Abstract
On-chip detection of low abundant protein biomarkers is of interest to enable point-of-care diagnostics. Using a simple form of integration, we have realized an integrated microfluidic platform for the detection of prostate specific antigen (PSA), directly in anti-coagulated whole blood. We combine acoustophoresis-based separation of plasma from undiluted whole blood with a miniaturized immunoassay system in a polymer manifold, demonstrating improved assay speed on our Integrated Acoustic Immunoaffinity-capture (IAI) platform.
The IAI platform separates plasma from undiluted whole blood by means of acoustophoresis and provides cell free plasma of clinical quality at a rate of 10 uL/min for an online immunoaffinity-capture of PSA on a porous silicon antibody microarray. Whole blood input (hematocrit 38-40%) rate was 50μl/min giving a plasma volume fraction yield of ≈33%.
PSA was immunoaffinity-captured directly from spiked female whole blood samples at clinically significant levels of 1.7-100ng/ml within 15 minutes and was subsequently detected via fluorescence readout, showing a linear response over the entire range with a coefficient of variation of 13%.
Introduction
On-chip detection of low abundant protein biomarkers is of interest to enable point-of-care (POC) diagnostics. However, detecting the biomarkers of interest directly from whole blood samples presents a high degree of complexity1 such as interference or non-specific binding from the cellular elements in blood.2 In addition, biomarker targets may only be present at low abundance, are rapidly degraded or eliminated by other mechanisms in vitro. Still, blood serum and plasma remain the most widely used biofluid in clinical diagnostics due to the fact that the blood/plasma biomarker profile reflects physiological and pathological changes relating to disease.3, 4 Although conventional immunoassays/antibody-validation assays, such as ELISA, have been the golden standard in clinical diagnostics, improvements with regards to time, sample/reagent consumption, portability and throughput are imperative. To implement a total LOC platform, the challenge lies not only in dealing with the sample complexity but also the sensitivity and specificity of the subsequent diagnostic assay, which needs to be simultaneously tackled. With regards to the needs of realizing a total LOC platform, developments using microfluidic technology has opened new possibilities for the detection of disease-correlated biomarkers from complex biological samples such as blood/plasma5-8 and urine.9, 10 Much of the recent advancements in microfluidic-based lab on a chip approaches that target POC settings11, 12 involves efforts towards full system integration, increased throughput, multiplexing, cost-efficiency, rapid ‘sample to answer’, miniaturized immunoassay systems. Reports on microfluidic immunoassay platforms using diffusion13, 14, surface/beads-immobilized15, 16, centrifugal17, 18 and other separation-based approaches19, 20, demonstrate a rapid progress in miniaturizing conventional immunoassays, taking into account the complexity of the unprocessed, whole blood samples.
Reduction of the sample complexity due to interference from blood cells in the whole blood sample is crucial to ensure low limit of detection for the biomarkers of interest. In this case, separation-based microfluidics relies on the capability of the system to perform efficient on-chip separation of plasma by removing the blood cells prior to immunoaffinity-capturing of the targeted biomarkers. For example, several studies specifically utilizing membrane filters21, 22, Zweifach-Fung effect19, 23 (bifurcation law) and acoustophoresis-based separations24 have shown successful combinations of on-chip separations and immunoassays. Being able to handle high cellular contents e.g. undiluted blood, these approaches offer a potential for the development of fully-integrated microfluidic whole-blood immunoassay platforms.
We here present an integrated microfluidic platform that uses acoustophoresis to extract plasma from whole blood and performs simultaneous immunoaffinity-capturing of a prostate cancer biomarker within 15 minutes. We have previously reported on an acoustophoresis-based microchip, which was capable of generating diagnostic quality anti-coagulated plasma from undiluted whole blood samples.24 It was further linked to a potential clinical application by measuring prostate-specific antigen (PSA) off-line on a porous silicon sandwich antibody microarray chip. In this work, we have proceeded to develop an integrated device where the porous silicon microarray chip was connected directly to the outlet of the plasmapheresis chip, namely, the Integrated Acoustic Immunoaffinity-capture (IAI) platform.
Materials and Methods
Proteins and Reagents
Prostate Specific Antigen (PSA) was obtained from Sigma. Anti-PSA monoclonal mouse antibodies 2E9 and H117 were produced and characterized as previously described.25, 26 2E9 monoclonal antibody was labelled with fluorescein isothiocyanate (FITC) isomer I-Celite (Sigma St. Louis, MO, USA) and separated on a PD10 column (Amersham, Uppsala, Sweden).
