Abstract
Mechanisms associated with type 1 diabetes (T1D) development remain incompletely defined. Using a sensitive array-based bioassay where patient plasma is used to induce transcriptional responses in healthy leukocytes, we previously reported disease-specific, partially interleukin (IL)-1−dependent signatures associated with preonset and recent onset (RO) T1D relative to unrelated healthy control subjects (uHC). To better understand inherited susceptibility in T1D families, we conducted cross-sectional and longitudinal analyses of healthy autoantibody-negative (AA−) high HLA−risk siblings (HRS) (DR3 and/or DR4) and AA− low HLA−risk siblings (LRS) (non-DR3/non-DR4). Signatures, scored with a novel ontology-based algorithm, and confirmatory studies differentiated the RO T1D, uHC, HRS, and LRS plasma milieus. Relative to uHC, T1D family members exhibited an elevated inflammatory state, consistent with innate receptor ligation that was independent of HLA, AA, or disease status and included elevated plasma IL-1α, IL-12p40, CCL2, CCL3, and CCL4 levels. Longitudinally, signatures of T1D progressors exhibited increasing inflammatory bias. Conversely, HRS possessing decreasing AA titers revealed emergence of an IL-10/transforming growth factor-β−mediated regulatory state that paralleled temporal increases in peripheral activated CD4+/CD45RA−/FoxP3high regulatory T-cell frequencies. In AA− HRS, the familial innate inflammatory state also was temporally supplanted by immunoregulatory processes, suggesting a mechanism underlying the decline in T1D susceptibility with age.
Introduction
Type 1 diabetes (T1D) arises through autoimmunity toward the insulin-producing pancreatic β-cells. It is a complex disease, developing through the interaction of an incompletely defined combination of genetic susceptibilities (1) and environmental factors (2,3). Inherited risk for T1D is largely conveyed by the HLA locus (4); however, changing environmental stressors that potentiate β-cell autoimmunity (viruses, diet, and/or altered gut microbiome) may account for the increasing incidence observed in most developed countries (5–7). Supporting this possibility is the decreased proportion of “high-risk” HLA genotypes among present new-onset T1D patients (8).
T1D pathogenesis involves innate immune activity (9–11) coupled with failures in central and peripheral tolerance mechanisms that enable expansion of disease-mediating autoreactive T cells. Other immune cell types are involved, including B cells, as evidenced by the development of autoantibodies (AAs) toward islet antigens that precede onset in ∼95% of patients (12–14). Chemokines and cytokines also are involved in T1D pathogenesis by influencing immunocyte activity, impairing β-cell function and inducing β-cell death (15).
T1D possesses a clinically silent preonset period that likely lasts years in many patients (16). This time of progressive β-cell loss represents a window for interventions aimed at preserving islet function and delaying and/or preventing T1D. Detection of multiple AA specificities is presently the best predictive biomarker (17,18), although it is thought that AAs do not directly mediate T1D development (19–21). Altered cytokine milieus represent a potential biomarker for T1D; however, they are generally dilute and difficult to directly measure in the periphery. Furthermore, measurement of a single or few cytokines may be uninformative as combinatorial effects are likely important.
Measures are needed that can detect alterations in the immune state associated with T1D progression or nonprogression. Toward this goal, we have used a sensitive and comprehensive genomics-based assay whereby serum- or plasma-borne mediators are used to drive transcription in a well-controlled “reporter” peripheral blood mononuclear cell (PBMC) population (10,22–25). Plasma of recently diagnosed (recent onset [RO]) T1D patients when cultured with unrelated healthy PBMCs induces a disease-specific signature compared with plasma of unrelated healthy control subjects (uHC). This signature includes upregulation of interleukin (IL)-1 cytokine family members, chemokines, and immune receptors and signaling molecules, as well as downregulation of genes involved with immunoregulatory processes, suggesting that RO T1D plasma possesses higher levels of proinflammatory mediators and lower levels of anti-inflammatory factors. Importantly, this signature has been detected years prior to T1D onset, before the appearance of AAs (10,24).
By using cross-sectional and longitudinal studies of progressors and nonprogressors to T1D, a novel quantitative signature scoring algorithm and targeted follow-up studies, we herein report the presence of an innate inflammatory state in T1D families that is independent of AA status, HLA haplotypes, or T1D progression. Importantly, among healthy siblings of probands possessing high-risk HLA haplotypes, we have identified age-dependent, regulatory processes that parallel temporal increases in peripheral activated regulatory T cell (Treg) frequencies. This observation offers mechanistic insight to the age-dependent decline in T1D susceptibility.
Research Design and Methods
Study Subjects
Subjects were recruited through Children’s Hospital of Wisconsin (CHW) and diagnosis of T1D was defined per World Health Organization criteria (26). All RO T1D subjects (n = 47) were positive for >1 AA. To avoid inducing transcription due to factors related to hyperglycemia, samples were collected 2–7 months after diagnosis from subjects with histories of good glycemic control. Control subjects, free of known infection at sample collection, belonged to three groups: AA− siblings of probands possessing either high-risk HLA genotypes (DR3 and/or DR4, termed high-risk siblings [HRS] n = 30), lower risk HLA genotypes (non-DR3/DR4, termed low-risk siblings [LRS] n = 42), and uHC (n = 44) possessing no family history of T1D. Details of the studied subjects are provided in Supplementary Table 1. The study was approved by the institutional review board of CHW (IRB 01–15) and written informed consent was obtained from subjects or their parents/legal guardians.
Peripheral blood was drawn into acid citrate dextrose solution A or K+EDTA anticoagulant. Ficoll Histopaque (Sigma-Aldrich, St. Louis, MO) density gradient centrifugation was used to separate blood components immediately postdraw, and plasma was stored at −80°C until use. Measurements of AAs targeting GAD, IA2, insulin, and zinc transporter 8 (ZnT8) were conducted as described (27). Subjects were genotyped for HLA-DQB1 and haplotypes were inferred using Caucasian haplotype frequencies as described (24,28).
Plasma-Induced Signature Analysis
Transcription induced in cryopreserved PBMCs (UPN727, Cellular Technology Ltd., Shaker Heights, OH) after coculture with 40% subject plasma in RPMI 1640 medium was measured using the GeneChip Human Genome U133 plus 2.0 array (Affymetrix, Santa Clara, CA) as previously described (24). The statistical significance of differentially induced transcription was assessed though ANOVA and false discovery rates (FDR) using Genomics Suite 6.5 (Partek, St. Louis, MO). Data sets were analyzed by the Database for Annotation, Visualization and Integrated Discovery (DAVID) (29) and Ingenuity Pathway Analysis (IPA) package (Ingenuity Systems, Redwood City, CA). Hierarchical clustering was conducted with Genesis (30). Data files are available through the National Center for Biotechnology Information Gene Expression Omnibus (31) (accession numbers GSE52724 and GSE35725).
25-Plex ELISA
Plasma cytokine levels were assayed with the Beadlyte cytokine assay kit (Millipore, Billerica, MA) and a Bio-Plex Luminex 100 XYP instrument. Concentrations were calculated with the Bio-Plex Manager 4.1 software; a five-parameter curve-fitting algorithm was applied for standard curve calculations.
Flow Cytometry Studies
PBMCs were stained with the fixable Live/Dead Violet dye (Life Technologies, Grand Island, NY) for 30 min on ice, followed by surface staining for anti-CD4 (clone RPA-T4), anti-CD25 (clone M-A251), anti-CD45RO (clone UCHL1), anti-CD45RA (clone HI100), and anti-CD127 (clone HIR-7R-M21) (BD Bioscience, San Jose, CA) on ice for 30 min, followed by intracellular staining with anti-FOXP3 (clone PCH101) (eBioscience, San Diego, CA). Each analysis included fluorescence minus one controls to ensure correct gating. Stained cells were analyzed on a LSR II flow cytometer (BD Bioscience).
Results
Cross-sectional Plasma-Induced Signatures of Healthy Siblings of T1D Patients
Siblings of probands have ∼6% probability of developing T1D, and most of those that progress to disease possess a DR3 and/or DR4 HLA haplotype (1,32). To better understand the inherited susceptibility present in T1D families, we examined plasma-induced signatures of RO T1D patients, uHC, and healthy AA− siblings of probands possessing or lacking DR3 and/or DR4 alleles (abbreviated HRS and LRS, respectively).
