Abstract
PiT-1 protein is a transmembrane sodium-dependent phosphate (Pi) transporter. PiT-1 knock out (KO) embryos die from largely unknown causes by embryonic day (E) 12.5. We tested the hypothesis that PiT-1 is required for endocytosis in the embryonic yolk sac (YS) visceral endoderm (VE). Here we present data supporting that PiT-1 KO results in a YS remodeling defect and decreased endocytosis in the YS VE. The remodeling defect is not due to an upstream cardiomyocyte requirement for PiT-1, as SM22αCre-specific KO of PiT-1 in the developing heart and the YS mesodermal layer (ME) does not recapitulate the PiT-1 global KO phenotype. Furthermore, we find that high levels of PiT-1 protein localize to the YS VE apical membrane. Together these data support that PiT-1 is likely required in YS VE. During normal development maternal immunoglobulin (IgG) is endocytosed into YS VE and accumulates in the apical side of the VE in a specialized lysosome termed the apical vacuole (AV). We have identified a reduction in PiT-1 KO VE cell height and a striking loss of IgG accumulation in the PiT-1 KO VE. The endocytosis genes Tfeb, Lamtor2 and Snx2 are increased at the RNA level. Lysotracker Red staining reveals a loss of distinct AVs, and yolk sacs incubated ex vivo with phRODO Green Dextran for Endocytosis demonstrate a functional loss of endocytosis. As yolk sac endocytosis is controlled in part by microautophagy, but expression of LC3 had not been examined, we investigated LC3 expression during yolk sac development and found stage-specific LC3 RNA expression that is predominantly from the YS VE layer at E9.5. Normalized LC3-II protein levels are decreased in the PiT-1 KO YS, supporting a requirement for PiT-1 in autophagy in the YS. Therefore, we propose the novel idea that PiT-1 is central to the regulation of endocytosis and autophagy in the YS VE.
Keywords: Slc20a1, PiT-1, yolk sac development, phosphate transport, endocytosis, microautophagy
1. Introduction
PiT-1 (Slc20a1) protein is a sodium-dependent phosphate (Pi) transporter that contains 12 transmembrane domains (Farrell et al., 2009; O'Hara et al., 1990). PiT-1 was first identified as the gibbon-ape leukemia virus receptor (Glvr1), and found to function as a sodium-dependent phosphate transporter (Johann et al., 1993; Kavanaugh and Kabat, 1996; Kavanaugh et al., 1994; Olah et al., 1994; Virkki et al., 2007). Inorganic phosphate (Pi) is required in high abundance for numerous biological processes including nucleic acid synthesis, mineralization, oxidative phosphorylation and many others. PiT-1 facilitates Pi uptake against the electrochemical gradient via Na-coupled exchange. PiT-1 has also been implicated in terminal B-cell and erythrocyte cell differentiation, as well as cell cycle progression (Beck et al., 2009; Byskov et al., 2012; Forand et al., 2013; Liu et al., 2013). PiT-1 KO embryos are embryonic lethal and display gross defects in yolk sac (YS) vascular development and hematopoiesis (Beck et al., 2009; Festing et al., 2009).
Mammalian embryonic development is highly dependent upon regulated maternal-fetal exchange. Initially, the growth of the embryo is largely self-sustaining. However, as development proceeds further, growth of the embryo requires formation of the YS. The YS isolates the embryo from the uterine lumen and facilitates uptake of maternal factors, including immunoglobulins, LDL, transferrin, and other identified and unidentified molecules by both endocytosis, in which molecules are delivered to a specialized lysosome called the apical vacuole (AV), as well as trancytosis in which molecules are transferred across cells. Presumably maternal Pi is included in these processes, but this remains to be clearly demonstrated in mammals. The YS contains two tissue layers: the mesodermal layer (ME), and the visceral endoderm (VE) layer which is a transporting epithelium that serves nutritive and metabolic functions (Jollie 1990, Palis 2005). The YS VE layer is derived from the primitive endoderm of the blastocyst. As the embryo develops, distal primitive endoderm gives rise to the embryonic visceral endoderm layer (emVE) and proximal primitive endoderm differentiates into the extraembryonic visceral endoderm layer (exVE) that undergoes transcytosis at embryonic day (E) 5.25-E6.5 (Viotti et al. 2012). The exVE then gives rise to the YS VE layer that undergoes yolk sac trafficking after E7.5 (Viotti et al. 2012). The ME layer is generated from cells in the posterior primitive streak during gastrulation after ~E6.5 (Viotti et al. 2012).
Several key players in YS endocytosis are known, but the inductive factors and many mechanistic steps are yet to be discovered. Recently published work indicates that endocytosis in the VE occurs at least in part via microautophagy in which microvesicles (MVs) containing maternal factors are endocytosed (Kawamura et al., 2012; Wada et al., 2013). The resulting intracellular double membrane organelle fuses with the AV and the MV is delivered into the AV lumen. Finally, the MV membrane and the contents of the MV are hydrolyzed (Kawamura et al., 2012; Wada et al., 2013).
A fully developed YS vasculature is absolutely required for survivability of the embryo. YS development is initiated during gastrulation. First, blood islands are formed in the mesodermal tissue adjacent to the VE in the proximal, extraembryonic region of the gastrulating mouse embryo by ~E7.5 (Palis et al. 1995). Next, the blood islands fuse via vasculogenesis to generate a network of primitive vessels known as the vascular plexus by E8.0 (Ema and Rossant, 2003 and Palis 2005). The vascular plexus is then “remodeled” via angiogenesis into a mature vasculature by E10.0 (Palis 2005). Although the VE directly mediates maternal-fetal communication, the underlying mesodermal layer (ME) is the site of YS vascular development. The complex morphogenetic vascular development that occurs within the ME is highly dependent on the presence and normal activity of the VE (Palis et al., 1995). Disruption of the remodeling process frequently results from loss of molecular signaling pathways that promote normal endothelial and hematopoietic cell differentiation and morphogenesis intrinsic to the YS tissue, including paracrine VE to ME signaling (Ema and Rossant, 2003; Goldie et al., 2008; Ueno and Weissman, 2010). Alternatively, a loss of mechanical stimulation/hemodynamic force due to upstream aberrant cardiac function also leads to defective YS remodeling (Culver and Dickinson, 2010; Lucitti et al., 2007). Lethality associated with abnormal yolk sac vascular development commonly occurs between E9.5 and E12.5. The role of Pi in the development and function of the YS has not yet been elucidated.
PiT-1 knock out (KO) embryos die by E12.5 from largely unknown causes, indicating that PiT-1 is necessary for embryonic development (Beck et al., 2009; Festing et al., 2009). Additionally, the time frame of PiT-1 KO embryonic death corresponds with that of yolk sac malfunction mortalities. However, the specific role of PiT-1 in the YS remains unknown. We have found that vascular smooth muscle cell (VSMC) knock down (KD) of PiT-1 in vitro decreases several processes, including Pi uptake, mineralization, and osteochondrogenic phenotype change in high Pi conditions (Li et al., 2006). Furthermore, others have recently shown that PiT-1 is required for homeostatic levels of autophagy; vesicular shedding in VSMCs is thought to be counteracted by Pi-induced ROS production that promotes autophagy in non-disease conditions (Dai et al., 2013). These in vitro analyses suggest that there may be an important role for PiT-1 in autophagy in vivo. In order to elucidate the in vivo roles of PiT-1, we have taken advantage of complete and tissue specific PiT-1 KO mouse lines. Here we present data supporting that PiT-1 is required for Pi-mediated endocytosis via microautophagy in the yolk sac visceral endoderm.
2. Experimental Procedures
2.1 Mice: Embryo production, Genotyping and Yolk Sac Tissue Layer Collection
For production of PiT-1 KO and control WT animals, heterozygous PiT-1 mice were mated. For production of SM22α-KO animals, PiT-1 fl/fl females and PiT-1 fl/fl, SM22αCre males were mated. SM22αCre mice were kindly provided by Dr. David Dichek (University of Washington). Timing of embryonic development was determined by the presence of a vaginal plug the morning after mating and designated as E0.5. At the time of embryonic collection, female mice were euthanized and embryos and extraembryonic tissues were collected by mechanical dissection in PBS.
