Abstract
Summary: ThunderSTORM is an open-source, interactive and modular plug-in for ImageJ designed for automated processing, analysis and visualization of data acquired by single-molecule localization microscopy methods such as photo-activated localization microscopy and stochastic optical reconstruction microscopy. ThunderSTORM offers an extensive collection of processing and post-processing methods so that users can easily adapt the process of analysis to their data. ThunderSTORM also offers a set of tools for creation of simulated data and quantitative performance evaluation of localization algorithms using Monte Carlo simulations.
Availability and implementation: ThunderSTORM and the online documentation are both freely accessible at https://code.google.com/p/thunder-storm/
Contact: guy.hagen@lf1.cuni.cz
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Single-molecule localization microscopy (SMLM) methods such as stochastic optical reconstruction microscopy (STORM; Rust et al., 2006) and photo-activated localization microscopy (PALM; Betzig et al., 2006) have recently emerged to overcome the diffraction barrier, offering ∼10 times higher lateral resolution and the possibility of 3D imaging by various approaches. In SMLM, a super-resolution image is reconstructed from a sequence of diffraction-limited images of sparsely distributed single photoswitchable molecules. As the sequence is usually long (thousands of images) and the positions of the molecules have to be estimated systematically with sub-diffraction precision, it is crucial to use specialized software for processing the data.
We present ThunderSTORM, an open-source, interactive, modular and platform-independent software, which provides a complete set of tools for automated processing, analysis and visualization of data acquired by SMLM methods. Our philosophy in developing ThunderSTORM has been to offer an extensive collection of processing and post-processing methods, which were developed based on extensive testing with both real and simulated data. We also provide a detailed description of the implemented methods and algorithms (Supplementary Note), as well as a user’s guide.
2 FEATURES AND METHODS
Most software tools currently available for SMLM data processing typically use only one particular algorithm for detection and localization of molecules. ThunderSTORM offers many different processing and post-processing methods so that users can adapt the analysis to their data. This approach can lead to higher quality results than existing solutions. Experienced users may use any combination of the available methods; however, we have designed the software’s default settings to produce good results on many of the datasets we have experimented with.
2.1 Raw data processing
Approximate molecular positions can be determined, in combination with a variety of feature-enhancing low-pass and band-pass filters (Křížek et al., 2011; Izeddin et al., 2012), by detection of local maxima, non-maximum suppression or calculation of the centroid of connected components of segmented objects. A feature exclusively unique to ThunderSTORM is the possibility of specifying the threshold for detection of molecules using a mathematical expression with quantities based on raw or filtered images. This allows computing the threshold value systematically for unknown input images with, for example, low signal to noise ratio, or where the global intensity slowly fluctuates. ThunderSTORM also offers a preview function to help visualize the detected molecules with the chosen combination of data processing settings.
Sub-diffraction localization of molecules is accomplished by computing the centroid of local neighborhoods, by a radial symmetry approach (Parthasarathy, 2012), or by fitting a suitable PSF model using standard or weighted non-linear least-squares methods, or using maximum-likelihood estimation (Mortensen et al., 2010). Users may also choose not to use any of the methods, thereby using the approximate localizations from the previous step. The uncertainty of the localization of molecules is calculated according to Thompson et al. (2002), or according to Quan et al. (2010) if EMCCD cameras are used.
Super-resolution 3D imaging is accomplished by an astigmatism approach (Huang et al., 2008). An integral part of this feature is the software’s calibration tool, in which a Z-stack of astigmatic images of sub-diffraction fluorescent beads is used to establish parameters for determining the axial position of each molecule.
Efforts to accelerate the acquisition process in SMLM have involved increasing the density of photoactivated fluorophores. In this case, ThunderSTORM uses an algorithm based on fitting of multiple emitters (Huang et al., 2011).
2.2 Post-processing and visualization
Post-processing routines offered by ThunderSTORM can eliminate molecules with poor localization or other user-defined criteria, merge molecules reappearing in subsequent frames, remove duplicated molecules obtained in multiple emitter analysis (Huang et al., 2011), correct molecular positions for lateral drift of the sample using fiducial markers or using cross-correlation methods (Mlodzianoski et al., 2011) and correct the absolute axial position of the molecules when the data were acquired in multiple Z-stage positions (Huang et al., 2008). Users can also select a region of interest to export only the localized molecules and their parameters from the region. Post-processing includes a live preview.
Visualization involves creation of a new high-resolution image based on the previously obtained sub-diffraction molecular coordinates. Several methods have been implemented for visualization such as Gaussian rendering and a 2D histogram with an option of jittering (Křížek et al., 2011). ThunderSTORM also introduces a new visualization method based on an average shifted histogram approach (Scott, 1985). This method provides similar results as the Gaussian rendering, but is orders of magnitude faster.