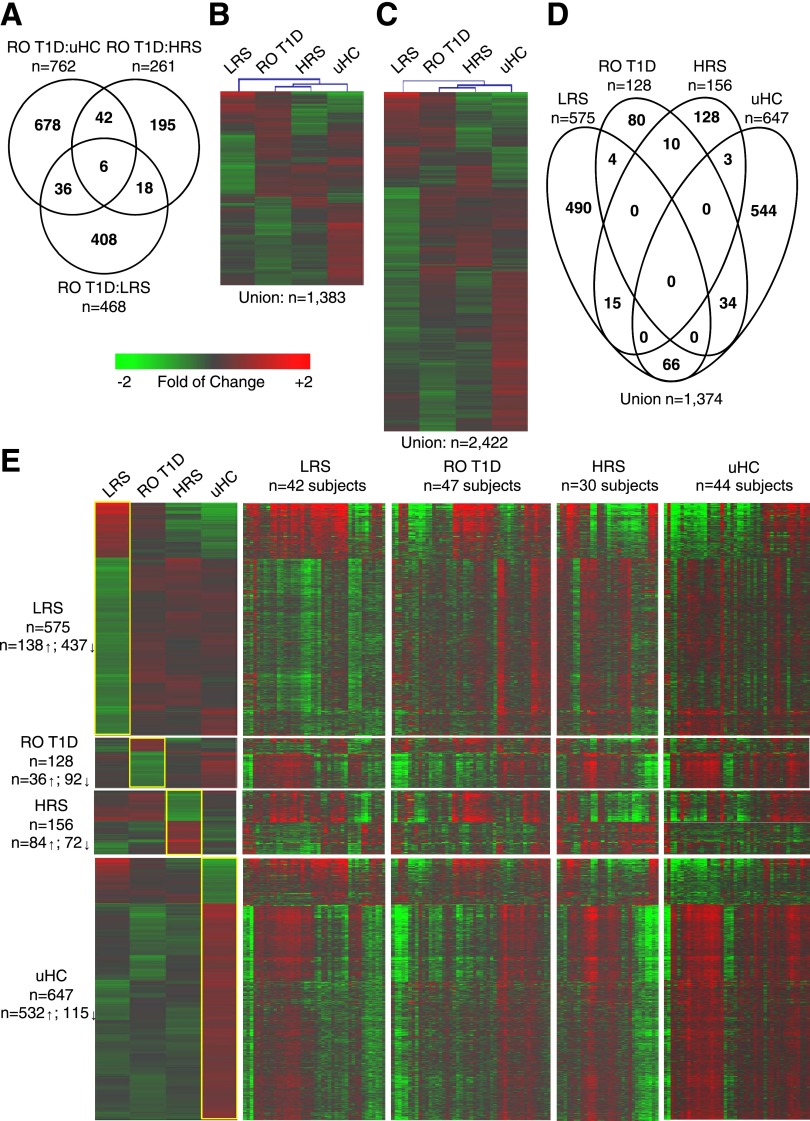
The comparison of transcripts induced by plasma of RO T1D patients to those induced by each of the healthy cohorts identified three distinct signatures, where the number of regulated probe sets was inversely related to the genetic risk possessed by the cohort (Fig. 1A). Hierarchical clustering of the 1,383 probe set union encompassed by these three comparisons found the RO T1D and HRS groups most similar. Unexpectedly, these exhibited intermediate relatedness to the LRS and uHC cohorts, which were most distinct from one another (Fig. 1B). A union of 2,422 regulated probe sets was identified among all six possible pairwise comparisons (Supplementary Table 2). The additional probe sets captured by this larger analysis did not alter the hierarchical clustering of the cohorts (Fig. 1C), confirming that the LRS cohort was indeed most distinct from that of the uHC cohort.
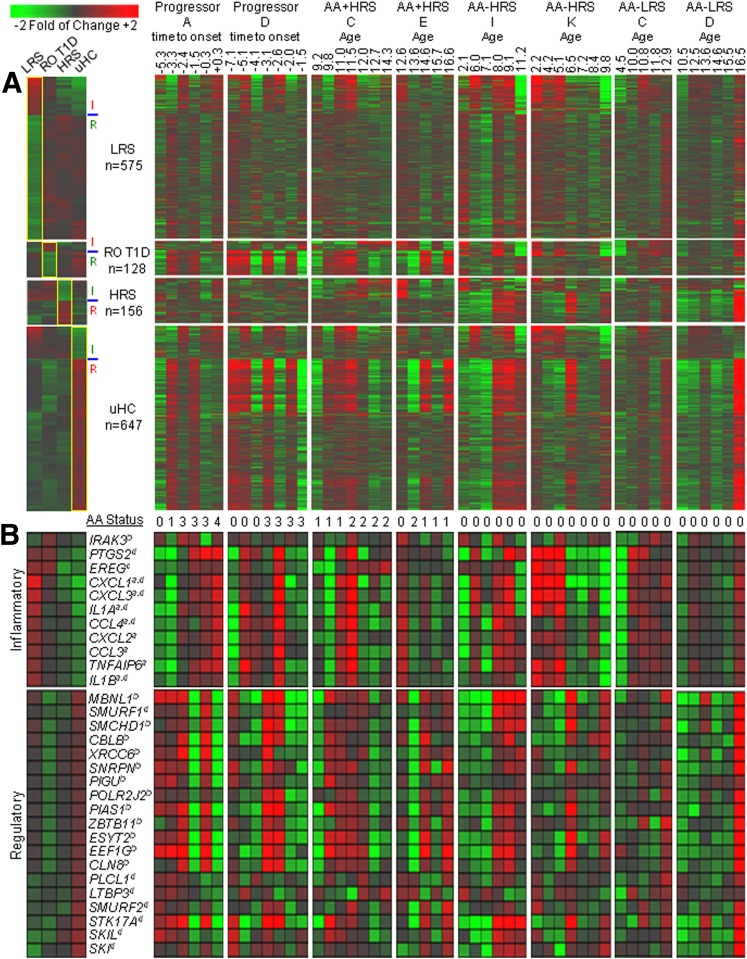
Figure 1.
Cross-sectional comparisons of the plasma-induced signatures. The analysis included RO T1D patients (n = 47, age 10.0 ± 2.9 years, blood glucose 155.0 ± 73.6 mg/dL, HbA1c 7.5 ± 1.2% [58 ± 9.3 mmol/mol]), LRS (n = 42, age 8.4 ± 2.0 years, blood glucose 93.5 ± 16.2 mg/dL), HRS (n = 30, age 8.6 ± 1.9 years, blood glucose 92.7 ± 11.7 mg/dL), and uHC (n = 44, age 15.0 ± 4.1 years, blood glucose 92.2 ± 13.2 mg/dL). A: Venn diagram illustrating the relationship of the 1,383 probe sets regulated to thresholds (|log2 ratio| > 0.263, 1.2-fold; FDR < 0.2; ANOVA P < 0.036) in the RO T1D:HRS (n = 261), RO T1D:LRS (n = 468), and RO T1D:uHC (n = 762) comparisons. B: Two-way hierarchical clustering (probe sets and cohorts) for the 1,383 regulated probe sets identified when comparing the RO T1D patients to the three healthy control cohorts. C: Two-way hierarchical clustering (probe sets and cohorts) for the 2,422 regulated probe sets identified among all possible pairwise comparisons between the RO T1D, LRS, HRS, and uHC cohorts (n = 6 comparisons: RO T1D:HRS [n = 261], RO T1D:LRS [n = 468], RO T1D:uHC [n = 762], LRS:uHC [n = 1,339], LRS:HRS [n = 612], and HRS:uHC [n = 635]). D: Venn diagram illustrating the relationship of the 1,374/2,422 probe sets remaining after identification of the most distinctive probe sets for each cohort. This was accomplished by filtering the 2,422 probe sets to retain those probe sets where the difference in the mean induction for one group was > |1.125-fold| relative to the mean of each of the other three groups. E: Heat maps illustrating the mean expression levels of probe sets that distinguish each cohort (leftmost panel); shown are the expression levels for these genes by the individual subjects comprising each cohort (rightmost panels, as indicated). Expression levels illustrated in the heat map are normalized against the mean expression of the four cohorts.
To focus the ontological studies on transcripts most distinctive of each group, we filtered the 2,422 probe set union, retaining for each cohort transcripts exhibiting the highest and lowest induced expression relative to each of the three other groups (Fig. 1D and E). These cohort-associated probe sets were then independently subjected to pathway analysis.
