Abstract
Mcl-1 is a unique antiapoptotic Bcl2 family member with a short half-life due to its rapid turnover through ubiquitination. We discovered that Ku70, a DNA double-strand break repair protein, functions as a deubiquitinase to stabilize Mcl-1. Ku70 knockout in mouse embryonic fibroblast (MEF) cells or depletion from human lung cancer H1299 cells leads to the accumulation of polyubiquitinated Mcl-1 and a reduction in its half-life and protein expression. Conversely, expression of exogenous Ku70 in Ku70−/− MEF cells restores Mcl-1 expression. Subcellular fractionation indicates that Ku70 extensively colocalizes with Mcl-1 in mitochondria, endoplasmic reticulum and nucleus in H1299 cells. Ku70 directly interacts with Mcl-1 via its C terminus (that is, aa 536–609), which is required and sufficient for deubiquitination and stabilization of Mcl-1, leading to suppression of apoptosis. Purified Ku70 protein directly deubiquitinates Mcl-1 by removing K48-linked polyubiquitin chains. Ku70 knockdown not only promotes Mcl-1 turnover but also enhances antitumor efficacy of the BH3-mimetic ABT-737 in human lung cancer xenografts. These findings identify Ku70 as a novel Mcl-1 deubiquitinase that could be a potential target for cancer therapy by manipulating Mcl-1 deubiquitination.
Mcl-1 is an antiapoptotic molecule that is overexpressed in various types of cancers, including lung cancer,1 leukemia,2 lymphoma,3 hepatocellular carcinoma4 and so on. In addition to its antiapoptotic function, Mcl-1 is also an oncoprotein that promotes the development of cancer.5 In contrast to other Bcl2 family members such as Bcl2 and Bcl-XL, Mcl-1 is unique in its short half-life (30 min–3 h) and short-term prosurvival function, which probably relates to the presence of a long proline-, glutamic acid-, serine- and threonine-rich (PEST) region upstream of the Bcl2 homology (BH) domain.1 The mechanism(s) that stabilizes the Mcl-1 protein are critical for its long-term survival function. Mcl-1 protein can be phosphorylated at multiple sites that distinctly regulate Mcl-1 protein turnover. For example, extracellular signal-regulated kinase 1/2-mediated T163 site phosphorylation enhances the half-life and antiapoptotic function of Mcl-1.1, 6 In contrast, S159 phosphorylation by GSK-3β facilitates Mcl-1 ubiquitination and degradation to reduce its survival activity.7
Ubiquitination and deubiquitination are two reversible processes that can control protein stability. E3 ligases and deubiquitinases (deubiquitinating enzymes (DUBs)) are two groups of regulatory enzymes that orchestrate the ubiquitination levels of target proteins in eukaryotic cells.8 Recently, Mule and FBW7 have been identified as Mcl-1 ubiquitin E3 ligases that can directly induce polyubiquitination and degradation of Mcl-1.9, 10 Inversely, USP9X has been demonstrated as the Mcl-1 deubiquitinase that removes the Lys 48-linked polyubiquitin chains that normally mark Mcl-1 for proteasomal degradation, leading to stabilization of Mcl-1.3 Therefore, the stability of Mcl-1 in cells is tightly regulated by its E3 ligases and deubiquitinase, which is dependent on Mcl-1 phosphorylation status.3, 11
Ku70 is a protein that binds to DNA double-strand break (DSB) ends and is required for the non-homologous end-joining pathway of DSB repair.12, 13, 14, 15 The Ku70 protein consists of three structural domains, including the N-terminal, central (that is, DNA binding) and C-terminal domains.16, 17 Ku70 usually heterodimerizes with Ku86, which forms a functional complex for DSB repair. By forming a bridge between the broken DNA ends, the Ku70/Ku86 heterodimer acts to structurally support and align the DNA ends, to protect them from degradation and to prevent promiscuous binding to unbroken DNA. Ku70/Ku86 effectively aligns the DNA, while still allowing access of polymerases, nucleases and ligases to the broken DNA ends to promote end joining.18 In some cases, a fourth domain is present at the C terminus of Ku86, which binds to the DNA-dependent protein kinase catalytic subunit.19 Importantly, Ku70 also regulates apoptosis independent of its DSB repair activity. For example, a recent report revealed that Ku70 regulates the proapoptotic function of Bax by sequestering Bax from the mitochondria and mediating Bax deubiquitylation.20 Here we discovered that Ku70 functions as a novel Mcl-1 deubiquitinase that directly removes polyubiquitin chains from Mcl-1 protein, leading to reduced Mcl-1 ubiquitination/degradation, enhanced stability and suppression of apoptosis.