At the time of dissection posterior embryonic trunk and tail were taken for genotyping. DNA extraction was performed using Proteinase K (Sigma P6556). WT and KO embryos were genotyped with established methods (Festing et al., 2009), and PiT-1 fl/fl and fl/fl,cre (SM22α-KO) embryos were genotyped with established methods (Crouthamel et al., 2013). Primer sequences and PCR conditions are listed in Supplemental Table 1.
The yolk sac layers were separated using the trypsin/pancreatin method as described in (Rhee et al., 2013). Briefly, yolk sacs were collected and incubated in Tyrode Ringers Solution (Fisher S25512) containing 0.005g/mL Trypsin (Fisher S25618) and 0.025g/mL Pancreatin (Fisher S25456) for 4h at 4°C with gentle shaking. Yolk sacs were then washed in DMEM (Life Technologies 11995-065) and tissue layers were mechanically separated and stored at −80°C until further processing.
2.2 Gene Expression: RNA Extraction, cDNA Synthesis, RT-PCR and qPCR
Embryonic tissues, whole yolk sacs or yolk sac layers were dissected and stored at −80°C. Total RNA was extracted using the RNeasy Mini Kit (Qiagen 74104) and DNaseI (Qiagen 1023460). cDNA synthesis was performed with 500 ng of whole yolk sac RNA, 250 ng of specific yolk sac tissue layer RNA or 250 ng of embryonic heart RNA. For reverse transcription the Omniscript RT Kit (Qiagen 205113) was used with both oligodT primers (Promega C110A) and random hexamers (Qiagen 79236) according to the manufacturer's instructions. RT-PCR was performed as in (Crouthamel et al., 2013) using GoTaq Flexi DNA Polymerase (Promega M829B) and 5× Green GoTaq Buffer Flexi (Promega M891A). Primer sequences and PCR conditions are listed in Supplemental Table 1. Gapdh was used as an internal control for all RT-PCR. The student's t-test was used to test the null hypothesis that there are no differences between WT and KO samples. At least three embryos per genotype were analyzed for each gene, and littermate controls (WT or fl/fl as indicated) were used in all cases.
Quantitative RT-PCR (qPCR) was performed using the primers listed in Supplemental Table 1, Taqman probes from ABI against PiT-1: 5’-ccgtaaggcagatcc-3’ and PiT-2: 5’-catggttggttcagctg-3’, and Taqman Universal PCR Master Mix (Roche 4304437). Assays were multiplexed with VIC labeled 18S probe (ABI 4308329). qPCR reactions were performed on an Applied Biosystems ABI Prism 7000 Sequence Detection System with Sequence Detection Software Version 1.2.3 and recommended cycling conditions. The student's t-test was used to test the null hypothesis that there are no differences between WT and KO samples. At least three embryos per genotype were analyzed for each gene, and littermate controls (WT or fl/fl as indicated) were used in all cases.
2.3 Histology
2.3.1 Fixation, Embedding and Sectioning
Embryonic tissue was processed as in (Trask et al., 2012). Samples were dissected and fixed in 4% paraformaldehyde (PFA)/PBS overnight at 4°C. They were then dehydrated through a series of methanol washes, treated with xylenes, embedded in paraffin and sectioned at 7.5 micrometers with a Leica RM2135.
2.3.2 Immunostaining
Immunofluorescence was performed as in (Wallingford et al., 2013), but treated with a serum block as opposed to a milk block. Heat mediated antigen retrieval was performed with 0.01M Tris Base pH10.0. The following antibodies and antibody concentrations were used: rabbit anti-CDH1 1:200 (Abcam ab53033); goat anti-SM22α 1:100 (Abcam 10135); mouse anti-PCNA 1:100 (Abcam ab29). All secondary antibodies were used at 1:400 and included: donkey anti-rabbit Dylight 549 (Jackson ImmunoResearch 711-505-152); donkey anti-goat Dylight (Jackson ImmunoResearch 488 705-485-147); and donkey anti-mouse Alexa Fluor 488 (Jackson ImmunoResearch 715-545-151).
PiT-1 immunohistochemistry was performed as in (Crouthamel et al., 2013) with chicken anti-rat PiT-1 primary antibody (provided by Dr. M. Levi, University of Colorado), Tris Base heat-mediated antigen retrieval and donkey serum block. Slides were counterstained with Harris Hematoxylin (Sigma HHS32).
Whole mount Pecam1 staining was performed with rat anti-CD31 (BD PharMingen 550274), 4% BSA block, and goat anti-rat secondary antibody (Jackson ImmunoResearch 712-035-153).
2.3.3 H&E
Slides were stained with Harris Hematoxylin (Sigma HHS32) and counterstained with Eosin Y (Sigma E4382).
2.4 Image Analysis
Digital images of whole mount embryos were captured on a Nikon SMZ-1500 stereomicroscope equipped with a Nikon D5100 DSLR camera. Digital images of sectioned embryos were taken with a Nikon Eclipse E800 inverted fluorescence microscope and RS Photometrics Coolsnap color digital camera, imaged with Metamorph Version 6.3r7 and converted with Irfanview Version 4.2.3. The Image-J version 1.46r (http://imagej.nih.gov/ij) ROI Manager Tool and the Image-J “Cell Counter” plug-in was used to assist with cell counting and cell height measurements. For cell height measurements, images of H&E stained yolk sac sections where used and were assessed blindly; the images had coded names and the measurer had no knowledge of the sample genotypes or any previous experience with the images. Twenty cells from each of three images were counted for three different yolk sacs for each genotype (180 cells total per genotype). For maximum artery width quantification, the three largest arteries on three images per yolk sac were measured with ImageJ; a total of three yolk sacs per genotype were examined. In order to determine the extent of endocytosis of phRODO Dextran, ImageJ was used to blindly determine the internal density value of ten cells for each of ten images/yolk sac; the normalization value was taken by averaging three readings and normalized internal density values for each yolk sac were calculated prior to genotyping. The student's t-test was used to test the null hypothesis that there are no differences between control and KO samples.
2.5 Western Blotting
For western blotting, yolk sac lysates were collected and treated with a protease inhibitor cocktail (Roche). Lysates were denatured in Laemli Buffer containing β-Mercaptoethanol and run in a 10% SDS Page gel. LC3A antibody (Cell Signaling Technology D50G8 XP) was used according to the manufacturers instructions. The western blot was stripped with Restore Plus Western Blot Stripping Buffer (Thermo Scientific PI-46430). β−Actin (Abcam ab8227) was used to determine a loading control. Densitometry was performed with ImageJ. PiT-1 antibody (See section 2.3) was used to confirm loss of PiT-1 protein.
2.6 ex vivo Yolk Sac Assays
Optimal dilutions of both Lysotracker Red (Life Technologies DND99) and phRODO Green Dextran for Endocytosis (Life Technologies P35368) were determined empirically (Supplemental Figures 2 and 3). Yolk sacs were dissected in PBS. For visualization of vacuoles, the lysosome stain Lysotracker Red was incubated at 1:1000 in PBS for 5 minutes at 37°C. Upon receipt, phRODO Dextran was resuspended according the manufacturer's instructions and stored at −20°C. Working solutions of phRODO Dextran were made by diluting stock solution in prewarmed DMEM with 10%FBS at 1:100 and yolk sacs were incubated for 2 hours at 37°C. Yolk sacs were immediately mounted in Prolong Gold (Life Technologies P36930) and imaged.
