Abstract
Lignin biosynthesis occurs via radical coupling of guaiacyl and syringyl hydroxycinnamyl alcohol monomers (i.e., “monolignols”) through chemical condensation with the growing lignin polymer. With each chain-extension step, monolignols invariably couple at their β-positions, generating chiral centers. Here, we report on activities of bacterial glutathione-S-transferase (GST) enzymes that cleave β-aryl ether bonds in lignin dimers that are composed of different monomeric units. Our data reveal that these sequence-related enzymes from Novosphingobium sp. strain PP1Y, Novosphingobium aromaticivorans strain DSM12444, and Sphingobium sp. strain SYK-6 have conserved functions as β-etherases, catalyzing cleavage of each of the four dimeric α-keto-β-aryl ether-linked substrates (i.e., guaiacyl-β-guaiacyl, guaiacyl-β-syringyl, syringyl-β-guaiacyl, and syringyl-β-syringyl). Although each β-etherase cleaves β-guaiacyl and β-syringyl substrates, we have found that each is stereospecific for a given β-enantiomer in a racemic substrate; LigE and LigP β-etherase homologues exhibited stereospecificity toward β(R)-enantiomers whereas LigF and its homologues exhibited β(S)-stereospecificity. Given the diversity of lignin’s monomeric units and the racemic nature of lignin polymers, we propose that bacterial catabolic pathways have overcome the existence of diverse lignin-derived substrates in nature by evolving multiple enzymes with broad substrate specificities. Thus, each bacterial β-etherase is able to cleave β-guaiacyl and β-syringyl ether-linked compounds while retaining either β(R)- or β(S)-stereospecificity.
Introduction
Lignin, a major component of plant cell walls, is a recalcitrant polymer composed of monomeric units (i.e., components derived from guaiacyl and syringyl monomers),1−3 providing plants with both pathogenic resistance and structural integrity.4,5 The β-O-4′-ether (hereafter termed β-ether) is the most prevalent type of intermolecular bond through which the guaiacyl (monomethoxylated) and syringyl (dimethoxylated) aromatic units are linked.6 Thus, the development of methodologies for β-ether cleavage and depolymerization of the lignin backbone may reveal novel aspects of catalysis and lead to lignin-derived products of high economic value.7−10
The formation of lignin polymers by radical coupling of monomeric units generates a racemic product containing both β(R)- and β(S)-ether bonds. Here, we report on enzyme activity with a set of newly analyzed substrates for a group of sequence-related bacterial β-etherases that are glutathione-S-transferase (GST) superfamily member enzymes, each of which catalyzes cleavage of β-ether bonds that are characteristically found in lignin polymers. Specifically, we reveal that each of these enzymes has activity with guaiacyl- and syringyl-containing substrates and that each enzyme exhibits stereospecifity for cleavage of either β(R)- or β(S)-ether-linked enantiomers.
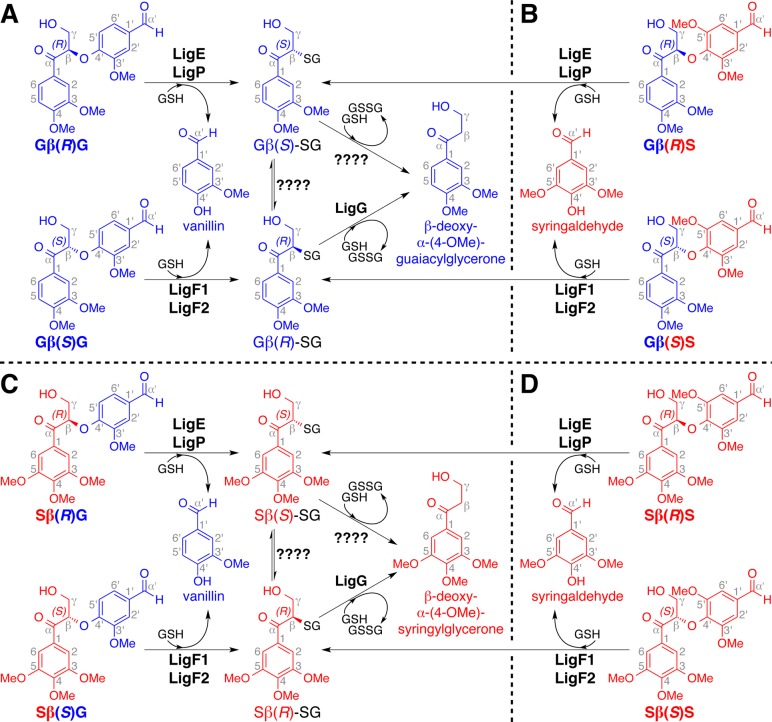
