Abstract
Many neurodegenerative diseases, such as Alzheimer's disease and Parkinson's disease, are characterized by abnormal accumulations of aggregated proteins. Brains in these diseases also show accumulation of autophagic vesicles in the neuronal cytoplasm, suggesting impairment of the autophagic process. As autophagy involves de novo membrane production and vesicle fusion, extensive changes in lipid molecules are necessary. However, the involvement of signaling lipid-modifying enzymes in autophagy and their roles in neurodegenerative diseases are not clear. Using specific inhibitor, we show that loss of phospholipase D1 (PLD1) activity resulted in an accumulation of microtubule-associated protein light chain 3 (LC3), p62, and polyubiquitinated proteins, signs representing malfunction in autophagic flux. Fluorescence and electron microscopic analyses demonstrated impaired fusion of autophagosomes with lysosomes, resulting in accumulation of autophagosomes. Within the cells with impaired autophagic flux, α-synuclein aggregates accumulated in autophagosomes. Knockdown of PLD1 expression using small interfering RNA also resulted in impaired autophagic flux and accumulation of α-synuclein aggregates in autophagosomes. Neuronal toxicity caused by α-synuclein accumulation was rescued by overexpression of PLD1; however, expression of activity-deficient mutant, PLD1-KRM, showed reduced rescue effects. Finally, we demonstrated that both PLD activity and expression levels were reduced in brain tissues of dementia with Lewy bodies (DLB) patients, whereas the amounts of α-synuclein and p62 were increased in the same tissue samples. Collectively, these results suggest that insufficient PLD activity, and therefore, the changes in phospholipid compositions within membranes, might be an important contributor to impaired autophagic process and protein accumulation in Lewy body diseases.
Macroautophagy is the best-characterized autophagy pathway that mediates the lysosomal degradation of the cytoplasmic organelles and proteins.1 In this paper, macroautophagy will be simply referred to as autophagy. Autophagy may be characterized by nonspecific sequestration and degradation of the bulk cytoplasm, a process that recycles essential building blocks for production of macromolecules under conditions where nutrients are limited. Autophagy may also occur to selectively degrade polyubiquitinated targets, and this is often referred to as the quality control autophagy.2 Many long-lived proteins and perhaps protein aggregates may be the substrates of the quality control autophagy.
As being an essential process for macromolecular metabolism, perturbation of autophagy has been linked to various human diseases, such as neurodegenerative diseases, cancer, and infectious diseases.3 Autophagic dysfunction in neurons, in particular, causes accumulation of aggregation-prone proteins and neurodegeneration that are associated with various neurodegenerative diseases, including Alzheimer's disease (AD), Parkinson's disease (PD), and Huntington's disease (HD).4 Recently, genetic mutations that are linked to some of the major neurodegenerative diseases have turned out to reside in the genes that are involved in multiple steps in the autophagic pathways,4 implicating the therapeutic potential of controlling autophagy.
Autophagy involves sequestration of cytoplasmic substrates by a double-membraned compartment known as the autophagosome (AP).1 Autophagy process initiates with the formation of a distinct structure referred to as the phagophore that extends its ends and seals in circle to form the AP. APs can fuse with various endosomal vesicles, forming amphisomes, and eventually fuse with lysosomes to form autolysosomes, where degradation of contents takes place. Autophagy involves a wide range of changes in membrane structures, such as de novo membrane biogenesis and membrane fusion. Therefore, lipid molecules and lipid-metabolizing enzymes must play essential roles in the autophagic process. A well-characterized such lipid enzyme is the class III phosphatidylinositol-3 kinase (PI3K), Vps34, that is essential for biogenesis of APs through interactions with various proteins.5 Other than Vps34, little has been known about the roles of lipid enzymes in autophagic process.
In the current study, we explored the role of phospholipase D1 (PLD1) in autophagy and clearance of α-synuclein aggregates, suspected culprit of PD.6 PLD1 generates phosphatidic acid (PA) from phosphatidylcholine and has been known to be involved in intracellular vesicle trafficking.7 Our results suggest that PLD1 is an important player for maintaining autophagic flux via regulating autolysosome formation. We also showed that enzymatic inhibition and reduction in expression of PLD1 resulted in impaired clearance of α-synuclein aggregates. Finally, our data showed that reduced expression, and thus activity, of PLD1 was associated with Lewy body diseases.
Results
Pharmacological inhibition of PLD1 interferes with autophagic flux
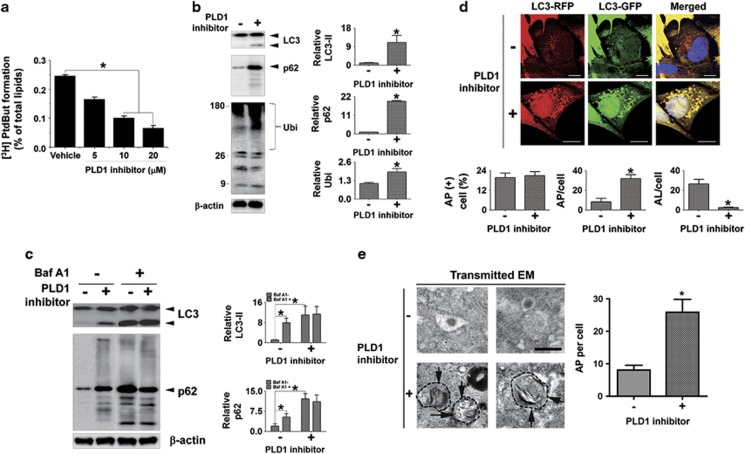
To investigate the roles of PLD1 in the induction and the maturation of autophagic vesicles, we examined the effects of a selective inhibitor of PLD1, VU0155069.8 Dose-dependent inhibition of PLD activity was confirmed in differentiated SH-SY5Y neuroblastoma cells (Figure 1a). We first analyzed the levels of endogenous LC3-II, a lipidated form of microtubule-associated protein light chain 3 (LC3) known as a marker of AP. Inhibition of PLD1 resulted in the accumulation of LC3-II (Figure 1b). Accumulation of LC3-II could indicate either increased formation of AP or decreased flux of AP to autolysosome, the latter reflecting impaired fusion between APs and lysosomes.9 Efficient flux of autophagy would result in a decrease in the substrates, such as polyubiquitinated proteins and p62, a polyubiquitin-binding protein. On the contrary, the levels of both polyubiquitinated proteins and p62 were elevated significantly in the cells treated with PLD1 inhibitor, suggesting failure of maturation of APs to autolysosomes (Figure 1b). To further evaluate the autophagic flux, we assessed the levels of LC3-II and p62 in the presence of bafilomycin A1 (Baf A1), a proton pump inhibitor that blocks the lysosomal acidification and the AP–lysosome fusion.10 In the presence of Baf A1, the PLD1 inhibitor did not alter the level of LC3-II or p62 (Figure 1c), indicating that the increases in these proteins upon PLD1 inhibition were due to the impaired autophagic flux. We next measured the fusion rate of APs to lysosomes by using LC3 tagged with GFP-RFP tandem fluorescent proteins (tfLC3). In APs where the internal pH is neutral, both GFP and RFP fluoresce, and hence it appears yellow under optical microscope. However, in autolysosomes where the internal pH becomes acidic, GFP no longer fluoresces, and hence only red fluorescence is visible.11 Therefore, tfLC3 allows for discrimination between AP and autolysosome. When PLD1 was inhibited, the total number of autophagic vesicles containing cells was not changed (Figure 1d). However, the average number of AP (red-positive/green-positive) per cell was elevated from 8 to 31 (Figure 1d). On the other hand, the average number of autolysosome (red-positive/green-negative) was reduced from 26 to 2.5 (Figure 1d). Furthermore, electron microscopy (EM) analysis verified an increase in the number of AP in cells treated with the PLD inhibitor (Figure 1e). These data indicate that inhibition of PLD1 blocks the fusion of AP to lysosome, thereby resulting in accumulation of APs.