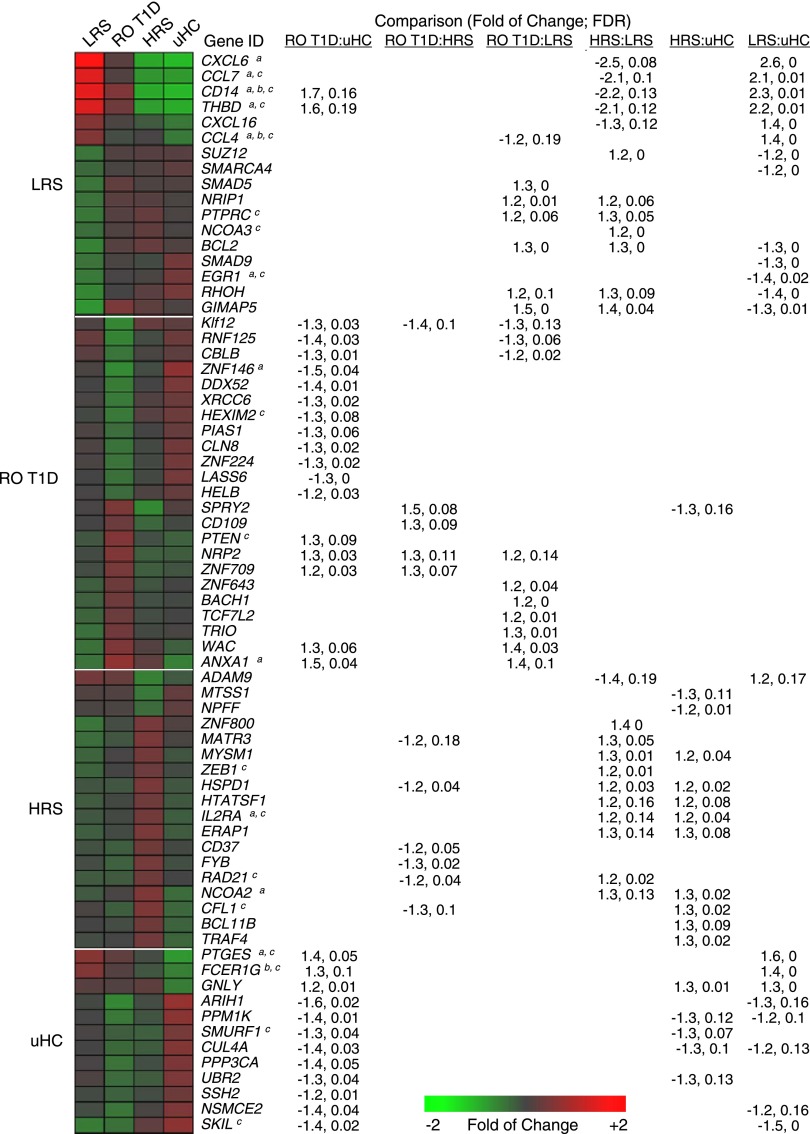
Consistent with a family history lacking autoimmunity, ontological analysis of the uHC-associated data set revealed the lowest induction of genes associated with immune activation (Fig. 2, additional annotations are shown in Fig. 3). These included IL-1 family members (IL1A, IL1B, IL1R1), cytokines (CXCL1, CXCL2, CXCL3, CCL4), immune receptors (CD93, FCER1G), and transcripts related to inflammatory mediator synthesis (PTGS2, PTGES). uHC plasma induced higher expression of genes implicated in regulating immune responses through protein phosphatase activity (PTPN4, SSH2, PPP3CA, PPP3CB) and protein ubiquitination (SMURF1, SMURF2, ITCH, UBR2).
Figure 2.
Expression levels and one-way hierarchical clustering (probe sets only) of selected probe sets regulated by LRS, RO T1D, HRS, and uHC plasma (among the 1,374 probe sets identified in Fig. 1D and E). Tabulated are the fold of change and FDR associated with each probe set for the six possible pairwise comparisons between the cohorts. a, b, and c denote regulation by IL-1, TGF-β, and IL-10, respectively, per IPA upstream regulator analysis. Expression levels illustrated in the heat map are normalized against the mean expression of the four cohorts.
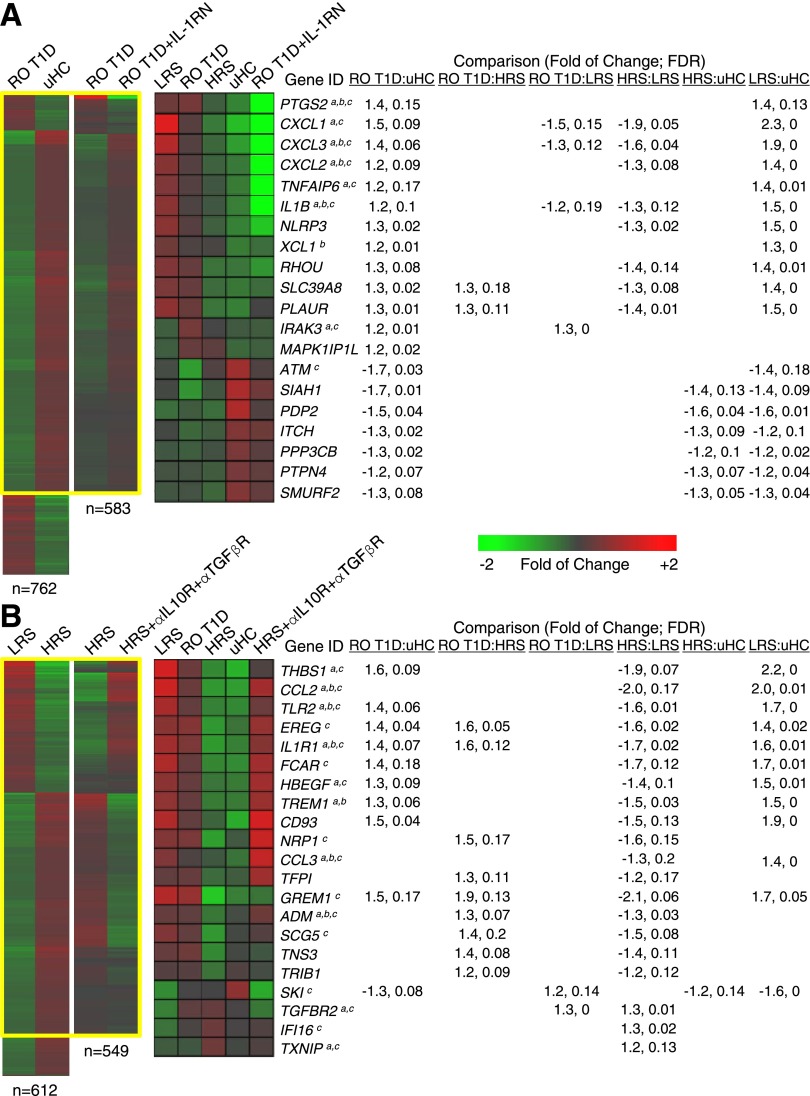
Figure 3.
Identification of ligands underlying plasma induced signatures. A: Adding 4 μg/mL IL-1RN to RO T1D cultures (n = 47 subjects) modulated IL-1–dependent components of the RO T1D:uHC signature, directionally altering expression of 583/762 genes (76.5%, χ2 P < 10E-6; yellow box). Notably, among the 2,422 union of probe sets regulated across the six possible pairwise cohort comparisons, 1,449/2,422 (59.8%, χ2 P < 1.13E-04) reversed the direction of induction when IL-1RN was introduced into the RO T1D coculture, further supporting an IL-1 bias in T1D families. B: A total of 612 significantly regulated genes were identified when directly comparing the LRS and HRS cohorts; 59.5% (n = 364) were among the 1,383 gene union in shown Fig. 1B. Adding 30 μg/mL α-TGF-βR– and 30 μg/mL α-IL-10R–neutralizing antibodies to HRS sera (n = 3 subjects) modulated transcription to mimic LRS sera, directionally altering expression of 549/612 genes (89.7%, χ2 P < 10E-6; yellow box). Tabulated are the fold of change and FDR associated with each probe set for the six possible pairwise comparisons between the cohorts. a, b, and c denote regulation by IL-1, TGF-β, and IL-10, respectively, per IPA upstream regulator analysis. Nonspecific isotypic control antibodies did not modulate expression (data not shown). Independent addition of α-TGF-βR– or α-IL-10R–neutralizing antibodies to HRS sera (n = 3 subjects) modulated fewer genes (511/612 and 425/612, respectively, χ2 P < 10E-3). Adding 2 ng/mL TGF-β and 5 ng/mL IL-10 to LRS sera (n = 3 subjects) modulated transcription to mimic HRS sera, directionally altering expression of 361/612 (59.0%, χ2 P = 0.048) genes. Independent addition of TGF-β or IL-10 to LRS sera modulated fewer genes (170/612 and 207/612, respectively, χ2 P > 0.05, data not shown).