The bacterium Sphingobium sp. strain SYK-6 possesses several metabolic enzymes that mediate metabolism of lignin-derived compounds.11 “Lig enzymes” that act in the proposed β-etherase pathway enable this organism to derive monoaromatic growth substrates from β-ether-linked α-keto diguaiacyl compounds such as α-(4-O-Me)-guaiacylglycerone-β-(1′-formyl)-guaiacyl ether (GβG). The β(R)- and β(S)-enantiomers of GβG (Gβ(R)G and Gβ(S)G, Figure 1A) arise as β-etherase pathway intermediates from the activities of nicotinamide adenine dinucleotide (NAD)-dependent Lig dehydrogenases, which oxidize the corresponding benzylic alcohols to α-ketones.12,13 It has been shown that, using glutathione (GSH) and GβG as cosubstrates, the β-etherases (LigE, LigP, and LigF1) cleave this aromatic dimer,14−17 producing vanillin and a GSH-conjugated guaiacyl monomer (Gβ-SG) as reaction products.18 Gβ-SG is further degraded by LigG (and other enzymes that have not yet been identified), yielding glutathione disulfide (GSSG) and the monoaromatic compound β-deoxy-α-(4-O-Me)-guaiacylglycerone (Figure 1A).15
Figure 1.
β-Etherase pathway-mediated conversion of β-enantiomers of substrates GβG, GβS, SβG, and SβS, in which vanillin and syringaldehyde are formed from cleavage of β-guaiacyl (in panels A and C) and β-syringyl (in panels B and D) ether-linked compounds. Compound names are displayed below each structure, and 3-methoxylated (i.e., guaiacyl) and 3,5-dimethoxylated (i.e., syringyl) units are shown in blue and red. Catabolism of (A) Gβ(R)G and Gβ(S)G, as well as (B) Gβ(R)S and Gβ(S)S, yields aromatic monomers Gβ(S)-SG, Gβ(R)-SG, and β-deoxy-α-(4-O-Me)-guaiacylglycerone as metabolic intermediates. Catabolism of (C) Sβ(R)G and Sβ(S)G, as well as (D) Sβ(R)S and Sβ(S)S, yields aromatic monomers Sβ(S)-SG, Sβ(R)-SG, and β-deoxy-α-(4-O-Me)-syringylglycerone as metabolic intermediates.
The racemic nature of the lignin backbone19−22 and the existence of both β(R)- and β(S)-configurations in lignin necessitate the ability to degrade both Gβ(R)G and Gβ(S)G enantiomers (Figure 1A). In Sphingobium sp. strain SYK-6, this is accomplished via the activities of multiple β-etherases with complementary stereochemical properties.12,15Sphingobium sp. strain SYK-6 LigE and LigP catalyze stereospecific cleavage of Gβ(R)G, and LigF1 exhibits stereospecificity for the Gβ(S)G enantiomer. In this organism, β-ether cleavage is coupled to GSH-conjugation, inversion of β-chirality, and stereoselective formation of Gβ(S)-SG (LigE and LigP) and Gβ(R)-SG (LigF1).18
The existence of guaiacyl and syringyl units in the lignin polymers of all land plants other than softwoods also necessitates the existence of enzymes that will cleave β-ether-linked units of different subunit composition (i.e., guaiacyl-β-guaiacyl (GβG), guaiacyl-β-syringyl (GβS), syringyl-β-guaiacyl (SβG), and syringyl-β-syringyl (SβS); Figure 1). Although the activities of Sphingobium sp. strain SYK-6 β-etherases have been shown to contribute to the stereospecific and stereoselective degradation of model compounds containing guaiacyl units, such as Gβ(R)G and Gβ(S)G,15,18,23,24 the role served by Lig enzymes in the catabolism of native lignin-derived compounds is largely unknown because (a) investigation of enzymes that might be involved in this pathway has been limited to those encoded in the genome of Sphingobium sp. strain SYK-6 and (b) the activities of β-etherase pathway enzymes have not been tested with the range of β-ether-containing oligomers composed of guaiacyl and syringyl subunits that are typically found in lignin (Figure 1B–D).
