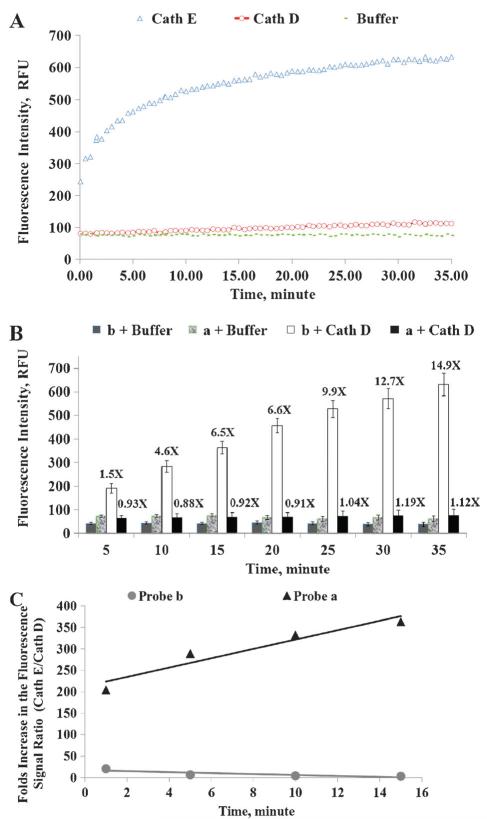
Cath E and Cath D are two closely related proteases, which share similar structure and substrate selectivity.32 Both of them favor hydrophobic residues at the scission sites.33,34 There was no specific peptide substrate, to the best of our knowledge, which could distinguish these two proteases until our recent development in identifying a selective peptide substrate for Cath E.25 In order to confirm that the image signal is truly generated by Cath E, a Cath D sensitive probe b was included in the study as a negative control. The major structural difference between these two probes lie in the natural hydrophobic amino acid residues at P1 and P1′ positions of the scissile bond. In Cath E selective probe a, Leu residue was selected as amino acid for the P1 position based on its hydrophobicity index. A less hydrophobic amino acid residue, Phe, was selected for the P1 position in probe b. A conformationally distinctive amino acid residue is inserted at the P1′ position in each probe. A structurally constrained Pro residue was selected for probe a while a less restricted Leu residue for probe b. Prior to in vivo validation, the enzyme selectivity of intramolecularly quenched imaging probes a and b was determined by measuring changes in fluorescence intensity with pure enzymes. A prominent increase of the fluorescence signal was observed instantaneously upon incubating imaging probe a with Cath E (Fig. 1A). While the fluorescence signal with Cath E continued to propagate over the monitoring period (15-fold in 35 min), the fluorescence signal with Cath D remained at the background level, comparable to that in buffer (Fig. 1A). Unlike Cath E selective probe a, probe b showed superior selectivity to Cath D (Fig. 1B). About 13-fold increase in fluorescence intensity was observed within the first 30 min. In contrast, less than 10% difference was observed for imaging probe a when treated with Cath D, similar to that of the no enzyme treated imaging probes (Fig. 1B). Statistical analysis using paired-samples t-test denoted the significant difference of fluorescence intensity, two-tailed P-values < 0.001. These results verified the pronounced selectivity difference between imaging probes a and b.
Fig. 1.
(A) Change in the fluorescence intensity of Cath E selective probe a (1 nmole) during incubation with 46 pmol of Cath E and Cath D in 50 mM NaOAc buffer of pH 4.0. Values represent the mean of at least three independent experiments; (B) change in the fluorescence intensity of Cath E selective probe a and control probe b (1 nmole, each) during the incubation with 46 pmole of Cath D in 50 mM NaOAc buffer of pH 4.0. Values lying above the bars denote the number of folds increase in the fluorescence signals relative to those of the nonenzyme treated probes. Values represent the mean of at least three independent experiments. Error bars represent the upper and lower values of the Standard Error Mean (SEM). (C) Profile of the folds ratio between the net fluorescence signal (Cath E/Cath D) of the Cath E selective probe a versus control probe b. Net fluorescence signals represent the signals after background signals correction.
To further confirm the inferences, imaging probes selectivity was calculated by the ratio of the folds change in the fluorescence signal of Cath E over Cath D (Fig. 1C). Imaging probe a demonstrated a significant increase, ~204-fold, at the initial time point and continued to elevate higher throughout the entire monitored time period. Comparatively, probe b showed ~3.3-fold at the end of 15 min (Fig. 1C). The results revealed the great potentiality of probe a in distinguishing the proteolytic activities of Cath E and Cath D.