Results
Disruption of Ku proteins downregulates Mcl-1 and sensitizes cells to staurosporine or Bcl2 inhibitor
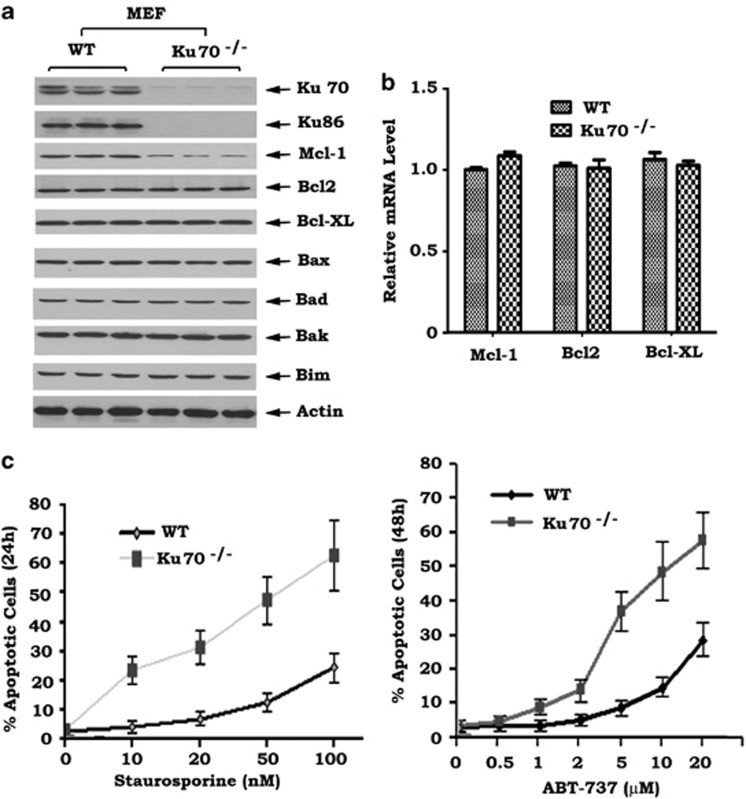
Ku70 and Ku86 usually exist in cells as functional heterodimers that are required for the repair of DSBs.12 Interestingly, no detectable levels of Ku86 could be observed in Ku70−/− mouse embryonic fibroblast (MEF) cells compared with wild-type (WT) MEF cells (Figure 1a). This supports the notion that Ku70 may stabilize Ku86 protein.21, 22 Ku proteins have been reported to support cell survival but the mechanism(s) involved is not fully understood.20, 23 To test whether Ku regulates apoptosis regulators, several Bcl2 family members were compared with WT MEF and Ku-deficient MEF cells. Significantly decreased levels of Mcl-1, but not Bcl2, Bcl-XL or other proapoptotic molecules, were observed in MEF Ku70−/− cells when compared with WT MEF cells (Figure 1a), suggesting that disruption of Ku70 may selectively reduce Mcl-1 expression. RT-PCR analysis provided evidence that the absence of Ku70 did not affect mRNA levels of Mcl-1, Bcl2 or Bcl-XL (Figure 1b). This indicates that downregulation of Mcl-1 mediated by Ku70 loss may not occur through a transcriptional mechanism. Functionally, decreased Mcl-1 by Ku70 loss significantly enhanced apoptosis following treatment of cells with staurosporine (Figure 1c, left panel). ABT-737 is a known inhibitor of Bcl2, Bcl-XL and Bcl-w but not of Mcl-1.24 Mcl-1 expression has been found to be associated with resistance of cells to ABT-737.25, 26 Our results show that the Ku70-deficient MEF cells are more sensitive to ABT-737 than WT MEF cells (Figure 1c, right panel), indicating that downregulation of Mcl-1 by disruption of Ku70 can sensitize cells to Bcl2 inhibition.
Figure 1.
Ku70 knockout downregulates Mcl-1 and enhances apoptosis following treatment with staurosporine or Bcl2 inhibitor. (a) Expression of Mcl-1, Ku70, Ku86 and other Bcl2 family members in wild type (WT) and Ku70−/− MEF cells. (b) mRNA levels of Bcl2, Bcl-XL and Mcl-1 in WT and Ku70−/− MEF cells. (c) Numbers of apoptotic cells in WT and Ku70−/− MEF cells treated with increasing concentrations of staurosporine or ABT-737 for 24 or 48 h
USP9X is a known Mcl-1 deubiquitinase that can stabilize Mcl-1 via deubiquitination.3 On the basis of our findings, Ku70 may have a similar role as USP9X in regulating Mcl-1 stability. To test this possibility, H1299 cells were transfected with Ku70 short hairpin RNA (shRNA), USP9X shRNA or their combination in the absence or presence of MG132. MG132 is a specific, potent, reversible and cell-permeable proteasome inhibitor that reduces the degradation of ubiquitin-conjugated proteins in mammalian cells.27 Silencing of Ku70, USP9X or both in H1299 cells resulted in decreased levels of Mcl-1, which could be reversed by MG132 (Figure 2a). Intriguingly, a greater degree of Mcl-1 reduction (that is, to an almost undetectable level) was observed in double-knockdown cells when compared with Ku70 shRNA or USP9X shRNA alone (Figure 2a). These data suggest that, in addition to USP9X, Ku70 may be another Mcl-1 deubiquitinase that stabilizes Mcl-1 through deubiquitination. Similar to Ku70−/− MEF cells, a significant decrease in Ku86 expression was also observed when Ku70 was knocked down by Ku70 shRNA in H1299 cells (Figure 2a). Because knockdown of Ku70 in H1299 cells did not affect mRNA levels of Mcl-1 (Figure 2b), this suggests that Ku70 may regulate Mcl-1 protein expression at a non-transcriptional level. Similar results were also obtained in HEK293T cells (Supplementary Figure S1). Functionally, silencing of Ku70 not only resulted in reduced levels of Mcl-1 but also enhanced the sensitivity of human lung cancer H1299 cells to the Bcl2 inhibitor ABT-737 (Figure 2c). Intriguingly, double knockdown of Ku70 and USP9X rendered H1299 cells more sensitive to ABT-737 when compared with knockdown of Ku70 or USP9X alone (Figure 2c).
Figure 2.
Ku70 depletion from human lung cancer H1299 cells reduces Mcl-1 and sensitizes cells to Bcl2 inhibition. (a) Expression of Mcl-1, Ku70, Ku86 and USP9X in H1299 cells transfected with Ku70 shRNA, USP9X shRNA or their combination, and after 24 h, treated with MG132 for 6 h. (b) mRNA levels of Bcl2, Bcl-XL and Mcl-1 in H1299 cells expressing Ku70 shRNA or control shRNA. (c) Numbers of apoptotic cells in H1299 cells expressing Ku70 shRNA, USP9X shRNA or their combination treated with increasing concentrations of ABT-737 for 48 h
Disruption of Ku enhances Mcl-1 ubiquitination in association with reduced half-life
To test whether Ku70 functions as a Mcl-1 deubiquitinase, Ku70 was depleted from H1299 cells by RNA interference using Ku70 shRNA. Mcl-1 ubiquitination was compared in H1299 cells expressing Ku70 shRNA, USP9X shRNA or control shRNA. Results revealed that significantly more Mcl-1 ubiquitination was observed in H1299 cells expressing Ku70 shRNA or USP9X shRNA when compared with control cells (Figure 3a), indicating that Ku70 may play a similar role as USP9X (a deubiquitinase) in regulating Mcl-1 ubiquitination in human lung cancer H1299 cells. Similarly, a greater level of Mcl-1 ubiquitination was also observed in Ku70−/− MEF cells when compared with WT MEF cells (Figure 3b). To further test the effect of Ku70 on Mcl-1 stability, the half-life of Mcl-1 was measured using cycloheximide-blocking methods as described.28 WT or Ku70 −/− MEF cells were treated with 100 μg/ml cycloheximide for various times as indicated. Levels of Mcl-1 were analyzed by western blot and further quantified by ImageJ software (National Institutes of Health, Bethesda, MD, USA) to calculate the half-life as described.29 Results indicated that knock out of Ku70 resulted in a significantly reduced half-life of Mcl-1 (that is, from 2.2 to 0.45 h) in MEF cells (Figure 3c).