In this work, we reveal the ability of β-etherases from Sphingobium sp. strain SYK-6 (SsLigE, SsLigP, and SsLigF1) to cleave model dimeric lignin compounds containing GβS, SβG, and SβS β-ether linkages, in addition to the previously reported GβG substrate.15,18 Further, we identify several additional sequence-related proteins with β-etherase activity from Novosphingobium aromaticivorans (N. aromaticivorans) DSM12444 (NaLigE, NaLigF1, and NaLigF2) and Novosphingobium sp. strain PP1Y (NsLigE). We demonstrate that each enzyme catalyzes cleavage of all four combinations of β-ether-linked substrates, GβG (Figure 1A), GβS (Figure 1B), SβG (Figure 1C), and SβS (Figure 1D), where each LigE/LigP β-etherase homologue has the conserved function of degrading β(R)-enantiomers whereas each LigF1/LigF2 β-etherase homologue exhibits stereospecificity for the β(S)-enantiomers. Thus, we show that several bacteria possess β-etherases that have a previously unreported ability to cleave lignin dimers containing GβG, GβS, SβG, and SβS β-ether linkages. Our results also reveal that each of these enzymes exhibits similar stereospecifity to that previously described for the enzymes from Sphingobium sp. strain SYK-6.15,18,25 These observations reveal important features of a conserved class of bacterial enzymes that have utility in the conversion of lignin during either plant biomass processing or the potential production of valuable compounds from this abundant polymer.
Experimental Section
Gene Cloning and Enzyme Purification
DNA manipulation and transformation into Escherichia coli (E. coli) were carried out according to standard methods26 and as previously described (see Supporting Information (SI) for details).18 DNA primers and restriction enzymes were obtained from Integrated DNA Technologies (Coralville, IA, USA) and New England Biolabs (Ipswich, MA, USA). Plasmids containing genes encoding SsLigE (locus tag SLG_08660), SsLigP (SLG_32600), and SsLigF1 (SLG_08650) from Sphingobium sp. strain SYK-6, the gene encoding potential Lig enzyme NsLigE (PP1Y_AT11664) from Novosphingobium sp. strain PP1Y, and the Vibrio cholarae (V. cholarae) rtxA gene (Vch1786_I0951) were obtained from Invitrogen (Carlsbad, CA, USA) and were codon-optimized for expression in E. coli. Genes encoding potential Lig enzymes NaLigE (Saro_2405), NaLigF1 (Saro_2091), NaLigF2 (Saro_2865), and RpHypGST (RPA4340) were cloned from genomic DNA from N. aromaticivorans strain DSM12444 or Rhodopseudomonas palustris (R. palustris) strain CGA009, respectively.
Protein Expression and Purification
Each N-terminal (encoded on vector pVP302 K) and C-terminal (encoded on vector pVP202 K) octa-histidine affinity tagged (NHis8 and CHis8, respectively) enzyme was purified using nickel-nitrilotriacetic acid resin (Ni-NTA) affinity chromatography. NHis8 tags were cleaved using Tev protease,27 and CHis8 tags were cleaved by induction of the fused V. cholarae RtxA protease.28−30 A second round of Ni-NTA affinity chromatography removed cleaved tags from enzyme preparations that were subsequently purified by size-exclusion chromatography,18 and evaluated by SDS-PAGE (Figure S1 of the Supporting Information).
NMR Spectroscopy
1H and 13C NMR spectra were recorded on a Bruker Biospin (Billerica, MA, USA) AVANCE 700 MHz spectrometer fitted with a cryogenically cooled 5 mm TXI gradient probe with inverse geometry (proton coils closest to the sample). See Supporting Information for additional details.
Syntheses of β-Ether-Linked Dimeric Model Compounds
Syntheses of β-Brominated Intermediates
β-bromination of commercially available α-(4-O-Me)-guaiacylethanone produced crystalline β-bromo-α-(4-O-Me)-guaiacylethanone (for additional details, see SI and Figure S2). Similarly, commercially available α-(4-O-Me)-syringylethanone was brominated, yielding crystalline β-bromo-α-(4-O-Me)-syringylethanone.
Syntheses of Achiral β-Ether-Linked Intermediates
Four achiral β-ether-linked compounds were synthesized using the preceding β-bromides as starting materials. The phenolate ion of vanillin was used for SN2 displacement of the β-bromo-α-(4-O-Me)-guaiacylethanone bromide, yielding α-(4-O-Me)-guaiacylethanone-β-(1′-formyl)-guaiacyl ether (Figure S2A of the Supporting Information). Similarly, β-bromo-α-(4-O-Me)-guaiacylethanone and syringaldehyde were used to synthesize α-(4-O-Me)-guaiacylethanone-β-(1′-formyl)-syringyl ether (SI Figure S2B). The vanillin phenolate ion was used to displace the β-bromo-α-(4-O-Me)-syringylethanone bromide, yielding α-(4-O-Me)-syringylethanone-β-(1′-formyl)-guaiacyl ether (SI Figure S2C). Using β-bromo-α-(4-O-Me)-syringylethanone and syringaldehyde as starting materials under similar reaction conditions yielded α-(4-O-Me)-syringylethanone-β-(1′-formyl)-syringyl ether (SI Figure S2D).
