Abstract
Mouse 3T3-L1 preadipocytes differentiate into adipocytes when treated with 3-isobutyl-1-methylxanthine, dexamethasone, and insulin. Although mechanisms of adipogenesis, including transcriptional cascades, are understood, it is still unclear how clonally expanded cells eventually enter the terminal differentiation program. From gene expression profile studies, we identified G0/G1 switch gene 2 (G0s2) as a novel regulator of adipogenesis. The gene was found to be expressed at a higher level in white and brown adipose tissues, and it was induced in 3T3-L1 cells by hormonal treatment. Importantly, G0s2 expression was closely associated with the transition from mitotic clonal expansion to terminal differentiation. Knockdown of G0s2 expression with siRNA inhibited adipocyte differentiation, whereas constitutive overexpression of G0s2 accelerated differentiation of preadipocytes to mature adipocytes. Expression of G0s2 was found to be regulated by peroxisome proliferator-activated receptor γ (PPARγ), which is a well-known regulator of adipocyte differentiation. Absence of either PPARγ or G0s2 expression resulted in apoptotic pathway activation before terminal differentiation. To determine whether G0s2 has a role in vivo, G0s2-knockout mice were generated. The knockout mice were normal in appearance, but they had less adipose mass than wild-type littermates. Mouse embryonic fibroblast cells from G0s2-deficient mice exhibited impaired adipogenesis and contained unusually small intracellular lipid droplets, suggesting that G0s2 has a role in lipid droplet formation. Our studies demonstrate that G0s2 has an important role in adipogenesis and accumulation of triacylglycerol.
Obesity is a serious health problem and a contributing risk factor for several diseases, such as hypertension, cancer, diabetes, and atherosclerosis.1 As a major cellular component of adipose tissue, adipocytes have a central role in normal physiology.2 Obesity results from excessive accumulation of white adipose tissue, accompanied by increased size and number of adipocytes.3 Although a number of molecular mechanisms regulating adipocyte development have been elucidated, there is a clear need to fully define the complex molecular processes that control adipose tissue formation, in an effort to regulate its accretion.
Adipogenesis is the process of formation of new adipocytes from preadipocyte precursors, and an understanding of this process is important for control of obesity.4 The preadipocyte murine cell line 3T3-L1 is a useful model for studying molecular mechanisms underlying adipocyte differentiation.5 Upon reaching confluence, proliferating preadipocytes become growth arrested by contact inhibition. When postconfluent 3T3-L1 cells are exposed to adipogenic agents, growth-arrested preadipocytes re-enter the cell cycle as part of mitotic clonal expansion (MCE), after which the cells finally undergo terminal differentiation.6, 7, 8 This process is transcriptionally controlled mainly by CCAAT/enhancer-binding proteins (C/EBPs) and peroxisome proliferator-activated receptor γ (PPARγ).9 After hormonal induction, cells immediately express C/EBPβ and C/EBPδ, and these in turn induce expression of C/EBPα and PPARγ, which are the key regulators of the adipose-specific phenotype.10, 11 C/EBPα and PPARγ act synergistically to govern the terminal differentiation process12 that ends in formation of mature adipocytes.13
MCE is required for 3T3-L1 adipogenesis, and selective inhibition of each cell cycle step is sufficient to totally block adipogenesis.7 Upon hormonal stimulation, cells begin to express C/EBPβ; this protein is then serially phosphorylated by mitogen-activated protein kinase and glycogen synthase kinase β, resulting in active C/EBPβ with full DNA-binding capacity.14 Activation of C/EBPβ is clearly associated with MCE, as cells without C/EBPβ cannot complete clonal expansion.15 Those studies revealed a role of MCE in adipogenesis, and they explain why C/EBPβ is needed for entering the terminal differentiation process. However, it remains largely unknown which factors or events required for cell entry into terminal differentiation are actually set in place during MCE.
The aim of the present work is to identify factors that may be involved in the control of progression of adipogenesis, with particular focus on the period between MCE and terminal differentiation. The G0/G1 switch gene 2 (G0s2) was found to be a strong candidate factor. Originally, G0s2 was recognized by its transient induction in lymphocytes.16 It was shown that murine G0s2 is predominantly expressed in adipose tissue, and the gene is upregulated by PPARγ in 3T3-L1 cells.17 More recently, it has been suggested that G0s2 is involved in lipid metabolism as an inhibitor of adipose triglyceride lipase (ATGL)-mediated lipolysis;18 however, a G0s2-related activity in adipogenesis has not been investigated so far. In this report, we present data supporting G0s2 as a novel regulator of the adipogenic process. Adipogenesis was found to be blocked in the absence of G0s2 expression, and G0s2-knockout mice contain reduced levels of fat tissue. These observations suggest that G0s2 is required for adipogenesis, where it has an important role in the transition from MCE to terminal differentiation.
Results
Conditioned medium (CM) stimulates early expression of G0s2, accelerating 3T3-L1 cell differentiation
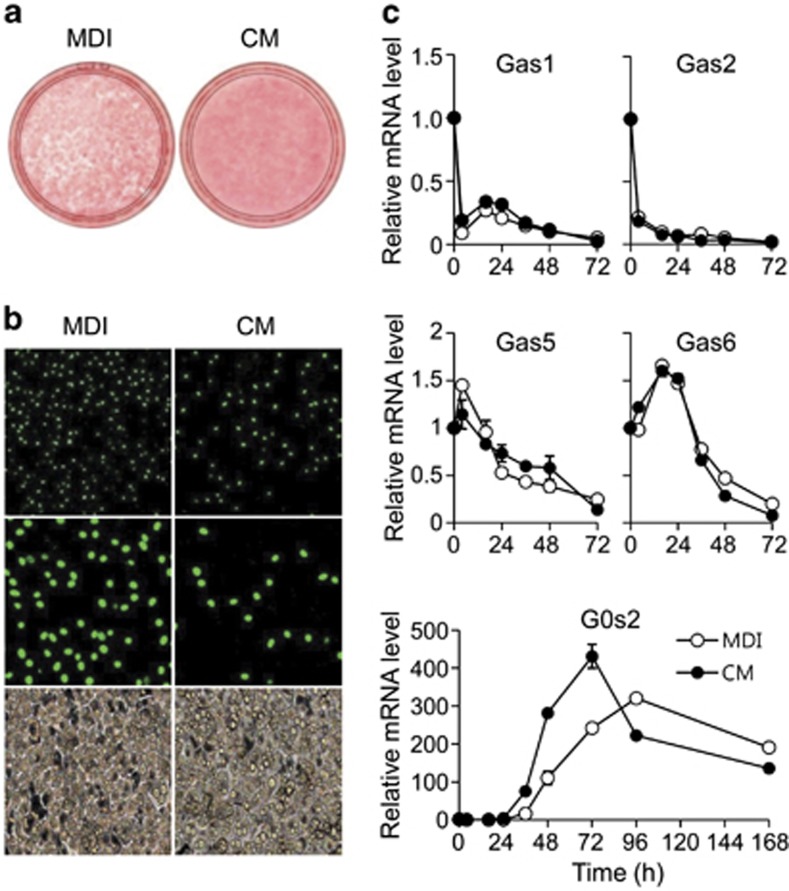
We assumed that MCE is followed by a critical adipocyte-specific event, necessary for the progression of terminal differentiation. Our first approach was to examine culture medium conditioned by cells undergoing clonal expansion, in search of a terminal differentiation-inducing activity. CM was collected from differentiating 3T3-L1 cultures after 48 h of hormone induction (Supplementary Figure S1A). When fresh 3T3-L1 preadipocytes were treated with the CM, adipocyte differentiation was clearly enhanced compared with control treatment with fresh medium containing 3-isobutyl-1-methylxanthine, dexamethasone, and insulin (MDI medium) (Figure 1a). Expression of adipocyte-related genes, such as C/EBPα, PPARγ, and fatty acid synthase (FAS), appeared earlier in CM- versus control-treated cells (Supplementary Figure S1B). In contrast, C/EBPβ expression decreased after 16 h of CM induction (Supplementary Figure S1B). Because C/EBPβ is required for MCE and is a key regulator of C/EBPα and PPARγ, this finding indicates that CM accelerates terminal differentiation by altered response of MCE.