Figure 1.
Inhibition of autophagic flux by pharmacological inhibition of PLD1. (a) The alteration in the PLD1 activity by pharmacological inhibition of PLD1. Differentiated SH-SY5Y cells were treated with 0, 5, 10, and 20 μM of PLD1 inhibitor for 2 days. N=3, *P<0.05. (b) The accumulation of autophagic substrates. Differentiated SH-SY5Y cells were treated with 20 μM of the PLD1 inhibitor for 2 days. The levels of LC3-II, p62, and polyubiquitinated proteins were normalized with β-actin. N=3, *P<0.05. (c) Cotreatment with PLD1 inhibitor and Baf A1. Differentiated SH-SY5Y cells were treated with the PLD1 inhibitor as above before treatment with 50 nM of Baf A1 for 24 h. The levels of LC3-II and p62 were normalized with β-actin. N=3, *P<0.05. (d) Accumulation of AP by PLD1 inhibition. Differentiated SH-SY5Y cells were transfected with tfLC3 and incubated with PLD1 inhibitor as above. Fluorescence images were analyzed using ImageJ software. A total of 300 cells were analyzed from three independent experiments (100 cells per experiment). Scale bar: 20 μm, *P<0.05. (e) EM of the accumulated APs in cells treated with PLD1 inhibitor. N=3 (20 cells per experiment). *P<0.05
PLD1 inhibition results in accumulation of α-synuclein aggregates in the AP
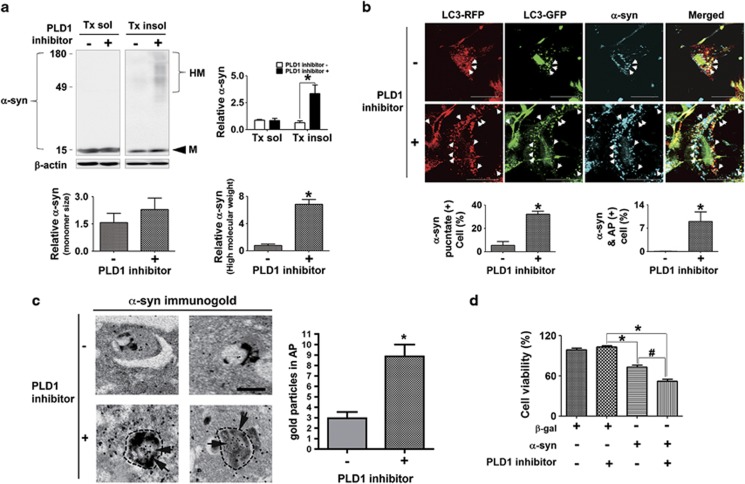
We and others have previously demonstrated that α-synuclein aggregates were degraded via the autophagic–lysosomal degradation pathway.12, 13 Furthermore, it has been reported that the autophagy pathway generally plays a major role in clearance of cytoplasmic protein aggregates.14 Therefore, we examined the effects of PLD1 inhibition on the cytoplasmic accumulation of α-synuclein. Differentiated human neuroblastoma SH-SY5Y cells overexpressing human α-synuclein were incubated with PLD1 inhibitor for 2 days. Inhibition of PLD1 significantly increased the accumulation of high-molecular-weight α-synuclein aggregates in the Triton-insoluble fraction (Figure 2a). On the other hand, the monomer in the same fraction showed a slight increase in the PLD1 inhibitor-treated group, but the change was not statistically significant. To examine the intracellular localization of α-synuclein accumulation under the same condition, we analyzed the cells co-transfected with human α-synuclein and tfLC3. Inhibition of PLD1 increased the number of α-synuclein puncta in the cytoplasm. A large portion of α-synuclein puncta colocalized with AP (red-positive/green-positive puncta) in the presence of PLD1 inhibitor (Figure 2b). Consistently, immuno-EM analysis showed increased number of immuno-gold labels for α-synuclein in APs in cells treated with PLD1 inhibitor (Figure 2c). Therefore, PLD1 inhibition resulted in the accumulation of α-synuclein aggregates in the APs, suggesting that their degradation was blocked because of impaired autophagic flux.
Figure 2.