Syntheses of Racemic β-Etherase Substrates
In parallel, each of the four racemic β-ether-linked intermediates was condensed with formaldehyde,31,32 yielding racemic β-aryl ether-linked dimeric model compounds that served as substrates for β-etherase enzyme assays. Accordingly, α-(4-O-Me)-guaiacylglycerone-β-(1′-formyl)-guaiacyl ether (GβG) was derived from α-(4-O-Me)-guaiacylethanone-β-(1′-formyl)-guaiacyl ether (SI Figure S2A), GβS was derived from α-(4-O-Me)-guaiacylethanone-β-(1′-formyl)-syringyl ether (SI Figure S2B), α-(4-O-Me)-syringylglycerone-β-(1′-formyl)-guaiacyl ether (SβG) was derived from α-(4-O-Me)-syringylethanone-β-(1′-formyl)-guaiacyl ether (SI Figure S2C), and SβG was derived from α-(4-O-Me)-syringylethanone-β-(1′-formyl)-syringyl ether (SIFigure S2D). Additional details on the syntheses can be found in the Supporting Information.
β-Etherase Enzyme Assays
Parallel 5 mL β-etherase reactions were conducted (assay buffer: 10 mM HEPES, 60 mM NaCl, 100 μM TCEP, 5% acetone, 2 mM GSH, pH 7.5) in which individual proteins NaLigE, NsLigE, SsLigE, SsLigP, NaLigF1, NaLigF2, or SsLigF1 (0.25 mg mL–1) were individually incubated with GSH and one of the racemic β-ethers GβG, GβS, SβG, or SβS (1.0 mM) as cosubstrates. Aliquots (2.5 mL) were collected prior to protein addition (0 h sample), and again after 1 h of incubation with each of the putative β-etherases. Each 2.5 mL sample was extracted six times with ethyl acetate, partitioning residual β-ether-linked enantiomers, and aromatic aldehydes to the organic phase and glutathione-conjugated products, Gβ-SG and Sβ-SG, to the aqueous layer. Ethyl acetate was then dried in vacuo, yielding residues containing the hydrophobic reaction products and residual substrate enantiomers. Residues from each sample were dissolved in 0.1 mL ethanol and analyzed by chiral chromatography.
Chiral Chromatography
Analytical Separation of GβG and GβS Enantiomers
Analyses of GβG- and GβS-derived β-etherase reaction products and residual substrates were conducted via chiral chromatographic separation using a Diacel Chemical Industries CHIRALPAK AD-H column (4.6 mm × 250 mm). A mobile phase of 3/2 hexane/ethanol was used at a flow rate of 1.0 mL min.–1 Vanillin, Gβ(S)G, and Gβ(R)G were detected in enzymatic reaction samples when racem-GβG was used as the substrate, with each eluting after tR = 4.8, 16.6, and 20.2 min, respectively. Absolute configurations of GβG enantiomers were determined previously.33 Syringaldehyde, Gβ(S)S, and Gβ(R)S were detected in reaction samples when racem-GβS was used as the substrate, eluting after tR = 6.3, 16.0, and 18.1 min.
Analytical Separation of SβG and SβS Enantiomers
Analyses of SβG- and SβS-derived β-etherase reaction products and residual substrate enantiomers were conducted via chiral chromatographic separation using a Diacel Chemical Industries CHIRALPAK AY-H column (10 mm × 250 mm). A mobile phase of 1/1 hexane/ethanol was used at a flow rate of 2.5 mL min.–1 Vanillin, Sβ(S)G, and Sβ(R)G were detected in reaction samples when racem-SβG was used as the substrate, eluting after tR = 6.9, 16.7, and 19.5 min, respectively. Syringaldehyde, Sβ(R)S, and Sβ(S)S were detected in reaction samples when racem-SβS was used as the substrate, eluting after tR = 8.0, 18.4, and 24.2 min, respectively.