Blood samples
Citrated blood samples from healthy female blood donors were obtained from Blood Center Skåne (Lund University Hospital, Sweden). The hematocrit level of the whole blood samples were measured by a hematocrit centrifuge (Hematocrit 210, Hettich, Tuttlingen, Germany). To determine the red blood cell content of the plasma samples after acoustic separation, a Coulter Counter (Multisizer 3, Beckman Coulter Inc., Fullerton, CA) was used to count cells in the range of 4-8 μm.
For analysis, a titration series of PSA-spiked female whole-blood samples was made in the range of 1-100 ng/ml. As a reference, fractions of the spiked whole blood samples were centrifuged for plasma separation prior to determination of total PSA concentration, using DELFIA Prostatus PSA free/total assay, a commercially available quantitative time-resolved fluoroimmunoassay targeting both free and total PSA (Perkin- Elmer, Turku, Finland).27
IAI platform
The IAI platform has two major components:
the acoustic plasmapheresis microchip
the porous silicon microarray for on-chip immunoaffinity-capturing of PSA.
Both components were integrated in a polymer manifold encompassing both the plasmapheresis microchip and a flow cell holding the porous silicon PSA microarray.
Acoustic Plasmapheresis microchip fabrication
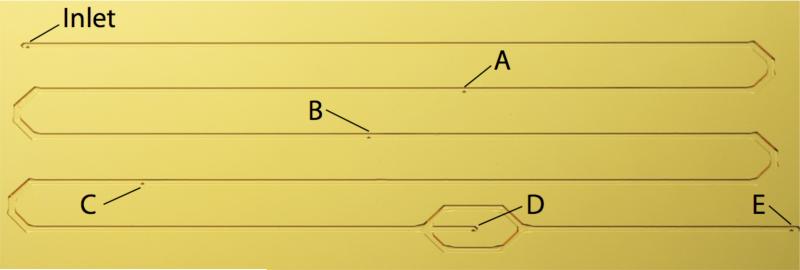
Briefly, the acoustic plasmapheresis microchip consists of a meander microchannel with a whole blood inlet, four blood cell waste outlets (outlets A-D, Figure 1) and one plasma outlet (outlet E, Figure 1). The four waste outlets were centred at the bottom along the microchannel. A more detailed description of the acoustic plasmapheresis chip has been described by Lenshof et al.24
Figure 1.
The acoustic plasmapheresis microchip with multiple outlet configuration (A-D) along the meander shaped separation microchannel for RBC removal and (E) extraction of pure plasma.
The immunoaffinity-capture region: Porous Silicon Microarray fabrication
Porous silicon chips were obtained by anodic dissolution of a p-type monocrystalline silicon wafer. A detailed description of the fabrication process has been described by Jaeras et al.28
Monoclonal mouse anti-PSA capture antibody H117 (0.5mg/ml in PBS) was arrayed onto the porous silicon chips using an in-house developed piezoelectric microdispenser29,30 forming an array of 600 antibody spots (100 pl/spot), at a 150 μm centre to centre distance. The chips were washed with a 3 time washing step in PBS-tween (0.05% tween 20 in PBS) to remove loosely bound capture antibodies. Finally, blocking with 5% non-fat dry milk was done prior to the insertion of the porous silicon chips to the IAI platform.
Microarray analysis
To perform analysis on the microarray, a confocal microscope setup (BX51W1, Olympus), oil immersion 20× objective, an ion laser (IMA101010BOS, Melles Griot Laser Group), with an excitation wavelength of 488nm was used for fluorescence detection. Microarray image analysis was performed using Fluoview 300 software (Fluoview, Olympus). The Fluoview 300 circle method was used to quantify the total intensity of each spot that was detected on the microarray. Background intensity was similarly measured and then subtracted from the total intensity of the spot. Nine spots and their backgrounds were measured for each image analysis, generating the mean spot intensities presented in the figures.