3. Results
3.1 Na-dependent Phosphate Transporter Gene Expression
Our lab previously identified that PiT-1 KO embryos display embryonic lethality and gross vascular abnormalities at E12.5 (Festing et al., 2009). We began to follow up on this research by assessing gene expression of PiT-1 and PiT-2 in WT and PiT-1 KO yolk sacs by qPCR (Figure 1). We observed a lack of WT PiT-1 mRNA in PiT-1 KO embryos as compared to WT embryos (p-value=0.000405) (Figure 1A). Unlike other PiT-1 KO models, PiT-2 expression was not significantly increased in the PiT-1 null YS (p-value=0.065123) (Figure 1B). PiT-1 and PiT-2 expression in the developing yolk sac were previously examined by RT-PCR and Southern blotting with conflicting results (Festing et al., 2009; Richardson et al., 1996). In order to address this issue we examined PiT-1 and PiT-2 expression during yolk sac development by qPCR. We found that both genes are expressed throughout development in the yolk sac. PiT-1 expression peaked at E9.5, and PiT-2 expression increased linearly with development (Figure 1C,D). Finally, PiT-1 protein was strongly localized to the apical VE in E9.5 YS (Figure 1E,F).
Figure 1. PiT Gene Expression.
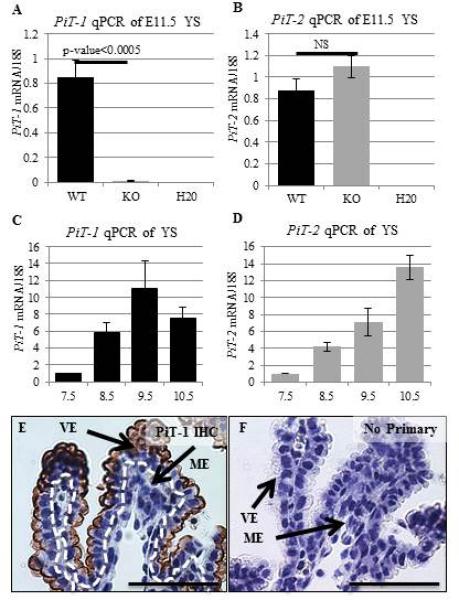
At E11.5 PiT-1 mRNA is absent from the KO YS (A) and there is no significant difference in PiT-2 expression (B) by qPCR (N=3/genotype). Developmental profiles of PiT mRNA during YS development collected by qPCR show a peak in PiT-1 expression at E9.5 (C) and a linear increase of PiT-2 expression (D) (N=3/embryonic stage). PiT-1 protein localizes to the apical membrane of YS VE at E11.5 (E) compared to a no primary control (F). Scale bars=25um.
3.2 PiT-1 KO results in a yolk sac remodeling defect
At E9.5 both WT and PiT-1 KO embryos and yolk sacs look largely normal (Figure 2A,D). Both display a beating heart and subsequent blood flow (data not shown). Primitive erythropoietic progenitors can be seen within the yolk sac vasculature in whole mount samples (circles in Figure 2A,D) and in H&E stained yolk sac sections (arrows in Figure 2B,E). However, whole mount Pecam1/CD31 staining at E11.5 reveals a characteristic remodeling defect in PiT-1 KO YS, evidenced by the failure of the hatched vascular plexus (asterisk in Figure 2C) to develop into a mature branching vasculature (asterisk in Figure 2F) with large arteries (arrows in Figure 2C,F). Maximum artery width measurements further support the failure of normal vascular development in the PiT-1 KO YS (Figure 2 G-I). In order to test whether developmental signaling pathways were disrupted in the PiT-1 KO YS, RT-PCR with WT and KO E11.5 YS RNA was performed and we found that expression was maintained for all VE to ME signaling pathways examined (Supplemental Figure 1). VE genes Hnf4α (Chen et al., 1994; Duncan et al., 1994), Tgfβ1 (Akhurst et al., 1990) and Vegfa (Breier et al., 1995) were increased in PiT-1 KO YS, but most ME genes examined, including Acta2 (Mahlapuu et al., 2001), Tagln (Li et al., 1996), TgfβrII (Wang et al., 1995), Flk1 (Nishikawa et al., 1998), and Tie2 (Mahlapuu et al., 2001) were largely unaltered (Supplemental Figure 1).
Figure 2. PiT-1 KO Vascular Remodeling Defect.
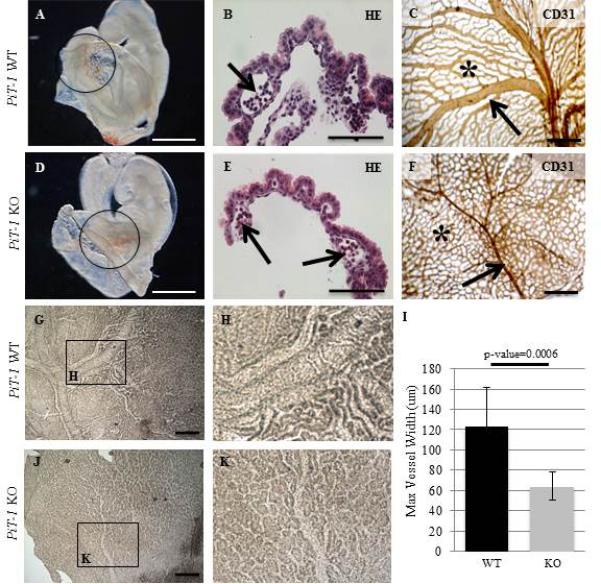
At E9.5 PiT-1 KO embryos are largely normal. Yolk sac vasculature is visible (circles in A, D) and H&E stained YS sections reveal presence of nucleated erythrocytes (arrows in B, E). However, whole mount IHC of Pecam1 at E11.5 shows a clear yolk sac remodeling defect, including absence of a mature branching vasculature (asterisks in C,F) and large arteries (arrows in C,F). Quantification of this by measurement of maximum artery width (described in experimental methods section) revealed a significant difference between the average WT artery width (120.9um ± 39.1um) and the average PiT-1 KO artery width (64.2um ± 14um) (G-I) (N=3/genotype). Scale bars: A and D=1mm; B and E=100um; C,F,G and J= 500um.
3.3 Deletion of PiT-1 in SM22α expressing cells of the developing heart does not result in embryonic lethality
Defects in YS angiogenesis or remodeling are commonly attributed to an upstream cardiac defect resulting in a lack of hemodynamic pressure, or defective developmental signaling. In order to determine whether there could be an upstream cardiac requirement for PiT-1 we turned to a tissue-specific knock out system. Our lab previously produced mice in which PiT-1 exons 3 and 4 are floxed by loxp sites (PiT-1 fl/fl) (Festing et al., 2009). We then bred PiT-1 fl/fl mice to SM22αCre mice to produce KO of PiT-1 in SM22α expressing cells; these mice are viable, reproduce normally and do not show heart abnormalities when analyzed by ultrasound (Crouthamel et al., 2013).
SM22αCre activity was shown previously in detail through the use of R26R mice; at E8.5 SM22αCre is active in a small number of cells in the primitive heart and ME layer of the YS, and at E9.5 SM22αCre is active throughout the developing heart and the ME layer of the YS (French et al., 2008). In order to determine the extent of Cre mediated loss of PiT-1, we collected E9.5 and E10.5 hearts and examined expression of SM22α, Cre and PiT-1 mRNA (Figure 3C, D). SM22α expression confirmed isolation of target tissue. Cre expression was only observed in the SM22α-KO heart RNA and not the fl/fl control heart RNA. As expected, PiT-1 expression was largely reduced in the SM22α-KO developing hearts. We then isolated E11.5 control PiT-1 fl/fl embryos and PiT-1 fl/fl, SM22αCre (SM22α-KO) embryos to examine YS vasculature and found that the SM22α-KO develop a mature YS vasculature with large hierarchical vessels by E11.5 (Figure 3A,B).
Figure 3. PiT-1 SM22α-KO Viability.