Figure 1.
The effect of treatment with differentiation medium or CM on differentiation of 3T3-L1 preadipocytes. (a) Post-confluent 3T3-L1 preadipocytes cells were treated with differentiation medium containing 3-isobutyl-1-methylxanthine, dexamethasone, and insulin (MDI) or CM. Cells were then stained with Oil red-O on day 5. (b) Immunofluorescence analysis of bromodeoxyuridine (BrdU)-labeled adipocytes. 3T3-L1 cells were induced by MDI or CM and then BrdU was pulsed for 2 h (from 16 to 18 h after initiation of differentiation). The time period of BrdU pulse corresponds to the S phase in cells induced by the standard protocol. Representative images of fluorescence microscopy with magnification × 20 (upper), × 40 (middle), and of light microscopy with magnification × 20 (lower). (c) Preadipocyte 3T3-L1 cells were induced with MDI or CM. Cells were harvested at 0, 4, 16, 24, 36, 48, 72, 96, and 168 h after induction. Total RNA was isolated, and the expression levels of Gas1, Gas2, Gas5, Gas6, and G0s2 were determined by quantitative PCR. All reactions were performed in triplicate. Data represent the mean±S.D.
We examined whether MCE occurred in these cells by detecting dividing cells that incorporate BrdU. After 16 h of induction, most of the MDI-treated control cells were labeled with BrdU; in contrast, very few CM-treated cells were labeled (Figure 1b). Consistently, induction of 3T3-L1 preadipocytes with MDI led to a three-fold increase in cell number at day 2, but cell counts of cultures induced with CM increased only two-fold (Supplementary Figure S1C), indicating that differentiation in CM is not dependent on clonal expansion. This finding was also confirmed by FACS analysis. After 16 h of MDI induction, 3T3-L1 cells re-entered the cell cycle and synthesized DNA (S phase), resulting in increase of the G2/M subpopulation seen after 24 h of induction. However, CM-treated preadipocytes did not show any increase in S phase (Supplementary Figure S1D). This result suggests that MCE is not critical for CM-induced adipocyte differentiation.
Using material from the CM experiments, we identified the G0s2 gene from results of a preliminary microarray analysis. Because G0s2 is known to be associated with cell cycle progression, we expected that G0s2 may have a role in the transition between MCE and terminal differentiation. We confirmed G0s2 expression by real-time qPCR, along with that of other genes that might affect clonal expansion, such as growth arrest-specific genes (Gas). We found that CM strongly accelerates the expression of G0s2 mRNA compared with MDI medium, but the expression patterns of Gas genes were not changed (Figure 1c). This suggests that G0s2 is associated with the transition into terminal differentiation.
G0s2 overexpression enhances adipocyte differentiation
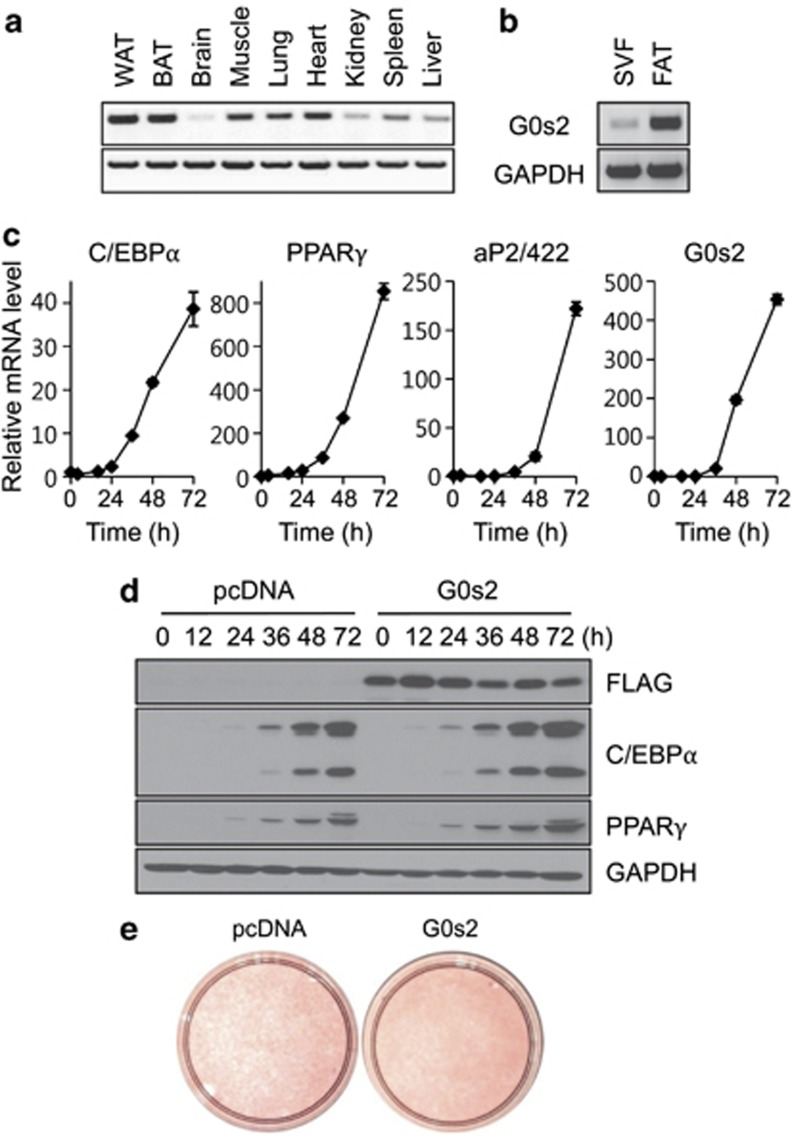
Tissue distribution of G0s2 mRNA determined by RT-PCR reveals that G0s2 expression is significantly high in adipose tissue (Figure 2a). In adipose tissue, expression of G0s2 was approximately 15-fold greater in the fat-cell fraction than stromal vascular fractions, indicating that G0s2 is expressed at higher levels in mature fat cells than in preadipocytes (Figure 2b). During 3T3-L1 cell differentiation, G0s2 mRNA is induced in a pattern similar to that of adipocyte markers, C/EBPα, PPARγ, and aP2/422 (Figure 2c). Given these results, we sought to determine whether G0s2 can enhance terminal differentiation. It was found that G0s2 overexpression caused slightly increased lipid droplet formation in 3T3-L1 adipocytes, accompanied by accelerated expression of C/EBPα and PPARγ (Figures 2d and e). This result suggests that G0s2 may be involved in adipocyte differentiation and lipid accumulation.
Figure 2.