Results
Identification of a Conserved Class of Putative β-Etherases
Given what is known about the β-etherase pathway in Sphingobium sp. strain SYK-6,12,15,18 we sought to investigate whether or not this pathway could be utilized for β-ether catabolism by other sphingomonads (bacteria from genera: Novosphingobium, Sphingobium, Sphingomonas, and Sphingopyxis),34 organisms that are often associated with the biodegradation of aromatic compounds in the environment.35−37 At the onset of this study, BLASTP searches38 querying the amino acid sequences of Lig enzymes from Sphingobium sp. strain SYK-639 revealed the existence of genes for putative LigE and LigF enzymes in two additional organisms for which full genome sequences were available: Novosphingobium sp. strain PP1Y and N. aromaticivorans strain DSM12444.40 Further, two homologues of each enzyme (LigE/LigP and LigF1/LigF2, respectively) were identified in strain SYK-6. We also found that both Novosphingobium strains encoded homologues of the NAD-dependent dehydrogenases that are essential for forming the α-ketones that undergo β-ether cleavage in strain SYK-6. In addition to the characterized β-etherase Lig enzymes (SsLigE, SsLigP, and SsLigF1), we expressed and purified homologous proteins encoded in the genomes of Novosphingobium sp. strain PP1Y (NsLigE) and N. aromaticivorans strain DSM12444 (NaLigE, NaLigF1, and NaLigF2). In sum, seven Lig homologues were tested for β-etherase activity with substrates GβG, GβS, SβG, and SβS; amino acid similarity to SsLigE or SsLigF1 is given in parentheses: SsLigE (100%), SsLigP (62%), NsLigE (78%), NaLigE (61%), SsLigF1 (100%), NaLigF1 (60%), and NaLigF2 (40%).
Cleavage of GβG Aromatic β-Ethers
Previously, it was reported that SsLigE-, SsLigP-, and SsLigF1-catalyzed stereospecific cleavage of a racemic diguaiacyl β-ether-linked substrate having a similar structure to that of GβG,15 but which contained an α′-H (rather than an α′-aldehyde) and a 4-OH (rather than a 4-OMe) (Figure 1A). We hypothesized that the Lig β-etherase homologues from each strain would catalyze the same reactions. In addition, we thought that it was likely, based on previously published data with diguaiacyl substrates,15,18 that neither the α′-aldehyde, nor the 4-O-Me moieties of GβG would inhibit β-etherase activity. Also, if each LigE/LigP enzyme had activity similar to that of the enzymes from Sphingobium sp. strain SYK-6, then they would each catalyze stereospecific degradation of Gβ(R)G whereas the Gβ(S)G enantiomer would be cleaved stereospecifically by the LigF1/LigF2 homologues. To test these predictions, we synthesized racem-GβG (Figure S2A) and performed β-etherase assays with each of the seven recombinant putative Lig enzymes using GSH and racem-GβG as cosubstrates. In comparing the chiral chromatogram of a sample containing substrates without protein (Figure 2A) with those representing materials from enzymatic assays that had been incubated with a homologue of LigE (Figure 2B–E), we found that, in each case, the LigE homologues released the expected product vanillin (tR = 4.8 min) and degraded the high-tR enantiomer (20.2 min) of GβG. Conversely, we found that each LigF homologue (Figure 2F–H) yielded vanillin as a reaction product while degrading the low-tR enantiomer (16.6 min). Given that previous work has shown that SsLigE and SsLigP, when incubated with substrate analogues of GβG, show β(R)-stereospecificity whereas SsLigF1 exhibits β(S)-stereospecificity,15,33 we propose that the high-tR compound degraded by the LigE homologues (Figure 2B–E) was Gβ(R)G and the low-tR compound degraded by the LigF homologues (Figure 2F–H) was Gβ(S)G. Further, analysis of the aqueous layers from these reactions18 confirmed that SsLigE- and SsLigP-catalyzed formation of Gβ(S)-SG whereas Gβ(R)-SG was formed as a product of SsLigF-catalyzed reactions, demonstrating that β-etherase catalysis involves formation of β-thioether compounds (Figure 1A). We therefore conclude that LigE homologues have the conserved function of β(R)-etherase activity with substrate GβG and, similarly, that LigF homologues in the sphingomonads each have the conserved function of catalyzing β(S)-ether cleavage.
Figure 2.