Results & discussion
Optimisation of flow based microarray assay
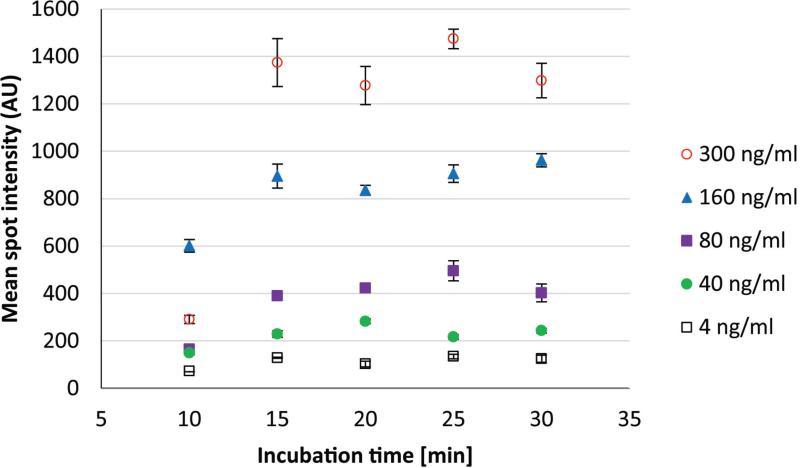
The continuous flow-based incubation of the microarray on the IAI platform offers different conditions for the plasma biomarkers to reach the immobilized antibodies on the porous silicon surface as compared to the previously reported microtitre plate-based porous silicon microarray assay.31 In order to optimize the sample incubation process in the IAI platform, PSA immunoaffinity-capturing was performed at incubation times ranging from 10-30 minutes. Obtained data showed that 15 minutes of PSA-spiked plasma perfusion was sufficient for PSA to bind to the immobilized anti-PSA H117 antibody in the continuous flow system, in the range of clinically relevant levels (4-300 ng/ml), Figure 2.
Figure 2.
Fluorescence detection results for PSA-spiked female plasma sample. PSA-spiked plasma perfused a flow cell holding the porous silicon antibody microarray for 10, 15, 20, 25 and 30 minutes (X-axis) to optimize the continuous flow PSA assay. Mean spot intensities (Y-axis) and standard deviation (error bars) were calculated from spots (N=9) obtained from microarray image analysis. Note that incubation time of more than 15 minutes did not result in a significantly higher mean spot intensity, thus this time was selected for the subsequent IAI protocol.
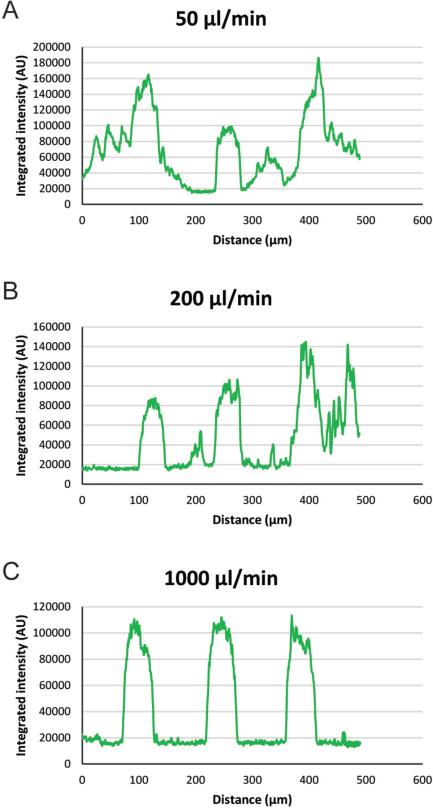
To optimize the washing steps in the PSA assay, 5 minute washing, after the plasmaincubation, at flow rates of 50, 100, 200, 500 and 1000 μl/min were investigated. Based on spot intensity profiles obtained by microarray image analysis, results show that a flow rate of 1000 μl/min (0.05 % Tween 20 in PBS) was sufficient to reduce background (Figure 3).
Figure 3.
Spot intensity profiles obtained via microarray image analysis. Washing buffer (0.05 % Tween 20 in PBS) was aspirated through the microarray flow cell at various flow rates for 5 minutes. Representative spot profiles for 50, 200 and 1000 are shown.
Whole blood analysis
One significant step towards more advanced POC diagnostics includes miniaturization of the conventional immunoassays. In this perspective we evaluated the application of the IAI platform to assay PSA, using female plasma samples spiked with PSA.