G0s2 is predominantly expressed in fat tissues and its overexpression enhances adipocyte differentiation. (a) Tissue distribution of G0s2 expression by reverse transcriptase-PCR analysis. (b) G0s2 expression in stromal vascular fraction (SVF) and fat cell fraction (FAT). (c) G0s2 expression during adipogenesis was validated by quantitative real-time PCR. C/EBPα, PPARγ, and aP2/422 were used as adipocyte differentiation markers. (d) 3T3-L1 cells expressing control vector (pcDNA) or G0s2-FLAG expression vector were transfected by electroporation and then induced to differentiate. Protein collected at indicated times after induction was used to assay levels of G0s2, C/EBPα, and PPARγ expression. GAPDH was used as loading control. (e) Oil red-O staining after complete differentiation (day 4)
Knockdown of G0s2 inhibits adipocyte differentiation and stimulates apoptosis in 3T3-L1 cells
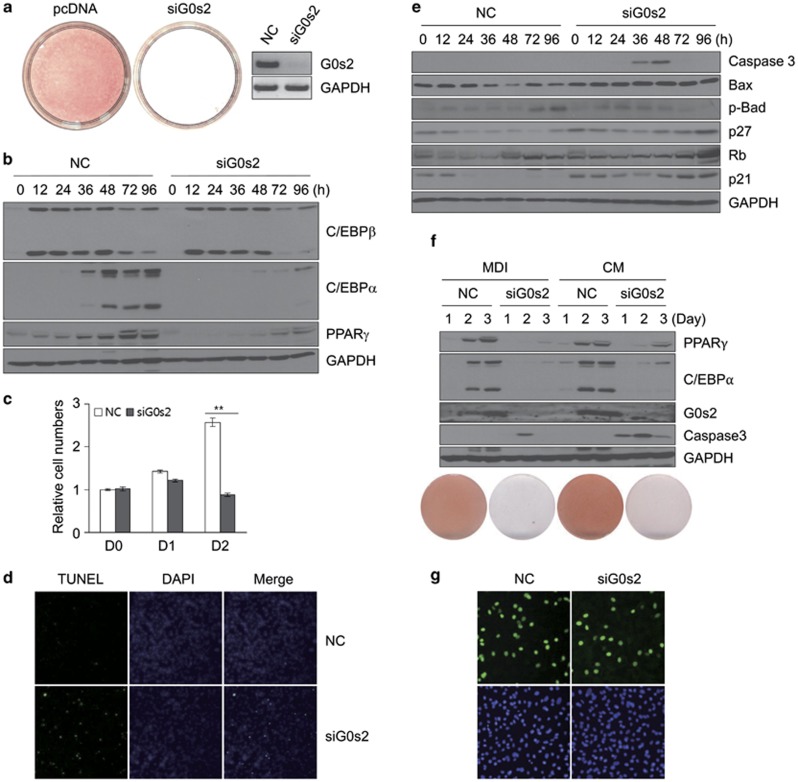
To examine the function of G0s2 in adipogenesis, we knocked down the expression of G0s2 in 3T3-L1 cells using siRNA. Knockdown of G0s2 resulted in inhibition of differentiation, as shown by Oil red-O staining (Figure 3a). Levels of C/EBPα and PPARγ proteins were reduced in the G0s2-knockdown cells; whereas, levels of C/EBPβ protein were unaffected (Figure 3b). This suggests that G0s2 affects adipogenesis at a time point between C/EBPβ and C/EBPα expression. Interestingly, G0s2 knockdown results in decreased cell counts at day 2 (Figure 3c). Because G0s2 expression during 3T3-L1 differentiation begins after around 36 h of induction, we assumed that G0s2 knockdown should not affect the initiation of MCE. With this in mind, we wondered whether inhibition of G0s2 may stimulate apoptosis during adipogenesis. To determine whether the reduction in cell number was caused by apoptosis, we performed a terminal deoxynucleotidyl transferase-mediated dUTP end labeling (TUNEL) assay. Those results clearly show increased labeling in G0s2-knockdown cells compared with negative controls (Figure 3d). Caspase 3, known as a marker of apoptosis,19 was activated between 36 and 48 h of induction, and this coincides with the point of transition from MCE to terminal differentiation. In addition, levels of the pro-apoptotic protein Bax20 were greatly enhanced in cells transfected with G0s2-specific siRNA (Figure 3e). Interestingly, phosphorylated form of Bad protein was decreased after siG0s2 transfection (Figure 3e). These changes of apoptosis-related proteins are thought to be associated with G0s2 function during adipogenesis.
Figure 3.
Knockdown of G0s2 by siRNA inhibits adipocyte differentiation and leads to apoptosis. (a) Knockdown of G0s2 abrogates 3T3-L1 cell differentiation. Oil red-O staining of adipocytes with negative control (NC) and G0s2-knockdown (siG0s2) at day 5. The knockdown efficiency by siRNA was assessed by reverse transcriptase-PCR at day 2 after differentiation. (b) C/EBPβ, C/EBPα, and PPARγ protein levels were measured in the NC and siG0s2 cells by western blotting analysis, using GAPDH as loading control. (c) Cell counts were determined at days 0, 1, and 2 after induction. Cells were induced with differentiation medium containing 3-isobutyl-1-methylxanthine, dexamethasone, and insulin (MDI) after transfection of siRNA, and the cells were counted. **P<0.01. Data represent the mean±S.D. (d) Cells treated with negative control (NC) or knockdown of G0s2 (siG0s2) were fixed 48 h after induction and then stained with DAPI (4,6-diamidino-2-phenylindole). They were analyzed by fluorescence microscopy or for DNA fragmentation by TUNEL. (e) Cells were harvested at the indicated times, and western blotting analysis of caspase 3, Bax, p-Bad, p27, Rb, or p21 expression levels during 3T3-L1 differentiation was performed using GAPDH as loading control. (f) Knockdown of G0s2 upon CM treatment did not induce 3T3-L1 differentiation. Cells treated with negative control (NC) or knockdown of G0s2 (siG0s2) were induced either by MDI or CM and were analyzed by western blotting and Oil red-O staining. (g) Bromodeoxyuridine (BrdU) incorporation analysis in the cells treated with siG0s2. BrdU was pulsed for 2 h (from 16 to 18 h after initiation of differentiation), and cells were observed by fluorescence microscopy with magnification × 40
Does CM-induced acceleration of adipogenesis depend on G0s2 expression? To assess this question, we attempted G0s2 knockdown with CM treatment in 3T3-L1 cells. As shown in Figure 3f, CM did not induce 3T3-L1 differentiation without G0s2. Moreover, CM treatment with G0s2 siRNA caused even earlier and more extensive expression of caspase 3. This result supports the conclusion that CM accelerates adipocyte differentiation by inducing G0s2 at an earlier time point than MDI and that G0s2 is closely associated with protecting cells from apoptosis. In addition, the BrdU incorporation assay clearly showed that G0s2 siRNA cells enter MCE to almost the same extent as the control cells (Figure 3g). This result suggests that the major effect of G0s2 knockdown on adipogenesis is not due to blocking the initiation of MCE but is due to inhibiting the apoptotic pathway after MCE. Taken together, these data indicate that G0s2 is required for terminal differentiation, and without G0s2 expression during these events, the cells will undergo apoptosis upon strong mitotic signal.
Anti-apoptotic activity of PPARγ is mediated by G0s2 in 3T3-L1 cells
Previously, it has been reported that G0s2 expression is upregulated by PPAR family proteins in hepatocytes, as well as in 3T3-L1 cells.17 In adipocytes, PPARγ has a central role both in adipocyte differentiation and maintenance of the adipocyte phenotype. However, it has not been clearly established whether PPARγ has anti-proliferative or anti-apoptotic activity during adipogenesis. In cells treated with rosiglitazone, a synthetic PPARγ agonist, higher levels of C/EBPα, as well as G0s2, were seen (Supplementary Figure S2A). In addition, loss of G0s2 expression resulted in induction of apoptosis, indicated by caspase 3 levels, even in the presence of rosiglitazone (Supplementary Figure S2B). This indicates that G0s2 is a target gene of PPARγ, and it is required for the maintenance of the terminal differentiation program.