Fluorescence imaging
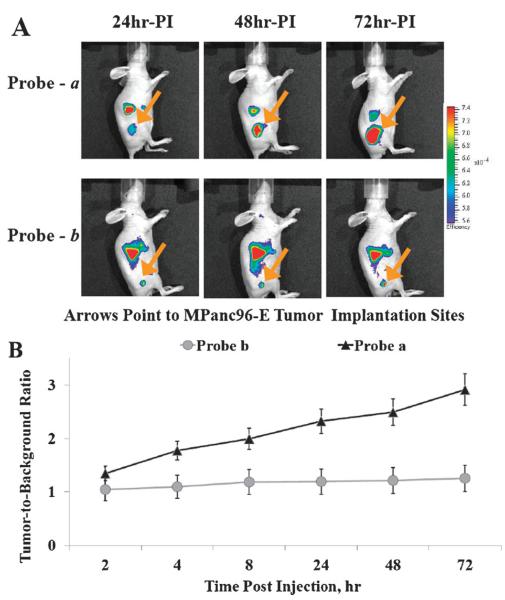
The capability of Cath E selective probe a to detect Cath E-positive MPanc96-E human tumor xenografts in vivo was investigated using NIRF imaging. After IV injection of probe a, Cath E-positive MPanc96-E tumors were clearly seen at 24 h (Fig. 2A). The signal continued to increase and the boundaries of tumor xenografts were much clearer at 48 and 72 h. Alongside, the relative abdominal fluorescence signals were reduced significantly at 72 h. Conversely, imaging probe b failed to report the tumors associated Cath E-proteolytic activity at all time points. Furthermore, high fluorescence signals were detected constantly in the upper quadrant of the abdominal cavity (Fig. 2A).
Fig. 2.
(A) In vivo fluorescence image of Cath E-positive MPanc96-E tumor xenografts in Nu/Nu mice at various time points following administration of 2 nmole of Cath E selective probe a and control probe b; (B) target-to-background fluorescence ratio of probes a and b in Cath E-positive MPanc96 tumor-bearing Nu/Nu mice. Images were collected and corrected for flat field and cosmic fluorescence. Radiance scale of the color bar represents the fluorescence emission normalized to the illumination intensity. The tumor associated fluorescence (target) to non-tumor associated fluorescence (non-target) was quantified using identical size regions of interest (ROI) around the area of interest on the animal image. The controls ROI represent an average of the pixels everywhere in the in vivo animal image except within the pixels of interest in the field of an identical size of the tumor image.
The tumor to background ratio of imaging probe a showed steady increase that achieves as high as ~3 at 72 h PI (Fig. 2B). The fluorescent signal in Cath E expressing tumor was much higher than that in non-Cath E expressing tissues in the near vicinity. In contrast, the imaging probe b showed minimum changes in the tumor-to-background ratios with a maximum value of ~1.2 at 72 h PI. This result demonstrated the superiority of Cath E selective imaging probe a in detecting the Cath E expressing tumor.
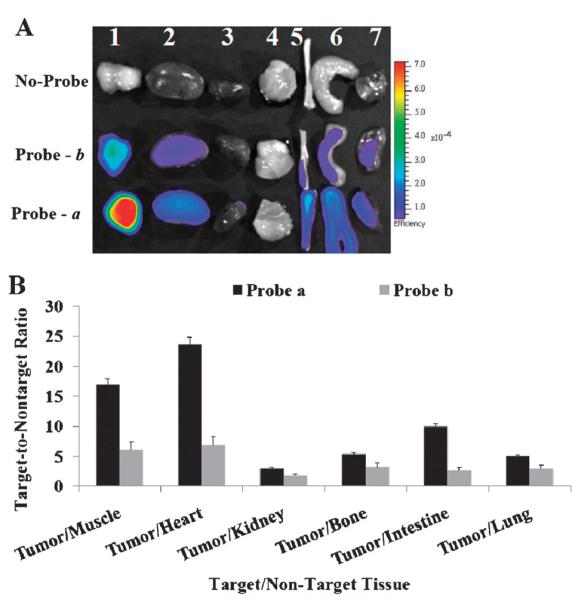
To further validate the specificity of the imaging probes to tumors associated Cath E, organs/tissues from two sets of animals were harvested and subjected to a side-by-side comparison. The ex vivo image of selected tissues/organs revealed that the highest fluorescence signal was from Cath E-positive MPanc96-E tumor with probe a (Fig. 3A). A significantly high tumor-to-nontumor fluorescence ratio of Cath E selective imaging probe a was achieved in most of the investigated tissues (Fig. 3B). The tumor-to-nontumor fluorescence ratios to muscle and heart were up to ~16 and 23, respectively. In contrast, fluorescence signal from control probe b in tumor/non-tumor was relatively low (Fig. 3B). High ex vivo fluorescent signals were associated with the liver images (data not shown). This is most probably due to the liver’s distinct capability of retaining high molecular weight identities, including probes, along with its conventional extraordinarily elevated enzymatic activity. The elevated ex vivo fluorescence signal associated with liver might explain the constantly in vivo detected high fluorescence signals within the upper quadrant of the abdominal cavity.