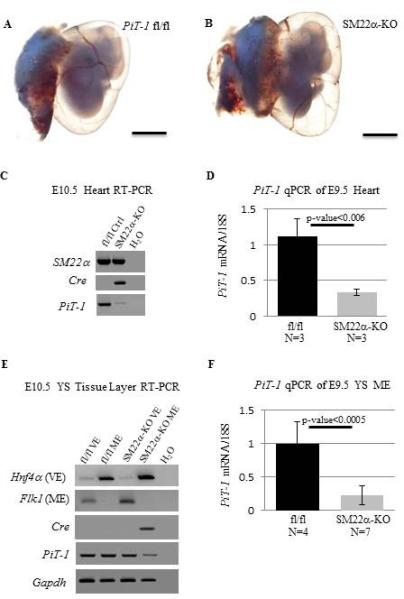
Tissue specific KO of loxp flanked PiT-1 (fl/fl) allele by SM22αCre does not disrupt YS remodeling (A, B). RT-PCR of E10.5 heart RNA (C, N=1/genotype) and qPCR of E9.5 heart RNA (D, N=3/genotype) confirm loss of PiT-1. Similarly, RT-PCR of E10.5 yolk sac mesoderm RNA (E, N=1 fl/fl and N=2 fl/fl, cre) and qPCR of E9.5 yolk sac mesoderm RNA (F, N=4 fl/fl and N=7 fl/fl, cre) confirm loss of PiT-1. Scale bars=1mm.
We next examined gene expression in E9.5 and E10.5 yolk sac VE and ME tissue layers that had been separated by enzymatic and mechanical methods as outlined in the Experimental Methods. Expression of the ME marker Flk1 and the VE marker Hnf4α were used to confirm effective tissue separation. Again, Cre expression and reduced PiT-1 levels were specifically observed in the expected tissue (SM22α-KO ME) at E9.5 and E10.5 (Figure 3E, F).
3.4 Altered endocytosis gene expression and IgG accumulation in PiT-1 KO YS
To identify the upstream cause of the PiT-1 KO YS vascular development phenotype, we examined the gross morphology of E9.5 WT and KO YS by analyzing H&E stained sections. We identified a focal disruption of normal nuclear localization relative to the apical and basal cell membranes in the PiT-1 KO YS VE. To assess proliferation, PCNA staining and DAPI counterstaining of WT and PiT-1 KO E9.5 YS sections was performed and did not reveal a gross difference in PCNA levels (data not shown). In epithelial cells E-Cadherin (CDH1) is absent from the apical membrane (Jenkins et al., 2013). We examined CDH1 localization in WT and PiT-1 KO E9.5 YS sections; VE/ME cells were delineated with the help of the ME marker SM22α (Figure 4). Both WT and PiT-1 KO VE apical cell membranes were found to be E-Cadherin negative, suggesting normal cell polarity (arrows in Figure 4).
Figure 4. Cell Polarity.
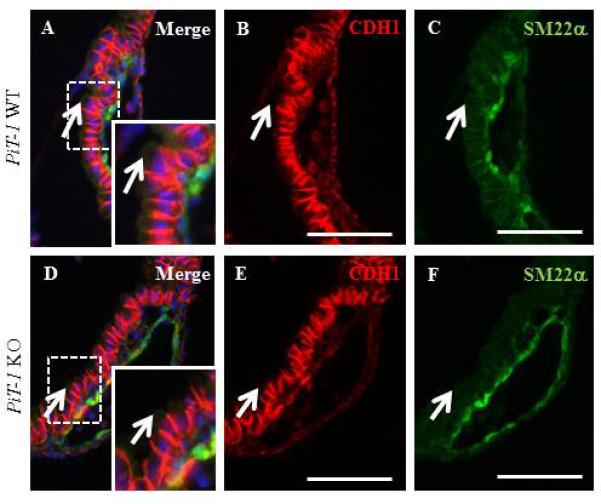
Appropriate localization of E-Cadherin (Red) and SM22α (Green) in PiT-1 KO VE (D-F) compared to WT controls (A-C). Insets: magnified image of area enclosed by dashed boxes. Scale bars=100um.
We then hypothesized that the nuclei appeared more centrally located due to a decrease in the height of the VE cells. Height and nuclear localization in WT and PiT-1 KO YS were quantified by performing blind measurements of total cell height and the cell height superficial to the nucleus in three different H&E stained sections from three different WT and KO yolk sacs for a total of 180 cells/genotype (Figure 5 A-C). The average height of E9.5 WT VE cells was significantly larger than the PiT-1 KO cells (WT=17.03um vs KO=14.82um; p-value=0.0018), as was the apical height (WT=7.12um vs KO=5.12um; p-value=0.00014) (Figure 5B). Together these data indicate that the total height is shorter in the PiT-1 KO VE, and this difference is due to a decrease in the height of the apical space of the cell.
Figure 5. Abnormal Morphology and IgG Localization in PiT-1 KO YS VE.
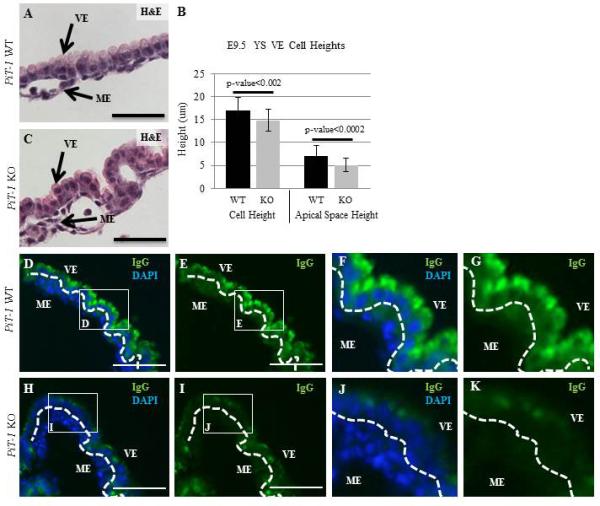
Quantification of cell size and nuclear localization relative to the apical membrane reveals that PiT-1 KO VE cells are smaller due to a decrease in the apical space (A-C). There is a large reduction in IgG accumulation in the PiT-1 KO YS VE (H-K), compared to controls (D-G). Scale bars=100um.
In WT YS VE the apical side of the cell contains specialized lysosomes termed apical vacuoles (AV). Maternal IgG and other molecules are normally taken up by the VE and delivered to the AV. We examined uptake of maternal IgG in WT and KO yolk sacs and observed a striking reduction in IgG staining on the PiT-1 KO YS (compare Figure 5D-G to H-K). This loss of IgG localization suggests a functional endocytosis defect and/or a structural AV defect.
In order to assess whether endocytosis and/or microautophagy was disrupted, we examined YS layer-specific expression of several genes related to endocytosis or autophagy including LC3, Tfeb, Lamtor2 and Snx2 (Kabeya et al., 2000; Griffin et al., 2005; Sardiello et al., 2009; Schwarz et al., 2002; Settembre et al., 2011; Sparber et al., 2013; Teis et al., 2006). As expected we found that the expression of these genes was largely restricted to the VE layer (Figure 6A). Tfeb, a master transcriptional regulator of both endocytosis and autophagy genes was expressed through out YS development and the important autophagy-related gene LC3 was expressed in a stage specific manner during YS development (Figure 6B). Tfeb, LC3, Lamtor2 and Snx2 were upregulated in the PiT-1 KO YS VE at the RNA level (Figure 6C1, C2). In order to quantify the level of autophagy in the YS, we examined expression of LC3 protein in the yolk sac. LC3-I is the unmodified protein that is found in the cytosol, and LC3-II is cleaved protein that localizes to the autophagosome and is representative of the degree of autophagy. The level of autophagy determined by LC3-II/LC3-I ratios was significantly decreased in the PiT-1 KO YS (p-value=0.037), and the overall LC3 protein levels normalized to β-Actin were modestly decreased (p-value=0.06) (Figure 6D1, quantified by densitometry in D2).
Figure 6. Endocytosis and Autophagy Gene Expression in the YS.