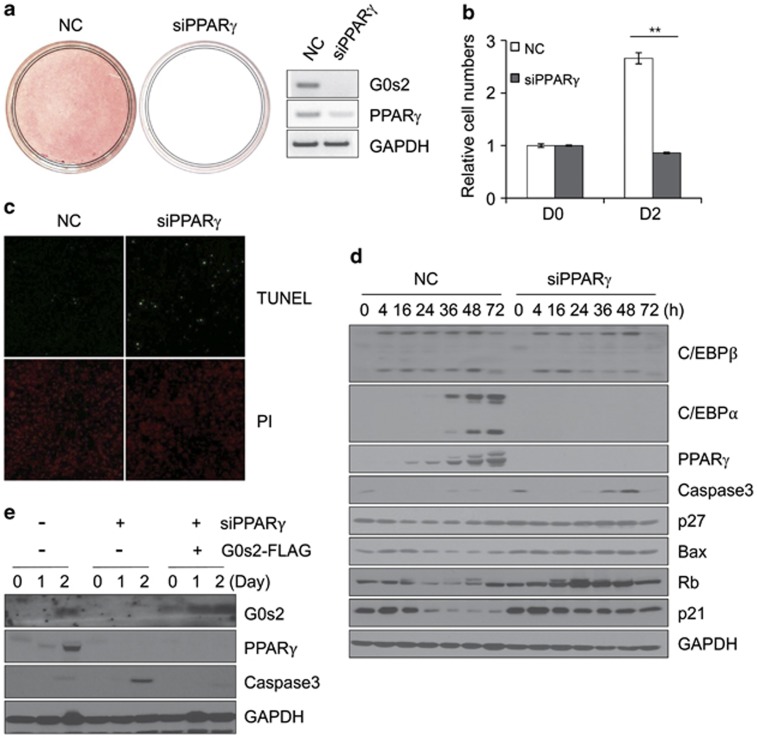
To examine the relationship between function of PPARγ and G0s2 in apoptosis, we depleted PPARγ by RNA interference. As expected, PPARγ-knockdown cells do not express G0s2, and they are unable to differentiate into adipocytes (Figure 4a). Knockdown of PPARγ also causes decreased cell number at day 2 after differentiation (Figure 4b). Moreover, the pattern of appearance of apoptosis after induction of differentiation is similar in PPARγ- and G0s2-knockdown cells (Figure 4c). Expression of C/EBPα is also markedly reduced, but caspase 3, Bax, and p27 levels were increased in PPARγ-knockdown cells (Figure 4d). To confirm that these effects on apoptosis are mainly mediated by G0s2, G0s2-overexpressing 3T3-L1 cells were constructed by retroviral transduction of G0s2 cDNA. We then knocked down expression of PPARγ with siRNA in G0s2-overexpressing 3T3-L1 cells. Whereas PPARγ knockdown resulted in significantly increased expression of caspase 3 compared with control cells, G0s2-overexpressing cells did not express caspase 3 even in the absence of PPARγ (Figure 4e). These data indicate that PPARγ is required not only for adipose-specific gene expression but also for protecting clonally expanded cells from apoptosis. In this regard, G0s2 is thought to have a major role in this protection, because G0s2 expression in the absence of PPARγ is seen to prevent caspase 3 activation. Taken together, these results indicate that G0s2 acts downstream of PPARγ as a regulator both of apoptosis and terminal differentiation.
Figure 4.
Knockdown of PPARγ by siRNA inhibits differentiation of 3T3-L1 preadipocytes and induces apoptosis during adipogenesis. (a) Effects of PPARγ knockdown was assessed by Oil red-O staining and reverse transcriptase-PCR. NC, Negative control, siPPARγ, PPARγ-knockdown. (b) Cell numbers were determined on days 0 and 2 after differentiation. **P<0.01. Data represent the mean±S.D. (c) Cells with negative control (NC) and knockdown of PPARγ (siPPARγ) were fixed 48 h after induction and then stained with DAPI (4,6-diamidino-2-phenylindole). They were analyzed by fluorescence microscopy or for DNA fragmentation by TUNEL. (d) Protein expression of the adipogenesis-associated targets C/EBPβ, C/EBPα, and PPARγ and apoptosis-related genes caspase 3, Bax, p27, and p21. (e) 3T3-L1 cells stably expressing control vector or G0s2-FLAG were transfected with siPPARγ, and effects on protein expression were examined
The function of G0s2 in adipogenesis is independent of ATGL
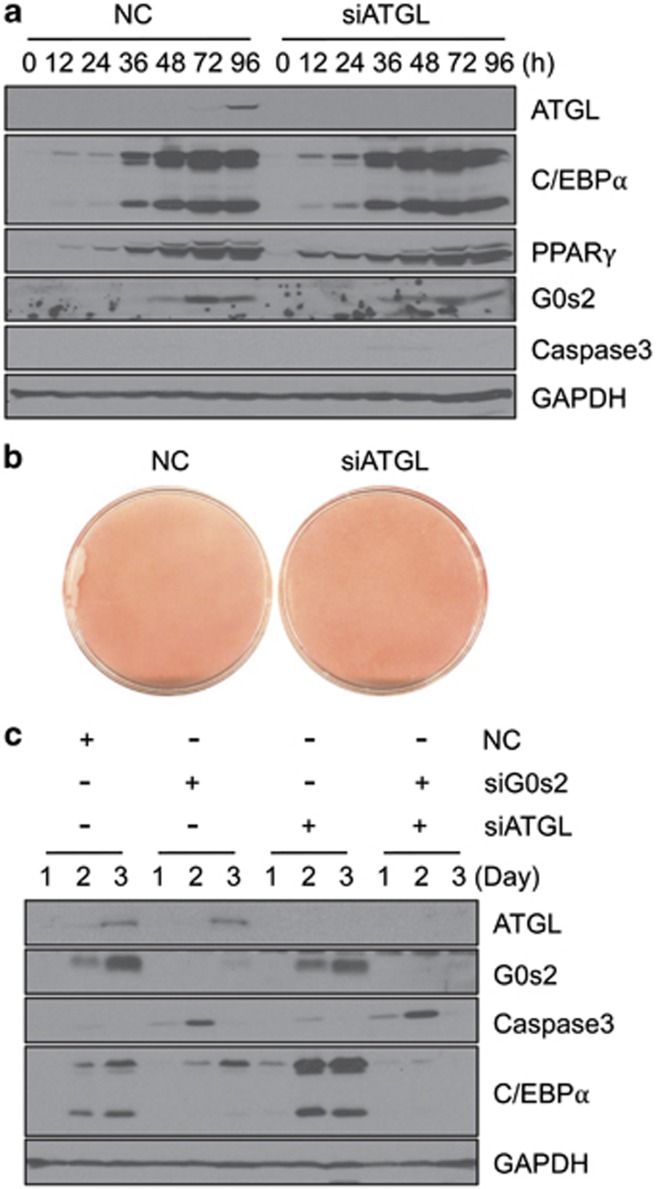
Previous reports indicate that G0s2 specifically interacts with ATGL, and G0s2 functions to attenuate ATGL activity.18 To examine whether the function of G0s2 in the terminal differentiation program is dependent on ATGL, we performed knockdown of ATGL expression in 3T3-L1 cells. As shown in Figures 5a and b, knockdown of ATGL resulted in slightly enhanced lipid accumulation without any significant effect on the expression levels of C/EBPα, PPARγ, G0s2, or caspase 3. We then knocked down both G0s2 and ATGL in 3T3-L1 cells (Figure 5c). As seen with G0s2 knockdown, the double G0s2 and ATGL knockdown also leads to induced caspase 3 expression at day 2. These results indicate that G0s2-depleted cells undergo apoptosis regardless of ATGL expression. Thus, G0s2 appears to have dual functions: (1) inhibition of ATGL-induced lipolysis in mature adipocytes; and (2) anti-apoptotic activity, which is ATGL-independent, during terminal adipocyte differentiation.