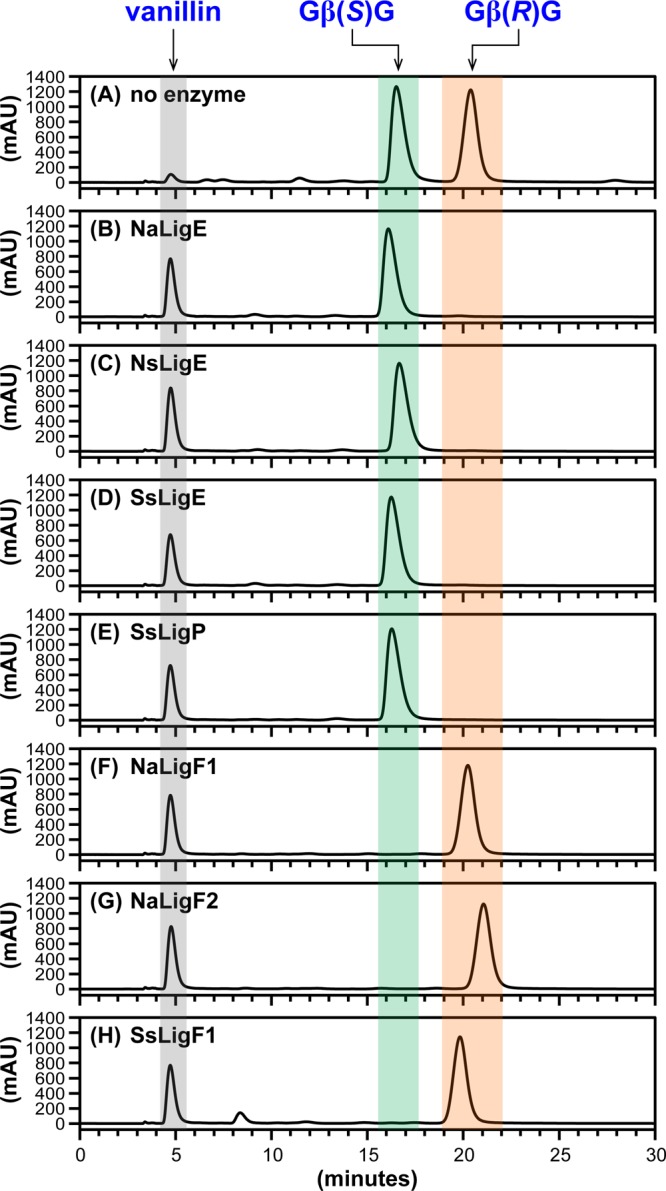
HPLC chromatographic traces (CHIRALPAK AD-H column, λ = 280 nm) of β-etherase enzyme assay samples from cosubstrates racem-GβG and glutathione. Chromatographic regions for vanillin (gray), Gβ(S)G (green), and Gβ(R)G (orange) peak elution times are highlighted by shading. (A) No enzyme added, 0 h sample, where the ratio of peak area integrals of Gβ(S)G to Gβ(R)G was ∼1:1. After 1 h incubation with either enzymatic catalyst: (B) NaLigE, (C) NsLigE, (D) SsLigE, (E) SsLigP, (F) NaLigF1, (G) NaLigF2, or (H) SsLigF1. Structures of vanillin, Gβ(S)G, and Gβ(R)G are shown in Figure 1A. Abbreviations: Na, N. aromaticivorans strain DSM12444; Ns, Novosphingobium sp. strain PP1Y; Ss, Sphingobium sp. strain SYK-6. See Experimental Section for details.
Cleavage of GβS Aromatic β-Ethers
To date, diguaiacyl compounds have been the only type of β-ether-linked lignin compound tested as a substrate of the Sphingobium sp. SYK-6 β-etherase pathway enzymes.15,25,33 We hypothesized that β-ether cleavage would occur with substrates containing additional methoxy groups on the aromatic rings, i.e., syringyl units. To test this hypothesis, we conducted additional β-etherase assays with each putative Lig enzyme using model β-ether compounds that contained either one or two syringyl units (Figure 1B–D) as substrates. The resulting data from assays in which GSH and racem-GβS (Figure 1B) were used as cosubstrates revealed that each β-etherase homologue catalyzed cleavage. Chiral chromatography of the reaction samples (Figure 3) indicated that each of the seven putative Lig enzymes produced the expected product (Figure 1B) syringaldehyde (tR = 6.3 min), with each LigE homologue (Figure 3B–E) degrading only the high-tR enantiomer (18.1 min) and each LigF homologue (Figure 3F–H) degrading the low-tR enantiomer (16.0 min). We propose that the GβG cleavage stereospecificity exhibited by each enzyme is also observed with the degradation of the GβS enantiomers. From this, we conclude that the LigE/LigP homologues exhibited β(R)-etherase activity, degrading Gβ(R)S (tR = 18.1 min), whereas each LigF-catalyzed β(S)-ether cleavage of Gβ(S)S (tR = 16.0 min).