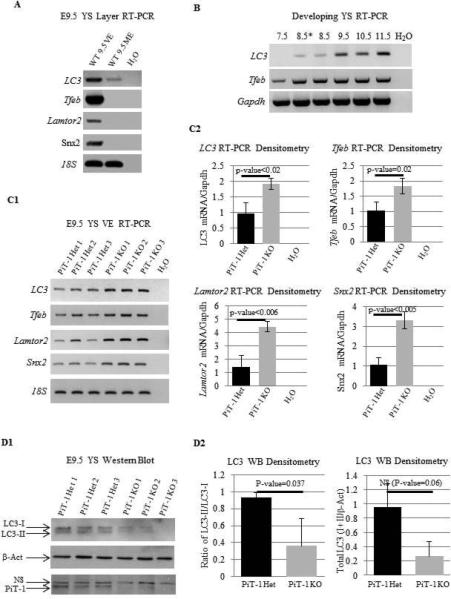
LC3, Tfeb, Lamtor2 and Snx2 are primarily expressed in the VE layer of the YS (A). LC3 is expressed in a stage specific manner during YS development and Tfeb is expressed at similar levels during YS development (B). YS VE RT-PCR of isolated YS VE mRNA reveals an upregulation of LC3, Tfeb, Lamtor2 and Snx2 in PiT-1 KO YS VE compared to heterozygous controls (C1, analyzed by densitometry in C2). Western blotting of LC3, B-Actin and PiT-1 reveals a slight decrease in total LC3 protein as well as a decrease in autophagy in PiT-1 KO yolk sacs identified by the ratio of LCII/LCI expression (D1, analyzed by densitometry in D2). Het: heterozygous. NS: nonspecific band. E8.5*: E8.5 allantois.
In order to test for vacuole presence we dissected fresh yolk sacs and incubated them with the lysosome stain Lysotracker Red according to the manufacturers instructions (Figure 7 A-D). This stain clearly demonstrated a loss of normal lysosome presence in the PiT-1 KO YS. We then examined endocytosis in dissected yolk sacs ex vivo with the use of the phRODO Green Dextran for Endocytosis. This functional assay demonstrated a significant decrease in endocytosis in the PiT-1 KO (Figure 7 E-I).
Figure 7. Presence of the Apical Vacuole and Endocytosis in the YS.
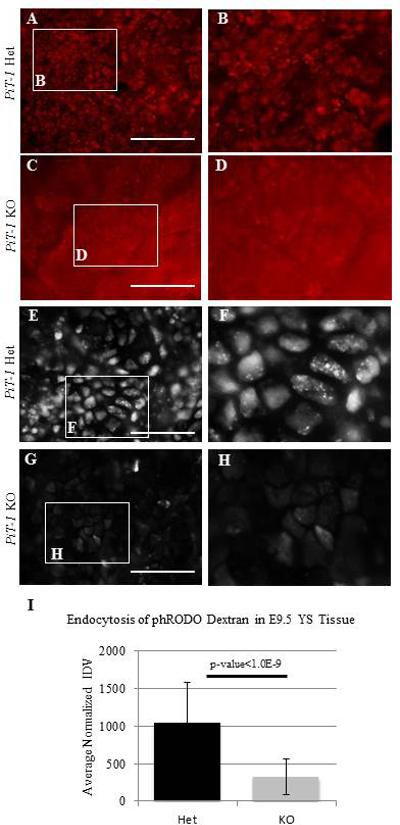
PiT-1 KO YS stained with the lysosome stain Lysotracker Red demonstrate a lack of normal AV maintenance (compare KO yolk sac in C, D to Het YS in A, B). PiT-1 KO YS incubated with phRODO Green Dextran for Endocytosis demonstrates a decrease in endocytosis (E-H). Dextran uptake was quantified by normalized internal density values; ten cells were measured on each of ten images for each yolk sac and averaged prior to genotyping. Normalization values were found by averaging three measurements per image. IDV: Internal density value. Het: heterozygous. Scale bars: AD=100um; E-H=50um.
4. Discussion
Our results shed light on the requirement for PiT-1 in yolk sac endocytosis. A loss of global PiT-1 results in abnormal VE gross morphology, decreased maternal-fetal IgG uptake, upregulation of endocytosis and autophagy gene transcription, disruption of AV maintenance and decreased endocytosis ex vivo. This data supports that normal endocytosis in the YS requires PiT-1. We hypothesize that the absence of PiT-1 leads to a decrease in endocytosis and lysosomal activity that in turn results in a failure of the VE to appropriately support the ME layer during yolk sac angiogenesis (Figure 8). In conjunction with published literature our data prompts additional questions about PiT-1 activity in both developmental and disease models.
Figure 8. Proposed Model of PiT-1-Dependent Microautophagy.
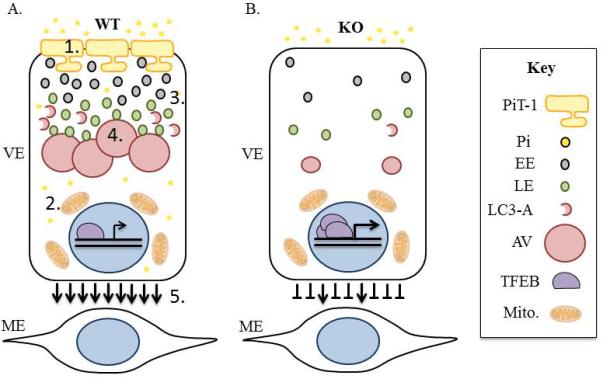
A.) In WT animals PiT-1 is present on the apical VE membrane, phosphate is taken into the cell (1), high Pi induces mitochondrial production of ROS (2) and vesicular uptake (3). As a result nutrients are delivered to the apical vacuole via microautophagy (4), and healthy VE promotes vascular development in the mesodermal layer (5). B.) In the PiT-1 KO Pi levels are not maintained, IgG does not properly accumulate in the AV, Snx2, Lamtor2, LC3 and TFEB are upregulated at the transcriptional level, LC3 protein is lost, and impaired VE endocytosis inhibits appropriate ME vascular development. Pi: inorganic phosphate. EE: early endosome. LE: late endosome. LC3-A: LC3-II associated autophagosome. AV: apical vacuole. Mito: mitochondria.
4.1 Working model of Pi-mediated endocytosis
The data presented here begs the question of whether PiT-1 is required for endocytosis via microautophagy. High Pi and reactive oxygen species (ROS) induce homeostatic levels of autophagy that are believed to counteract vesicle shedding in cultured human vascular smooth muscle cells (HA-VSMCs); as would be predicted, HA-VSMC PiT-1 KD in vitro results in decreased autophagy (Dai et al., 2013). Similarly, we found that KO of PiT-1 results in decreased autophagy in the PiT-1 KO YS. This novel regulation of endocytosis by a phosphate transporter highlights the importance of Pi localization during development. However, the dynamics of uptake and endocytic cycling of Pi in the YS remains unknown.
The data supports that endocytic cycling in disrupted in the PiT-1 KO VE and manifests in altered epithelial cell morphology, endocytosis and autophagy. This implies that despite the expression increase of both Tgfβ1 and Vegfa RNA in the PiT-1 KO VE, these important paracrine signaling molecules Tgfβ1 and Vegfa do not successfully reach their respective receptors that are localized to cells in the ME layer. Therefore, the increased expression does not have an effect on the ME layer, and it is instead the lack of normal endocytic processes that impact yolk sac development. This work is the first to suggest connections between Pi uptake, endocytosis, and paracrine support of vascular development. Pi uptake in this case may be directly through PiT-1, or indirectly through a currently unidentified mechanism. Additionally, it is possible that both the abnormal cellular morphology and the endocytosis failure involve Pi-dependent defects in the cytoskeletal network. Further research will elucidate the specific role of Pi uptake in this system, as well as the direct impact of Pi uptake and Pi deprivation on endosome trafficking and the cytoskeletal network.
In the absence of PiT-1 endocytosis and autophagy genes are upregulated in the VE, possibly through a compensatory mechanism. Although RNA levels are high, it is likely that proteins are not functioning normally. In support of this idea, we found a decrease in LC3 protein by western blotting, despite an increase in LC3 RNA. This indicates that the increased RNA levels may indeed reflect a compensatory mechanism attempting to counteract the loss of endocytosis and autophagy in the PiT-1 KO YS. Snx2 shuttles CIMPR in yeast and other cell types. Of note, the Snx1/2 double KO is lethal due to YS malfunction that includes VE AV defects, yet Snx2 does not shuttle CIMPR in the YS as it does in yeast – the specific role of Snx2 in the YS remains enigmatic (Griffin et al., 2005; Schwarz et al., 2002). Therefore, it is possible that Snx2 shuttles PiT-1 or a microautophagy-related protein between the apical cell membrane, endosomes, apical vacuoles and mitochondria.