Fig. 3.
(A) Ex vivo fluorescence image of excised representative organs and tissues 48 h post injection; 1, Cath E-positive MPanc96-E tumor; 2, kidney; 3, heart; 4, muscle; 5, bone; 6, small intestine; 7, lung. (B) Target-to-nontarget fluorescence ratio of probes a and b in MPanc96-E tumor and representative organs and tissues 48 h post injection. The controls ROI represent an average of the pixels every-where in the ex vivo organs image except within the pixels of interest in the field of an identical size of the tumor image.
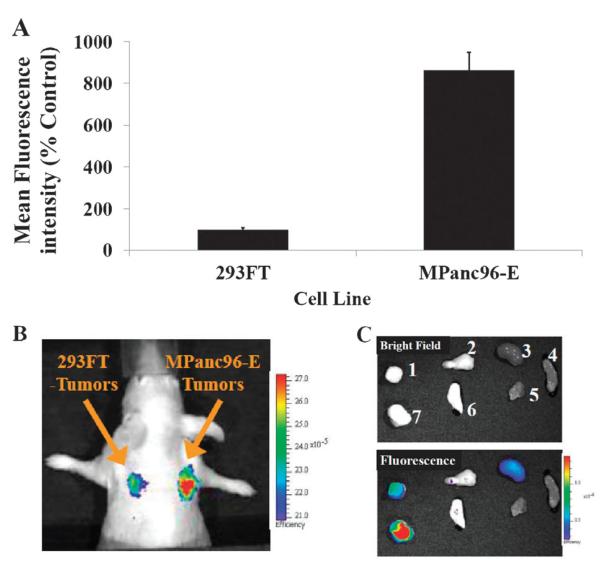
To explore the detection sensitivity and to further confirm the confined specificity of imaging probe a to Cath E, 293FT cells with lower expression levels of Cath E relative to that of MPanc96-E cells were used as control xenografts. FACS analysis showed that the in vitro expression levels of Cath E in MPanc96-E were ~8 times higher than that in 293FT cells (Fig. 4A). An in vivo image of probe a in nude mice bearing dual tumors, MPanc96-E and 293FT, showed considerable difference between these tumors. High fluorescence signal was originated from the Cath E-positive (MPanc96-E) tumor, 2.5-fold higher than the signal in 293FT tumor (Fig. 4B). An ex vivo image of selected tissues and organs also illustrated that probe a fluorescence signal was mainly from Cath E-positive (MPanc96-E) tumor while very low signal was derived from 293FT tumor, 3.6-fold difference (Fig. 4C). Ex vivo quantitative analysis showed that considerably high fluorescence ratio (~8-15) of probe a in Cath E-positive MPanc96-E tumor to most investigated non-tumoral tissues was achieved. Both ex vivo and in vivo results were consistent with Cath E differential expression in MPanc96-E and 293FT tumors.
Fig. 4.
(A) Relative expression of Cath E in 293FT and MPanc96-E tumor cell lines; (B) in vivo fluorescence image of Cath E-positive 293FT/MPanc96-E tumor xenografts in Nu/Nu mice after 48 h of 2 nmole administration of Cath E selective probe a; (C) ex vivo bright and fluorescence image of excised representative organs and tissues 48 h post injection; 1, 293FT-tumor; 2, pancreas; 3, kidney; 4, small intestine; 5, muscle; 6, lung; 7, MPanc96-E pancreatic tumor.
Discussion
The current imaging modalities for early detection of tumor lesions and accurate distinguishing of tumors from inflammations are limited. For decisive diagnosis of cancers, most patients are required to be subjected to invasive biopsies. Molecular targeting agents possessing distinctive capabilities to target certain upregulated tumor biomarkers are crucial for both early diagnosis and subsequent therapeutic regimen. Recent findings on Cath E upregulation in tumors suggest that the proteolytic activity of Cath E could be an ideal biomarker for early- and late-stage tumor diagnosis.14–16 However, a specific Cath E imaging probe is an essential tool to be able to validate this hypothesis.
In this study, we aimed to evaluate the potentiality of a Cath E sensitive molecular imaging probe a for Cath E activity imaging. Our prior study suggested that a peptide substrate possessing -Leu-Pro- residues at the scissile site could be activated selectively by Cath E, but not by Cath D.25 Since Cath E and Cath D are two closely related proteases which have similar enzymatic substrate specificity, it is critical to confirm the protease selectivity of the developed probes. To better confirm the activation mechanism, a Cath D activatable peptide b was, thus, included in the study.