Effects of PLD1 inhibition on the accumulation of α-synuclein aggregates. (a) PLD1 inhibition induced accumulation of α-synuclein. Differentiated SH-SY5Y cells overexpressing human α-synuclein were treated with 20 μM of the PLD1 inhibitor for 2 days. The levels of α-synuclein were normalized with β-actin. HM, high molecular species; M, Monomeric size. N=3, *P<0.05. (b) α-Synuclein accumulation in APs. Differentiated SH-SY5Y cells were co-transfected with human α-synuclein and tfLC3 and incubated with the PLD1 inhibitor as above. Cells were permeabilized with 40 μM of digitonin before immunofluorescence staining. A total of 300 cells were analyzed from three independent experiments (100 cells per experiment). Scale bar: 20 μm, *P<0.05. (c) Immunogold EM of α-synuclein in APs in cells treated with PLD1 inhibitor. N=3 (15 cells per experiment). *P<0.05. (d) Cytotoxicity analysis. Control cells and cells overexpressing human α-synuclein were cultured for 1 day and treated with either DMSO or PLD1 inhibitor for 2 days. N=3, *P<0.05, #P<0.05
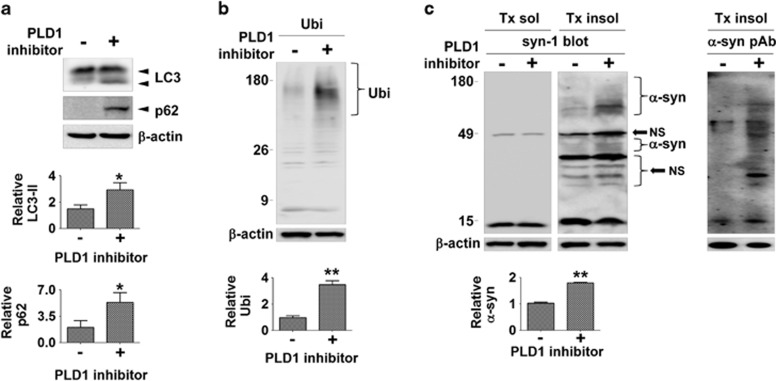
The effects of PLD inhibition on autophagic flux and α-synuclein clearance has also been validated in rat primary cortical neurons. Treatment of neurons with the PLD1 inhibitor resulted in the accumulation of LC3-II, p62, and polyubiquitinated proteins, as well as the high-molecular-weight α-synuclein aggregates in the Triton-insoluble fraction (Figure 3).
Figure 3.
PLD1 inhibitor induced accumulation of α-synuclein in primary cortical neurons. (a and b) The accumulation of autophagic substrates by PLD1 inhibitor in rat primary cortical neurons. Primary cortical neurons were treated with 10 μM of PLD1 inhibitor for 2 days. The levels of LC3-II, p62, and polyubiquitinated proteins were normalized with β-actin. N=3, *P<0.05, **P<0.01. (c) The PLD1 inhibition induced accumulation of α-synuclein in primary rat cortical neurons. The levels of α-synuclein were normalized with β-actin, NS, nonspecific band, N=3, **P<0.01
We then assessed whether PLD1 inhibition, and hence the blockade of autophagic flux, modifies α-synuclein toxicity. It has previously been shown that overexpression of α-synuclein by itself could cause neuronal cell death in differentiated SH-SY5Ycells.12 This was confirmed in the current study where overexpression of wild-type α-synuclein caused reduction of cell viability by 27% in 3 days (Figure 2d). When these cells were treated with PLD1 inhibitor, cell viability was further reduced to 52% (Figure 2d). On the other hand, expression of control protein, β-galactosidase, did not cause cell death by itself or in combination with PLD1 inhibitor treatment. This experiment was conducted under the condition where PLD1 inhibitor itself did not cause cell death (Figure 2d), and therefore the data indicate that the toxicity is specific to α-synuclein expression and that PLD1 inhibition potentiates the toxic effects of α-synuclein. Collectively, inhibition of PLD1 activity blocks normal autophagic flux by interfering with fusion between AP and lysosome, causing accumulation of α-synuclein aggregates within APs, and thereby increasing α-synuclein toxicity.
Knockdown of PLD1 expression inhibits the autophagic flux and clearance of α-synuclein aggregates
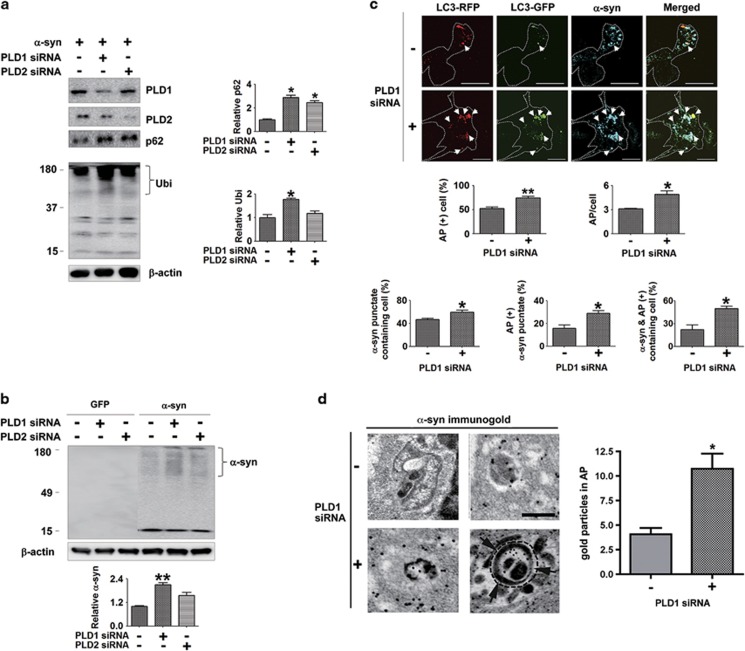
To further validate the roles of PLD1 in autophagy and α-synuclein accumulation, we knocked down the expression of PLD1 and PLD2 using RNAi. Expression of siRNAs against PLD1 and PLD2 specifically decreased the levels of the endogenously expressed proteins in SH-SY5Y cells (Figure 4a). Consistent with the inhibitor study, knockdown of PLD1 resulted in an increase in p62, polyubiquitinated proteins, and detergent-insoluble high-molecular-weight α-synuclein aggregates (Figures 4a and b). Knocking down of PLD2 also caused an increase in these proteins, but the extent of increase was not as great as PLD1 knockdown; some of the changes were not statistically significant under the current condition (Figures 4a and b). Next, we evaluated the maturation of APs in the cells that were co-transfected with human α-synuclein, tfLC3, and siRNA for PLD1. Confocal microscopy analysis showed that APs (red-positive/green-positive) were significantly increased after PLD1 knockdown. This was assessed by both the number of AP-containing cells and the number of APs per cell (Figure 4c). Similarly, knockdown of PLD1 also increased the number of cells containing α-synuclein puncta (Figure 4c). Some of α-synuclein puncta colocalized with APs in basal condition, and knockdown of PLD1 resulted in increased colocalization of APs and α-synuclein puncta (Figure 4c). As a result, the number of cells that contain APs with α-synuclein inside also increased (Figure 4c). To verify this result, we investigated the accumulation of α-synuclein in APs with immuno-EM. Knockdown of PLD1 resulted in an increase in the number of immuno-gold labels of α-synuclein in APs (Figure 4d).