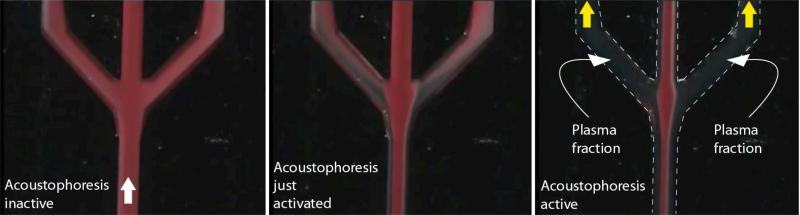
The plasmapheresis microchip has been modified relative our earlier generation to improve the plasma separation/generation and enable integration with the microarray flow cell. The plasmapheresis chip was slightly elongated with a separation channel length of 238 mm (compared to 224 mm in earlier design), yet having the same number of outlets. The microchip was placed in a manifold to couple the plasma microchannel outlet into the flow cell holding the porous silicon microarray chip. PSA-spiked undiluted whole blood was drawn through the acoustic plasmapheresis chip by software controlled syringe pumps (neMESYS, Cetoni, GmbH, Germany). The acoustophoresis chip was actuated according to the previously described protocol,24 and hence a half wave length ultrasonic standing wave was used to accomplish on-chip separation of anti-coagulated plasma from red and white blood cells.32-34 As seen in Figure 4, the primary acoustic radiation force focus the blood cells into the pressure node of the standing wave field, moving them to the centre of the microfluidic channel, while a cell free plasma emerges along the channel sidewalls. Lower flow rates were applied in all waste outlets (outlet A-D) as compared to the earlier design, see Table 1. This contributed to a longer retention time for acoustic focusing and hence improved the plasma separation efficiency. The lower total flow rate of the acoustic plasmapheresis microchip also resulted in a lower consumption of whole blood sample.
Figure 4.
Sequence showing the starting phase of plasma production where A) ultrasound is inactive, B) the acoustophoresis is beginning to focus the RBC in the channel centre and C) continuous phase of plasma production where the final fraction of RBC are removed via the central outlet.
Table 1.
Flow rates for inlet and outlets A-E, plasma yield and the amount of cells/L of the generated plasma for the improved acoustic plasmapheresis microchip as compared to our previously reported microchip design.
| Inlet (μl/min) | Waste outlet A (μl/min) | Waste outlet B (μl/min) | Waste outlet C (μmin) | Waste outlet D (μl/min) | Plasma outlet E (μl/min) | Fraction of plasma from whole blood | Erythrocytes/L in produced plasma | |
|---|---|---|---|---|---|---|---|---|
| Previous design | 80 | 20 | 20 | 15 | 15 | 10 | 12.5 % | 3.7×109 |
| Improved design | 50 | 10 | 10 | 10 | 10 | 10 | 20% | 4.2×109 |
To deplete the focused blood cells from the plasma, these were aspirated through the waste outlet A-D located in the bottom centre of the microchannel, Figure 1. The sequential removal of focused blood cells via multiple outlets along the bottom center of the microfluidic channel gradually reduced the hematocrit level. The trifurcation at the end of the microchannel provided the final cell separation, yielding high quality plasma with a low cellular content through the side outlets. The plasma was generated at a rate of 10 ul/min from undiluted whole blood (38-40 % hematocrit) flow rate of 50 μl/min as compared to 80 μl/min in earlier version. A higher plasma yield of 33 % of the total plasma volume was achieved as compared to the 21 % in the previous generation. Flow rate settings, plasma yield and plasma cell count are given in Table 1. The plasma generated in the new set-up showed a slightly higher cell content than previous version, which can be attributed to the fact that a larger fraction of the total plasma volume is extracted for the diagnostic step. It should, however, be commented that the cell background was still well within the criterions < 6*10^9 cells/L, as recommended by the Council of Europe.35
Increasing the extracted plasma fraction inherently increases the risk of having a carry over of cells. It should also be noted that the entire plasma volume in a blood sample is not accessible in the acoustophoresis based approach described herein since the blood cells are not exposed to forces of the same magnitude as in a centrifugation step and hence not packed as densely. By exposing the blood sample to the acoustic force field for a longer time it may be possible to concentrate the blood cells more and thus, accomplish a higher plasma fraction yield but then on the account of either a longer separation channel, higher acoustic input power or lower flow rate.