Unexpectedly, transcripts upregulated by LRS plasma were consistent with an inflammatory response, cytokine activity, and exposure to bacterial antigens (Fig. 2). Many proinflammatory annotations that exhibited the lowest induction by uHC plasma were most highly induced by LRS plasma. These included chemokines (CCL2, CCL3, CCL4), pattern recognition receptors (CD14, TLR2), as well as immune receptors, growth factors, signaling molecules, and transcription factors involved in promoting immune responses (FCAR, PLAUR, TREM1, NLRP3). Transcription downregulated by LRS plasma relative to the other cohorts was related to transforming growth factor (TGF)-β signaling (TGFBR2, SMAD9, SMAD5, SKI, SKIL), transcriptional regulation/repression (NRIP1, NCOA3, TBL1XR1, SUZ12), and regulation of immune activation (PTPRC).
Among the three T1D family cohorts, hierarchical clustering found the RO T1D intermediate to the LRS and HRS. While RO T1D plasma induced transcription consistent with innate immune activation, it was to a lesser degree than that of LRS plasma (Fig. 2). Previously (24), we identified a 762 probe set signature when comparing transcription induced by RO T1D and uHC plasma. Given its intermediate positioning in this analysis, only 128 probe sets were exclusively associated with the RO T1D cohort after filtering for the ontological studies. RO T1D plasma induced increased expression related to regulation of apoptosis (PTEN, TCF7L2), regulation of protein kinase activity (IRAK3, SPRY2), transcriptional regulation (BACH1, ZNF643, ZNF709), and adhesion and inflammation (NRP2, CD109, ANTXR2). RO T1D plasma induced underexpression of transcripts related to regulation of immune responses through ubiquitin mediated proteolysis (CBLB, PIAS1, SIAH1, RNF125), transcriptional regulators (KLF12, SLC11A1, ZNF224), and ATP-dependent helicase activity (DDX52, XRCC6, HELB).
Subjects within the HRS group have the greatest probability of T1D progression among the three healthy cohorts, yet LRS plasma induced more robust transcription related to innate inflammation. Notably, ontological analysis of probe sets uniquely upregulated by HRS plasma identified activities related to regulation of T-cell/immune cell activation (IL2RA, ARHGDIB), transcriptional regulation (UBTF, EIF2C1, TXNIP, ANP32A), transcriptional repression (ZEB1, NCOA2, BCL11B), and cell growth/survival (CD37, TRAF4, CFL1, IFI16), many of which are involved in the attenuation of immune responses (Fig. 2). HRS plasma downregulated transcripts associated with cell adhesion and migration (ADAM9, GREM1, ITGA2, NRP1), positive regulation of cell proliferation (EREG, ADM, TNS3), and signal transduction (MTSS1, MITF, NPFF).
The cross-sectional profiling suggests that an inflammatory state exists in T1D families. This is supported by the distinct underinduction of inflammatory transcripts by uHC relative to LRS, RO T1D, and HRS plasma. Unexpectedly, LRS plasma induced the most robust induction of proinflammatory transcripts, consistent with the inheritance of risk not related to the HLA. The data also suggest that in HRS, where inheritance of high-risk HLA haplotypes increases the likelihood of adaptive autoimmune responses, the underlying inflammatory state is actively attenuated.
Mediators Underlying Cohort Signatures
Upstream regulator analysis was performed on the cross-sectional data sets (Table 1). The IL-1 pathway was activated when analyzing probe sets of the RO T1D:uHC and LRS:uHC comparisons (Z scores >2.8) and was inhibited when analyzing probe sets of the HRS:LRS comparison (Z score <−2.7). Analysis of the uHC-associated probe sets also detected inhibition of the IL-1 pathway (Z score <−2.4), suggesting that IL-1 contributes to the inflammatory bias observed in T1D families relative to uHC. The partial IL-1 dependence of the RO T1D:uHC signature was confirmed by introducing IL-1 receptor antagonist (IL-1RN) to cocultures (Fig. 3A), which modulated IL-1−dependent components of the RO T1D:uHC signature by directionally altering expression of 583/762 probe sets (76.5%, χ2 P < 10E-6).
Table 1.
Upstream regulator analysis using IPA
| Upstream regulator |
||||
|---|---|---|---|---|
| n | IL-1 | IL-10 | TGF-β | |
| RO T1D:uHC | 762 | 3.2E-6 (↑) | 1.3E-9 (↓) | 1.3E-7 (↓) |
| RO T1D:HRS | 261 | 1.7E-2 (↑) | ns | 6.3E-7 (↓) |
| RO T1D:LRS | 468 | 4.4E-2 (↓) | 5.0E-2 (↑) | ns |
| HRS:LRS | 621 | 5.8E-5 (↓) | 4.0E-5 (↑) | 3.6E-9 (↑) |
| HRS:uHC | 635 | ns | ns | ns |
| LRS:uHC | 1,339 | 1.6E-12 (↑) | 1.8E-22 (↓) | 5.7E-17 (↓) |
| LRS | 575 | 3.6E-8 (↑) | 7.5E-14 (↓) | 2.6E-8 (↓) |
| RO T1D | 128 | ns | ns | ns |
| HRS | 156 | ns | ns | 9.7E-3 (↑) |
| uHC | 647 | 6.8E-5 (↓) | 5.8E-6 (↑) | 1.3E-4 (↑) |
Analysis was performed on each of the six pairwise comparisons among the four cohorts, as well as for the probe sets most distinctly regulated by each cohort. Indicated are the number of regulated probe sets in the analysis and the upstream regulator. The P value (determined with a Fisher exact test) reflects the significance of the overlap between the regulated probe sets within the data set and genes regulated by the transcriptional regulator. ns, not significant P > 0.05. Arrow-up or arrow-down denotes the direction of enrichment in the comparison. Values < 0.01 are considered significant.
Consistent with the failure of LRS plasma to induce genes related to immunoregulatory processes, upstream regulator analysis identified IL-10 and TGF-β as potential mediators, showing the most significance when analyzing the LRS:uHC comparison (P = 1.8E-22 and 5.7E-17, respectively). Among the T1D family cohorts, IL-10 and TGF-β were most significantly identified as potential regulators when analyzing the HRS:LRS comparison (P = 4.0E-5 and 3.6E-9, respectively), consistent with the induction of immunoregulatory genes by HRS plasma. The dependence of the HRS signature on IL-10 and TGF-β was confirmed by introducing α-IL-10R and α-TGF-βR antibodies to cocultures (Fig. 3B) which directionally altered expression of 549/612 genes (89.7%, χ2 P < 10E-6) of the HRS:LRS signature.