Figure 3.
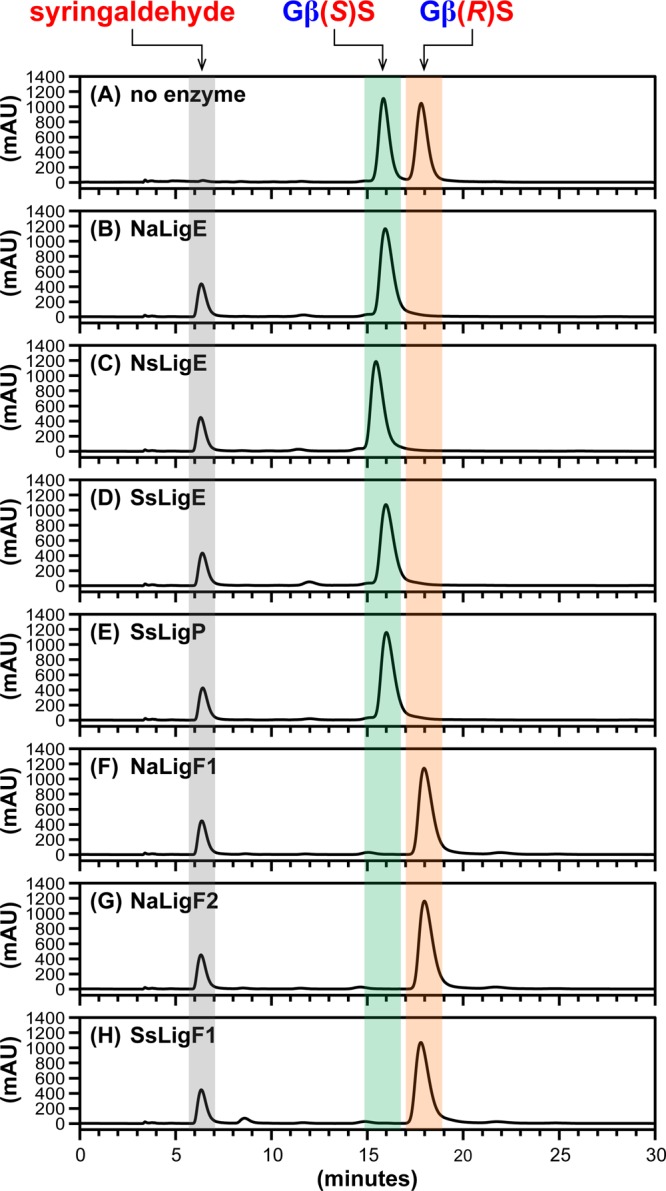
HPLC chromatographic traces (CHIRALPAK AD-H column, λ = 280 nm) of β-etherase enzyme assay samples from cosubstrates racem-GβS and glutathione. Chromatographic regions for syringaldehyde (gray), Gβ(S)S (green), and Gβ(R)S (orange) peak elution times are highlighted by shading. (A) No enzyme added, 0 h sample, where the ratio of peak area integrals of Gβ(S)S to Gβ(R)S was ∼1:1. After 1 h incubation with either enzymatic catalyst: (B) NaLigE, (C) NsLigE, (D) SsLigE, (E) SsLigP, (F) NaLigF1, (G) NaLigF2, or (H) SsLigF1. Structures of syringaldehyde, Gβ(S)S, and Gβ(R)S are shown in Figure 1B. Abbreviations: Na, N. aromaticivorans strain DSM12444; Ns, Novosphingobium sp. strain PP1Y; Ss, Sphingobium sp. strain SYK-6. See Experimental Section for details.
Cleavage of SβG Aromatic β-Ethers
To test whether Lig β-etherases catalyze cleavage of the geometric isomer containing its syringyl and guaiacyl units in the opposite bonding orientation of GβS, we assayed β-etherase activity of each putative Lig enzyme using GSH and racem-SβG as cosubstrates and found that each catalyzed cleavage. An alignment of the chiral chromatograms (Figure 4) reveals that each of the seven enzymes cleaved a single SβG enantiomer, yielding the expected product (Figure 1C), vanillin (tR = 6.9 min). We also found that each putative LigE/LigP enzyme (Figure 4B–E) catalyzed stereospecific cleavage of Sβ(R)G (tR = 19.5 min), whereas the LigF homologues (Figure 4F–H) exhibited stereospecificity toward Sβ(S)G (tR = 16.7 min).