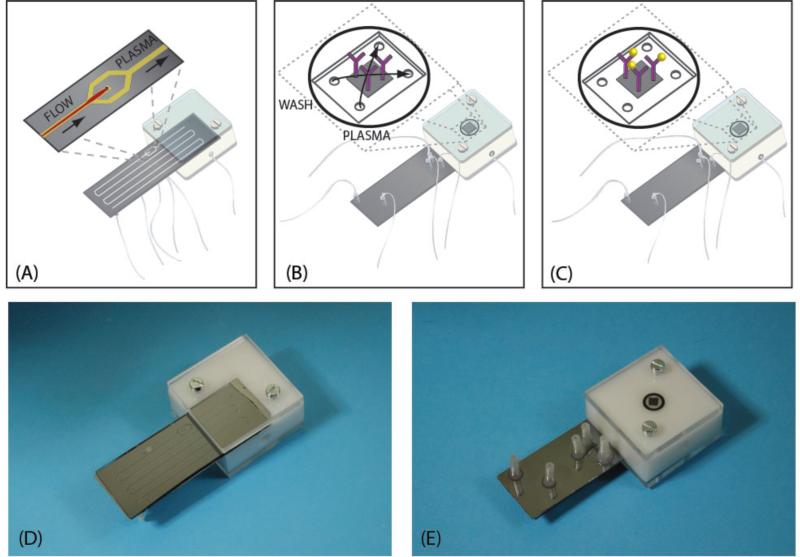
Figure 5 A-E, shows the acoustic plasmapheresis microchip mounted in the manifold that connects the plasma microchannel outlet to the flow cell holding the immunoaffinity-capture microarray (Figure 5B-C). Because of the high surface area attributed by the porous silicon 3D morphology, spots with high antibody density and spot quality were obtained.36, 37 Via outlet E, the clean plasma continuously perfused the microarray flow cell. After 15 minutes of continuous flow plasma incubation, the washing buffer (0.05% tween 20 in PBS) was subsequently aspirated through the flow cell. After incubation the PSA microarray was subjected to off-line 10μl FITC-labeled secondary antibody incubation and fluorescence detection. The on-line processing and the short (15 minutes) PSA immunoaffinity-capture time could reduce the risk of plasma protein degradation induced by the standard blood pre-processing and extended time before analysis.38 Hence, this approach opens the route to more accurate assaying of other more delicate biomarkers.
Figure 5.
The Integrated Acoustic Immunoaffinity-capture (IAI) platform. The plasma microchannel outlet is connected to the flow cell containing the porous silicon immunoaffinity-capture region which is perfused by the acoustophoretically generated plasma for 15 minutes. A) The acoustophoresis-based plasma separation region. B) The porous silicon immunoaffinity-capture microarray region at the back side of the IAI manifold containing the microarrayed anti-PSA H117 antibody (in purple). C) Immunoaffinity-capture of PSA (in yellow) by the anti-PSA H117 antibody. D) Photo of the IAI manifold. E) Photo of the IAI manifold (back side).
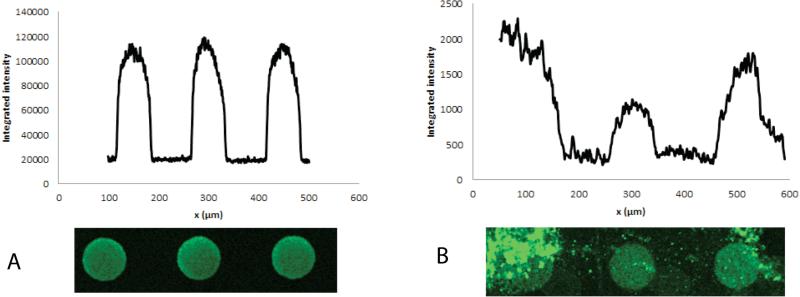
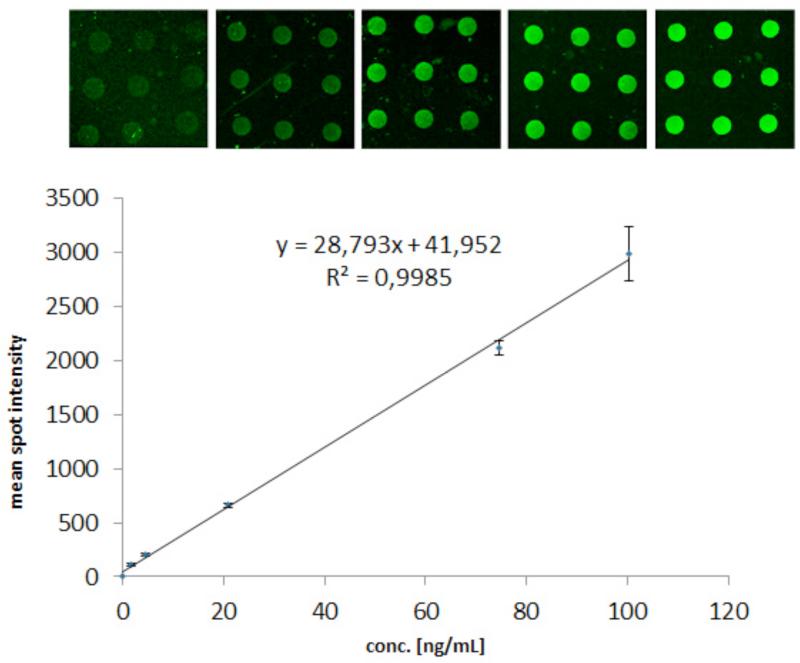
The IAI protocol for whole blood samples spiked with PSA generated high quality/homogeneous spots (Table 2). An example of a spot intensity profile obtained from the porous silicon PSA microarray can be seen in Figure 6A. Figure 6B shows the effect of non-optimal manual washing steps that now has been successfully eliminated via automated and reproducible aspiration of washing buffer on the IAI platform. The mean spot intensities and coefficient of variation (CV) of Table 2 were calculated from the spots of the insert images of Figure 7. CVs ~13% was in agreement with previously published work.28 The graph in Figure 7 shows mean spot intensity versus PSA concentration as determined by the DELFIA reference assay. This corresponded to a coefficient of determination of R2 > 0.99, indicating a good linearity within the studied concentration range. The error bars show the standard deviations calculated from the spot intensities. Based on duplicate runs of the IAI protocol, PSA was detectable