2.3 Simulation engine and performance evaluation
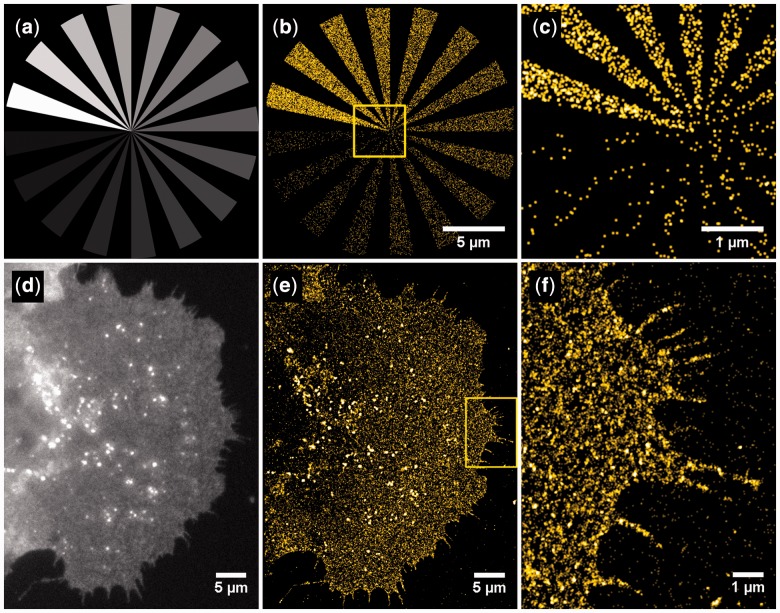
ThunderSTORM is capable of generating realistic sequences of SMLM-like images in which the ground-truth positions of the molecules are known. A grayscale mask can be used to vary the spatial density of molecules [Fig. 1(a–c)]. When the localization data and the ground-truth positions of molecules are available, ThunderSTORM can quantitatively evaluate the performance of localization algorithms (see Supplementary Note Sections 8 and 9). This allows users to perform sophisticated Monte Carlo simulations (Křížek et al., 2011) (see User’s Guide Sections 8–10).
Fig. 1.
Simulations and SMLM reconstruction with ThunderSTORM. (a) Example of a mask used for generating simulated SMLM data. The gray-scale intensity values are interpreted as molecular densities within a user-specified range. (b) SMLM reconstruction of a simulated dataset. (c) Detail of b. (d) Widefield fluorescence image of an A431 epidermoid carcinoma cell expressing the membrane protein mCitrine-erbB3. (e) SMLM reconstruction using the default settings. (f) Detail of e. SMLM imaging was performed as previously described (Křížek et al., 2011)
3 SUMMARY
ThunderSTORM introduces several new features and concepts for 2D and 3D SMLM data analysis. The software combines several algorithms for SMLM analysis into one comprehensive environment. One of the main features is the ability to process the data using any combination of the implemented feature-enhancing, spot detection and fitting methods. An important feature in ThunderSTORM is the possibility of specifying the threshold for detection of molecules using mathematical expressions. This allows users to systematically maximize the efficiency of molecule detection in the raw data by searching for the optimum combination, which may vary from experiment to experiment. ThunderSTORM also offers a much higher degree of user interactivity during data post-processing compared with other SMLM software packages, and introduces a new and fast visualization method that creates high-quality results. A realistic data generator within ThunderSTORM allows users to run multidimensional Monte Carlo simulations to evaluate the performance of localization methods. We have found ThunderSTORM’s flexibility and performance to be of critical importance when analyzing data with low molecular brightness, which we encountered when imaging A431 cells expressing mCitrine-erbB33 (Křížek et al., 2011) [Fig. 1(d–f)].
Funding: This work was supported by the Czech Science Foundation [P304/09/1047, P205/12/P392, P302/12/G157, 14-15272P]; Charles University [Prvouk/1LF/1, UNCE 204022]; the European Regional Development Fund [OPPK CZ.2.16/3.1.00/24010, BIOCEV CZ.1.05/1.1.00/02.0109]; and the European Social Fund [OPVK CZ.1.07/2.3.00/30.0030].
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Huang B, et al. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, et al. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed. Opt. Express. 2011;2:1377–1393. doi: 10.1364/BOE.2.001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, et al. Wavelet analysis for single molecule localization microscopy. Opt. Express. 2012;20:2081–2095. doi: 10.1364/OE.20.002081. [DOI] [PubMed] [Google Scholar]
- Křížek P, et al. Minimizing detection errors in single molecule localization microscopy. Opt. Express. 2011;19:3226–3235. doi: 10.1364/OE.19.003226. [DOI] [PubMed] [Google Scholar]
- Mlodzianoski MJ, et al. Sample drift correction in 3D fluorescence photoactivation localization microscopy. Opt. Express. 2011;19:15009–15019. doi: 10.1364/OE.19.015009. [DOI] [PubMed] [Google Scholar]
- Mortensen KI, et al. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods. 2010;7:377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R. Rapid, accurate particle tracking by calculation of radial symmetry centers. Nat. Methods. 2012;9:724–726. doi: 10.1038/nmeth.2071. [DOI] [PubMed] [Google Scholar]
- Quan T, et al. Localization capability and limitation of electron-multiplying charge-coupled, scientific complementary metal-oxide semiconductor, and charge-coupled devices for superresolution imaging. J. Biomed. Opt. 2010;15:066005. doi: 10.1117/1.3505017. [DOI] [PubMed] [Google Scholar]
- Rust MJ, et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DW. Averaged shifted histograms: effective nonparametric density estimators in several dimensions. Ann. Stat. 1985;13:1024–1040. [Google Scholar]
- Thompson RE, et al. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.