Figure 3.
Ku70 disruption results in accumulation of ubiquitinated Mcl-1 and reduced Mcl-1 half-life. (a) Mcl-1 ubiquitination analyzed by anti-HA western blot following anti-Mcl-1 co-IP in H1299 cells expressing Ku70 shRNA, USP9X shRNA or control shRNA 48 h after transfection with HA-tagged ubiquitin (HA-Ub) constructs. (b) Mcl-1 ubiquitination analyzed as in (a) in WT or Ku70−/− MEF cells transfected with HA-Ub constructs. (c) Mcl-1 expression in WT or Ku70−/− MEF cells treated with 100 mg/ml cycloheximide 5 min before starting the indicated time course. Mcl-1 levels were quantified by ImageJ software to calculate the half-life
Expression of exogenous Ku70 in Ku70−/− MEF cells restores Mcl-1 expression and prolongs cell survival following stress
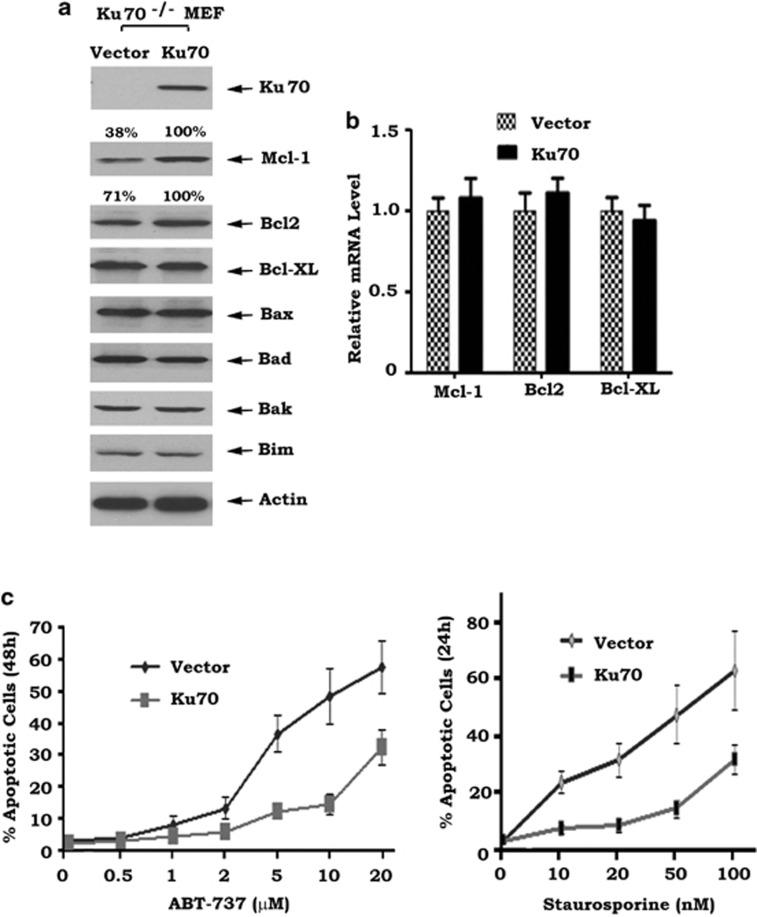
Conversely, to demonstrate whether Ku70 functions as a deubiquitinase to stabilize Mcl-1, Ku70 was exogenously expressed in Ku70−/− MEF cells. Intriguingly, restoration of Ku70 significantly upregulated Mcl-1 protein but had no effect on Bcl-XL, Bax, Bad, Bak or Bim (Figure 4a). Ku70 did not affect Mcl-1 mRNA level, suggesting that Ku70-mediated upregulation of Mcl-1 does not result from a transcriptional mechanism (Figure 4b). Similar experiments were also performed in HEK293T or HCT116 cells. Consistently, overexpression of Ku70 in HEK293T cells resulted in increased Mcl-1 protein level in association with reduced Mcl-1 ubiquitination and prolonged half-life of Mcl-1 (that is, from 1.54 to 3.87 h) (Supplementary Figure S2). Importantly, Ku70-enhanced Mcl-1 reduced the sensitivity of MEF cells to ABT-737 or staurosporine (Figure 4c). It seems that expression of exogenous Ku70 also slightly increased Bcl2 level in Ku70−/− MEF cells (Figure 4a), but overexpression of exogenous Ku70 in HEK293T cells did not enhance Bcl2 level (Supplementary Figure S2a). Further work may be required to determine whether Ku70 may also regulate Bcl2 in various cell systems.
Figure 4.