at clinically significant levels of 1.7-100 ng/ml after 15 minutes of immunoaffinity-capturing at 50 μl/min total flow rate of whole blood via fluorescence readout. Although the lowest concentration measured in blood samples in this study was 1.7 ng/ml as lowest, we have previously shown that the porous silicon sandwich assay used herein has a limit of detection of 0.14 ng/ml PSA28. Unspiked human female whole blood sample was used as a negative control in our experiments. No unspecific binding from plasma proteins was found (data not shown).
Table 2.
PSA concentrations (DELFIA), mean spot intensities, and the corresponding standard deviations and coefficients of variation.
| Concentration, ng/ml | Mean spot intensity, AU | Standard deviation, AU (n=9) | CV% (n=9) |
|---|---|---|---|
| 100 | 2983 | 507 | 17 |
| 75 | 2116 | 133 | 6.3 |
| 21 | 660 | 42 | 6.3 |
| 4 | 200 | 26 | 13.1 |
| 1.7 | 110 | 22 | 20.2 |
Figure 6.
A) An example of spots and spot intensity profile obtained for PSA detection via fluorescence readout on the IAI platform. B) Effect of non-optimal washing steps on the spot profile.
Figure 7.
Fluorescence readout of the titrated series of PSA-spiked female whole blood obtained via the IAI protocol. The insert images show microarray images of the corresponding concentration ranges. Mean spot intensities (Y-axis) and standard deviations (error bars) were calculated from the spots (N=9) in the images captured via a 20× lens. The PSA concentrations on the X-axis were obtained by the DELFIA reference assay.
Performed as an integrated microfluidic assay, the IAI platform has minimized the number of conventional assay steps resulting in a reduced total assay time and consumption of sample/reagents. Immunoaffinity- capturing of PSA from whole blood, with optimized flow conditions and incubation times, was performed in 15 minutes as compared to 60 minutes on our previous PSA microarray platform. The assay steps performed on the IAI (15 minutes of immunoaffinity-capturing of PSA from whole blood and 5 minutes washing) lasted for a total of 20 minutes which should be compared with at least 75 minutes for the corresponding assay steps performed on our previous PSA microarray platform.
Conclusion & outlook
The IAI platform, in its simplest form of integration, has shown proof of principle for whole blood sample input and biomarker quantitation output. The platform integrates multiple functions i.e. 1) on-chip plasma separation, 2) immunoaffinity-capture and 3) steps of loading samples/reagents/washing buffers. In this paper, we show for the first time microchip integration of acoustically driven plasmapheresis and microarray-based protein biomarker detection, which provides reduced assay times and enables automated microfluidic sample processing.