Figure 4.
Inhibition of autophagic flux and α-synuclein accumulation in APs by PLD1 RNAi. (a) Effects of PLD1 RNAi on the accumulation of autophagic substrates. Differentiated SH-SY5Y cells overexpressing human α-synuclein were transfected with control, PLD1 siRNA, or PLD2 siRNA. The levels of p62 and polyubiquitinated proteins were normalized with β-actin. N=3, *P<0.05. (b) The accumulation of α-synuclein by PLD1 RNAi. The level of α-synuclein was normalized with β-actin. N=3, **P<0.01. (c) Deposition of α-synuclein in APs by PLD1 RNAi. Differentiated SH-SY5Y cells overexpressing human α-synuclein were co-transfected with tfLC3 and PLD1 siRNA. Cells were permeabilized with 40 μM of digitonin before immunofluorescence staining. A total of 300 cells were analyzed from three independent experiments (100 cells per experiment). Scale bar: 20 μm, *P<0.05, **P<0.01. (d) Immunogold-EM of α-synuclein in APs after PLD1 knockdown. N=3 (15 cells per experiment). *P<0.05
Inhibition of the activity and reduction of the expression of PLD1 collectively showed that PLD1 plays a role in maturation of APs to autolysosomes and in the clearance of α-synuclein aggregates.
Ectopic expression of PLD1 reduced protein aggregate accumulation and cytotoxicity
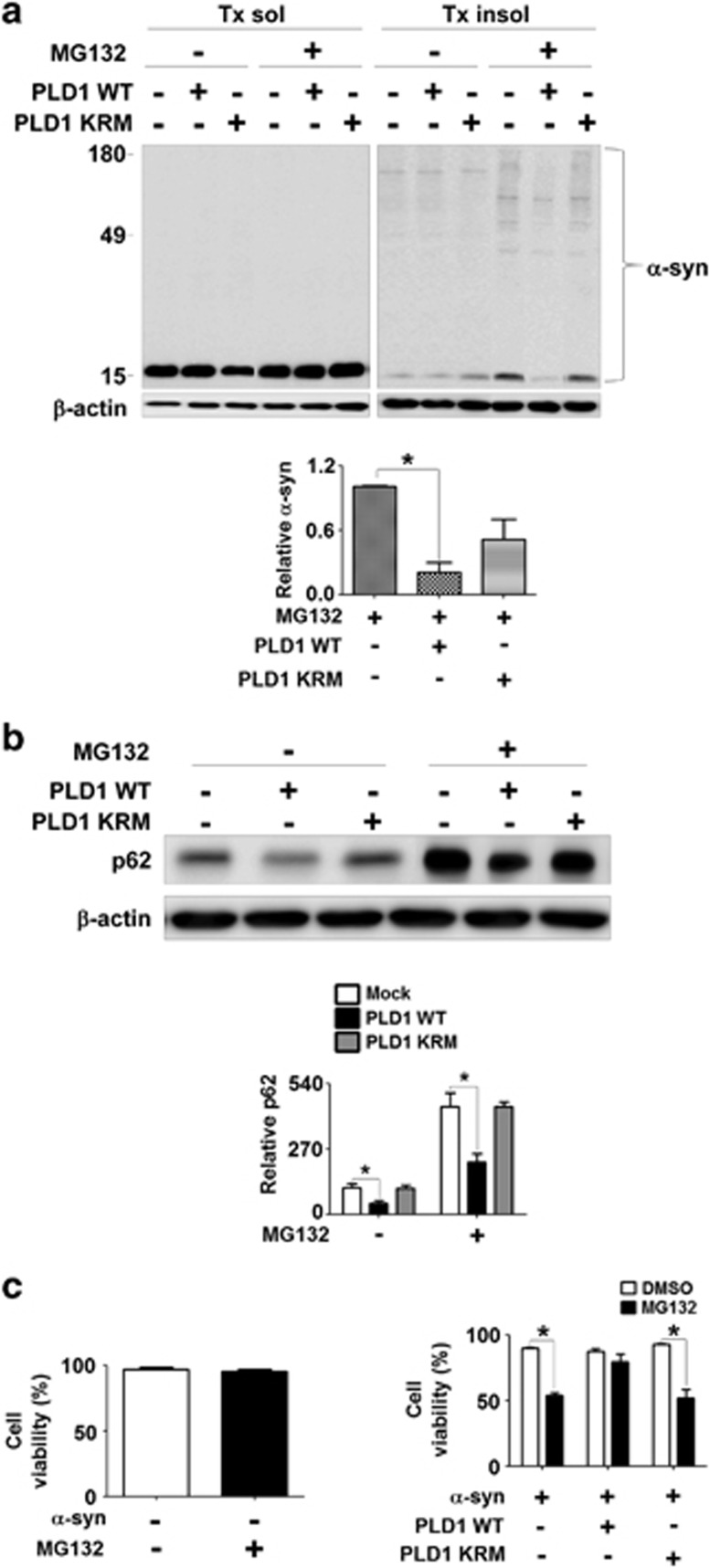
As our results suggest that PLD1 is required for autophagic flux and clearance of α-synuclein aggregates, we reasoned that ectopic overexpression of PLD1 might reverse the cytotoxic effects of α-synuclein aggregates. To rapidly induce aggregation of α-synuclein, differentiated SH-SY5Y cells overexpressing human α-synuclein were treated with 40 μM of MG132 for 24 h. Treatment of MG132 resulted in the increase of α-synuclein in the Triton-insoluble fraction (Figure 5a) and reduced cell viability (Figure 5c). We conducted these experiments under the condition that MG132 alone did not cause cytotoxicity but only potentiated the toxicity of α-synuclein (Figure 5c). When these cells were transfected with the wild-type PLD1, both the MG132-induced Triton-insoluble α-synuclein aggregates and cytotoxicity were significantly reduced (Figure 5). In contrast, expression of catalytically inactive form of PLD1 (mutation of lysine to arginine in the catalytic region of PLD1 (PLD1 KRM)) resulted in much less reduction in α-synuclein deposition (Figure 5a) and no effects on cytotoxicity (Figure 5c). Next, to verify the normal flux of autophagy, we examined the alterations in the levels of p62. In the cells transfected with wild-type PLD1, the levels of p62 were significantly reduced (Figure 5b), suggesting increased autophagic clearance, whereas expression of PLD1KRM did not affect the levels of p62. These results suggest that overexpression of PLD1 rescue the cells from both aggregated protein accumulation and aggregate-induced cytotoxicity by enhancing autophagic degradation.