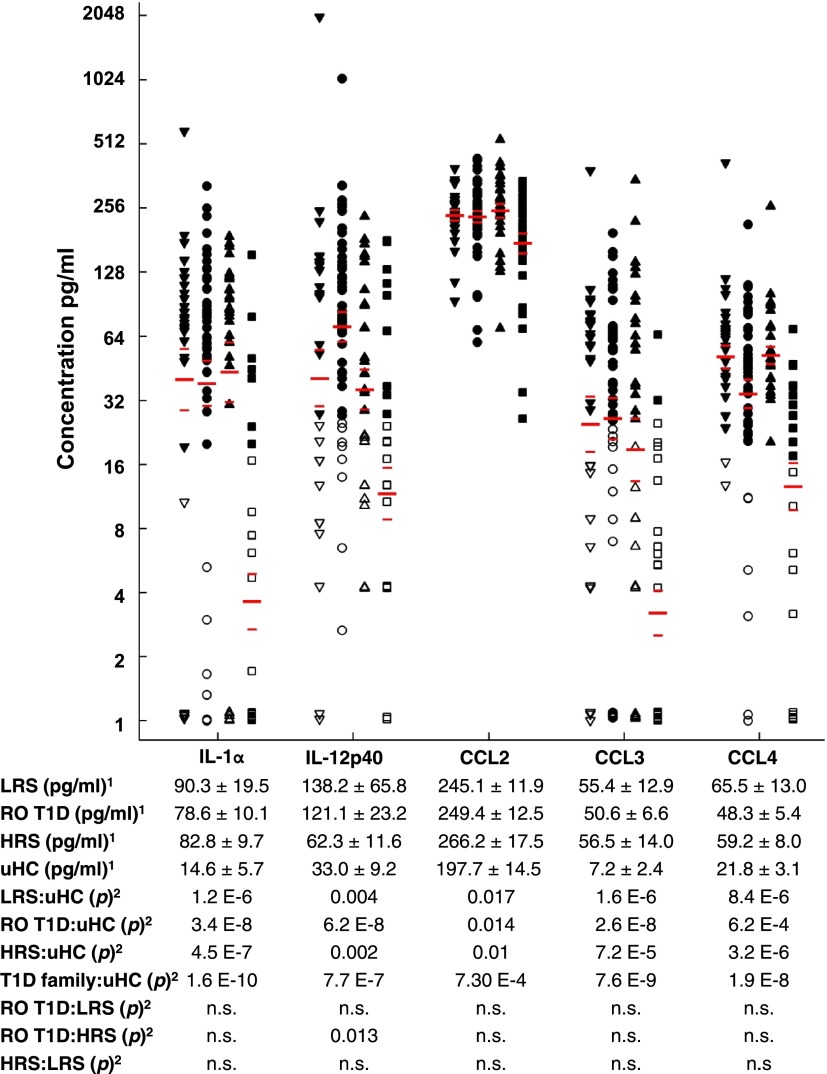
Plasma cytokine levels of the four cohorts were investigated (Fig. 4). Significantly higher levels of IL-12p40, IL-1α, CCL2, CCL3, and CCL4 were detected in RO T1D compared with uHC plasma. These cytokines were also significantly elevated in the LRS and/or HRS cohorts relative to uHC, further indicating the presence of an inflammatory state in T1D families that is independent of AA and HLA status and disease progression. Unlike the signature studies, direct cytokine measurements did not differentiate the T1D family cohorts.
Figure 4.
Plasma IL-1α, IL-12p40, CCL2, CCL3, and CCL4 levels in the cross-sectional cohorts. A total of 30 LRS, 48 RO T1D, 30 HRS, and 31 uHC subjects were assayed in duplicate; respectively 28, 31, 28, and 22 of these subjects were analyzed in the expression studies. 1Tabulated data are expressed as cohort mean values ± SE in pg/mL. 2P value for two sample t test. LRS (closed downward triangles), RO T1D (closed circles), HRS (closed upward triangles), and uHC (closed squares); open symbols are below assay detection limit. Among the LRS, RO T1D, HRS, and uHC cohorts, respectively: 80.0%, 81.3%, 83.3%, and 22.6% of subjects had IL-1α levels >16 pg/mL detection limit; 56.7%, 81.3%, 60.0%, and 29.0% of subjects had IL-12p40 levels >25 pg/mL detection limit; all subjects had CCL2 levels >16 pg/mL detection limit; 63.3%, 66.7%, 56.7%, and 6.5% of subjects had CCL3 levels >25 pg/mL detection limit; and 93.3%, 87.5%, 100%, and 64.5% of subjects had CCL4 levels >16 pg/mL detection limit. The average blood glucose measurements (nonfasting) for these LRS, RO T1D, HRS, and uHC cohorts were 93.1 ± 12.0 mg/dL, 166.4 ± 69.9 mg/dL, 91.6 ± 10.6 mg/dL, and 89.0 ± 10.3 mg/dL, respectively. While blood glucose levels were higher in the RO T1D cohort, these cytokines also were significantly elevated in the LRS and/or HRS cohorts, suggesting that the elevated cytokine levels are not related to hyperglycemia. Eotaxin, GMCSF, IFNα2, IFNγ, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p70, IL-13, IL-15, IL-17A, IP-10, TNF-α, and TNF-β were included in the 25-plex panel, but statistically significant differences between the cohorts were not detected.
Longitudinal Analyses of Progressors and Nonprogressors
Plasma-induced signatures were used to longitudinally study five T1D progressors. As we have previously observed (10,24,25), crescendos in induced proinflammatory transcription with coincident reductions in induced regulatory transcription were associated with T1D progression (Fig. 5). This pattern was evident in the DR3/DR4-negative “Progressor A” at −5.3 years and the DR3/DR4-positive “Progressor D” at −4.1 years prior to T1D onset, which was prior to the emergence of AAs in these two subjects. This pattern was observed in all five T1D progressors examined (Supplementary Figure 1).
Figure 5.
Longitudinal analyses of T1D progressors and siblings of T1D probands. In all cases ≥4 samplings per subject were available, spanning a time period of 2.6–10 years. A: Analysis from the perspective of the 1,374 regulated transcripts identified in cross-sectional studies. Inflammatory activity was associated with probe sets upregulated by LRS and RO T1D plasma and downregulated by HRS and uHC plasma, while regulatory activity was associated with probe sets downregulated by LRS and RO T1D plasma and upregulated by HRS and uHC plasma. These inflammatory and regulatory probe sets are indicated by I and R, respectively. Presented are the mean responses for each cross-sectional cohort (leftmost panel) and eight longitudinal data sets: T1D progressors A and D, AA+ HRS C and E, AA− HRS I and K, and AA− LRS C and D (as labeled). Indicated for the T1D progressors is the time of sample collection relative to T1D onset. Progressor A and D were diagnosed at the ages of 21 and 17, respectively. For nonprogressors, the age of sample collection is indicated. B: Expression levels of 30 well-annotated regulated transcripts having inflammatory and regulatory roles. The number of detected AA specificities is indicated (0–4). The cross-sectional data set to which each probe set is associated (LRS, RO T1D, HRS, uHC) is indicated (a, b, c, d, respectively).
Plasma-induced signatures were used to longitudinally study 27 nonprogressors (9 AA+ HRS, 12 AA− HRS, and 6 AA− LRS; all shown in Supplementary Figure 1). In AA+ HRS, greater inflammatory bias was evident immediately prior to, or associated with, increases in the number of AA specificities detected. The opposite also was observed, with greater regulatory bias being associated with decreases in the number of detected AA specificities (Fig. 5, subjects AA+ HRS C and AA+ HRS E). As a group, the AA− HRS exhibited reduced temporal induction of inflammatory components and increased temporal induction of regulatory components of the 1,374 cross-sectional data set. This overall pattern was less evident among AA− LRS (Fig. 5, subjects AA− HRS I and AA− HRS K).
Overall, these analyses support that plasma-induced signatures captured temporal alterations in immunological balance within T1D family members. The data also indicate that a single transcript may not be informative, but combinations of transcripts may comprehensively capture the nature of the immune state.
Ontology-Based Quantitative Scoring of Plasma-Induced Signatures
As defined by the ontological analyses and illustrated in Fig. 5, the regulated probe sets identified in the cross-sectional studies can be broadly considered as “inflammatory” or “regulatory.” Therefore, signatures were quantitatively scored with a composite inflammatory index (I.I.com) (formulas provided in Supplementary Data) determined by calculating an average ratio between the mean log intensity of the inflammatory genes versus the mean log intensity of the regulatory genes of the four cross-sectional data subsets.
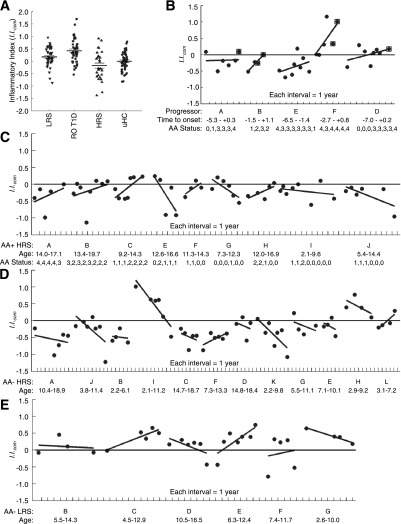
The mean I.I.com of RO T1D patients was significantly higher than that of LRS, HRS, and uHC, and with the exception of the HRS:uHC comparison, it was significantly different between comparison of any of the cohorts (Fig. 6A). All five longitudinally monitored T1D progressors possessed regressions of I.I.com versus time that exhibited positive slopes (Fig. 6B). The I.I.com for all HRS AA+ nonprogressors that persistently possessed titers for >1 AA specificity also exhibited positive slopes (Fig. 6C, subjects A, B, and C), whereas most AA+ HRS subjects possessing transient or decreasing titers exhibited temporally decreasing I.I.com (Fig. 6C, subjects E, G, I, and J). Notably, AA+ HRS subjects F and H exhibited positive slopes, indicative of increasing inflammatory bias despite decreases in AA titers. The I.I.com plots of most longitudinally monitored AA− HRS subjects (10/12, 83%) exhibited negative slopes indicative of decreasing inflammatory bias over time; this pattern was not observed in the longitudinally monitored AA− LRS that overall exhibited higher I.I.com.