Lastly, the mechanistic step between Pi uptake and microautophagy induction is not clear. Mitochondria are well-known sites of the electron transport chain and oxidative phosphorylation. Interestingly, Vamp1 (also known as Synaptobrevin1) may play a role in Pi-dependent endocytosis. Vamp1 is expressed in the YS and closely parallels expression of PiT-2 in the brain (da Silva et al., 2013). Vamp1 splice-isoforms vary in sequence and function. Vamp1a targets the plasma membrane and mediates exocytosis in neuronal cells and cardiomyocytes. Importantly, the splice-isoform Vamp1b targets the mitochondria (Isenmann et al., 1998). We found that Vamp1b expression exceeds Vamp1a expression in the YS, and increased at a higher rate during YS development (data not shown). This novel data leads us to speculate that Vamp1b could potentially mediate delivery of Pi or other vesicle contents to mitochondria in PiT expressing cells. Together these data suggest a working model of endocytosis in which PiT-1 promotes Pi delivery to mitochondria, thus inducing ROS production and microautophagy. Future studies examining Pi dynamics, ROS levels and Vamp1 activity in PiT-1 KO YS will help differentiate these possibilities.
4.2 Tissue-Specific Transporter Function
Previously, our lab found that PiT-2 is increased in PiT-1 null VSMCs (Crouthamel et al. 2013). Additionally, PiT-1 compensation has been suggested as a potential pathogenic mechanism for development of Familial Idiopathic Basal Ganglia Calcification (FIBGC) in human cases associated with mutated PiT-2 sequences (da Silva et al., 2013; Hsu et al., 2013; Wang et al., 2012; Zhang et al., 2013). For these reasons, the mechanisms controlling compensatory PiT expression are of interest. Notably, we have found that the PiT-1 global KO did not cause a significant increase in PiT-2 RNA in whole YS lysates. Of note, PiT-2 RNA is expressed, indicating a PiT-1 specific role at this stage of embryonic development. This data suggests that compensatory PiT-1 and PiT-2 gene expression is cell type-dependent, and further suggests that PiT-1 plays a specific role in yolk sac development for which PiT-2 is not capable of compensating. Conversely, SM22α-KO mice are viable (Crouthamel et al., 2013) despite the altered PiT-1 expression in the developing heart and ME layer. This suggests that either of the type III transporters may be sufficient to fulfill requirements for type III sodium-dependent phosphate transporters in SM22α positive lineages. Importantly, a low level of PiT-1 expression was observed in the SM22α-KO hearts and YS ME layer. This expression most likely originates from the developing endocardium of the heart, and the endothelium and primitive blood cells of the YS ME layer; the work presented here does not distinguish between these possibilities. Examination of Pi-uptake dynamics as well as tissue specific single and double PiT-1 and PiT-2 KO will yield valuable information about the complex interplay of these two genes and their potential roles in vascular development and disease.
4.3 PiT-1 and Vascular Disorders
The PiT-1 global KO phenotype we have observed, coupled with the contrasting viability of PiT-1 SM22α-KO mice suggests that PiT-1 may be required in the YS VE, but either PiT-1 or PiT-2 may subserve type III transporter requirements in SM22αCre expressing tissues. Work aimed at understanding mechanisms of vascular calcification has revealed a compensatory increase in PiT-2 gene expression in mature vasculature of the adult. PiT-1 is believed to play a role in vascular calcification in patients with Chronic Kidney Disease (CKD), in which ectopic hydroxyapatite formation may lead to vessel stiffening and mortality. Elevated serum Pi is a key risk factor for vascular calcification (Dhingra et al., 2007; Kestenbaum et al., 2011; Tonelli et al., 2009) and when VSMCs are exposed to high Pi, calcification is observed (Jono et al., 2000; Lomashvili, 2004). This data suggests that Pi-driven mechanisms may control ectopic calcification observed in patients with CKD (Lau et al., 2010; Shanahan et al., 2011). KO of PiT-1 in human cells alleviated calcification (Li et al., 2006). However, we found that KO of PiT-1 in mouse cells failed to act similarly and instead resulted in an increase of PiT-2 expression both in vitro and in vivo in the aorta of CKD mouse models (Crouthamel et al., 2013). Furthermore, in vitro expression of PiT-2 in PiT-1 null cells revealed that PiT-2 was capable of compensating for PiT-1 with respect to Pi uptake and calcification (Crouthamel et al., 2013). As PiT-2 was able to compensate for Pi transport in PiT-1 KD human VSMC, both type III transporters may play critical roles in ectopic mineralization. Currently, the specific mechanistic steps remain enigmatic. Furthermore, abnormal levels of PiT gene expression may promote FIBGC. Large subsets of patients with FIBGC were recently found to contain deleterious mutations in PiT-2 (da Silva et al., 2013; Hsu et al., 2013; Wang et al., 2012; Zhang et al., 2013), which has been suggested to result in compensatory PiT-1 overexpression and subsequent disease-related calcification. Interestingly, PiT-1 is not capable of compensating for loss of PiT-2 in FIBGC. This indicates a PiT-2 specific requirement and further supports the presence of distinct roles for each type III transporter.
4.4 Conclusions
We have found that global PiT-1 KO results in a YS remodeling/angiogenesis defect early in development. However, Sm22αCre mediated tissue-specific PiT-1 KO mice are viable and display high decreased levels of PiT-1 loss in the developing heart and YS ME layer. The viability of PiT-1 SM22α-KO mice supports that the PiT-1 global KO YS remodeling defect observed at E11.5 is not due to an upstream cardiac requirement for PiT-1. At E9.5, prior to disruption of normal developmental gene expression, maternal IgG accumulation in the AV is lost and mRNA of several endocytosis and autophagy genes is increased in the PiT-1 KO VE, likely reflecting a compensatory mechanism. LC3 protein levels are decreased, and Lysotracker Red confirms that AV maintenance is disrupted. Lastly, we find that uptake of fluorescent dextran is disrupted in an ex vivo functional endocytosis assay performed with dissected yolk sacs. Together, this data supports the novel idea that PiT-1 is required for endocytosis in the yolk sac. Additionally, this work suggests that there are tissue-specific differences in the roles of PiT-1 and PiT-2 in vivo.
Supplementary Material
Supplemental Figure 1. Whole YS mRNA RT-PCR analyzed by densitometry suggests minor gene expression changes in the E11.5 YS PiT-1 KO VE. A: Genes expressed in the VE. B: Genes expressed in the ME. C: Genes expressed in endothelial cells. NS: not significant. #: 0.050012.
Supplemental Figure 2. Lysotracker Red diluted at 1:1000 in PBS and incubated for 5 minutes at 37°C resulted in effective staining of WT lysosomes (compare A-D). Comparison of fluorescence images to light microscopy images confirms localization of the staining to within cells (C Expanded).
Supplemental Figure 3. phRODO Green Dextran for Endocytosis diluted at 1:100 in DMEM with 10% FBS and incubated for 2h at 37°C resulted in uptake of fluorescent dextran (compare A-D). E: Magnified area indicated in C.
Supplemental Table 1. Oligomer sequences (5′-3′) and sources for all genotyping and gene expression primer pairs used.
Highlights.
Global PiT-1 KO results in a yolk sac remodeling/angiogenesis defect.
Yolk sac remodeling failure is not due to a cardiomyocyte requirement for PiT-1.
Maternal IgG accumulation in the apical vacuole is defective in PiT-1 KO.
Apical vacuole maintenance, endocytosis and autophagy in the YS require PiT-1.