Equivalent size and depth tumor xenografts in nude mice were used in the in vivo fluorescence imaging studies. The fluorescence signal emitted from Cath E expressing MPanc96-E tumors was much higher than that from the low Cath E expressing 293FT tumors and other tissues, a direct reflection of the superior selectivity of imaging probe a for in vivo imaging of Cath E proteolytic activity. As expected, control imaging probe b failed to achieve comparable fluorescent signal enhancement within the Cath E-positive tumor xenografts, supporting that the tumor signals were protease dependent. Another significant difference between images obtained using these two probes was the fluorescence signal located in the upper quadrant of the abdominal cavity. While the fluorescence signal associated with probe a in this particular area was diminished quickly, the signal associated with probe b remained high throughout the monitored period. The ex vivo strong fluorescent signals associated with liver relative to that of other organs suggest that the detected in vivo high fluorescence signal in the upper quadrant of the abdominal cavity might be due to the excessive Cath D enzymatic activity within the liver,35,36 along with the preferred substrate selectivity of probe b.
In order to selectively image tumor, it is preferred to augment tumor specific signals than the overall signals. Widely used targeting probes, which without a ‘turn-off” mechanism, raise signals in tumors and entire non-tumor tissues after injection. The tumor-to-background ratio relies heavily on the wash-out effect. A high tumor-to-background ratio could be expected only if the applied imaging probes were retained in targeted tissue while unbound probes were eliminated quickly from circulation. In contrast, the described Cath E activatable probe is injected in its quenching state, so that the background signal was not a concern. The tumor-to-background ratio depends on the activation of the probes by locally upregulated proteases. Our results show that polymeric-based imaging probe a was able to target-passively and uptake-efficiently in tumor tissues by virtue of an enhanced permeability and retention (EPR) effect. Only the up-taken probe a turned on selectively within the tumor tissues, by the discerning proteolytic activity of Cath E, but not in surrounding tissues lacking the regional expression of Cath E. With this incisive activation character, imaging probe a was able to reveal tumor location clearly and achieve high tumor-to-background ratio. In addition, the extended retention and extensive exposure to the proteolytic activities within the target tissue might help in detecting the preneoplastic lesions and/or predisseminated cells possessing adequate Cath E expression, as opposed to the instant delivery and brief exposure by simple passive targeting.
Here we demonstrated that the molecular differences between tissues could be selectively detected using specifically designed imaging probes. The developed Cath E sensitive molecular imaging probe was activated selectively by tumor associated Cath E in whole animals. Potential Cath E-selective probes, such as the described probe a, could be extremely useful in detecting Cath E-positive tumors.11–13
Materials and methods
Materials
All solvents used were of analytical or HPLC grade. Dichloromethane (DCM), N-methylpyrrolidone (NMP) and methanol (MeOH) were purchased from Fisher (Fair Lawn, NJ, USA). N,N-Dimethylformamide (DMF), diethylether, acetonitrile (MeCN), diisopropylethylamine (DIPEA), dl-dithiothreitol (DTT), triethylacetate (TEA), dimethylsulfoxide (DMSO), iodoacetic anhydride and piperidine were purchased from Sigma-Aldrich (St. Louis, MO, USA). Triisopropylsilane (TIS), triethylamine (TEA) and 1,2-ethanedithiol (EDT) and Kaiser test kit were purchased from Sigma-Aldrich (Milwaukee, WI, USA). HOBt and HBTU were purchased from Applied Biosystems (Foster City, CA, USA). Fmoc protected amino acids were purchased from AnaSpec (Fremont, CA, USA). Fmoc-Rink Amide MBHA-resin was purchased from Novabiochem (La Jolla, CA, USA). Cy5.5 mono NHS ester was purchased from GE Healthcare (Buckinghamshire, UK). d-Polylysine (44 KDa) and polyethylene glycol (5 KDa) grafted copolymer (DPGC), ~28% PEGylation and ~60 substituted lysines, were custom synthesized by VisEn Medical (Bedford, MA, USA). Polyacrylamide gel (Bio-Gel P-10 Gel) was purchased from Bio-Rad (Hercules, CA, USA).
Probe synthesis
Cath E peptide substrate a, Ala-Gly-Phe-Ser-Leu-Pro-Ala-Gly-Cys-CONH2 and control peptide b, Gly-Ser-Pro-Ala-Phe-Leu-Ala-Gly-Cys-CONH2, were synthesized by solid-phase peptide synthesis (SPPS) on Rink amide Novagel resin (0.1 mmole, 0.61 μmol mg −1) using standard Fmoc chemistry with HBTU/HOBT coupling on an automatic synthesizer (ABI-433A, Applied Biosystems). The peptides were purified by HPLC and then Cy5.5-NHS ester was coupled to the N-termini in anhydrous DMSO and TEA overnight in the dark. After HPLC purification, the identity of Cy5.5 labeled peptides was confirmed by LC-MS using a Thermo Finnigan LCQ Fleet mass spectrometer (Thermo-Fisher Scientific, West Palm Beach, FL, USA).