Expression of exogenous Ku70 in Ku70−/− MEF cells restores Mcl-1 expression and suppresses apoptosis following stress. (a) Expression of Ku70, Mcl-1 and other Bcl2 family members in Ku70−/− MEF cells transfected with WT Ku70 or vector-only control. Mcl-1 and Bcl2 levels were quantified by ImageJ software. (b) mRNA levels of Bcl2, Bcl-XL and Mcl-1 in Ku−/− MEF cells expressing exogenous WT Ku70 or vector-only control. (c) Numbers of apoptotic cells in Ku70−/− MEF cells expressing exogenous WT Ku70 or vector-only control treated with increasing concentrations of ABT-737 or staurosporine for 48 or 24 h
Ku70 partially colocalizes and directly interacts with Mcl-1 via its C terminus
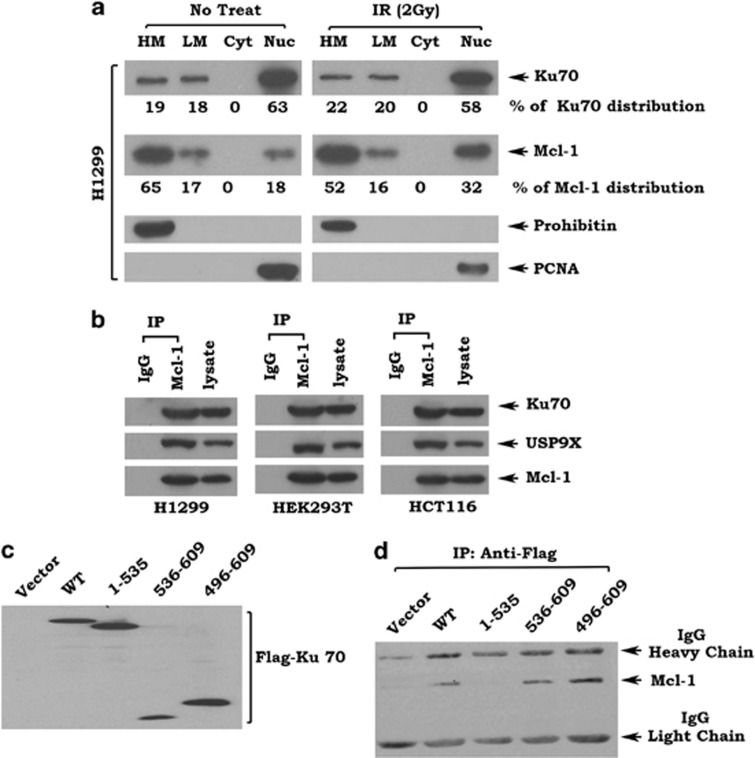
To further uncover the mechanism by which Ku70 regulates the stability of Mcl-1, localization of Ku70 and Mcl-1 was analyzed by subcellular fractionation as we described in ‘Materials and Methods'.30, 31, 32 As shown in Figure 5a, left panel, Ku70 was colocalized with Mcl-1 in heavy membrane (HM) containing mitochondria, light membrane (LH) containing endoplasmic reticulum (ER) and in nuclear (Nuc) fractions in H1299 cells. Quantification analysis of Ku70 and Mcl-1 using ImageJ software revealed that 19% of Ku70 and 65% of Mcl-1 were localized in mitochondria. Eighteen percent of Ku70 and 17% of Mcl-1 were localized in the ER. Sixty-three percent of Ku70 and 18% of Mcl-1 were localized in the nucleus (Figure 5a, left panel). On the basis of these findings, 19% of Ku70 molecules may be able to stabilize 65% of Mcl-1 in mitochondria. Obviously, there are sufficient Ku70 molecules to stabilize Mcl-1 in the ER and nucleus as well. Intriguingly, ionizing radiation (IR) enhanced the nuclear localization of Mcl-1 (that is, from 18 to 32% Figure 5a), suggesting that radiation may enhance Mcl-1 colocalization with Ku70 in the nucleus. In addition, radiation also slightly enhanced Ku70 levels in the mitochondria (that is, from 19 to 22%) or ER (that is, from 18 to 20%) (Figure 5a). To verify the purity of the subcellular fractions obtained, fraction-specific proteins were assessed by probing the same filters. Prohibitin, an exclusively mitochondrial protein,33 was detected only in the mitochondrial fraction, whereas proliferating cell nuclear antigen (PCNA), a nuclear marker,34 was detected exclusively in the nuclear fraction (Nuc) (Figure 5a). These data reveal that the fractionation procedure does not cause cross-contamination between the fractions.
Figure 5.
Ku70 colocalizes with and interacts with Mcl-1 via its C terminus. (a) Subcellular fractionation was performed to isolate heavy membrane (HM), light membrane (LM), cytosol (Cyt) and nuclear (Nuc) fractions as described in ‘Materials and Methods'. Western blot analysis of subcellular fractions was performed to detect Ku70 and Mcl-1. Levels of Ku70 and Mcl-1 were quantified by ImageJ software. (b) Mcl-1-associated Ku70 or USP9X and Mcl-1 analyzed by western blot following anti-Mcl-1 co-IP in H1299, HEK293 or HCT116 cells. (c) Expression of exogenous Ku70 analyzed by anti-Flag western blot in HEK293 cells transfected with Flag-tagged WT and each Ku70 deletion mutant. (d) Ku70-associated Mcl-1 analyzed by anti-Mcl-1 western blot following anti-Flag co-IP in HEK293T cells expressing exogenous Ku70 WT or each deletion mutant
It is known that USP9X as a Mcl-1 deubiquitinase directly interacts with Mcl-1.3 To further test whether Ku70 interacts with Mcl-1, co-immunoprecipitation (co-IP) experiments were performed using Mcl-1 antibody in H1299, HEK293T and HCT116 cells. Results revealed that both Ku70 and USP9X interacted with Mcl-1 in various types of cells (Figure 5b). To test whether the Ku70/Mcl-1 interaction is regulated in response to DNA damage induced by IR and whether Ku70 and USP9X interact with Mcl-1 in a competitive manner, H1299 cells were treated with increasing doses of IR. Results indicated that IR promoted Ku70 to interact with Mcl-1 in association with decreased USP9X/Mcl-1 binding in a dose-dependent manner (Supplementary Figure S3). To further demonstrate the binding site of Mcl-1 on Ku70 protein, a series of Flag-tagged Ku70 deletion mutants were exogenously expressed in Ku70−/− MEF cells (Figure 5c). Co-IP experiments revealed that Mcl-1 interacted with Ku70 WT, aa 536–609 and aa 496–609, but failed to bind the aa 1–535 mutant protein (Figure 5d). These findings indicate that Mcl-1 may bind to Ku70 at its C-terminal tail (aa 536–609). It was previously demonstrated that USP9X interacts with Mule.35 The Ku70/Mule association was also analyzed in H1299 cells by co-IP experiments. Results indicate that, in addition to USP9X, Ku70 also interacted with Mule (Supplementary Figure S4).
The C terminus is essential for Ku70 to deubiquitinate and stabilize Mcl-1
To test the role of Ku70/Mcl-1 binding in regulating Mcl-1 ubiquitination and stability, WT and each Ku70 deletion mutant were overexpressed in Ku70-deficient cells (Figure 6a). Intriguingly, expression of WT or the aa 536–609 or aa 496–609 Ku70 mutants not only significantly enhanced Mcl-1 level but also reduced Mcl-1 ubiquitination (Figures 6a and b). In contrast, the Mcl-1-binding-deficient Ku70 mutant (aa 1–535) failed to reduce ubiquitination and had no effect on Mcl-1 protein level (Figures 6a and b and Supplementary Figure S2d). These findings indicate that the C terminus of Ku70 (aa 536–609) is required and sufficient to stabilize Mcl-1 via deubiquitination. Functionally, expression of exogenous WT, aa 536–609 or 496–609 but not the Mcl-1-binding-deficient mutant (aa 1–535) suppressed ABT-737-induced apoptosis in Ku70-deficient MEF cells (Figure 6c).
Figure 6.