Figure 6.
Ontology-based scoring of cross-sectional and longitudinal samples (≥4 samplings/subject). As indicated in Fig. 5, inflammatory activity was associated with probe sets upregulated by LRS and RO T1D plasma and downregulated by HRS and uHC plasma, while regulatory activity was associated with probe sets downregulated by LRS and RO T1D plasma and upregulated by HRS and uHC plasma. This formed the basis of I.I.com, determined by calculating an average ratio between the mean log intensity of the inflammatory genes vs. the mean log intensity of the regulatory genes of the four cross-sectional data subsets. Given their relevance, higher weight was ascribed to probe sets of the RO T1D (13.0) and HRS (11.5) cohorts. A: The mean composite I.I.com of the 47 cross-sectional RO T1D subjects (mean ± SE 0.42 ± 0.07) was significantly higher than that observed for the 42 LRS (0.17 ± 0.06), 30 HRS (−0.18 ± 0.10), and 44 uHC (0.00 ± 0.06) subjects (P = 0.007, 2.6E-06, and 7.8E-06, respectively). The mean I.I.com of the LRS vs. HRS and LRS vs. uHC cohorts also were significantly different (P = 0.002 and 0.036, respectively). The mean I.I.com of the HRS vs. uHC was not significantly different (P = 0.11). B–E: Each point is a sampling and each line is a longitudinal series. B: Longitudinal analysis of progressors to T1D (n = 5); subjects A, B, E, F, and D were diagnosed at the ages of 21.7, 9.0, 19.3, 6.9, and 17.4, respectively. Indicated are the time of sampling relative to onset and the number of detected AA specificities (0–4). Postonset time points are boxed. C: Longitudinal analysis of presently nondiabetic, AA+ HRS (n = 9). Indicated are the age and AA status at each sampling. D: Longitudinal analysis of healthy AA− HRS (n = 12). Indicated is the age at each sampling. E: Longitudinal analysis of healthy AA− LRS (n = 6). Indicated is the age at each sampling. Analysis and regressions were conducted in MatLab.
The data support that the scored signatures captured temporal dynamics of the inflammatory state, as among T1D progressors (5/5) and HRS showing increases in the number of detected AA specificities (3/3), there are concordant increases in I.I.com. Regulatory activity also was captured as the majority of AA+ HRS showing transient AA positivity or decreasing numbers of detected AA specificities and most AA− HRS exhibited concordant decreases in I.I.com.. This observation supports the hypothesis that among sibling nonprogressors possessing high-risk HLA haplotypes immunoregulation becomes temporally more robust.
Increased Activated Treg Percentages Are Associated With Immunoregulation Temporally Measured in Plasma-Induced Signatures
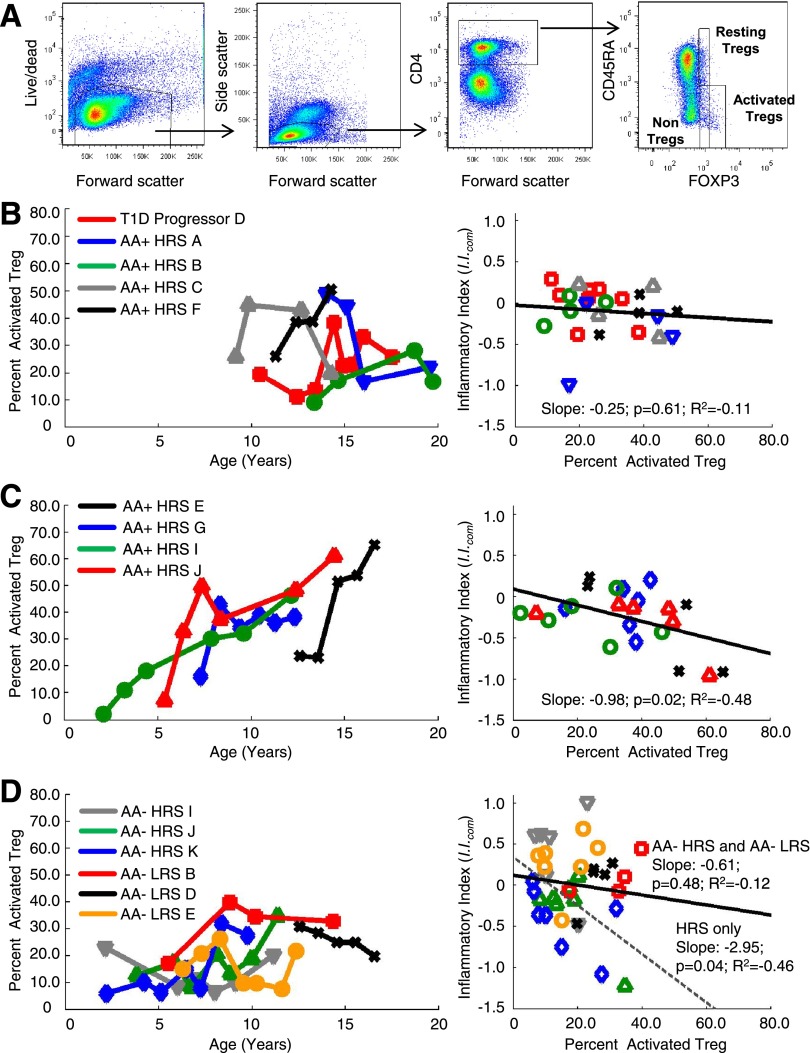
Human CD4+FoxP3+ T cells can be divided into three functionally distinct subpopulations based on expression of CD45RA and the level of FOXP3. CD4+/CD45RA−/FoxP3high cells are activated Tregs with high suppressive capacity, CD4+/CD45RA+/FoxP3low cells are resting Tregs also possessing suppressive function, and CD4+/CD45RA−/FoxP3low identify nonsuppressive FoxP3+ T cells that can produce proinflammatory cytokines (33). For subjects where cryopreserved PBMCs were available, we longitudinally examined the frequency of activated and resting Tregs in the periphery of T1D progressors, AA+ HRS, AA− HRS, and AA− LRS subjects that had been analyzed by plasma-induced signature analysis (Fig. 7A).
Figure 7.
Longitudinal analysis of activated and resting Treg cell abundances in peripheral blood of T1D family members. A: Representative flow cytometry profiles showing the gating strategy for resting and activated Treg populations. Resting and activated CD4+ Tregs were respectively defined as CD45RA+/FoxP3low and CD45RA−/FoxP3high. The expression CD45RO and CD25 confirmed the phenotype of resting (CD45RO−/CD25+) and activated (CD45RO+/CD25high) Tregs. Both populations were mostly CD127low, further excluding them as effector T cells. B: Analysis of a T1D progressor and AA+ HRS possessing temporally measured I.I.com possessing positive slopes. C: Analysis of AA+ HRS possessing temporally measured I.I.com possessing negative slopes. D: Analysis of AA− HRS and AA− LRS. B–D: The left panel plots the percentage of CD4+/CD45RA−/FoxP3high Treg among total Treg vs. time for each subject; the right panel plots I.I.com vs. the percentage of CD4+/CD45RA−/FoxP3high Treg among total Treg for each subject. The regression is plotted for all samples in the analysis (solid line); the dotted line (D) shows the regression for only the AA− HRS subjects I, J, and K.
T1D progressor D and AA+ HRS subjects A, B, and C, whose temporally plotted I.I.com exhibited positive slopes indicating increasing inflammatory bias, showed percentages of activated Tregs that had a flat or downward trend over time (Fig. 7B). AA+ HRS subject F, sampled annually over ∼4 years, did not fit this pattern, exhibiting a maximal I.I.com at time point 3, but with transient positivity for AA, consistent with increasing activated Treg percentages during the third through fourth visits (Fig. 7B).