Figure 5.
Reversal of MG132-induced α-synuclein accumulation and cytotoxicity by PLD1 overexpression. (a and b) Cells overexpressing human α-synuclein were transfected with either the wild-type PLD1 or PLD1 KRM (catalytic inactive mutant). The levels of α-synuclein (a) and p62 (b) in Triton-insoluble fraction were normalized with β-actin. The left and right panels in (a) were derived from the same gel. (c) Cell viability was analyzed using an automatic cell counter. N=3, *P<0.05
Reduction in PLD1 is associated with autophagic dysfunction and α-synuclein accumulation in Lewy body diseases
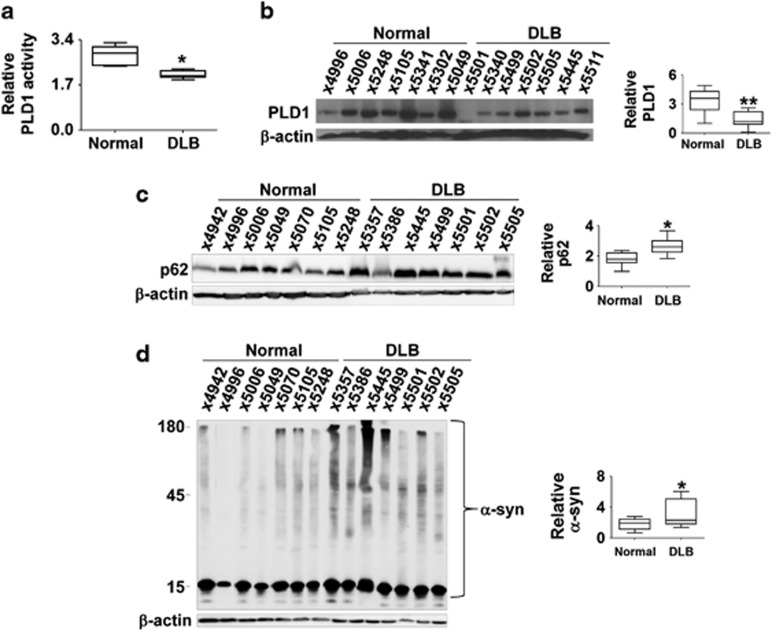
Based on our results, we postulated that the normal function of PLD1 might be compromised in Lewy body diseases, where autophagic dysfunction and α-synuclein accumulation have been well documented.4 First, to confirm the PLD1 expression in human neurons, we have performed immunohistochemistry with brain sections from control and dementia with Lewy bodies (DLB) patients. The data clearly show PLD1 immunoreactivities in neurons with vesicular patterns (Supplementary Figure 2). In DLB cases, PLD1 also showed vesicular patterns in neurons, but the levels of PLD1 were decreased (Supplementary Figure 2b). These data are consistent with the previous study showing that PLD1 is expressed in neurons in humans and is associated in neurite outgrowth.15 We then compared the enzymatic activities and expression levels of PLD1 in post-mortem brain samples obtained from the control subjects and DLB patients. There was no correlation between post-mortem delay and PLD activity (Supplementary Figure 1). We found that both enzymatic activities and expression levels of PLD1 were significantly lower in DLB brain samples than in control samples (Figures 6a and b). To assess autophagic dysfunction, we examined the levels of p62, and these were significantly increased in DLB samples than in control (Figure 6c). Western analysis also showed that the detergent-insoluble α-synuclein was drastically increased in DLB samples compared with the control (Figure 6d). These results suggest that reduced expression and activity of PLD1 is associated with autophagic dysfunction and α-synuclein accumulation in Lewy body diseases.
Figure 6.
The levels of PLD1 and autophagy substrates in DLB brain samples. (a and b) Relative activities (a) and levels (b) of PLD1 in the brains of control subjects and DLB patients. N=7 for each group. *P<0.05, **P<0.01. (c and d) Relative levels of p62 (c) and α-synuclein (d) in the brain of control subjects and DLB patients. N=7, *P<0.05
Discussion
In the current study, we showed that reduced PLD activity and PLD1 expression resulted in impairment in autophagic flux, probably at the stage of AP–lysosome fusion, thereby causing accumulation of α-synuclein aggregates in the AP. This accumulation of α-synuclein aggregates caused cell death that could be rescued by ectopic expression of PLD1. We also found that PLD activity and PLD1 expression levels were reduced in the brain tissues of DLB patients with concomitant increases in α-synuclein aggregates. These results provide significant implications of phospholipid metabolism in the regulation of autophagic process, and therefore in the clearance of pathogenic protein aggregates in neurodegenerative diseases.
Our study suggests that PLD1 is necessary for the maturation of APs to autolysosomes. Inhibition of PLD1 activity and reduction in PLD1 expression resulted in excessive accumulation of APs with substrates such as p62, polyubiquitinated proteins, and α-synuclein aggregates inside, whereas autolysosomes were reduced under the same conditions. Our findings are consistent with the previous study by Dall'Armi et al.,16 where PLD1 was predominantly associated with endosomes in normal growth medium and translocated to the limiting membranes of the amphisomes during starvation-induced autophagy.
Although involvement of PLD1 in late stages of autophagy is now clear, the mechanism has not been clearly addressed. Several studies suggested the role of PLD1 in the fusion of secretory vesicles with the plasma membrane in various mammalian cells (reviewed in Bader and Vitale17). In yeast, PLD ortholog, Spo14, was shown to be necessary for membrane fusion during the formation of prospore membranes.18 More recently, in yeast strain Pichia pastoris, Spo14 has been implicated in unconventional exocytosis of Acb1, and the process involves the fusion of APs with the plasma membrane.19 PLD catalyzes the removal of choline from PC, generating PA, that could then serve as a precursor for the production of diacylglycerol (DAG). When locally concentrated, both PA and DAG favor negative curvature in membranes, lowering the energy barrier to fusion.7 Our data showed that inhibition of PLD1 led to accumulation of preacidic autophagic compartments with simultaneous reduction in autolysosomes. Considering the previous studies pointing to the role of PLD in membrane fusion, we are inclined to suggest that PLD1 is directly involved in the fusion of AP/amphisome with lysosomes.