Ku70 C terminus is essential for Mcl-1 deubiquitination and stabilization. (a) Expression of Ku70 and Mcl-1 in Ku70−/− MEF cells transfected with Flag-tagged WT and each Ku70 deletion mutant. (b) Mcl-1 ubiquitination analyzed as in (Figure 3a) in Ku70−/− MEF cells expressing exogenous WT or each deletion mutant transfected with HA-ubiquitin constructs. (c) Percentage of apoptotic cells in Ku70−/− MEF cells expressing exogenous WT or each Ku70 deletion mutant treated with increasing concentrations of ABT-737 for 48 h
Direct deubiquitination of Mcl-1 by Ku70 in vitro
To test whether Ku70 protein can directly remove polyubiquitins from Mcl-1 protein, first, Flag-Ku70 and Flag-Mcl-1 were individually overexpressed in HEK293T cells. Flag-Ku70 and Flag-Mcl-1 proteins were then purified from HEK293T cells as described.3 To assess whether Ku70 protein directly deubiquitinates Mcl-1 in vitro, purified Ku70 and Mcl-1 were mixed under conditions supporting Mcl-1 deubiquitination. Purified USP9X and BSA were used as positive and negative controls, respectively. Results revealed that Ku70 directly deubiquitinated Mcl-1 in a dose-dependent manner in vitro (Figure 7a). In addition, purified recombinant Ku70 protein hydrolyzed polyubiquitin chains into monoubiquitin units (Supplementary Figure S5).These findings indicate that Ku70 protein has potent intrinsic deubiquitination enzymatic activity. As only WT, aa 536–609 and aa 496–609 but not the Mcl-1-binding-deficient aa 1–535 Ku70 mutant protein directly deubiquitinated Mcl-1 (Figure 7b), this suggests that the C terminus of Ku70 is required and sufficient to remove polyubiquitins from Mcl-1 protein in vitro. Western blot analysis revealed that Mcl-1 protein purified from MG132-treated HEK293T cells was only detected by a Lys 48 linkage-specific but not a Lys 63 linkage-specific polyubiquitin antibody (Figure 7c). Ku70 may stabilize Mcl-1 by removing its degradative Lys 48-linked polyubiquitin chains.
Figure 7.
Purified Ku70 protein directly deubiquitinates Mcl-1 protein in vitro. (a and b) Mcl-1 ubiquitination following incubation of purified ubiquitinated Mcl-1 protein with (a) purified Ku70, USP9X or BSA control, (b) purified WT or each Ku70 deletion mutant. (c) Flag-Mcl-1 purified from denatured HEK293T cell lysates was western blotted for ubiquitin, K48-linked polyubiquitin, K63-linked polyubiquitin and FLAG. The smear represents polyubiquitinated Flag-Mcl-1
Downregulation of Mcl-1 by silencing of Ku70 enhances antitumor efficacy of ABT-737 in lung cancer xenografts
Our findings show that depletion of Ku70 from human lung cancer H1299 cells using Ku70 shRNA not only downregulates Mcl-1 but also sensitizes cells to Bcl2 inhibitor (that is, ABT-737) (Figures 2 and 8a). To further test whether reduction of Mcl-1 by silencing of Ku70 enhances the antitumor efficacy of ABT-737 in an animal model, we generated lung cancer xenografts using H1299 cells expressing Ku70 shRNA or control shRNA. Mice with xenografts were treated with ABT-737 (50 mg/kg/day) for 12 days. Results indicated that silencing of Ku70 synergizes with ABT-737 in suppression of lung cancer growth (Figure 8b). To assess whether ABT-737 induced tumor growth regression via apoptosis in vivo, representative samples from harvested tumor tissues were analyzed by immunohistochemistry (IHC) for active caspase 3 as described.24 Silencing of Ku70 significantly enhanced apoptosis in tumor tissues after ABT-737 treatment (Figure 8c).
Figure 8.
Ku70 depletion sensitizes lung cancer to ABT-737 in vivo. (a) Cell growth in H1299 cells expressing Ku70 shRNA or control shRNA treated with ABT-737. (b) Tumor growth of Ku70 shRNA- or control shRNA-H1299 xenografts treated with ABT-737 (50 mg/kg/day) for 12 days. (c) Active caspase 3 was analyzed in tumor tissues at the end of experiments by IHC staining and quantified as described in ‘Materials and Methods'
Discussion
Protein ubiquitination is a reversible process involving both ubiquitin-ligating enzymes (E3 ligases) and DUBs. DUBs cleave the isopeptide bond at the C terminus of ubiquitin.36 Ubiquitin is a highly conserved 76-amino-acid protein that undergoes covalent attachment via isopeptide linkage to lysine residues in target proteins. The ubiquitin conjugation system is composed of three classes of enzymes, a Ub-activating enzyme (E1) that hydrolyzes ATP and adenylates ubiquitin, a Ub-conjugating enzyme (E2) that catalyzes the covalent attachment of ubiquitin to lysine residues in target proteins and a Ub-ligating enzyme (E3) that confers substrate specificity. Ubiquitin has seven internal lysine residues, any of which can be polyubiquitinated to create ubiquitin chains of distinct linkages. A ubiquitin code exists whereby proteins tagged with K6-, K11-, K27-, K29- and K48-linked polyubiquitin chains are targeted for proteasome degradation, whereas K63-linked chains are preferentially involved in the lysosomal pathway.37 Mcl-1 differs from other Bcl2 family members in its high turnover rate. Mcl-1 contains a PEST sequence that can act as a transferrable degradation signal.6, 38 The expression level of Mcl-1 is tightly regulated by ubiquitinating (that is, Mule, FBW7 and β-TrCP)9, 10, 39 and deubiquitinating (USP9X) enzymes.3, 40 Here we found that in the absence of Ku70, Mcl-1 is heavily modified by ubiquitination in association with a shorter half-life and decreased protein level in various types of cells (Figures 1, 2, 3 and Supplementary Figure S1). Conversely, overexpression of Ku70 enhances Mcl-1 level by reduction of ubiquitination (Figures 4, 6 and Supplementary Figure S2). These findings suggest that Ku70, in addition to its DSB repair activity, may also function as a deubiquitinase to remove K48-linked polyubiquitin chains from the ubiquitinated Mcl-1. Our in vitro deubiquitination assay further demonstrated that purified Ku70 not only directly deubiquitinated Mcl-1 protein but also hydrolyzed polyubiquitin chains into monoubiquitin units (Figure 7 and Supplementary Figure S5). These findings strongly support that Ku70 possesses intrinsic deubiquitination enzymatic activity that is responsible for deubiquitination and stabilization of Mcl-1. In addition to Mcl-1, a slight decrease in Bax expression was observed in Ku70−/− MEF cells (Figure 1a), which supports a previous report showing that Ku70 may also function as Bax deubiquitinase.20 However, the effect of Ku70 on Bax is relatively weak in our system.