Figure 5.
ATGL gene knockdown does not induce apoptosis in 3T3-L1 preadipocytes. (a) ATGL expression was suppressed in 3T3-L1 preadipocytes by siRNA, and then cells were differentiated into adipocytes. Cells were harvested at the indicated times, followed by western blotting analysis of ATGL, G0s2, C/EBPα, PPARγ, caspase 3, and GAPDH expression. (b) Oil red-O staining after differentiation (day 5). (c) 3T3-L1 cells were transfected with siRNAs targeting G0s2 and/or ATGL, followed by differentiation into adipocytes. Levels of ATGL, G0s2, caspase 3, C/EBPα, and GAPDH proteins were analyzed in immunoblots at the indicated days
G0s2-knockout mice have decreased adiposity
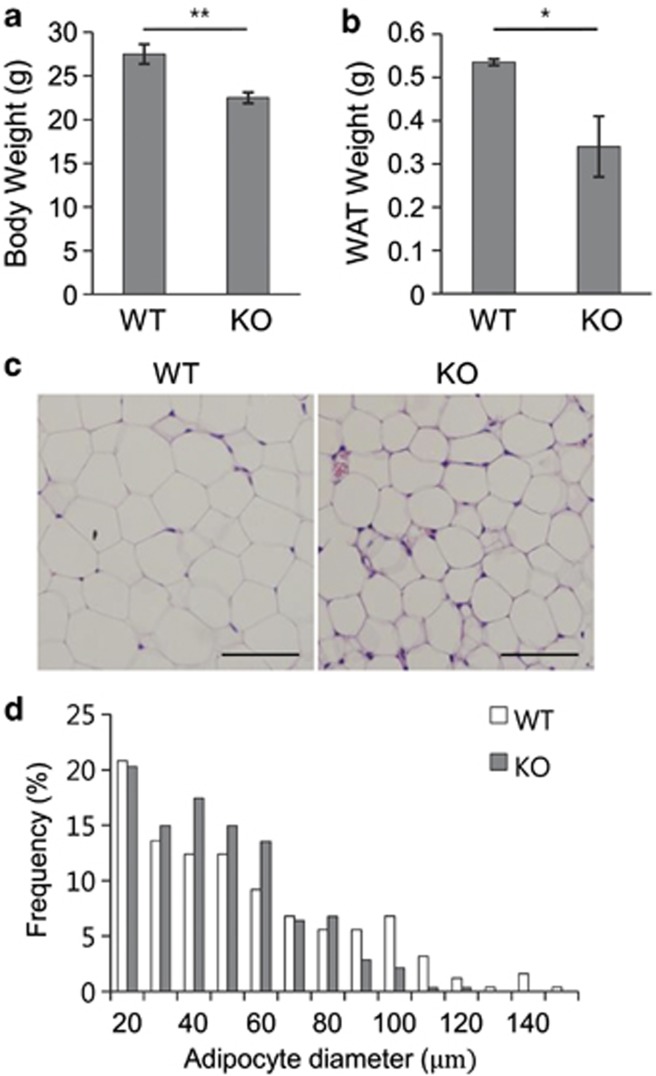
To elucidate the physiological role of G0s2 in vivo, we generated G0s2-knockout mice. Successful ablation was confirmed by the virtual absence of G0s2 gene expression (Supplementary Figures S3A and S3B). Analysis of adipose stores from 12-week-old animals showed a 15% reduction in body weight in G0s2-knockout versus wild-type mice, owing to a 30% reduction in adipose content (Figures 6a and b). No significant difference in liver weight was seen (Supplementary Figure S3C). A decrease in adipose tissue mass could result from a reduction in adipocyte size, as well as adipocyte number, with impaired differentiation. Histological analysis of epididymal fat pads revealed that adipocytes from G0s2-knockout mice were smaller than those from wild type (Figures 6c and d). These data suggest that G0s2 has a role in promoting fat tissue formation in vivo.
Figure 6.
Fat development is impaired in G0s2-knockout mice. (a) Body weight and (b) epididymal fat pad from white adipose tissue (WAT) were evaluated in G0s2-knockout mice and wild-type control mice at the age of 12 weeks. Data represent the mean±S.D. *P<0.05, **P<0.01. (b) Hematoxylin/eosin-stained WAT sections from wild-type and G0s2-knockout mice. Scale bar=100 μm. (c) Distribution of adipocyte size in epididymal WAT from wild-type or G0s2-knockout mice on normal diet (n=3 per group). (d) The diameter of epididymal white adipocytes was determined by ImageJ, and >500 adipocytes were examined for each group. All data are expressed as means±S.D.
G0s2-knockout mouse embryo fibroblasts (MEFs) show impaired adipogenesis
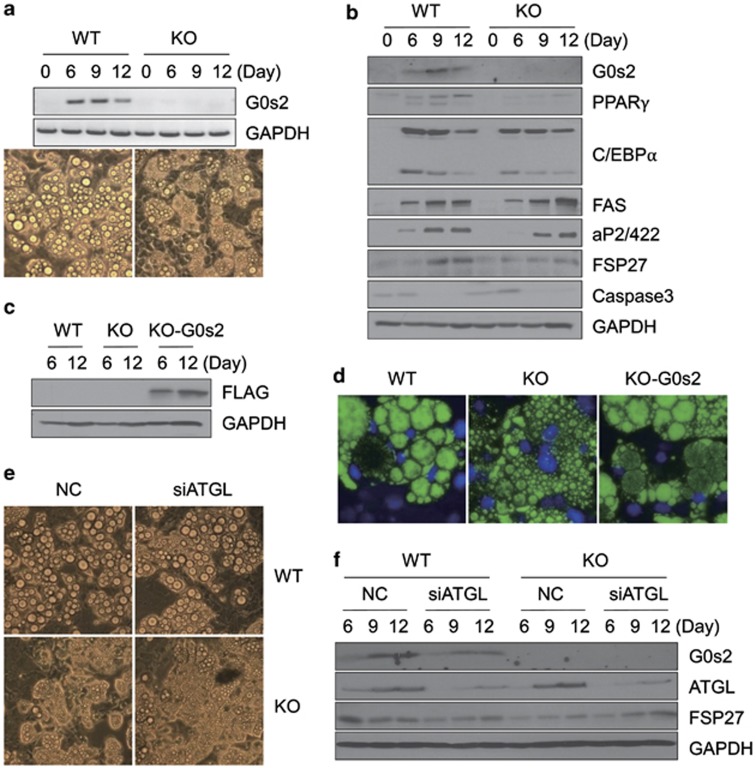
We explored specific action of G0s2 on the adipocyte differentiation process per se in MEFs isolated from G0s2-knockout versus wild-type littermate mouse embryos. After induction by adipogenic agents, less accumulation of lipid droplets was seen in knockout versus wild-type MEFs (Figure 7a). Unlike the pattern observed in G0s2-knockdown 3T3-L1 cells, G0s2-knockout MEF cells accumulate many small lipid droplets during adipogenesis, but these do not mature into the larger lipid droplets observed in wild-type MEFs. Immunoblot analysis of adipocytes at days 6, 9, and 12 revealed that, to some extent, expression levels of PPARγ and its regulatory genes, aP2/422, and fat-specific protein 2721 were reduced in G0s2-knockout MEFs (Figure 7b). The reason why MEFs from G0s2-knockout mice do not exhibit increased caspase 3, as seen in 3T3-L1 cells, is not clear. However, this phenomenon is probably due to differences between 3T3-L1 and MEF cells, as MCE apparently has not been observed in MEF cells.8 Nevertheless, G0s2-knockout MEFs show impaired adipogenesis compared with wild-type MEFs, suggesting that G0s2 also has a major role in normal development of fat pad in vivo.