The probe preparation protocol is similar to the previously reported protocol for other protease sensitive probes.37,38 Briefly, iodoacetylated-DPGC was prepared by reacting iodoacetic anhydride with DPGC (1 mg, MW = 375 KDa) in basic conditions, and then the prepared iodoacetylated-DPGC was reacted with Cy5.5-peptide substrates (1 mg) in sodium acetate buffer (300 μl, 50 mM, pH 6.5) overnight at rt in the dark. The formed imaging probes a and b were collected by membrane cutoff filtration and quantified by measuring Cy5.5 molar absorbance at 681 nm. Peptide loadings on DPGC were calculated using the relative mole ratio of fluoro-chrome to DPGC. On average, 21–23 peptides were attached to each DPGC molecule. The quenching efficiency of the probes was quantified by measuring the fluorescence signal intensity of imaging probes to that of free equal-moles of Cy5.5 dye. More than 95% of fluorescent quenching was obtained.
In vitro validation of probe activation
Imaging probes a and b (20–25 μl, 1 nmole) were incubated with 46 pmole of Cath E or Cath D (Calbiochem, La Jolla, CA) in 50 mM sodium acetate buffer, pH 4, containing 150 mM NaCl, and the total volume was brought to 100 μL using the same buffer. All assays were performed in triplicate in 96-well black walls, clear-bottom plates (Corning, NY, USA). The change in the fluorescence intensity was monitored over time using a fluorescence spectrophotometer (SpectraMax.M2e, Molecular Devices, Sunnyvale, CA, USA) at an excitation wavelength (λex) of 645 nm and an emission wavelength (λem) of 695 nm at 27 °C. Control experiments were performed simultaneously by replacing the enzyme with assay buffer.
Cell lines
Two cell lines with different Cath E expressions were used for this study. Cath E expressing pancreatic cancer cell line (Mpanc96)39 and FG30 lentiviral plasmid40 were kindly provided by Dr. Timothy J. Eberlein (St. Louis, MO) and Dr. Xiao-Feng Qin (M. D. Anderson Cancer Center), respectively. The Cath E expression was augmented in Mpanc96 cells by infection with recombinant non-replicative FG30 lentiviral plasmid containing human Cath E cDNA. The Cath E vector was cotransfected with packaging constructs pRSVREV, pMDLg/pRRE, and VSV-G expression plasmid pHCMVG. Lentivirus was produced in 293T cells using the LipofectAMINE reagent (Invitrogen, Carlsbad, CA). MPanc96 cells were infected with lentivirus (25 μl supernatant per ml medium) mixed with polybrene (4 μg ml −1). The 293FT cell line, with lower levels of Cath E expression, was selected to explore the detection sensitivity limits of the developed probe. It is a fast-growing highly transfectable variant of the 293 cell line derived from human embryonic kidney cells transformed with sheared human adenovirus type 5 DNA and stably expressing SV40 large T antigen (Invitrogen, Carlsbad, CA). Both Cath E-positive (Mpanc96-E) and 293FT Cell lines were cultured routinely in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) with 10% FBS. All cells were maintained at 37 °C in a humidified atmosphere of 5% CO2.
Quantitation of Cath E expression
Fluorescence-activated cell sorting was used to quantify Cath E expression. Suspended cells were fixed with 2% paraformaldehyde, permeabilized with 100% cold methanol, blocked for 1 h with 1 × Dulbecco’s PBS (pH 7.4) (Gibco, Carlsbad, CA) containing 250 μg ml−1 heat inactivated BSA (Dulbecco’s PBS/BSA), incubated with primary antibody Cath E H-40 rabbit polyclonal sc-30055 (Santa Cruz Biotechnologies, Santa Cruz, CA) or rabbit (DA1E) mAb IgG XP isotype control (Cell Signaling Technology, Danvers, MA) for 1 hour at 4 °C and washed three times with Dulbecco’s PBS/BSA. Cells were then incubated with anti-rabbit IgG, F(ab′)2 fragment (Alexa fluor® 647 Conjugate), antibody (Cell Signaling Technology, Danvers, MA) and analyzed on a Dako CyAn ADP analyzer (Beckman Coulter, Brea, CA). Relative fluorescence values from the IgG only samples were subtracted from the samples treated with primary antibody.