The Ku70 protein contains several functional domains, including the Ku86-binding sites (aa 1–115 and aa 378–482), DNA-binding domain (aa 277–341) and the C-terminal portion (aa 536–609).16, 17 The N-terminal portion (aa 1–535) is important for Ku70 to heterodimerize with Ku86 and bind to DNA to mediate its DSB repair capacity. The C terminus of Ku70 is mainly responsible for its antiapoptotic function,20 through mechanism(s) that were not fully understood. Our studies here demonstrated that Ku70 interacts with Mcl-1 via its C terminus (aa 536–609) (Figure 5). Functionally, the C terminus of Ku70 is required and sufficient not only for deubiquitination and stabilization of Mcl-1 but also for its antiapoptotic activity (Figures 6 and 7). In contrast, the N-terminal portion (aa 1–535) of Ku70, which contains the DSB repair capacity, has no deubiquitination enzymatic activity and fails to deubiquitinate Mcl-1 protein in vitro and in cells (Figures 6 and 7). On the basis of these findings, we propose that the antiapoptotic activity of Ku70 may occur, at least in part, through deubiquitination and stabilization of Mcl-1, which is independent of its DNA repair function.
Functionally, knockout of Ku70 increases cellular sensitivity to apoptotic stress (that is, staurosporine) (Figure 1), and overexpression of Ku70 suppresses staurosporine-induced apoptosis (Figure 4). As staurosporine is a pan-kinase inhibitor and it does not induce apoptosis through double-strand DNA breakage, this further supports the notion that the antiapoptotic activity of Ku70 is independent of its known activity in double-strand DNA break repair. ABT-737 is a known BH3-mimetic Bcl2 inhibitor.24 However, overexpression of Mcl-1 may mediate cancer cell resistance to ABT-737.26 Our data reveal that downregulation of Mcl-1 by disrupting Ku70 via RNAi significantly enhances sensitivity of human lung cancer cells to ABT-737 treatment in vitro and in lung cancer xenografts (Figures 2 and 8), indicating that inhibition of Mcl-1 deubiquitination can enhance the anti-cancer efficacy of the BH3-mimetic ABT-737.
In summary, our findings demonstrate that the absence of Ku70 leads to decreased levels of Mcl-1, mediated by increased polyubiquitination and degradation. Ku70 protein has DUB enzymatic activity that stabilizes Mcl-1 by directly removing degradative Lys 48-linked polyubiquitin chains. Ku70 directly interacts with Mcl-1 via its C-terminal tail, which is essential for Ku70 to deubiquitinate Mcl-1 and suppress apoptosis. Thus, inhibition of Mcl-1 deubiquitination by disrupting Ku70 DUB enzymatic activity should represent a new strategy for cancer therapy.
Materials and Methods
Materials
Ku70, Ku86, Mcl-1 and HA antibodies as well as ABT-737 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Bcl-XL and Bak antibodies were purchased from Epitomics (Burlingame, CA, USA). Mule antibody was obtained from Bethyl Laboratories Inc. (Montgomery, TX, USA). USP9X antibody was purchased from Abnova (Walnut, CA, USA). Bcl2 antibody was obtained from Calbiochem (Darmstadt, Germany). The pCMV-Flag-hMcl-1 plasmid was obtained from Addgene (Cambridge, MA, USA). Ku70 shRNA and USP9 shRNA were purchased from Santa Cruz Biotechnology. MG132 was purchased from EMD chemicals, Inc. (Darmstadt, Germany). Flag-tagged Ku70 WT and its mutants (that is, aa 1–535, aa 496–609 and aa 536–609) in pCMV2B vector were obtained from Dr. Shigemi Matsuyama (Case Western Reserve University). K48- or K63-linked polyubiquitin chains were purchased from Boston Biochem (Boston, MA, USA). Anti-ubiquitin-, anti-K48- or anti-K63-specific ubiquitin antibodies were obtained from Cell Signaling (Danvers, MA, USA). All of the reagents used were obtained from commercial sources unless otherwise stated.
Cell lines, cell culture and plasmid transfection
WT and Ku70−/− MEF cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% FBS and 4 mM L-glutamine. HCT116 and human lung cancer H1299 cells were maintained in RPMI-1640 with 10% serum. HEK293T cells were grown in DMEM with 10% FBS. Transfections of HA-tagged ubiquitin, Flag-Mcl-1 and Flag-Ku70 plasmids were performed using NanoJuice transfection Kit (Novagen, San Diego, CA, USA) according to the manufacturer's instructions.41, 42, 43
Preparation of cell lysates
Cells were washed with 1 × PBS and resuspended in ice-cold EBC buffer (0.5% Nonidet P-40 (Sigma, St. Louis, MO, USA), 50 mM Tris, pH 7.6, 120 mM NaCl, 1 mM EDTA, and 1 mM β-mercaptoethanol) containing protease inhibitor mixture. Cells were lysed by sonication and centrifuged at 14 000 × g for 10 min at 4 °C. The resulting supernatant was collected as the total cell lysate. Western blot was performed by loading 50 μg of protein per lane on an 8–12% SDS-PAGE, followed by protein transfer to nitrocellulose membrane for analysis of specific protein(s).44
Subcellular fractionation