Among the AA+ HRS subjects (E, G, I, J), whose temporally plotted I.I.com showed negative slopes indicating increasing regulatory bias, the percentages of activated Tregs were higher and exhibited a robust upward trend (Fig. 7C). A significant negative relationship between I.I.com and the percentage of activated Tregs for these subjects was observed (R2 = −0.48, P = 0.02).
The AA− HRS (I, J, K) also exhibited negative slopes to their temporally plotted I.I.com. Although the increase in activated Treg percentage was not as robust as that seen in the AA+ HRS (Fig. 7C), again a significant negative relationship between I.I.com and the percentage of activated Tregs for these subjects was observed (Fig. 7D, R2 = −0.46, P = 0.04). These results indicate that temporal decreases in I.I.com are associated with increased percentages of activated Tregs in the periphery. This relationship was not observed for the LRS subjects (B, D, E) analyzed.
Discussion
Through analyses of LRS, RO T1D, HRS, and uHC subjects, our findings suggest that an innate inflammatory state exists in T1D families that is independent of HLA type, AA status, and disease progression. Using novel ontology-based signature scoring, temporal increases in inflammatory processes with concurrent reductions in immunoregulatory activity were measured in T1D progressors. Among AA− HRS and AA+ HRS showing decreases in the number of detected AA specificities, temporal induction of an IL-10/TGF-β–dependent signature was concomitant with increased percentages of activated peripheral Tregs. These data suggest that in the presence of high-risk HLA haplotypes the innate inflammatory state common to T1D family members becomes more actively regulated in an age-dependent manner, possibly providing insight as to why T1D susceptibility declines with age.
The bioinformatic strategies, receptor modulations, and cytokine measurements support a decreasing inflammatory bias (involving at least IL-1) and an increasing regulatory bias (involving at least IL-10 and TGF-β) when examining the LRS→RO T1D→HRS→uHC continuum (Fig. 1). There is inconsistent literature describing plasma cytokine levels in T1D. This stems from, but is not limited to, differences in the analytes measured and methods and reagents used. Furthermore, few studies have included both related and unrelated AA− healthy control subjects. Consistent with studies that referenced uHC (34,35), we observed elevated cytokine levels in T1D families. Specifically, increased plasma IL-1α, IL-12p40, CCL2, CCL3, and CCL4 levels were detected in RO T1D, HRS, and LRS relative to uHC. Like Svensson et al. (36), we found differences in plasma cytokine levels between RO T1D patients and their healthy AA− siblings negligible. Consistent with the signature analysis, plasma cytokine levels also indicated an inflammatory state in T1D families. However, unlike the signature analysis, which reflects a response to the sum of plasma-borne factors, the cytokine panel tested could not differentiate the T1D family cohorts.
The inflammatory state in T1D families may arise from inheritance of potentiating genetic variants in immune pathways. This is supported by the association of polymorphisms in PTPN22, CTLA4, IL2RA, IFIH1, SH2B3, and other non-MHC loci with T1D (1). Another potential contributor is intestinal hyperpermeability, which could result in translocation of bacteria and/or pattern recognition receptor ligands and an elevated inflammatory state. Studies have linked intestinal permeability and intestinal ultra-structure alterations with T1D (37–40). Furthermore, zonulin, a protein that modulates intestinal permeability was found to be elevated and associated with increased intestinal permeability in T1D patients and their healthy relatives (41). Our seminal report (10) focused on IL-1 as a mediator of the RO T1D signature, a conclusion supported by this report. However, Toll-like receptor signaling also could contribute to inflammatory components of the RO T1D, HRS, and LRS signatures. While measurable endotoxin levels were not detected in the T1D family cohorts (data not shown) we cannot exclude this possibility.
The DR3 and DR4 HLA haplotypes are associated with many autoimmune diseases (42). This stems from the evolutionarily selected capacity of the DR3/DQ2 and DR4/DQ8 peptides to present a broad range of pathogen-derived peptides to T cells, which is associated with an increased propensity to present self-peptides to autoreactive T cells (43). Previous studies did not detect altered Treg frequencies in T1D patients (44). In this study, among AA+ HRS subjects showing robust induction of the IL-10− and TGF-β−dependent HRS signature, we observed temporal increases in the percentage of activated Tregs associated with decreases in the number of detected AA specificities. Negative slopes in I.I.com were observed in 83% of AA− HRS, where increased regulatory activity also was associated with increased activated Treg percentages in peripheral blood. Our findings support the hypothesis that an innate inflammatory state exists in T1D families that is actively regulated in the context of high-risk HLA haplotypes through increased activity and frequency of immunoregulatory cells over time. Exposure of Tregs to inflammatory inputs (e.g., endotoxin, IL-1β) can impair their suppressive capacities by promoting FOXP3 protein proteosomal degradation (45,46). It is therefore possible to envision that the addition of inflammatory stimuli, such as viral infection, to this preexisting inflammatory environment might overwhelm an emerging state of regulated susceptibility, promoting a break in tolerance and autoimmunity.
Besides AA+ relatives of T1D patients that never develop overt diabetes, studies of human subjects and rodent models support the existence of mechanisms that control progression of autoimmunity in the context of high genetic risk. AA− first-degree relatives of T1D patients have been reported to have a balance of islet antigen-specific inflammatory and regulatory T cells intermediate to that of RO T1D patients and uHC (47). Furthermore, IL-10−secreting, islet autoantigen−specific Tregs have been isolated from subjects with no family history of T1D (48). Similar to the regulated susceptibility observed in HRS, we have identified temporal induction of an immunoregulatory state in the BioBreeding (BB) diabetes-resistant (BBDR) rat that supplants a preexisting innate inflammatory state (25). Importantly, establishment of this regulated state coincides with the failure of Kilham rat virus to induce T1D in older BBDR rats, again suggesting a mechanism for why T1D susceptibility declines with age. As observed in the BB rat (49), pharmacological strategies targeting the preexisting inflammatory state in T1D families may be useful as a component of preventative or interventional immunotherapy. Such approaches in younger at-risk subjects may only be temporarily necessary until endogenous regulatory mechanisms become sufficiently established.
This study extends our use of plasma-induced transcription as a tool for understanding and as a biomarker of processes associated with T1D pathogenesis. We identified an inflammatory state in T1D families and the emergence of an age-dependent immunoregulated state in HRS. In many respects, the RO T1D signature represents an intermediate between the LRS and HRS signatures. As the majority of siblings that progress to T1D possess DR3 and/or DR4 haplotypes, this intermediate signature raises the question of whether T1D arises from a loss of the HRS regulatory state or a failure to initially establish it. It is from this perspective that we are extending this modestly sized analysis to larger-scale human longitudinal studies aimed at assessing the inflammatory state associated with diabetes susceptibility and disease progression.
Supplementary Material
Article Information
Acknowledgments. The authors thank the patients and subjects who participated in this study as well as the physicians, nurses, and staff of Children’s Hospital of Wisconsin and The Max McGee National Research Center for Juvenile Diabetes who assisted in subject recruitment and sample collection and processing.
Funding. This work was supported by JDRF International (grants 1-2008-1026, 5-2012-220, 17-2012-621 to M.J.H.), American Diabetes Association (grant 7-12-BS-075 to M.J.H.), National Institutes of Health (grants R00DK077443 to Y.-G.C., R01DK080100 to X.W., R01AI078713 to M.J.H., DP3DK098161 to C.J.G. and M.J.H., and 1-UL1-RR031973 to the Clinical and Translational Science Institute of Southeast Wisconsin), and the Children’s Hospital of Wisconsin Foundation.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Y.-G.C. designed experiments, researched data, and wrote, reviewed, and edited the manuscript. S.M.C. and J.E.W. designed experiments, researched data, and reviewed the manuscript. S.J. and S.C. developed and tested algorithms, conducted data analysis, and reviewed the manuscript. M.L.K., J.K., R.G., and M.F.R. recruited subjects, processed samples, researched data, and reviewed the manuscript. C.J.G. wrote, reviewed, and edited the manuscript. X.W. designed, supervised, and reviewed the data analysis and reviewed the manuscript. M.J.H. designed experiments, analyzed data, and wrote, reviewed, and edited the manuscript. M.J.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-0214/-/DC1.
See accompanying article, p. 3581.