Figure 7.
Adipocyte differentiation is inhibited in G0s2-knockout MEF cells, and ectopic expression of G0s2 in G0s2-knockout MEF restores adipogenesis. (a) MEFs from wild-type (WT) or G0s2-knockout (KO) mice were induced for adipocyte differentiation. Expression of G0s2 was analyzed by reverse transcriptase-PCR, and microscopy was done at day 12. (b) Western blotting analysis of G0s2 and adipocyte-specific proteins in WT and KO MEFs. (c) KO MEFs were transfected with G0s2-FLAG-overexpression vector. Western blotting analysis of expression in WT containing empty vector (WT), G0s2-KO containing empty vector (KO), or rescued G0s2-KO (KO-G0s2) was performed with anti-FLAG antibody. (d) BODIPY staining of WT, G0s2-KO, and KO-G0s2 MEF cells. The fluorescent dyes BODIPY and DAPI (4,6-diamidino-2-phenylindole) were used to stain the lipids contained in droplets of differentiated fat cells and nuclei of existing cells. (e) WT or KO MEFs were transfected with siRNA directed against ATGL, and microscopy ( × 20) was performed. (f) Western blotting analysis was carried out in WT or KO MEFs after ATGL siRNA transfection
Finally, we rescued G0s2 gene expression in G0s2-knockout MEFs. Transfection of a vector expressing G0s2 into knockout cells led to reactivation of the adipocyte differentiation program, as shown by the appearance of lipid droplets visualized after BODIPY staining (Figures 7c and d). Furthermore, ATGL knockdown by siRNA merely affected the pattern of differentiation in both wild-type and G0s2-knockout MEFs (Figures 7e and f), suggesting that this impairment in fat development is not dependent on ATGL.
Discussion
Preadipocyte 3T3-L1 cells have been used as a primary model for the study of adipogenesis. It is thought that several proteins involved in regulation of growth and differentiation are secreted from differentiating 3T3-L1 cells. In this regard, candidate secreted proteins from 3T3-L1 cells have been identified by a systematic proteomic approach, based on selective blocking of secretion.22 Although secreted proteins during adipogenesis have been suggested to be important, their identity and physiological roles remain elusive. Other studies have revealed that an endogenous PPARγ ligand is generated during an early phase of the adipogenic process in 3T3-L1 cells.23 This suggests a hypothesis that some factors released during adipogenesis, particularly as a result of MCE, are essential for the transition of the cells into terminal differentiation. In this study, we have shown that CM accelerates differentiation into mature adipocytes, suggesting that additional events are required for the transition into terminal differentiation. More importantly, CM was seen to induce adipogenesis in proliferating preadipocytes without MCE. MCE is reported to be necessary for the progression through subsequent steps in the differentiation program;24 however, its role in adipocyte differentiation is not fully understood. From this point of view, it is surprising that CM treatment directly leads to the terminal differentiation program without MCE, which we nonetheless confirmed by FACS analysis and BrdU incorporation. This indicates that certain cell-specific events necessary for entry into terminal differentiation are truly associated with, and a result of, MCE and are mediated by some secreted factors. It remains to be discovered what proteins are secreted during MCE, which of these are active, and how they act on the cells for entry into terminal differentiation.
This study was designed to define events that are necessary and sufficient for 3T3-L1 cell differentiation. From screens to identify genes temporally regulated during transition from MCE to terminal differentiation in 3T3-L1 adipogenesis, we found G0s2 as a novel regulator of adipogenesis. Subsequent experimental results suggest that G0s2 is essentially involved in adipocyte differentiation and lipid accumulation. Overexpression of G0s2 was found to stimulate lipid droplet formation in 3T3-L1 adipocytes, and knockdown of endogenous G0s2, on the other hand, was seen to inhibit fat accumulation and stimulate apoptosis. In addition, CM-induced terminal differentiation appeared to require G0s2 expression, because both G0s2 siRNA 3T3-L1 cells (Figure 3f) and G0s2 KO MEF cells (Supplementary Figure S4) did not affect the differentiation pattern upon CM treatment.
The small, basic G0s2 protein is a multifaceted protein involved in proliferation, apoptosis, inflammation, metabolism, and carcinogenesis. In cultured human Simpson–Golabi–Behmel syndrome cells and 3T3-L1 cells, both G0s2 mRNA and protein steadily increase as cells enter the growth arrest stage preceding terminal differentiation.17 However, despite its postulated role as a cell cycle regulator, it has not yet been investigated whether G0s2 has any active role in the cell cycle withdrawal during adipogenic differentiation. The current study identifies a previously unrecognized G0s2 function. We demonstrated that G0s2 is mainly associated with the terminal differentiation program. Moreover, our data suggest that G0s2 functions to protect against apoptosis. This activity of G0s2 is thought to be independent of ATGL-lipolytic activity. Thus, it is likely that G0s2 has a dual mechanistic function, one being ATGL dependent25 and the other ATGL independent.
We also explored a unique role of PPARγ to protect differentiating cells from apoptosis. Knockdown of PPARγ by siRNA resulted in apoptosis after induction of differentiation, similar in pattern to that observed in G0s2-knockdown cells. The latter effect was shown to be attenuated by G0s2 restoration. It does not necessarily mean that PPARγ itself is an active apoptotic regulator. Rather, our result suggests that G0s2 and PPARγ are essential for a transition from the end of the cell cycle stage to terminal differentiation. In other words, cells expressing G0s2 and PPARγ are able to steadily enter and maintain the terminal differentiation process, but without G0s2, cells cannot withstand a strong mitotic signal, which then leads to apoptosis. Similarly, C/EBPα is well known to have anti-proliferative activity. Because C/EBPα and PPARγ reciprocally regulate each other during adipogenesis, we suggest that the anti-proliferative functions of C/EBPα or PPARγ might essentially be mediated by G0s2.
In the present study, we have demonstrated that G0s2 function has a strong impact on adipocyte formation. We found that adipocyte differentiation was reduced in cultured G0s2-knockout MEF cells. In MEF cells, in contrast to 3T3-L1 cells, G0s2 does not clearly affect C/EBPα and caspase 3 expression. This may simply reflect a biological difference between MEF and 3T3-L1 cells. Among the cell models of adipogenesis, 3T3-L1 and 3T3-F442A cells clearly undergo one or two rounds of cell division called MCE.26 However, this postconfluence mitosis has not been observed in mouse C3H10T1/2 or human preadipocytes.8, 27 It has been suggested that preadipocytes isolated from fat have already undergone the prerequisite clonal expansion in vivo, and thus further cell cycle events would not be observed in such cells.4 Nevertheless, it is clear that some of the checkpoint proteins for mitosis are involved in regulation of adipogenesis.8 Our data show that fat mass in G0s2-knockout mouse was significantly reduced in vivo. Given the phenotype of many small droplets found in G0s2-knockout adipocytes, the ultimate fat reduction might be a result of defects in droplet enlargement. But we cannot exclude the possibility that a deficiency of G0s2 in mice, as in 3T3-L1 cells, affects the integrity and maintenance of existing committed stem cells. It would be of interest to measure active proliferating cells by BrdU incorporation in the white fat pads of mice.