Although our study showed the involvement of PLD1 in later stages of autophagy, that is, the fusion between APs and lysosome, it does not necessarily rule out the possibility of PLD1 playing a role in early stages of AP formation. The source for AP membrane biogenesis appears to be diverse. Membranes derived from the Golgi, endoplasmic reticulum, mitochondria, and plasma membrane have been implicated as the source membranes.20, 21, 22, 23, 24, 25 It is possible that vesicles from multiple membrane sources may fuse heterotypically to generate APs. This early fusion steps may involve lipid-hydrolyzing enzymes, possibly PLD. Alternatively, PLD1 might be involved in the early stages of autophagy through regulation of the mTOR signaling pathway. It has been suggested that PA exerted its signaling actions as a second messenger through regulating mTOR and other proteins in the mTOR pathway.26, 27 An earlier study by Moreau et al.28 showed that a small G-protein, Arf6, facilitated AP biogenesis via inducing PLD activity.
Autophagy is the major mechanism for the clearance of intracellular protein aggregates that are associated with various human diseases, notably neurodegenerative diseases.4, 29 Genetic ablation of key autophagy genes in the nervous system caused neurodegenerative diseases with associated protein deposition pathology in mice.30, 31 Pharmacological induction of autophagy promoted clearance of protein aggregates and delayed neurodegeneration as well as behavioral symptoms in animal models of neurodegenerative diseases.14, 32, 33 We have recently shown that genetic and pharmacological ablation of autophagy promoted exocytosis and transcellular transmission of α-synuclein aggregates.34 Consistent with the role of autophagy in aggregate clearance, our data showed that impairment in autophagic flux upon PLD1 inhibition resulted in accumulation of α-synuclein in APs. Similarly, RNAi-mediated reduction in PLD1 expression caused the same pattern of α-synuclein accumulation. Our results are consistent with the previous study where pharmacological inhibition of PLD led to an increase in detergent-insoluble tau in brain slice culture.16 Our study further validated the importance of PLD1 by showing that accumulation of α-synuclein aggregates and concomitant cytotoxicity were reversed by ectopic expression of the wild-type PLD1, but not by the expression of an activity-deficient mutant PLD1. Therefore, maintaining the normal levels of PLD activity may be critical to cellular protein homeostasis, and thus to preventing neuronal cells from degeneration. The relevance of these findings to human neurodegenerative diseases has been verified in human autopsy samples. Both PLD activities and the expression levels of PLD1 were significantly reduced in brain tissue samples of DLB patients compared with the samples from neurologically normal individuals.
Why PLD1 expression is reduced in Lewy body diseases is not clear. Expression of PLD1 is known to be regulated in cell proliferation and differentiation;35 however, the mechanism underlying the regulation of PLD1 expression is not well understood. PLD1 levels and activities were reduced during the ceramide-induced apoptosis.36 Mitochondrial production of reactive oxygen species (ROS) by complex I plays an important role in ceramide-induced neuronal cell death.37 Therefore, we speculate that expression of PLD1 is regulated by ROS generated by the decline in complex I activity that is thought to be a critical event in dopaminergic degeneration in PD.
In conclusion, PLD1, a lipid-hydrolyzing enzyme previously implicated in intracellular signaling and vesicle trafficking, plays a critical roles in later stages of autophagy, most likely in fusion between AP/amphisome and lysosome to produce autolysosome. Through regulation of autophagic flux, PLD1 serves an important role in clearance of protein aggregates that are associated with neurodegenerative diseases. Consistently, both the activities and expression levels of PLD1 were reduced in brain tissues of Lewy body disease. Furthermore, accumulation of α-synuclein aggregates and its cytotoxic effects were reversed by ectopic expression of PLD1. Our results suggest that induction of PLD1 could be a strategy for restoring deficits in autophagic flux, and thereby delaying progression of neurological diseases. However, a recent study showed that genetic ablation of PLD2 ameliorated synaptic dysfunction and cognitive decline in models of AD.38 Therefore, understanding isotype-specific effects of PLD would be necessary for identifying effective therapeutic strategies targeting PLD enzymes.
Materials and Methods
Materials
The following antibodies were used in this study: α-synuclein monoclonal antibody (610787; BD Biosciences, San Diego, CA, USA),α-synuclein polyclonal antibody (2642; Cell Signaling Technology, Beverly, MA, USA), LC3 polyclonal antibody (ab48394; Abcam, Cambridge, MA, USA), p62 monoclonal antibody (c2384-0B; BD Transduction Laboratories, Swampscott, MA, USA), ubiquitin polyclonal antibody (Dako, Glostrup, Denmark), and β-actin monoclonal antibody (Sigma, St. Louis, MO, USA). Fluorescence dye-conjugated goat anti-rabbit IgG was purchased from Jackson Immunoresearch Laboratories (West Grove, PA, USA). PLD1 inhibitor (VU0155069) was purchased from Cayman Chemical (Ann Arbor, MI, USA). Bafilomycin A1 was purchased from Calbiochem (San Diego, CA, USA). Digitonin was obtained from Sigma.
Cell culture and transfection
SH-SY5Y cells were maintained and differentiated as previously described.12 For expression of human α-synuclein and tfLC3,11 SH-SY5Y cells were transfected by using electroporation as instructed in the manufacturer's protocol (Invitrogen Corp., Carlsbad, CA, USA). For knockdown of phospholipase D, SH-SY5Y cells were transfected with phospholipase D1 siRNA and phospholipase D2 siRNA using electroporation. For primary rat cortical neurons, cerebral cortices obtained from embryonic day 15 embryos of pregnant Sprague-Dawley rat were incubated with the papain solution (10 U/ml papain, 0.2 mg/ml cysteine, 0.5 mM ethylenediamine tetraacetic acid, 1 mM calcium chloride, and 0.003 M NaOH), followed by treatment with the serum inhibitor solution (minimum essential media with Earl's salts containing 5% fetal bovine serum, 2.5 mg/ml bovine serum albumin, and 2.5 mg/ml trypsin inhibitor). After dissociation by pipetting and centrifugation, cell pellets were resuspended and plated on the poly-D-lysine and laminin-coated culture dish. Cells were cultured in neurobasal medium (Invitrogen) containing 2% of B-27 supplement (Invitrogen) and 0.5 mM Glutamax-1 (Invitrogen). The medium was changed every 3 days.
Preparation of cell extracts
Cells were washed with PBS and lysed in extraction buffer (PBS, 1% Triton X-100, 1% (v/v) protease inhibitor cocktail (Sigma)) before incubation on ice for 10 min. Cell lysates were centrifuged at 16 000 × g for 10 min. Triton X-100-insoluble pellet was resuspended in the Laemmli sample buffer and sonicated briefly.