References
- 1.Steck AK, Rewers MJ. Genetics of type 1 diabetes. Clin Chem 2011;57:176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stene LC, Oikarinen S, Hyöty H, et al. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the Diabetes and Autoimmunity Study in the Young (DAISY). Diabetes 2010;59:3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hober D, Sauter P. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat Rev Endocrinol 2010;6:279–289 [DOI] [PubMed] [Google Scholar]
- 4.Erlich H, Valdes AM, Noble J, et al. Type 1 Diabetes Genetics Consortium . HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unwin N, Gan D, Whiting D. The IDF Diabetes Atlas: providing evidence, raising awareness and promoting action. Diabetes Res Clin Pract 2010;87:2–3 [DOI] [PubMed] [Google Scholar]
- 6.Vehik K, Dabelea D. The changing epidemiology of type 1 diabetes: why is it going through the roof? Diabetes Metab Res Rev 2011;27:3–13 [DOI] [PubMed] [Google Scholar]
- 7.Atkinson MA, Chervonsky A. Does the gut microbiota have a role in type 1 diabetes? Early evidence from humans and animal models of the disease. Diabetologia 2012;55:2868–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fourlanos S, Varney MD, Tait BD, et al. The rising incidence of type 1 diabetes is accounted for by cases with lower-risk human leukocyte antigen genotypes. Diabetes Care 2008;31:1546–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todd JA. Etiology of type 1 diabetes. Immunity 2010;32:457–467 [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol 2008;180:1929–1937 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Fillmore TL, Schepmoes AA, et al. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J Exp Med 2013;210:191−203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenbaum CJ, Palmer JP, Kuglin B, Kolb H. Insulin autoantibodies measured by radioimmunoassay methodology are more related to insulin-dependent diabetes mellitus than those measured by enzyme-linked immunosorbent assay: results of the Fourth International Workshop on the Standardization of Insulin Autoantibody Measurement. J Clin Endocrinol Metab 1992;74:1040–1044 [DOI] [PubMed] [Google Scholar]
- 13.Wenzlau JM, Juhl K, Yu L, et al. The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci USA 2007;104:17040–17045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gianani R, Rabin DU, Verge CF, et al. ICA512 autoantibody radioassay. Diabetes 1995;44:1340–1344 [DOI] [PubMed] [Google Scholar]
- 15.Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol 2009;5:219–226 [DOI] [PubMed] [Google Scholar]
- 16.Knip M. Natural course of preclinical type 1 diabetes. Horm Res 2002;57(Suppl. 1):6–11 [DOI] [PubMed] [Google Scholar]
- 17.Bingley PJ, Bonifacio E, Williams AJ, Genovese S, Bottazzo GF, Gale EA. Prediction of IDDM in the general population: strategies based on combinations of autoantibody markers. Diabetes 1997;46:1701–1710 [DOI] [PubMed] [Google Scholar]
- 18.Bingley PJ, Bonifacio E, Ziegler AG, Schatz DA, Atkinson MA, Eisenbarth GS, Immunology of Diabetes Society . Proposed guidelines on screening for risk of type 1 diabetes. Diabetes Care 2001;24:398. [DOI] [PubMed] [Google Scholar]
- 19.Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet 2001;358:221–229 [DOI] [PubMed] [Google Scholar]
- 20.Devendra D, Liu E, Eisenbarth GS. Type 1 diabetes: recent developments. BMJ 2004;328:750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skyler JS. Prediction and prevention of type 1 diabetes: progress, problems, and prospects. Clin Pharmacol Ther 2007;81:768–771 [DOI] [PubMed] [Google Scholar]
- 22.Kaldunski M, Jia S, Geoffrey R, et al. Identification of a serum-induced transcriptional signature associated with type 1 diabetes in the BioBreeding rat. Diabetes 2010;59:2375–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia S, Kaldunski M, Jailwala P, et al. Use of transcriptional signatures induced in lymphoid and myeloid cell lines as an inflammatory biomarker in type 1 diabetes. Physiol Genomics 2011;43:697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy H, Wang X, Kaldunski M, et al. Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes. Genes Immun 2012;13:593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YG, Mordes JP, Blankenhorn EP, et al. Temporal induction of immunoregulatory processes coincides with age-dependent resistance to viral-induced type 1 diabetes. Genes Immun 2013;14:387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 1998;15:539–553 [DOI] [PubMed] [Google Scholar]
- 27.Woo W, LaGasse JM, Zhou Z, et al. A novel high-throughput method for accurate, rapid, and economical measurement of multiple type 1 diabetes autoantibodies. J Immunol Methods 2000;244:91–103 [DOI] [PubMed] [Google Scholar]
- 28.Klitz W, Maiers M, Spellman S, et al. New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans. Tissue Antigens 2003;62:296–307 [DOI] [PubMed] [Google Scholar]
- 29.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 30.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics 2002;18:207–208 [DOI] [PubMed] [Google Scholar]
- 31.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aly TA, Ide A, Humphrey K, et al. Genetic prediction of autoimmunity: initial oligogenic prediction of anti-islet autoimmunity amongst DR3/DR4-DQ8 relatives of patients with type 1A diabetes. J Autoimmun 2005;25(Suppl.):40–45 [DOI] [PubMed] [Google Scholar]
- 33.Miyara M, Yoshioka Y, Kitoh A, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 2009;30:899–911 [DOI] [PubMed] [Google Scholar]
- 34.Chatzigeorgiou A, Harokopos V, Mylona-Karagianni C, Tsouvalas E, Aidinis V, Kamper EF. The pattern of inflammatory/anti-inflammatory cytokines and chemokines in type 1 diabetic patients over time. Ann Med 2010;42:426–438 [DOI] [PubMed] [Google Scholar]
- 35.Hussain MJ, Maher J, Warnock T, Vats A, Peakman M, Vergani D. Cytokine overproduction in healthy first degree relatives of patients with IDDM. Diabetologia 1998;41:343–349 [DOI] [PubMed] [Google Scholar]
- 36.Svensson J, Eising S, Hougaard DM, et al. Danish Childhood Diabetes Registry . Few differences in cytokines between patients newly diagnosed with type 1 diabetes and their healthy siblings. Hum Immunol 2012;73:1116–1126 [DOI] [PubMed] [Google Scholar]
- 37.Carratù R, Secondulfo M, de Magistris L, et al. Altered intestinal permeability to mannitol in diabetes mellitus type I. J Pediatr Gastroenterol Nutr 1999;28:264–269 [DOI] [PubMed] [Google Scholar]
- 38.Kuitunen M, Saukkonen T, Ilonen J, Akerblom HK, Savilahti E. Intestinal permeability to mannitol and lactulose in children with type 1 diabetes with the HLA-DQB1*02 allele. Autoimmunity 2002;35:365–368 [DOI] [PubMed] [Google Scholar]
- 39.Bosi E, Molteni L, Radaelli MG, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia 2006;49:2824–2827 [DOI] [PubMed] [Google Scholar]
- 40.Secondulfo M, Iafusco D, Carratù R, et al. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis 2004;36:35–45 [DOI] [PubMed] [Google Scholar]
- 41.Sapone A, de Magistris L, Pietzak M, et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes 2006;55:1443–1449 [DOI] [PubMed] [Google Scholar]
- 42.Gough SC, Simmonds MJ. The HLA region and autoimmune disease: associations and mechanisms of action. Curr Genomics 2007;8:453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol 2013;190:513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brusko T, Wasserfall C, McGrail K, et al. No alterations in the frequency of FOXP3+ regulatory T-cells in type 1 diabetes. Diabetes 2007;56:604–612 [DOI] [PubMed] [Google Scholar]
- 45.van Loosdregt J, Fleskens V, Fu J, et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 2013;39:259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Barbi J, Bu S, et al. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 2013;39:272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrich de Marquesini LG, Fu J, Connor KJ, et al. IFN-gamma and IL-10 islet-antigen-specific T cell responses in autoantibody-negative first-degree relatives of patients with type 1 diabetes. Diabetologia 2010;53:1451–1460 [DOI] [PubMed] [Google Scholar]
- 48.Tree TI, Lawson J, Edwards H, et al. Naturally arising human CD4 T-cells that recognize islet autoantigens and secrete interleukin-10 regulate proinflammatory T-cell responses via linked suppression. Diabetes 2010;59:1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C, Jurczyk A, diIorio P, et al. Salicylate prevents virus-induced type 1 diabetes in the BBDR rat. PLoS One 2013;8:e78050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.