Subcellular fractionation was performed to isolate HM, LM, cytosol and nuclear fractions as we described previously.32 Briefly, H1299 cells (2 × 107) were washed once with cold 1 × PBS and resuspended in isotonic mitochondrial buffer (210 mM mannitol, 70 mM sucrose, 1 mM EGTA, 10 mM Hepes, pH 7.5) containing protease inhibitor mixture. The resuspended cells were homogenized with a polytron homogenizer (Fisher Scientific, Pittsburgh, PA, USA) operating for four bursts of 10 s each at a setting of five and then centrifuged at 2000 × g for 3 min to pellet the nuclei. The supernatant was centrifuged at 13 000 × g for 10 min to pellet mitochondria. The supernatant was further centrifuged at 150 000 × g to pellet LMs. The resulting supernatant contained the cytosolic fraction. The mitochondria was washed with mitochondrial buffer twice, resuspended with 1% Nonidet P-40 lysis buffer, rocked for 60 min and then centrifuged at 17 530 × g for 10 min at 4 °C. The supernatant containing mitochondrial proteins was collected. For nuclear fractionation, the cells were washed with 1 × PBS and suspended in 2 ml of buffer A (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.03% Nonidet P-40 with fresh protease inhibitor mixture). The samples were incubated on ice until more than 95% of cells could be stained by trypan blue. The samples were then centrifuged at 500 × g at 4 °C for 5 min. The resulting nuclear pellet was washed with buffer B (50 mM NaCl, 10 mM Hepes, pH 8.0, 25% glycerol, 0.1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine) and then resuspended in 150 μl of buffer C (350 mM NaCl, 10 mM Hepes, pH 8.0, 25% glycerol, 0.1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine) and rocked at 4 °C for 30 min. After centrifugation (14 000 × g) at 4 °C, the supernatant (nuclear fraction) was collected. Protein (50 μg) from each fraction was subjected to SDS-PAGE and analyzed by western blot using antibody to Ku70 or Mcl-1. The purity of fractions was confirmed by assessing the localization of fraction-specific proteins including prohibitin (a mitochondrial Marker)33 and PCNA (a nuclear marker).34
Silencing of Ku70 or USP9X
Ku70 shRNA, USP9X shRNA and control shRNA were obtained from Santa Cruz Biotechnology. For pseudovirus production, Ku70 shRNA, USP9X shRNA or control shRNA were cotransfected into 293FT cells with a lentivector packaging plasmid mixture (System Biosciences, Mountain View, CA, USA) using the NanoJuice transfection kit (EMD Chemical, Inc.) as described.45 After 48 h, the virus-containing media were harvested by centrifugation at 20 000 × g. Cells were infected with the virus-containing media in the presence of polybrene (8 μg/ml) for 24 h. Stable positive clones were selected using 1 μg/ml puromycin.
In vitro deubiquitination
Flag-Mcl-1, Flag-Ku70 or Flag-USP9X in transfected HEK293T cells were purified with anti-Flag M2 affinity gel in RIPA buffer. High-salt (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% glycerol) and low-salt (20 mM Tris, pH 7.4, 300 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.1% NP-40) washes preceded elution with 500 μg ml−1 3 × Flag peptide. Purified ubiquitinated Mcl-1 protein was incubated with purified Ku70, USP9X or BSA control in 50 mM HEPES, pH 7.5, 10 mM 2-mercaptoethanol and 0.5 mM EDTA at 30 °C for 30 min. Mcl-1 ubiquitination was analyzed by western blot. For direct in vitro deubiquitylation activity assay, 40 ng of K48-linked tetraubiquitin chains was incubated with purified Ku70/80 complex or BSA (negative control) at 30 °C for 1 or 2 h as described.20 The reaction products were separated by SDS/PAGE, and the hydrolysis of ubiquitin chains into monoubiquitin units was detected by silver staining.
Measurement of apoptotic cells
Apoptotic cells were detected using an ApoAlert Annexin-V kit from Clontech (Mountain View, CA, USA) according to the manufacturer's instructions. The percentage of apoptotic cells was determined by fluorescence-activated cell sorter analysis.
Lung cancer xenografts and treatments
Animal experiments were approved by the Institutional Animal Care and Use Committee of Emory University. Six-week-old Nu/Nu nude mice were purchased from Harlan (Indianapolis, IN, USA) and housed under pathogen-free conditions in microisolator cages. Xenografts were raised by injecting 5 × 106 H1299 cells expressing control shRNA or Ku70 shRNA in a balanced salt solution into subcutaneous tissue at the flank region of nude mice. The tumors were allowed to grow to an average volume of 250 mm3 before initiation of therapy as described.24 H1299 xenografts were randomized into four groups (n=8 per group) as follows: (1) control shRNA; (2) Ku70 shRNA; (3) control shRNA+ABT-737 (50 mg/kg/day, i.p.); (4) Ku70 shRNA+ABT-737 (50 mg/kg/day, i.p.). Tumor volume (V) was measured by caliper measurements once every 2 days and calculated with the formula: V=L × W2/2 (L: length; W: width) as described.46 After 12 consecutive days of treatment, all mice were killed by inhaled CO2. The tumors were then removed, weighed and fixed with formalin for IHC.
IHC staining
Tumors were harvested, fixed in formalin and embedded in paraffin. Representative sections from paraffin-embedded tumor tissues were analyzed by IHC staining using an active caspase 3-specific antibody. Active caspase-positive cells in tumor tissues were scored at × 400 magnification. The average number of positive cells per 0.0625 mm2 area was determined from three separate fields in each of three independent tumor samples as described.24
Statistical analysis
Significant differences between two groups were analyzed using two-sided unpaired Student's t-test and P-value <0.05 was considered statistically significant.
Acknowledgments
We thank Anthea Hammond for editing of the manuscript. This work was supported by NCI, National Institutes of Health grants R01CA112183 and R01CA136534, and by Flight Attendant Medical Research Institute Clinical Innovator Award.