Acknowledgements
Dr. Giachelli's lab is supported by NIH grants HL62329, HL081785, and HL114611. Dr. Wallingford was supported by NHLBI T32HL007828. Chicken anti-rat PiT-1 primary antibody was provided by Dr. M. Levi (University of Colorado). We thank Dr. Maria Festing and Evan Narasimhan for Pecam-1 immunostaining and JiaJun Chia for blind cell height measurements.
Abbreviations
- AV
apical vacuole
- CKD
chronic kidney disease
- FIBGC
familial idiopathic basalganglia calcification
- IgG
immunoglobulin
- ME
mesodermal YS layer
- MV
microvesicle
- Pi
phosphate
- VE
visceral endoderm
- VSMC
vascular smooth muscle cell
- YS
yolk sac
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhurst RJ, Lehnert S. a, Faissner a, Duffie E. TGF beta in murine morphogenetic processes: the early embryo and cardiogenesis. Development. 1990;108:645–56. doi: 10.1242/dev.108.4.645. [DOI] [PubMed] [Google Scholar]
- Baldwin HS, Shen HM, Yan HC, DeLisser HM, Chung a, Mickanin C, Trask T, Kirschbaum NE, Newman PJ, Albelda SM. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31): alternatively spliced, functionally distinct isoforms expressed during mammalian cardiovascular development. Development. 1994;120:2539–53. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- Beck L, Leroy C, Beck-Cormier S, Forand A, Salaün C, Paris N, Bernier A, Ureña-Torres P, Prié D, Ollero M, Coulombel L, Friedlander G. The phosphate transporter PiT1 (Slc20a1) revealed as a new essential gene for mouse liver development. PLoS One. 2010;5:e9148. doi: 10.1371/journal.pone.0009148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck L, Leroy C, Salaün C, Margall-Ducos G, Desdouets C, Friedlander G. Identification of a novel function of PiT1 critical for cell proliferation and independent of its phosphate transport activity. J. Biol. Chem. 2009;284:31363–74. doi: 10.1074/jbc.M109.053132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisovska M, Zhao Y, Tsytsyura Y, Glyvuk N, Takamori S, Matti U, Rettig J, Südhof T, Bruns D. v-SNAREs control exocytosis of vesicles from priming to fusion. EMBO J. 2005;24:2114–26. doi: 10.1038/sj.emboj.7600696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier G, Clauss M, Risau W. Coordinate expression of vascular endothelial growth factor receptor-1 (flt-1) and its ligand suggests a paracrine regulation of murine vascular development. Dev. Dyn. 1995;204:228–39. doi: 10.1002/aja.1002040303. [DOI] [PubMed] [Google Scholar]
- Burgers PMJ. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009;284:4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byskov K, Jensen N, Kongsfelt IB, Wielsøe M, Pedersen LE, Haldrup C, Pedersen L. Regulation of cell proliferation and cell density by the inorganic phosphate transporter PiT1. Cell Div. 2012;7:7. doi: 10.1186/1747-1028-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan S. a, Plump a S., Prezioso VR, Bachvarova RF, Darnell JE. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Crouthamel MH, Lau WL, Leaf EM, Chavkin NW, Wallingford MC, Peterson DF, Li X, Liu Y, Chin MT, Levi M, Giachelli CM. Sodium-Dependent Phosphate Cotransporters and Phosphate-Induced Calcification of Vascular Smooth Muscle Cells: Redundant Roles for PiT-1 and PiT-2. Arterioscler. Thromb. Vasc. Biol. 2013;33:2625–32. doi: 10.1161/ATVBAHA.113.302249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver JC, Dickinson ME. The effects of hemodynamic force on embryonic development. Microcirculation. 2010;17:164–78. doi: 10.1111/j.1549-8719.2010.00025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva RJG, Pereira ICL, Oliveira JRM. Analysis of gene expression pattern and neuroanatomical correlates for SLC20A2 (PiT-2) shows a molecular network with potential impact in idiopathic basal ganglia calcification (“Fahr's disease”). J. Mol. Neurosci. 2013;50:280–3. doi: 10.1007/s12031-013-0001-0. [DOI] [PubMed] [Google Scholar]
- Dai X-Y, Zhao M-M, Cai Y, Guan Q-C, Zhao Y, Guan Y, Kong W, Zhu W-G, Xu M-J, Wang X. Phosphate-induced autophagy counteracts vascular calcification by reducing matrix vesicle release. Kidney Int. 2013;83:1042–51. doi: 10.1038/ki.2012.482. [DOI] [PubMed] [Google Scholar]
- Dhingra R, Sullivan LM, Fox CS, Wang TJ. Relations of serum phosphorus and calcium levels to the incidence of cardiovascular disease in the community. Arch Intern Med. 2007;167:879–885. doi: 10.1001/archinte.167.9.879. [DOI] [PubMed] [Google Scholar]
- Duncan S. a, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7598–602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Rossant J. Cell fate decisions in early blood vessel formation. Trends Cardiovasc. Med. 2003;13:254–9. doi: 10.1016/s1050-1738(03)00105-1. [DOI] [PubMed] [Google Scholar]
- Farrell KB, Tusnady GE, Eiden MV. New structural arrangement of the extracellular regions of the phosphate transporter SLC20A1, the receptor for gibbon ape leukemia virus. J. Biol. Chem. 2009;284:29979–29987. doi: 10.1074/jbc.M109.022566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing MH, Speer MY, Yang H-Y, Giachelli CM. Generation of mouse conditional and null alleles of the type III sodium-dependent phosphate cotransporter PiT-1. Genesis. 2009;47:858–863. doi: 10.1002/dvg.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forand A, Beck L, Leroy C, Rousseau A, Boitez V, Cohen I, Courtois G, Hermine O, Friedlander G. EKLF-driven PIT1 expression is critical for mouse erythroid maturation in vivo and in vitro. Blood. 2013;121:666–78. doi: 10.1182/blood-2012-05-427302. [DOI] [PubMed] [Google Scholar]
- French WJ, Creemers EE, Tallquist MD. Platelet-derived growth factor receptors direct vascular development independent of vascular smooth muscle cell function. Mol. Cell. Biol. 2008;28:5646–57. doi: 10.1128/MCB.00441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldie LC, Nix MK, Hirschi KK. Embryonic vasculogenesis and hematopoietic specification. Organogenesis. 2008;4:257–263. doi: 10.4161/org.4.4.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CT, Trejo J, Magnuson T. Genetic evidence for a mammalian retromer complex containing sorting nexins 1 and 2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15173–7. doi: 10.1073/pnas.0409558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenmann S, Khew-Goodall Y, Gamble J, Vadas M, Wattenberg BW. A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol. Biol. Cell. 1998;9:1649–60. doi: 10.1091/mbc.9.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PM, Vasavda C, Hostettler J, Davis JQ, Abdi K, Bennett V. E-cadherin polarity is determined by a multifunction motif mediating lateral membrane retention through ankyrin-G and apical-lateral transcytosis through clathrin. J. Biol. Chem. 2013;288:14018–31. doi: 10.1074/jbc.M113.454439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johann SV, van Zeijl M, Cekleniak J, O'Hara B. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J. Virol. 1993;67:6733–6. doi: 10.1128/jvi.67.11.6733-6736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jollie W. Development, morphology, and function of the yolk-sac placenta of laboratory rodents. Teratology. 1990;41:361–81. doi: 10.1002/tera.1420410403. [DOI] [PubMed] [Google Scholar]
- Jono S, McKee MD, Murry CE, Shioi a., Nishizawa Y, Mori K, Morii H, Giachelli CM. Phosphate Regulation of Vascular Smooth Muscle Cell Calcification. Circ. Res. 2000;87:e10–e17. doi: 10.1161/01.res.87.7.e10. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto a, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh MP, Kabat D. Identification and characterization of a widely expressed phosphate transporter/retrovirus receptor family. Kidney Int. 1996;49:959–963. doi: 10.1038/ki.1996.135. [DOI] [PubMed] [Google Scholar]