Animal preparation
All animal studies were performed in compliance with the approved animal protocols and guidelines of Institutional Animal Care and Use Committee at The Methodist Hospital Research Institute and MD Anderson Cancer Center. To prepare the tumor cells for inoculation, MPanc96-E human pancreatic cancer cells and 293FT cells were detached from the culture dish using 0.25% trypsin–EDTA (1×) and suspended in DMEM high glucose cultural media containing 10% FBS. After centrifugation and culture media aspiration, the resultant cell pellet was suspended in Dulbecco’s phosphate-buffered saline solution. Approximately 6 million MPanc96-E or 293FT tumor cells in 100 μl normal saline were implanted subcutaneously under aseptic conditions into both hind legs or shoulders of each immunocompromised nude (Nu/Nu) male mice (5–6 weeks old, average weight, 28 g, Charles River, Wilmington, MA, USA) under anesthesia, 1.5–3% isoflurane inhalation with supplemental oxygen (1–2 L min−1), via a nose cone connected to an Isoflurane vaporizer (VetEquip, Pleasanton, CA, USA). The mice were inspected daily until tumors were grown to 3–5 mm (3–4 weeks), as measured by calipers. Mice were fed with chlorophyll-free diet for optical imaging studies.
Fluorescence imaging studies
Two groups of MPanc96-E tumor-bearing mice (n = 6 per group) were injected a single dose of optical imaging probes a or b (2 nmol/100 μl PBS) intravenously through tail vein using a 30 gauge needle. The third group (n = 6) of 293FT/MPanc96-E double-tumor bearing nude mice was injected a single dose of imaging probe a (2 nmol/100 μl PBS) intravenously. An untreated control group of 4 tumor-bearing Nu/Nu mice were administrated a single dose of 100 μl normal saline. The anesthetized mice were placed on a prone position without any constrain or compression in a heated imaging platform of a small animal dedicated optical imaging system (IVIS-200/100, Xenogen/Caliper, Mountain View, CA). White light and NIRF images were acquired sequentially using Cy5.5 fluorescence filters (615–665 nm excitation filter and 695–770 nm emission filter). The mice were imaged before and immediately after the probe administration and latter sequentially at 2, 4, 8, 24, 48 and 72 h PI. Following the last image acquisition, mice were sacrificed by cervical dislocation and organs/tissues of interest were collected. Excised organs/tissues were rinsed with PBS and imaged for their associated NIR fluorescence. Fluorescence variations between different organs/tissues were corrected by subtracting the autofluorescence signals obtained from mice with no probe. General illumination setting and image acquisition parameters were: epi-illumination; 0.5 s exposure time; f/stop = 2; binning (HR) 4; field of view (FOV = 12.9 cm or 6.5 cm width and height). The mean fluorescence flux from each image was defined as photons per second per centimetre squared per steradian (p−1 s−1 cm−2 sr−1). Images were collected and corrected for flat field and cosmic fluorescence. Acquired images were analyzed using Living Image 3.1 software (Xenogen/Caliper, Alameda, CA). Fluorescence contrast was quantified by measuring the radiance within identical size regions-of-interest (ROI) on the animal image.
Statistical and image analyses
For quantitation, the value of each pixel in images was presented as a fractional ratio of the fluorescent emitted photons per incident excitation photon. This presentation reduces the effects of the illumination variations across the FOV of the imaging system, and minimizes the background autofluorescence originated from biological components other than the fluorophore of interest.41-43 Imaging data were presented as mean ±SD. Student’s paired-sample t-tests were used to determine whether a significant difference exists within and between different groups. P value ≤ 0.001 was considered to be significant. For tumor signal analysis, ROI were applied to measure the peak fluorescence efficiency. The target-to-non-target ratio was defined as the tumor fluorescence efficiency divided by the non-tumor tissue fluorescence efficiency.
Conclusions
The developed imaging probe a allows selective visualization of the molecular alternations in Cath E-positive tumor xenografts at the molecular levels through non-invasive detection of the proteolysis in vivo. The present study displayed the unique capability of the distinctive peptide substrate to specifically detect Cath E proteolytic activity in tissues. The main advantages of the developed protease-cleavable targeting probe are its ability to visualize the early enzymatic alternation in cancer cells in contrast to traditional late morphological remodeling. This molecular imaging approach could not only facilitate the early diagnosis of cancers but also benefit treatment and surgical procedures.
Supplementary Material
Acknowledgements
This research was supported, in part, by National Institute of Health (NIH) Grants by NIH CA135312 (CT) and by The Lockton Endowment to The University of Texas M. D. Anderson Cancer Center (CL).