The role of the G0s2 gene in obesity should be carefully accessed. Our data clearly show that G0s2 KO mice exhibit reduced body weights and adipose contents; however, previous studies of models of mouse and human obesity indicate decreased levels of G0s2 in white adipose fat pads.25 Because G0s2 also functions in the regulation of ATGL-dependent lipolysis, further investigations are necessary to examine whether reduced G0s2 in obesity has a causal relationship. It will also be interesting to analyze whether the regulation of cell differentiation involved in PPARγ/G0s2 function is associated with energy metabolism of ATGL-mediated lipolysis.
Materials and Methods
Cell culture and induction of differentiation
Preadipocyte 3T3-L1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 100 U/ml penicillin, 100 mg/ml streptomycin, and 8 mg/ml biotin and supplemented with 10% (v/v) heat-inactivated calf serum, at 37 °C in an atmosphere of 90% air and 10% CO2. To induce differentiation, 2-day postconfluent 3T3-L1 cells (designated day 0) were incubated in MDI medium, which consists of DMEM with 10% fetal bovine serum (FBS), 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, and 1 μg/ml of insulin, for 2 days. Cells were then cultured in DMEM containing 10% FBS and insulin for another 2 days, after which they were grown in DMEM containing 10% FBS. Cell numbers were determined on day 2. For preparation of CM, 2-day postconfluent 3T3-L1 cells were induced by MDI for 48 h, after which the used culture medium was harvested. Oil red-O staining was done at the indicated time points.
RNA isolation and PCR analysis
Total RNA was isolated using Trizol reagent according to the manufacturer's instructions. First-strand cDNA synthesis from 5 μg of total RNA was performed using SuperScript II reverse transcriptase primed by random hexamer primer. The transcripts of G0s2, PPARγ2, C/EBPα, and GAPDH were evaluated by PCR analysis. Amplification was performed using the forward and reverse primers, respectively: 5′-AAAGTGTGCAGGAGCTGATC-3′, 5′-GGACTGCTGTTCACACGCTT-3′ (G0s2); 5′-TATGGGTGAAACTCTGGGAG-3′, 5′-GCTGGAGAAATCAACTGTGG-3′ (PPARγ2); and 5′-ACCACAGTCCATGCCATCAC-3′, 5′-TCCACCACCCTGTTGCTGTA-3′ (GAPDH). Real-time qPCR was used to detect other transcripts, that is, Gas1, Gas2, Gas5, Gas6, G0s2, C/EBPα, PPARγ, and aP2/422. Real-time qPCR was performed using the SYBR Green Master mix in a total volume of 20 μl, as previously described.28 Transcripts were detected by real-time qPCR with a Step One instrument (Applied Biosystems, Foster City, CA, USA). A standard curve was used to calculate mRNA level relative to that of a control gene, ribosomal L32. Primer pairs for specific target genes were designed as listed in Supplementary Table S1. All reactions were performed in triplicate. Relative expression levels and S.D. values were calculated using the comparative method.
Western blotting analysis and antibodies
For protein analysis, cells were washed with cold PBS and then lysed directly in 60 mM Tris-HCl (pH 6.8) containing 1% (w/v) SDS. The lysate was heated at 100 °C for 10 min, centrifuged for 5 min at 13 000 rpm, and supernatants were then frozen until further analysis. For western blots, cell extracts were subjected to SDS-PAGE separation and then transferred to nitrocellulose membranes. Primary antibodies used for blotting were anti-C/EBPβ, anti-C/EBPα,29 anti-PPARγ (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-GAPDH, anti-caspase3, anti-Bax, anti-p-Bad, anti-p21, anti-p27, anti-FAS (Cell Signaling, Danvers, MA, USA), anti-FLAG (Sigma, St. Louis, MO, USA), and anti-Rb (BD Biosciences, Franklin Lakes, NJ, USA). After blocking, filters were immunoblotted with each primary antibody for 2 h at room temperature, followed by incubation with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) for 1 h. Target proteins were visualized by enhanced chemiluminescence detection system (Amersham, Boston, MA, USA). Western blotting results were analyzed to quantify signal intensities using Image J (National Institute of Health, Bethesda, MD, USA) after normalization to GAPDH levels (Supplementary Figure S5).
FACS analysis
3T3-L1 cells were trypsinized and collected by centrifugation, washed with PBS, and then fixed in 90% (v/v) cold methanol. Fixed cells were washed with PBS and stained with 50 μg/ml propidium iodide (PI) and 100 μg/ml RNase A in PBS for 30 min in the dark. Labeled cells were analyzed using a FACS caliber flow cytometry system, and data were analyzed using the ModFit software (BD Biosciences) as described.30
Cell labeling with BrdU and immunofluorescence analysis
Labeling with BrdU was done using the BrdU Labeling and Detection Kit I (Roche, Mannheim, Germany) according to the manufacturer's instructions. Briefly, cells were plated and grown on glass coverslips; at 2 days of postconfluence, they were induced to differentiate with differentiation medium. After 16 h, cells were incubated for 2 h with 10 μM BrdU labeling solution to label DNA synthesis. Cells on the coverslips were fixed in 70% (v/v) ethanol for 20 min at −15 °C to −25 °C for immunofluorescence analysis. Fixed cells were incubated with anti-BrdU primary antibody for 30 min at 37 °C. After washing, anti-mouse-IgG-fluorescein secondary antibody was added to coverslips and incubated for 30 min at 37 °C and then with 0.1 μg/ml DAPI for 5 min at room temperature, after which the coverslips were mounted for immunofluorescence microscopic analysis.
RNA interference (siRNA)
Preadipocyte 3T3-L1 cells were plated into 60-mm-diameter dishes 18–24 h before transfection. The following double-stranded stealth siRNA oligonucleotides (Invitrogen, Carlsbad, CA, USA) were used: mouse G0s2, sense 5′-AGGGACUGCUGUUCACACGCUUCCC-3′ and antisense 5′-GGGAAGCGUGUGAACAGCAGUCCCU-3′ mouse PPARγ, sense 5′-CAGAGCAAAGAGGUGGCCAUCCGAA-3′ and antisense 5′-UUCGGAUGGCCACCUCUUUGCUCUG-3′ and mouse ATGL, a set of three validated siRNA oligonucleotides (cat. no. 1320003). Control oligonucleotides with comparable GC content were also from Invitrogen. For knockdown, cells were transfected with control or gene-specific siRNA at 50 nM in OPTI-MEM medium using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's protocol. The next day, the medium was replaced with fresh DMEM containing 10% calf serum, and the cells were incubated for 24 h before the induction of differentiation. Oil red-O staining of G0s2 knockdown was performed at day 5.
Staining with Annexin-V and PI
The AnnexinV-FITC Apoptosis Detection Kit (BD Biosciences) was used to stain cells in Annexin binding buffer according to the manufacturer's instructions. Briefly, cells were induced to differentiate for 48 h, and then they were harvested by trypsinization and centrifugation, washed in PBS, and then resuspended in 1x binding buffer containing Annexin-V-FITC antibody and PI according to the manufacturer's protocol. Single-cell fluorescence was measured with a BD FACS caliber flow cytometry system (BD Biosciences).