Western blotting
Western blotting was performed as previously described.39 Images were obtained and quantified using the FUJIFILM Luminescent Image Analyzer LAS-3000 and Multi Gauge (v3.0) software (FUJIFILM, Tokyo, Japan).
Immunofluorescence staining and image analysis
The procedure for cell staining was performed as previously described.39 Briefly, cells differentiated on poly-L-Lysine-coated coverslips were fixed in 4% paraformaldehyde in PBS and permeabilized in 0.1% Triton X-100. Cells were incubated in blocking solution (5% BSA/3% goat serum in PBS) before addition of primary antibodies diluted in blocking solution. After washing, cells were incubated with fluorescent dye-conjugated secondary antibodies. Nuclei were stained with TOPRO-3 iodide (Invitrogen), and coverslips were mounted onto slide glasses using antifade reagent (Invitrogen). Olympus (Tokyo, Japan) FV1000 confocal laser scanning microscopy was used for observation of cells. Colocalization analysis was performed using the ImageJ software (NIH, Bethesda, MD, USA). In each cell, the colocalization of green and red fluorescence was calculated as the ratio between the area of the single channel and the area matching both channels.
Cell viability
To determine cell viability, the total number of cells and nonviable cells were counted by using the ADAM automatic cell counter (Digital Bio, Seoul, Korea) with propidium iodide staining with or without cell lysis, respectively.
In vitro PLD activity assay
In vitro PLD activity was measured using the Amplex Red PLD assay kit (Molecular Probes, Eugene, OR, USA) according to the manufacturer's protocol. The resulting fluorescence was detected using a fluorescence microplate reader at an excitation of 530 nm and an emission of 590 nm.
Electron microscopy
Briefly, cells were grown in glass bottom dishes (MatTek Corporation, Ashland, MA, USA) and then postfixed in 1% glutaraldehyde, treated with osmium tetraoxide, embedded in epon araldite, detached from the glass coverslips and sectioned with the ultramicrotome (Leica, Wetzlar, Germany). Grids were analyzed with a Zeiss (Jena, Germany) OM 10 electron microscope as previously described40 in order to evaluate the approximate numbers of APs per cell. For morphometric analysis of APs from each section, electron micrographs were obtained at a final magnification of × 12 000, and a total of 20 cells were analyzed for each experiment. Three independent experiments were performed. For immunogold labeling, sections of the epoxi embedded cells were mounted in nickel grids, etched, and incubated with antibodies against α-synuclein (Millipore, Temecula, CA, USA) followed by the corresponding secondary antibody and labeling with 10 nm Aurion ImmunoGold particles (1 : 50, Electron Microscopy Sciences, Fort Washington, PA, USA) with silver enhancement. A total of 15 cells were analyzed per condition. Cells were randomly acquired from 3 grids, and electron micrographs were obtained at a magnification of × 25 000.
Human specimens, neuropathological examination, and criteria for diagnosis
A total of 18 cases (n=9 nondemented controls; n=9 DLB cases) were included for the present study. Autopsy material was obtained from patients (Table 1) studied neurologically and psychometrically at the Alzheimer Disease Research Center/University of California, San Diego (ADRC/UCSD). The last neurobehavioral evaluation was performed within 12 months before death and included Blessed score, Mini Mental State Examination (MMSE), and dementia-rating scale (DRS).41, 42 Brains were processed and evaluated according to standard methods.43 At autopsy, brains were divided sagitally, the left hemibrain was fixed in formalin of 4% paraformaldehyde (PFA) for neuropathological analysis, and the right frozen at −70°C for subsequent neurochemical analysis. Paraffin sections from 10% buffered formalin-fixed neocortical, limbic system, and subcortical material stained with hematoxylin and eosin (H&E), thioflavine-S, ubiquitin (Dako, Carpinteria, CA, USA), and α-synuclein (Millipore) were used for routine neuropathological analysis that included assessment of plaques, tangles, Lewy bodies, and Braak stage.43 The diagnosis of DLB was based in the initial clinical presentation with dementia followed by parkinsonism and the presence of α-synuclein and ubiquitin-positive LBs in cortical and subcortical regions.44, 45 The diagnosis of PD was based on the initial presentation with parkinsonism and presence of α-synuclein and ubiquitin-positive LBs in subcortical regions.
Table 1. Human specimens, neuropathological evaluation, and diagnosis.
| Case number | Age | Sex | PM HOURS | Diagnosis | Braak_combo |
|---|---|---|---|---|---|
| X5501 | 86 | Female | 8 | DLB | 1 |
| X5505 | 67 | Female | 6 | DLB Alzheimer's changes | 2.2 |
| X5499 | 76 | Male | 6 | DLB Alzheimer's changes | 1.2 |
| X5386 | 74 | Male | 27 | DLB Alzheimer's changes | 1.1 |
| X5357 | 94 | Male | 12 | DLB Alzheimer's changes | 1.2 |
| X5340 | 84 | Male | 12 | DLB Alzheimer's changes | |
| X5502 | 83 | Female | DLB Alzheimer's disease | 5.2 | |
| X5445 | 86 | Male | 12 | DLB Alzheimer's disease | 2.2 |
| X5511 | 80 | Female | 24 | DLB Alzheimer's disease, mild lacunar infarct | 3.2 |
| X5302 | 83 | Female | 72 | Normal | 1.1 |
| X5341 | 77 | Female | 12 | Normal infarct, other; ischemic changes, acute | 0 |
| X5248 | 93 | Female | 18 | Normal | 1 |
| X5105 | 74 | Female | 8 | Normal lacunar infarct, chronic (old) infarct, other; ischemic changes | 0 |
| X5070 | 97 | Female | 12 | Normal | 1 |
| X5049 | 102 | Female | 9 | Normal | 1 |
| X5006 | 69 | Male | 24 | Normal | 0 |
| X4996 | 91 | Male | 6 | Normal | 3 |
| X4942 | 83 | Male | Normal | 0 |
Abbreviation: PM HOURS, postmortem delay hours
There are 18 tissue samples: 9 DLB cases and 9 controls. The tissues were taken from the frontal cortex
Preparation of brain tissue extracts
Tissue were lysed in extraction buffer (1% Triton X-100, 1% (v/v) protease inhibitor cocktail in PBS) with sonication. After centrifugation at 1000 × g for 10 min, the supernatant was obtained.