Glossary
- BH
Bcl2 homology
- DSB
DNA double-strand break
- HM
heavy membrane
- LH
light membrane
- ER
endoplasmic reticulum
- PCNA
proliferating cell nuclear antigen
- DUBs
deubiquitinating enzymes
- MEF
mouse embryonic fibroblast
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Zhao J, Xin M, Wang T, Zhang Y, Deng X. Nicotine enhances the antiapoptotic function of Mcl-1 through phosphorylation. Mol Cancer Res. 2009;7:1954–1961. doi: 10.1158/1541-7786.MCR-09-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgul C. Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell Mol Life Sci. 2009;66:1326–1336. doi: 10.1007/s00018-008-8637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- Liao M, Zhao J, Wang T, Duan J, Zhang Y, Deng X. Role of bile salt in regulating Mcl-1 phosphorylation and chemoresistance in hepatocellular carcinoma cells. Mol Cancer. 2011;10:44. doi: 10.1186/1476-4598-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Levy NB, Xie H, Qian L, Lee CY, Gascoyne RD, et al. MCL1 transgenic mice exhibit a high incidence of B-cell lymphoma manifested as a spectrum of histologic subtypes. Blood. 2001;97:3902–3909. doi: 10.1182/blood.v97.12.3902. [DOI] [PubMed] [Google Scholar]
- Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23:5301–5315. doi: 10.1038/sj.onc.1207692. [DOI] [PubMed] [Google Scholar]
- Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Hu HY. Editorial: protein ubiquitination and deubiquitination. Curr Protein Pept Sci. 2012;13:413. doi: 10.2174/138920312802430626. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Wang Q, Gao F, May WS, Zhang Y, Flagg T, Deng X. Bcl2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway. Mol Cell. 2008;29:488–498. doi: 10.1016/j.molcel.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Gao F, Wang T, Flagg T, Deng X. A nonhomologous end-joining pathway is required for protein phosphatase 2A promotion of DNA double-strand break repair. Neoplasia. 2009;11:1012–1021. doi: 10.1593/neo.09720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Owonikoko TK, Sun SY, Ramalingam SS, Doetsch PW, Xiao ZQ, et al. c-Myc suppression of DNA double-strand break repair. Neoplasia. 2012;14:1190–1202. doi: 10.1593/neo.121258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dong X, Myung K, Hendrickson EA, Reeves WH. Identification of two domains of the p70 Ku protein mediating dimerization with p80 and DNA binding. J Biol Chem. 1998;273:842–848. doi: 10.1074/jbc.273.2.842. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R, Esposito D, Sankar A, Maman JD, Hinks JA, Pearl LH, et al. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR) J Mol Biol. 2004;335:573–582. doi: 10.1016/j.jmb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- Amsel AD, Rathaus M, Kronman N, Cohen HY. Regulation of the proapoptotic factor Bax by Ku70-dependent deubiquitylation. Proc Natl Acad Sci USA. 2008;105:5117–5122. doi: 10.1073/pnas.0706700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Ye R, Veillette CJ, Lees-Miller SP. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry. 1999;38:1819–1828. doi: 10.1021/bi982584b. [DOI] [PubMed] [Google Scholar]
- Gama V, Gomez JA, Mayo LD, Jackson MW, Danielpour D, Song K, et al. Hdm2 is a ubiquitin ligase of Ku70-Akt promotes cell survival by inhibiting Hdm2-dependent Ku70 destabilization. Cell Death Differ. 2009;16:758–769. doi: 10.1038/cdd.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Peng Z. MG132, a proteasome inhibitor, induces apoptosis in tumor cells. Asia Pac J Clin Oncol. 2013;9:6–11. doi: 10.1111/j.1743-7563.2012.01535.x. [DOI] [PubMed] [Google Scholar]
- Arnold HK, Sears RC. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol. 2006;26:2832–2844. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejda B, Kidd M, Kazberouk A, Lawrence B, Pfragner R, Modlin IM. Limitations in small intestinal neuroendocrine tumor therapy by mTor kinase inhibition reflect growth factor-mediated PI3K feedback loop activation via ERK1/2 and AKT. Cancer. 2011;117:4141–4154. doi: 10.1002/cncr.26011. [DOI] [PubMed] [Google Scholar]
- Deng X, Ruvolo P, Carr B, May WS., Jr Survival function of ERK1/2 as IL-3-activated, staurosporine-resistant Bcl2 kinases. Proc Natl Acad Sci USA. 2000;97:1578–1583. doi: 10.1073/pnas.97.4.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Ito T, Carr B, Mumby M, May WS., Jr Reversible phosphorylation of Bcl2 following interleukin 3 or bryostatin 1 is mediated by direct interaction with protein phosphatase 2A. J Biol Chem. 1998;273:34157–34163. doi: 10.1074/jbc.273.51.34157. [DOI] [PubMed] [Google Scholar]
- Jin Z, Gao F, Flagg T, Deng X. Tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone promotes functional cooperation of Bcl2 and c-Myc through phosphorylation in regulating cell survival and proliferation. J Biol Chem. 2004;279:40209–40219. doi: 10.1074/jbc.M404056200. [DOI] [PubMed] [Google Scholar]
- Ikonen E, Fiedler K, Parton RG, Simons K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 1995;358:273–277. doi: 10.1016/0014-5793(94)01444-6. [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Gomez-Bougie P, Menoret E, Juin P, Dousset C, Pellat-Deceunynck C, Amiot M. Noxa controls Mule-dependent Mcl-1 ubiquitination through the regulation of the Mcl-1/USP9X interaction. Biochem Biophys Res Commun. 2011;413:460–464. doi: 10.1016/j.bbrc.2011.08.118. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- D'Arcy P, Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol. 2012;44:1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Yaglom J, Linskens MH, Sadis S, Rubin DM, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivigno D, Essmann F, Huber SM, Rudner J. Deubiquitinase USP9x confers radioresistance through stabilization of Mcl-1. Neoplasia. 2012;14:893–904. doi: 10.1593/neo.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Liu X, Yue P, Benbrook DM, Berlin KD, Khuri FR, et al. Involvement of c-FLIP and survivin down-regulation in flexible heteroarotinoid-induced apoptosis and enhancement of TRAIL-initiated apoptosis in lung cancer cells. Mol Cancer Ther. 2008;7:3556–3565. doi: 10.1158/1535-7163.MCT-08-0648. [DOI] [PubMed] [Google Scholar]
- Liu X, Yue P, Schonthal AH, Khuri FR, Sun SY. Cellular FLICE-inhibitory protein down-regulation contributes to celecoxib-induced apoptosis in human lung cancer cells. Cancer Res. 2006;66:11115–11119. doi: 10.1158/0008-5472.CAN-06-2471. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sun SY, Owonikoko TK, Sica GL, Curran WJ, Khuri FR, et al. Rapamycin induces Bad phosphorylation in association with its resistance to human lung cancer cells. Mol Cancer Ther. 2012;11:45–56. doi: 10.1158/1535-7163.MCT-11-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Deng X, Carr B, May WS. Bcl-2 phosphorylation required for anti-apoptosis function. J Biol Chem. 1997;272:11671–11673. doi: 10.1074/jbc.272.18.11671. [DOI] [PubMed] [Google Scholar]
- Huang S, Okumura K, Sinicrope FA. BH3 mimetic obatoclax enhances TRAIL-mediated apoptosis in human pancreatic cancer cells. Clin Cancer Res. 2009;15:150–159. doi: 10.1158/1078-0432.CCR-08-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, et al. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res. 2011;17:710–720. doi: 10.1158/1078-0432.CCR-10-0331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.