- Kavanaugh MP, Miller DG, Zhang W, Law W, Kozak SL, Kabat D, Miller AD. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N, Sun-Wada G-H, Aoyama M, Harada A, Takasuga S, Sasaki T, Wada Y. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 2012;3:1071. doi: 10.1038/ncomms2069. [DOI] [PubMed] [Google Scholar]
- Kestenbaum BR, Adeney KL, Boer IH, De Ix, J.H., Michael G, Siscovick DS. kidney disease[uni2009]: the Multi-Ethnic Study of Atherosclerosis. 2011;76:991–998. doi: 10.1038/ki.2009.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WL, Festing MH, Giachelli CM. Phosphate and vascular calcification: Emerging role of the sodium-dependent phosphate co-transporter PiT-1. Thromb. Haemost. 2010;104:464–70. doi: 10.1160/TH09-12-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Miano J, Cserjesi P, Olsen E. SM22 , a Marker of Adult Smooth Muscle, Is Expressed in Multiple Myogenic Lineages During Embryogenesis. Circ. Res. 1996;78:188–195. doi: 10.1161/01.res.78.2.188. [DOI] [PubMed] [Google Scholar]
- Li X, Yang H-Y, Giachelli CM. Role of the sodium-dependent phosphate cotransporter, Pit-1, in vascular smooth muscle cell calcification. Circ. Res. 2006;98:905–12. doi: 10.1161/01.RES.0000216409.20863.e7. [DOI] [PubMed] [Google Scholar]
- Liu L, Sánchez-Bonilla M, Crouthamel M, Giachelli C, Keel S. Mice lacking the sodium-dependent phosphate import protein, PiT1 (SLC20A1), have a severe defect in terminal erythroid differentiation and early B cell development. Exp. Hematol. 2013;41:432–443.e7.. doi: 10.1016/j.exphem.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomashvili K. a. Phosphate-Induced Vascular Calcification: Role of Pyrophosphate and Osteopontin. J. Am. Soc. Nephrol. 2004;15:1392–1401. doi: 10.1097/01.asn.0000128955.83129.9c. [DOI] [PubMed] [Google Scholar]
- Lucitti JL, Jones E. a V, Huang C, Chen J, Fraser SE, Dickinson ME. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development. 2007;134:3317–26. doi: 10.1242/dev.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 2003;116:3051–60. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerbäck S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–66. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Nishikawa SI, Nishikawa S, Kawamoto H, Yoshida H, Kizumoto M, Kataoka H, Katsura Y. In vitro generation of lymphohematopoietic cells from endothelial cells purified from murine embryos. Immunity. 1998;8:761–9. doi: 10.1016/s1074-7613(00)80581-6. [DOI] [PubMed] [Google Scholar]
- O'Hara B, Johann SV, Klinger HP, Blair DG, Rubinson H, Dunn KJ, Sass P, Vitek SM, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- Olah Z, Lehel C, Anderson WB, Eiden MV, Wilson CA. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J. Biol. Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- Palis J, McGrath KE, Kingsley PD. Initiation of hematopoiesis and vasculogenesis in murine yolk sac explants. Blood. 1995;86:156–63. [PubMed] [Google Scholar]
- Palis J. Hematopoietic Stem Cell Development. Yolk Sac Development in the Mouse. Landes Biosciences. 2005;62:2006. [Google Scholar]
- Rhee S, Guerrero-Zayas M-I, Wallingford MC, Ortiz-Pineda P, Mager J, Tremblay KD. Visceral endoderm expression of Yin-Yang1 (YY1) is required for VEGFA maintenance and yolk sac development. PLoS One. 2013;8:e58828. doi: 10.1371/journal.pone.0058828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Bank A, Richardson Christine, Bank Arthur. Developmental-stage-specific expression and regulation of an amphotropic retroviral receptor in hematopoietic cells . Developmental-Stage-Specific Expression and Regulation of an Amphotropic Retroviral Receptor in Hematopoietic Cells. 1996:16. doi: 10.1128/mcb.16.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Schwarz DG, Griffin CT, Schneider EA, Yee D, Magnuson T. Genetic Analysis of Sorting Nexins 1 and 2 Reveals a Redundant and Essential Function in Mice. 2002;13:3588–3600. doi: 10.1091/mbc.E02-03-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan CM, Crouthamel MH, Kapustin A, Giachelli CM. Arterial calcification in chronic kidney disease: key roles for calcium and phosphate. Circ. Res. 2011;109:697–711. doi: 10.1161/CIRCRESAHA.110.234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparber F, Scheffler JM, Amberg N, Tripp CH, Heib V, Hermann M, Zahner SP, Clausen BE, Reizis B, Huber L. a, Stoitzner P, Romani N. The late endosomal adaptor molecule p14 (LAMTOR2) represents a novel regulator of Langerhans cell homeostasis. Blood. 2013 doi: 10.1182/blood-2013-08-518555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011;107:1127–40. doi: 10.1093/aob/mcq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Taub N, Kurzbauer R, Hilber D, de Araujo ME, Erlacher M, Offterdinger M, Villunger A, Geley S, Bohn G, Klein C, Hess MW, Huber L. a. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. 2006;175:861–8. doi: 10.1083/jcb.200607025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli M, Curhan G, Pfeffer M, Sacks F, Thadhani R, Melamed ML, Wiebe N, Muntner P. Relation between alkaline phosphatase, serum phosphate, and all-cause or cardiovascular mortality. Circulation. 2009;120:1784–92. doi: 10.1161/CIRCULATIONAHA.109.851873. [DOI] [PubMed] [Google Scholar]
- Trask M, Tremblay KD, Mager J. Yin-Yang1 is required for epithelial-tomesenchymal transition and regulation of Nodal signaling during mammalian gastrulation. Dev. Biol. 2012 doi: 10.1016/j.ydbio.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Weissman IL. The origin and fate of yolk sac hematopoiesis: application of chimera analyses to developmental studies. Int. J. Dev. Biol. 2010;54:1019–31. doi: 10.1387/ijdb.093039hu. [DOI] [PubMed] [Google Scholar]
- Viotti M, Niu L, Shi S, Hadjantonakis A. Role in the gut endoderm in relaying left-right patterning in mice. PLoS One. 2012;10:e1001276. doi: 10.1371/journal.pbio.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virkki LV, Biber J, Murer H, Forster IC. Phosphate transporters: a tale of two solute carrier families. Am. J. Physiol. Renal Physiol. 2007;293:F643–F654. doi: 10.1152/ajprenal.00228.2007. [DOI] [PubMed] [Google Scholar]
- Wada Y, Sun-Wada G-H, Kawamura N. Microautophagy in the visceral endoderm is essential for mouse early development. Autophagy. 2013;9:252–4. doi: 10.4161/auto.22585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford MC, Angelo JR, Mager J. Morphogenetic analysis of peri-implantation development. Dev. Dyn. 2013;242:1110–20. doi: 10.1002/dvdy.23991. [DOI] [PubMed] [Google Scholar]
- Wang S, Sizeland A, Wang X, Sassoon D. Restricted expression of type-II TGF beta receptor in murine embryonic development suggests a central role in tissue modeling and CNS patterning. Mech Dev. 1995;52:275–89. doi: 10.1016/0925-4773(95)00408-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Whole YS mRNA RT-PCR analyzed by densitometry suggests minor gene expression changes in the E11.5 YS PiT-1 KO VE. A: Genes expressed in the VE. B: Genes expressed in the ME. C: Genes expressed in endothelial cells. NS: not significant. #: 0.050012.
Supplemental Figure 2. Lysotracker Red diluted at 1:1000 in PBS and incubated for 5 minutes at 37°C resulted in effective staining of WT lysosomes (compare A-D). Comparison of fluorescence images to light microscopy images confirms localization of the staining to within cells (C Expanded).
Supplemental Figure 3. phRODO Green Dextran for Endocytosis diluted at 1:100 in DMEM with 10% FBS and incubated for 2h at 37°C resulted in uptake of fluorescent dextran (compare A-D). E: Magnified area indicated in C.
Supplemental Table 1. Oligomer sequences (5′-3′) and sources for all genotyping and gene expression primer pairs used.