Statistical analysis
Values shown in the figures are presented as mean±S.E.M. P-values for determination of the statistical significance of differences were calculated using paired, two-tailed Student's t-test using GraphPad InStat version 3.05 software (San Diego, CA, USA).
Acknowledgments
This work was supported by the National Research Foundation (NRF) grant funded by the Korean Government (MEST) (no. 2010-0015188), and the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A111228).
Glossary
- AD
Alzheimer's disease
- AP
autophagosome
- Baf A1
bafilomycin A1
- DLB
dementia with Lewy bodies
- EM
electron microscopy
- HD
Huntington's disease
- LC3
microtubule-associated protein light chain 3
- PA
phosphatidic acid
- PI3K
phosphatidylinositol-3 kinase
- PLD1
phospholipase D1
- PLD1 KRM
mutation of lysine to arginine in the catalytic region of PLD1
- tfLC3
LC3 tagged with GFP-RFP tandem fluorescent proteins
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by N Bazan
Supplementary Material
References
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2012;8:108–117. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Lee S-J. Controlling the mass action of alpha-synuclein in Parkinson's disease. J Neurochem. 2008;107:303–316. doi: 10.1111/j.1471-4159.2008.05612.x. [DOI] [PubMed] [Google Scholar]
- Roth MG. Molecular mechanisms of PLD function in membrane traffic. Traffic. 2008;9:1233–1239. doi: 10.1111/j.1600-0854.2008.00742.x. [DOI] [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Lee H-J, Khoshaghideh F, Patel S, Lee S-J. Clearance of alpha-synuclein oligomeric intermediates via the lysosomal degradation pathway. J Neurosci. 2004;24:1888–1896. doi: 10.1523/JNEUROSCI.3809-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Yon C, Park SY, Oh DY, Han AH, Kim DS, et al. Role of phospholipase D1 in neurite outgrowth of neural stem cells. Biochem Biophys Res Commun. 2005;329:804–811. doi: 10.1016/j.bbrc.2005.02.087. [DOI] [PubMed] [Google Scholar]
- Dall'Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun. 2010;1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MF, Vitale N. Phospholipase D in calcium-regulated exocytosis: lessons from chromaffin cells. Biochim Biophys Acta. 2009;1791:936–941. doi: 10.1016/j.bbalip.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Morishita M, Schwartz CL, Coluccio A, Engebrecht J, Neiman AM. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J Cell Sci. 2006;119 (Pt 7:1406–1415. doi: 10.1242/jcs.02841. [DOI] [PubMed] [Google Scholar]
- Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy. 2010;6:301–303. doi: 10.4161/auto.6.2.11134. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DA. Regulation of mTOR by phosphatidic acid. Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, et al. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci USA. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Puri C, Rubinsztein DC. Arf6 promotes autophagosome formation via effects on phosphatidylinositol 4,5-bisphosphate and phospholipase D. J Cell Biol. 2012;196:483–496. doi: 10.1083/jcb.201110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Rose C, Menzies FM, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O, et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington's disease. Hum Mol Genet. 2010;19:2144–2153. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Cho ED, Lee KW, Kim JH, Cho SG, Lee SJ. Autophagic failure promotes the exocytosis and intercellular transfer of alpha-synuclein. Exp Mol Med. 2013;45:e22. doi: 10.1038/emm.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable ME, Obeid LM. Phospholipase D in cellular senescence. Biochim Biophys Acta. 1999;1439:291–298. doi: 10.1016/s1388-1981(99)00101-8. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Sakai H, Ohguchi K, Nakashima S, Banno Y, Nishimura Y, et al. Changes in the activity and mRNA levels of phospholipase D during ceramide-induced apoptosis in rat C6 glial cells. J Neurochem. 1997;69:713–720. doi: 10.1046/j.1471-4159.1997.69020713.x. [DOI] [PubMed] [Google Scholar]
- France-Lanord V, Brugg B, Michel PP, Agid Y, Ruberg M. Mitochondrial free radical signal in ceramide-dependent apoptosis: a putative mechanism for neuronal death in Parkinson's disease. J Neurochem. 1997;69:1612–1621. doi: 10.1046/j.1471-4159.1997.69041612.x. [DOI] [PubMed] [Google Scholar]
- Oliveira TG, Chan RB, Tian H, Laredo M, Shui G, Staniszewski A, et al. Phospholipase d2 ablation ameliorates Alzheimer's disease-linked synaptic dysfunction and cognitive deficits. J Neurosci. 2010;30:16419–16428. doi: 10.1523/JNEUROSCI.3317-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Lee S-J. Characterization of cytoplasmic alpha-synuclein aggregates. Fibril formation is tightly linked to the inclusion-forming process in cells. J Biol Chem. 2002;277:48976–48983. doi: 10.1074/jbc.M208192200. [DOI] [PubMed] [Google Scholar]
- Masliah E, Rockenstein E, Mante M, Crews L, Spencer B, Adame A, et al. Passive immunization reduces behavioral and neuropathological deficits in an alpha-synuclein transgenic model of Lewy body disease. PLoS One. 2011;6:e19338. doi: 10.1371/journal.pone.0019338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascovsky K, Salmon DP, Ho GJ, Galasko D, Peavy GM, Hansen LA, et al. Cognitive profiles differ in autopsy-confirmed frontotemporal dementia and AD. Neurology. 2002;58:1801–1808. doi: 10.1212/wnl.58.12.1801. [DOI] [PubMed] [Google Scholar]
- Salmon DP, Thal LJ, Butters N, Heindel WC. Longitudinal evaluation of dementia of the Alzheimer type: a comparison of 3 standardized mental status examinations. Neurology. 1990;40:1225–1230. doi: 10.1212/wnl.40.8.1225. [DOI] [PubMed] [Google Scholar]
- Hansen LA, Terry RD. Position paper on diagnostic criteria for Alzheimer disease. Neurobiol Aging. 1997;18 (4 Suppl:S71–S73. doi: 10.1016/s0197-4580(97)00073-0. [DOI] [PubMed] [Google Scholar]
- McKeith IG, Galasko D, Kosaka K, Perry EK, Dickson DW, Hansen LA, et al. Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): report of the consortium on DLB international workshop. Neurology. 1996;47:1113–1124. doi: 10.1212/wnl.47.5.1113. [DOI] [PubMed] [Google Scholar]
- Galasko D, Katzman R, Salmon DP, Hansen L. Clinical and neuropathological findings in Lewy body dementias. Brain Cogn. 1996;31:166–175. doi: 10.1006/brcg.1996.0040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.