Abbreviations list
- Cath E
Cathepsin E
- MPanc96
Human pancreatic cancer cells
- PDAC
Pancreatic ductal adenocarcinoma
- 293FT
Human embryonic kidney cells
- PET
Positron emission tomography
- DPGC
d-Polyethylene glycol protected grafted copolymer
- NIR
Near infrared
- PI
Post injection
- RP-HPLC
Reversed phase high performance liquid chromatography
- MS
Mass spectra
- UV
Ultraviolet
- TNBS
2,4,6-Trinitrobenzenesulfonic acid
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c1mb05215b
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Schneider G, Siveke JT, Eckel F, Schmid RM. Gastro-enterology. 2005;128:1606–1625. doi: 10.1053/j.gastro.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 2.von Burstin J, Eser S, Seidler B, Meining A, Bajbouj M, Mages J, Lang R, Kind AJ, Schnieke AE, Schmid RM, Schneider G, Saur D. Int. J. Cancer. 2008;123:2138–2147. doi: 10.1002/ijc.23780. [DOI] [PubMed] [Google Scholar]
- 3.Funovics M, Weissleder R, Tung CH. Anal. Bioanal. Chem. 2003;377:956–963. doi: 10.1007/s00216-003-2199-0. [DOI] [PubMed] [Google Scholar]
- 4.Reichert M, Saur D, Hamacher R, Schmid RM, Schneider G. Cancer Res. 2007;67:4149–4156. doi: 10.1158/0008-5472.CAN-06-4484. [DOI] [PubMed] [Google Scholar]
- 5.Hood JD, Cheresh DA. Nat. Rev. Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 6.Kelly KA, Bardeesy N, Anbazhagan R, Gurumurthy S, Berger J, Alencar H, Depinho RA, Mahmood U, Weissleder R. PLoS Med. 2008;5:e85. doi: 10.1371/journal.pmed.0050085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montet X, Weissleder R, Josephson L. Bioconjugate Chem. 2006;17:905–911. doi: 10.1021/bc060035+. [DOI] [PubMed] [Google Scholar]
- 8.Reubi JC, Waser B. Eur. J. Nucl. Med. Mol. Imaging. 2003;30:781–793. doi: 10.1007/s00259-003-1184-3. [DOI] [PubMed] [Google Scholar]
- 9.Reubi JC, Korner M, Waser B, Mazzucchelli L, Guillou L. Eur. J. Nucl. Med. Mol. Imaging. 2004;31:803–810. doi: 10.1007/s00259-004-1476-2. [DOI] [PubMed] [Google Scholar]
- 10.Korner M, Hayes GM, Rehmann R, Zimmermann A, Friess H, Miller LJ, Reubi JC. Am. J. Pathol. 2005;167:959–968. doi: 10.1016/S0002-9440(10)61186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuo K, Kobayashi I, Tsukuba T, Kiyoshima T, Ishibashi Y, Miyoshi A, Yamamoto K, Sakai H. Hum. Pathol. 1996;27:184–190. doi: 10.1016/s0046-8177(96)90373-1. [DOI] [PubMed] [Google Scholar]
- 12.Tenti P, Romagnoli S, Silini E, Zappatore R, Giunta P, Stella G, Carnevali L. Pathol., Res. Pract. 1994;190:342–349. doi: 10.1016/S0344-0338(11)80406-1. [DOI] [PubMed] [Google Scholar]
- 13.Lin CK, Lai KH, Lo GH, Cheng JS, Hsu PI, Mok KT, Tseng HH. Chin. Med. J. 2001;64:331–336. [PubMed] [Google Scholar]
- 14.Uno K, Azuma T, Nakajima M, Yasuda K, Hayakumo T, Mukai H, Sakai T, Kawai K. J. Gastroenterol. Hepatol. 2000;15:1333–1338. [PubMed] [Google Scholar]
- 15.Azuma T, Hirai M, Ito S, Yamamoto K, Taggart RT, Matsuba T, Yasukawa K, Uno K, Hayakumo T, Nakajima M. Int. J. Cancer. 1996;67:492–497. doi: 10.1002/(SICI)1097-0215(19960807)67:4<492::AID-IJC5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.Zaidi N, Kalbacher H. Biochem. Biophys. Res. Commun. 2008;367:517–522. doi: 10.1016/j.bbrc.2007.12.163. [DOI] [PubMed] [Google Scholar]
- 17.Luker GD, Luker KE. J. Nucl. Med. 2008;49:1–4. doi: 10.2967/jnumed.107.045799. [DOI] [PubMed] [Google Scholar]
- 18.Nichols MT, Russ PD, Chen YK. Pancreas. 2006;33:211–220. doi: 10.1097/01.mpa.0000227912.71202.2c. [DOI] [PubMed] [Google Scholar]
- 19.Wallace MB. Gastroenterology. 2007;132:484–487. doi: 10.1053/j.gastro.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Kaida S, Cabral H, Kumagai M, Kishimura A, Terada Y, Sekino M, Aoki I, Nishiyama N, Tani T, Kataoka K. Cancer Res. 2010;70:7031–7041. doi: 10.1158/0008-5472.CAN-10-0303. [DOI] [PubMed] [Google Scholar]