In oncoming work we anticipate enhancing the diagnostic value in the current PSA microarray by implementing a multiplex microarray holding complementary biomarkers and realizing further steps of integration and miniaturization.
In order to progress the IAI platform to a true sample-in-answer-out Point Of Care system the current off-line incubation of the detector antibody should be integrated in the microfluidic sequence, which is on-going work. The confocal fluorescence readout can be translated to a conventional bench top fluorescence microarray scanner.
Acknowledgements
Financial support is acknowledged from Swedish Research Council [VR 2009-5361 and VR/Vinnova/SSF MTBH 2006-7600 and K 2009-20095 (Medicine)], the Royal Physiographic Society, the Crafoord Foundation, the Carl Trygger Foundation, the SSF Strategic Research Centre (Create Health), funding (grant no. 3455) to H. Lilja from the Swedish Cancer Society, FiDiPro grant support to H. Lilja from TEKES, funds from David H. Koch, provided through the Prostate Cancer Foundation, and the Sidney Kimmel Center for Prostate and Urologic Cancers.
Footnotes
Conflict of interest
Dr Hans Lilja holds patents for free PSA, hK2 and intact PSA assays.
References
- 1.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Toner M, Irimia D. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lathrop JT, Anderson NL, Anderson NG, Hammond DJ. Curr Opin Mol Ther. 2003;5:250–257. [PubMed] [Google Scholar]
- 4.Drake RR, Cazares L, Semmes OJ. Proteom Clin Appl. 2007;1:758–768. doi: 10.1002/prca.200700175. [DOI] [PubMed] [Google Scholar]
- 5.Jiang H, Weng XA, Li DQ. Microfluid Nanofluid. 2011;10:941–964. [Google Scholar]
- 6.Stern E, Vacic A, Rajan NK, Criscione JM, Park J, Ilic BR, Mooney DJ, Reed MA, Fahmy TM. Nat Nanotechnol. 2010;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersaudy-Kerhoas M, Kavanagh DM, Dhariwal RS, Campbell CJ, Desmulliez MPY. Lab Chip. 2010;10:1587–1595. doi: 10.1039/b926834k. [DOI] [PubMed] [Google Scholar]
- 8.Chen WW, Li TS, He S, Liu DB, Wang Z, Zhang W, Jiang XY. Sci China Chem. 2011;54:1227–1232. [Google Scholar]
- 9.Wang J, Chatrathi MP. Anal. Chem. 2003;75:525–529. doi: 10.1021/ac020560b. [DOI] [PubMed] [Google Scholar]
- 10.Garcia CD, Henry CS. Analyst. 2004;129:579–584. doi: 10.1039/b403529a. [DOI] [PubMed] [Google Scholar]
- 11.Huckle D. Expert Rev Med Devic. 2006;3:421–426. doi: 10.1586/17434440.3.4.421. [DOI] [PubMed] [Google Scholar]
- 12.Huckle D. Expert Rev Mol Diagn. 2008;8:679–688. doi: 10.1586/14737159.8.6.679. [DOI] [PubMed] [Google Scholar]
- 13.Hosokawa K, Omata M, Maeda M. Anal. Chem. 2007;79:6000–6004. doi: 10.1021/ac070659o. [DOI] [PubMed] [Google Scholar]
- 14.Hatch A, Kamholz AE, Hawkins KR, Munson MS, Schilling EA, Weigl BH, Yager P. Nat Biotechnol. 2001;19:461–465. doi: 10.1038/88135. [DOI] [PubMed] [Google Scholar]
- 15.Lim TK, Ohta H, Matsunaga T. Anal. Chem. 2003;75:3316–3321. doi: 10.1021/ac020749n. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann O, Voirin G, Niedermann P, Manz A. Anal. Chem. 2002;74:5243–5250. doi: 10.1021/ac025777k. [DOI] [PubMed] [Google Scholar]
- 17.Schaff UY, Sommer GJ. Clinical Chemistry. 2011;57:753–761. doi: 10.1373/clinchem.2011.162206. [DOI] [PubMed] [Google Scholar]