TUNEL assay
TUNEL assay was used to measure cell apoptosis. Cells were seeded on glass coverslips in six-well dishes, and at 48 h after differentiation, staining of fragmented DNA from apoptotic cells was carried out using an in situ Cell Death Detection Kit Fluorescein (Roche). Briefly, cells were rinsed with PBS followed by fixation in 4% (w/v) paraformaldehyde for 1 h at room temperature. Fixed cells were permeabilized by incubation with 0.5% (v/v) Triton X-100 in PBS for 1 h at 37 °C. The cells were rinsed again with PBS and incubated with 50 μl TUNEL reaction mixture per sample for 1 h at 37 °C. Following washes with PBS, samples were mounted on glass slides with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA) and examined by inverted fluorescence confocal microscopy.
Transient transfection assay of G0s2-overexpressing cells
A G0s2-overexpression vector (pcDNA3.0-G0s2-FLAG) was constructed by inserting the whole open reading frame of mouse G0s2 with an N-terminal FLAG tag into pcDNA3.0. To maximize transfection efficiency, microliter-volume electroporation of 3T3-L1 preadipocytes was performed with OneDrop MicroPorator MP-100 (Digital Bio, Seoul, South Korea). The cells were trypsinized, washed with PBS, and finally resuspended in 10 μl of resuspension buffer R with 0.5 μg of plasmid at a concentration of 200 000 cells per pipette. The cells were then microporated at 1300 V, with a 20-ms pulse width, two pulses. Following microporation, the cells were seeded into 35-mm cell culture dishes and placed in a culture incubator at 37 °C.
Generation of G0s2-knockout mice and genotyping
G0s2-knockout mice were generated by homologous recombination and were obtained from Macrogene (Seoul, South Korea). Exons 1 and 2 of the G0s2 gene were replaced with a LacZ-neo cassette. ES cells were purchased from KOMP (https://www.komp.org/), in which gene trap-targeted G0s2-knockout ES cells were made on the original 129Sv founder background, and the knockout mice were generated on a C57BL/6J background. Mice were maintained on a 12-h light and 12-h dark cycle. All animal procedures were approved by the Institutional Animal Case Use Committee of the Yonsei University Health System. For genotyping of mice, genomic DNAs were isolated from tails of adults or heads of 13.5 days post conception (dpc) embryos by incubation at 55 °C overnight in 0.5 ml of 50 mM Tris-HCl (pH 6.8) containing 50 mM EDTA, 0.5% (w/v) SDS, and 0.2 mg proteinase K; this was followed by precipitation with isopropanol. Genotyping by PCR was performed using the following primers: Mutant, 5′-AACCGTGCATCTGCCAGTTT-3′ Wild type, 5′-AAAGTGTGCAGGAGCTGATC-3′ and Common, 5′-GCCTGTCTCCTTCTCTAATG-3′. Expected band size for G0s2 during gel analysis is 526 bp for the wild-type allele and 610 bp for the knockout allele.
Preparation of MEF cells
Wild-type and G0s2-knockout E13.5 embryos were isolated from a single heterozygous female that had been paired with a heterozygous male. Primary MEFs (passage 0 or 1) from the 13.5-day embryos were grown to confluence (day 0) in DMEM containing 10% FBS. Differentiation was initiated by treating cells with 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, 10 μg/ml insulin, and 2 μM rosiglitazone for 6 days. Subsequently, cells were maintained in DMEM supplemented with 10% FBS, 10 μg/ml insulin, and 2 μM rosiglitazone. After 3 days, the medium was replaced with maintenance medium, that is, DMEM supplemented with 10% FBS.
Staining of lipid droplets with BODIPY
MEFs were maintained at proper densities on glass cover slips placed in six-well dishes. After differentiation, cells were fixed in 4% formaldehyde for 30 min, followed by rinsing with PBS three times. To visualize lipid droplets, BODIPY 493/503 dye was added to a final concentration of 0.3 μg/ml and incubated for 20 min. Following three washes with PBS, samples were mounted on glass slides with Vectashield mounting medium (Vector Laboratories) containing DAPI and examined by inverted confocal fluorescence microscopy.
Acknowledgments
We thank Ms. Ji Eun Kim for technical assistance. This work was supported by the National Research Foundation of Korea (NRF) Grants 2011-0030086 and 2011-0015665, funded by the Korea government, Ministry of Science, ICT and Future Planning (MSIP).
Glossary
- G0s2
G0/G1 switch gene 2
- C/EBP
CCAAT/enhancer-binding protein
- PPAR
peroxisome proliferator-activated receptor
- MEF
mouse embryonic fibroblast
- MCE
mitotic clonal expansion
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by JA Cidlowski
Supplementary Material
References
- Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc Natl Acad Sci USA. 2003;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, et al. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, et al. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci USA. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci USA. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L, Forsdyke DR. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol. 1991;10:581–591. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- Zandbergen F, Mandard S, Escher P, Tan NS, Patsouris D, Jatkoe T, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J. 2005;392:313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lu X, Lombes M, Rha GB, Chi YI, Guerin TM, et al. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Litwack G, Alnemri ES. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- Finucane DM, Bossy-Wetzel E, Waterhouse NJ, Cotter TG, Green DR. Bax-induced caspase activation and apoptosis via cytochrome c release from mitochondria is inhibitable by Bcl-xL. J Biol Chem. 1999;274:2225–2233. doi: 10.1074/jbc.274.4.2225. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Ko EH, Kim JE, Kim E, Lee H, Choi H, et al. Nuclear receptor PPARgamma-regulated monoacylglycerol O-acyltransferase 1 (MGAT1) expression is responsible for the lipid accumulation in diet-induced hepatic steatosis. Proc Natl Acad Sci USA. 2012;109:13656–13661. doi: 10.1073/pnas.1203218109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Mariman E, Keijer J, Bouwman F, Noben JP, Robben J, et al. Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell Mol Life Sci. 2004;61:2405–2417. doi: 10.1007/s00018-004-4256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli I, Fang H, Ollero M, Shi H, Hamm JK, Kievit P, et al. Regulated production of a peroxisome proliferator-activated receptor-gamma ligand during an early phase of adipocyte differentiation in 3T3-L1 adipocytes. J Biol Chem. 2004;279:36093–36102. doi: 10.1074/jbc.M405346200. [DOI] [PubMed] [Google Scholar]
- Cornelius P, MacDougald OA, Lane MD. Regulation of adipocyte development. Annu Rev Nutr. 1994;14:99–129. doi: 10.1146/annurev.nu.14.070194.000531. [DOI] [PubMed] [Google Scholar]
- Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochim Biophys Acta. 2013;1831:276–281. doi: 10.1016/j.bbalip.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Lane MD. Adipogenesis: from stem cell to adipocyte. Annu Rev Biochem. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci USA. 2004;101:9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kim HJ, Lee YJ, Lee MY, Choi H, Kim JW. Kruppel-like factor KLF8 plays a critical role in adipocyte differentiation. PLoS One. 2012;7:e52474. doi: 10.1371/journal.pone.0052474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tang QQ, Li X, Lane MD. Effect of phosphorylation and S-S bond-induced dimerization on DNA binding and transcriptional activation by C/EBPbeta. Proc Natl Acad Sci USA. 2007;104:1800–1804. doi: 10.1073/pnas.0611137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lee YJ, Choi H, Ko EH, Kim JW. Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J Biol Chem. 2009;284:10601–10609. doi: 10.1074/jbc.M808742200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.