- 21.Pakzad F, Groves AM, Ell PJ. Semin. Nucl. Med. 2006;36:248–256. doi: 10.1053/j.semnuclmed.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Mertz HR, Sechopoulos P, Delbeke D, Leach SD. Gastrointest. Endosc. 2000;52:367–371. doi: 10.1067/mge.2000.107727. [DOI] [PubMed] [Google Scholar]
- 23.Ford EC, Herman J, Yorke E, Wahl RL. J. Nucl. Med. 2009;50:1655–1665. doi: 10.2967/jnumed.108.055780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schellenberg D, Goodman KA, Lee F, Chang S, Kuo T, Ford JM, Fisher GA, Quon A, Desser TS, Norton J, Greco R, Yang GP, Koong AC. Int. J. Radiat. Oncol., Biol., Phys. 2008;72:678–686. doi: 10.1016/j.ijrobp.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 25.Abd-Elgaliel WR, Tung CH. Biochim. Biophys. Acta. 2010;1800:1002–1008. doi: 10.1016/j.bbagen.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuda Y, Kohmura K, Kadowaki T, Tsukuba T, Yamamoto K. Biol. Chem. 2005;386:299–305. doi: 10.1515/BC.2005.036. [DOI] [PubMed] [Google Scholar]
- 27.Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K. J. Biochem. 1999;125:1137–1143. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]
- 28.Gulnik SV, Suvorov LI, Majer P, Collins J, Kane BP, Johnson DG, Erickson JW. FEBS Lett. 1997;413:379–384. doi: 10.1016/s0014-5793(97)00886-7. [DOI] [PubMed] [Google Scholar]
- 29.Baechle D, Cansier A, Fischer R, Brandenburg J, Burster T, Driessen C, Kalbacher H. J. Pept. Sci. 2005;11:166–174. doi: 10.1002/psc.607. [DOI] [PubMed] [Google Scholar]
- 30.Tung CH. Biopolymers. 2004;76:391–403. doi: 10.1002/bip.20139. [DOI] [PubMed] [Google Scholar]
- 31.Law B, Tung CH. Bioconjugate Chem. 2009;20:1683–1695. doi: 10.1021/bc800500a. [DOI] [PubMed] [Google Scholar]
- 32.Kageyama T, Takahashi K. J. Biochem. 1980;87:725–735. doi: 10.1093/oxfordjournals.jbchem.a132801. [DOI] [PubMed] [Google Scholar]
- 33.Scarborough PE, Guruprasad K, Topham C, Richo GR, Conner GE, Blundell TL, Dunn BM. Protein Sci. 1993;2:264–276. doi: 10.1002/pro.5560020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kageyama T. Methods Enzymol. 1995;248:120–136. doi: 10.1016/0076-6879(95)48010-2. [DOI] [PubMed] [Google Scholar]
- 35.Huang XF, Wang CM, Dai XW, Li ZJ, Pan BR, Yu LB, Qian B, Fang L. World J. Gastroenterol. 2000;6:693–698. doi: 10.3748/wjg.v6.i5.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moles A, Tarrats N, Fernandez-Checa JC, Mari M. Hepatology. 2009;49:1297–1307. doi: 10.1002/hep.22753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tung CH, Bredow S, Mahmood U, Weissleder R. Bioconjugate Chem. 1999;10:892–896. doi: 10.1021/bc990052h. [DOI] [PubMed] [Google Scholar]
- 38.Law B, Curino A, Bugge TH, Weissleder R, Tung CH. Chem. Biol. 2004;11:99–106. doi: 10.1016/j.chembiol.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Peiper M, Nagoshi M, Patel D, Fletcher JA, Goegebuure PS, Eberlein TJ. Int. J. Cancer. 1997;71:993–999. doi: 10.1002/(sici)1097-0215(19970611)71:6<993::aid-ijc15>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 40.Qin XF, An DS, Chen IS, Baltimore D. Proc. Natl. Acad. Sci. U. S. A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Mol. Imaging. 2004;3:9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 42.Mansfield JR, Gossage KW, Hoyt CC, Levenson RM. J. Biomed. Opt. 2005;10:41207. doi: 10.1117/1.2032458. [DOI] [PubMed] [Google Scholar]
- 43.Bogaards A, Sterenborg HJ, Trachtenberg J, Wilson BC, Lilge L. Lasers Surg. Med. 2007;39:605–613. doi: 10.1002/lsm.20525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.