- 18.Riegger L, Grumann M, Nann T, Riegler J, Ehlert O, Bessler W, Mittenbuehler K, Urban G, Pastewka L, Brenner T, Zengerle R, Ducree J. Sensor Actuat a-Phys. 2006;126:455–462. [Google Scholar]
- 19.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin LD, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Browne AW, Ramasamy L, Cripe TP, Ahn CH. Lab Chip. 2011;11:2440–2446. doi: 10.1039/c1lc20144a. [DOI] [PubMed] [Google Scholar]
- 21.Thorslund S, Klett O, Nikolajeff F, Markides K, Bergquist J. Biomed Microdevices. 2006;8:73–79. doi: 10.1007/s10544-006-6385-7. [DOI] [PubMed] [Google Scholar]
- 22.Stevens DY, Petri CR, Osborn JL, Spicar-Mihalic P, McKenzie KG, Yager P. Lab Chip. 2008;8:2038–2045. doi: 10.1039/b811158h. [DOI] [PubMed] [Google Scholar]
- 23.Qin LD, Vermesh O, Shi QH, Heath JR. Lab Chip. 2009;9:2016–2020. doi: 10.1039/b821247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenshof A, Ahmad-Tajudin A, Jaras K, Sward-Nilsson AM, Aberg L, Marko-Varga G, Malm J, Lilja H, Laurell T. Anal. Chem. 2009;81:6030–6037. doi: 10.1021/ac9013572. [DOI] [PubMed] [Google Scholar]
- 25.Pettersson K, Piironen T, Seppala M, Liukkonen L, Christensson A, Matikainen MT, Suonpaa M, Lovgren T, Lilja H. Clinical Chemistry. 1995;41:1480–1488. [PubMed] [Google Scholar]
- 26.Lilja H, Christensson A, Dahlen U, Matikainen MT, Nilsson O, Pettersson K, Lovgren T. Clinical Chemistry. 1991;37:1618–1625. [PubMed] [Google Scholar]
- 27.Mitrunen K, Pettersson K, Piironen T, Bjork T, Lilja H, Lovgren T. Clinical Chemistry. 1995;41:1115–1120. [PubMed] [Google Scholar]
- 28.Jaeras K, Ressine A, Nilsson E, Malm J, Marko-Varga G, Lilja H, Laurell T. Anal. Chem. 2007;79:5817–5825. doi: 10.1021/ac0709955. [DOI] [PubMed] [Google Scholar]
- 29.Laurell T, Wallman L, Nilsson J. Journal of Micromechanics and Microengineering. 1999;9:369–376. [Google Scholar]
- 30.Onnerfjord P, Nilsson J, Wallman L, Laurell T, Marko-Varga G. Analytical Chemistry. 1998;70:4755–4760. doi: 10.1021/ac980207z. [DOI] [PubMed] [Google Scholar]
- 31.Jaras K, Adler B, Tojo A, Malm J, Marko-Varga G, Lilja H, Laurell T. Clin Chim Acta. 2012;414:76–+. doi: 10.1016/j.cca.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson A, Petersson F, Jonsson H, Laurell T. Lab Chip. 2004;4:131–135. doi: 10.1039/b313493h. [DOI] [PubMed] [Google Scholar]
- 33.Hawkes JJ, Barber RW, Emerson DR, Coakley WT. Lab Chip. 2004;4:446–452. doi: 10.1039/b408045a. [DOI] [PubMed] [Google Scholar]
- 34.Harris NR, Hill M, Beeby S, Shen Y, White NM, Hawkes JJ, Coakley WT. Sensor Actuat B-Chem. 2003;95:425–434. [Google Scholar]
- 35.Cardigan R. Guide to the Preparation, Use and Quality Assurance of Blood Components. 13th edn 2007. [Google Scholar]
- 36.Ressine A, Ekstrom S, Marko-Varga G, Laurell T. Anal. Chem. 2003;75:6968–6974. doi: 10.1021/ac034425q. [DOI] [PubMed] [Google Scholar]
- 37.Steinhauer C, Ressine A, Marko-Varga G, Laurell T, Borrebaeck CAK, Wingren C. Anal Biochem. 2005;341:204–213. doi: 10.1016/j.ab.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh SY, Chen RK, Pan YH, Lee HL. Proteomics. 2006;6:3189–3198. doi: 10.1002/pmic.200500535. [DOI] [PubMed] [Google Scholar]