Target audience
Scientists and clinicians studying large lesions or pathologies with magnetic resonance spectroscopy.
Purpose
Potentially useful clinical MRS chemical shift imaging (CSI) acquisitions are often precluded by long study times. The newly-proposed Spectroscopy with Linear Algebraic Modeling (SLAM) method1 can provide compartmental-average spectra many-fold faster than CSI and at little cost to signal-to-noise ratio, by incorporating information from scout MRI1,3. Prior work demonstrated quantitative agreement in 31P 1D CSI heart studies1. Here, SLAM with a 6 to14-fold speed-up is extended to 2D 1H MRS studies of patients with brain tumors. Quantitative results are compared with CSI.
Methods
Proactive SLAM is implemented as follows: (i) Acquire MRI; (ii) Apply a fraction of the central k-space CSI phase encodes based on the number of compartments and speed-up factor; (iii) Co-register the CSI grid with the MRI; (iv) Segment the CSI voxels into the desired MRI-based compartments; (iv) Reconstruct compartmental spectra using SLAM. SLAM is tested retroactively on CSI data, by using only the central k-space phase-encoded signals in Step (ii).
SLAM reconstruction was implemented on1H CSI data from 15 brain tumor patients undergoing clinical/research MRI and MRS in a 3T Philips scanner, using 1/6 of the acquired (central k-space) CSI data. Five compartments—tumor, contralateral, brain (excluding tumor and contralateral), scalp and background— were segmented for SLAM reconstruction. Cho/Cr ratios were quantified in compartment spectra and compared with the compartmental average CSI spectra.
The proactive SLAM protocol was applied to volunteers after CSI protocol, using a SLAM speedup factor of ~14. Four compartments—a user-defined “lesion”, brain, scalp and background—were assigned and results compared to CSI.
Results
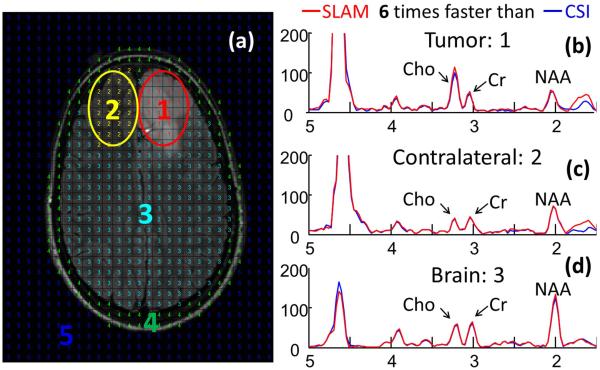
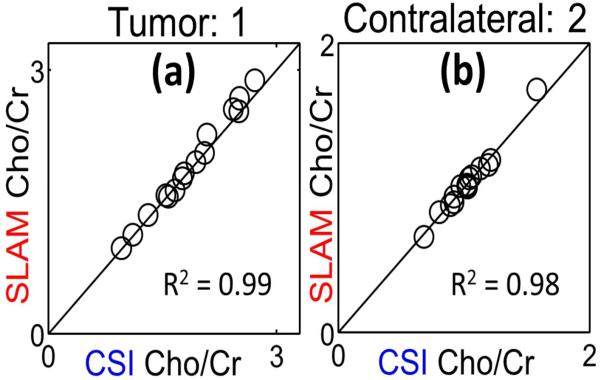
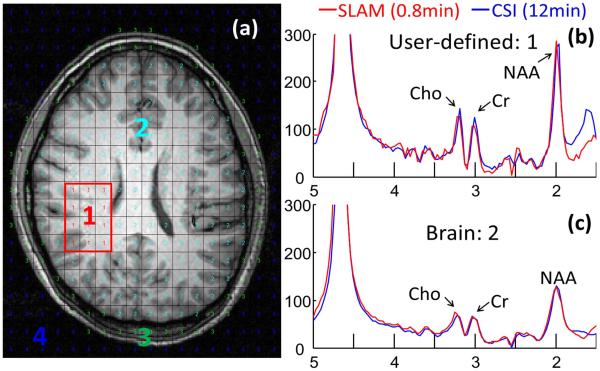
Fig. 1 shows segmentation in a brain tumor study (a), and the corresponding spectra (b-d) reconstructed from CSI (blue) and SLAM (red). Fig. 2 shows that SLAM Cho/Cr in tumor (a) and contralateral (b) compartments agree with CSI(vertical) in 15 patients. Fig. 3 shows proactive implementation of SLAM on a normal volunteer.
Fig. 1.
Retroactive SLAM using central 12×13 phase encodes out of original 27×33 CSI phase encodes. (a) Brain segmentation into 5 compartments. SLAM (red) spectra overlaid on CSI (blue) spectra for the tumor compartment #1 (b), the contralateral compartment #2 (c) and brain compartment #3 (d).
Fig. 2.
SLAM vs. CSI choline/creatine (Cho/Cr) ratios in 15 brain tumor patients. SLAM used 1/6 of CSI data.
Fig. 3.
(a) Brain segmentation from normal volunteer with 4 compartments. (b) Proactive 0.8 min SLAM spectrum (red) from compartment #1 compared to 12 min CSI acquisition (blue). (c) 0.8 min SLAM and 12 min CSI spectra from brain compartment #2 (excluding compartment #1).
Conclusion
SLAM was successfully applied retroactively to patients with brain tumors. It is also implemented proactively for brain studies. SLAM yielded spectra and quantitative results almost identical to CSI, but much faster (≤14 times). SLAM could enable whole-brain MRS in clinical studies that were otherwise precluded by long MRS acquisition times.
Acknowledgments
Grant support: EB007829, HL61912, EB009731
References
- [1].Zhang Y,, et al. JMR. 2012;218:66–76. doi: 10.1016/j.jmr.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brown TR,, et al. PNAS. 1982;79:3523–3526. doi: 10.1073/pnas.79.11.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhang Y,, et al. Proc. ISMRM. 2012:0709. [Google Scholar]