Figure 4.
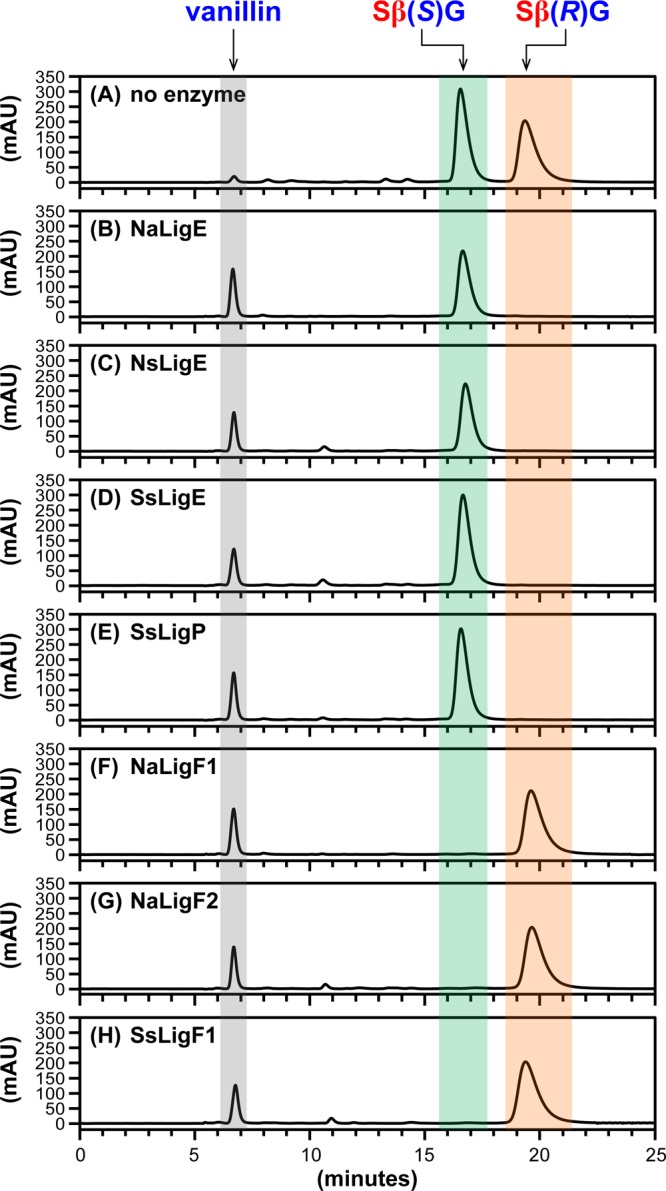
HPLC chromatographic traces (CHIRALPAK AY-H column, λ = 280 nm) of β-etherase enzyme assay samples from cosubstrates racem-SβG and glutathione. Chromatographic regions for vanillin (gray), Sβ(S)G (green), and Sβ(R)G (orange) peak elution times are highlighted by shading. (A) No enzyme added, 0 h sample, where the ratio of peak area integrals of Sβ(S)G to Sβ(R)G was ∼1:1. After 1 h incubation with either enzymatic catalyst: (B) NaLigE, (C) NsLigE, (D) SsLigE, (E) SsLigP, (F) NaLigF1, (G) NaLigF2, or (H) SsLigF1. Structures of vanillin, Sβ(S)G, and Sβ(R)G are shown in Figure 1C. Abbreviations: Na, N. aromaticivorans strain DSM12444; Ns, Novosphingobium sp. strain PP1Y; Ss, Sphingobium sp. strain SYK-6. See Experimental Section for details.
Cleavage of SβS Aromatic β-Ethers
To test for activity with a lignin compound composed of two syringyl units, we assayed for β-etherase activity with each putative Lig enzyme using GSH and racem-SβS as cosubstrates. Chiral chromatography (Figure 5) revealed that each enzyme degraded a single SβS enantiomer, resulting in the release of the expected product (Figure 1D), syringaldehyde (tR = 8.0 min). In contrast with chromatogram alignments from GβG (Figure 2), GβS (Figure 3), and SβG assay samples (Figure 4), where LigE/LigP homologues degraded the high-tR isomer and LigF homologues cleaved the low-tR enantiomer, we found that the putative LigE/LigP enzymes (Figure 5B–E) catalyzed degradation of the low-tR SβS isomer (tR = 18.4 min) and the LigF homologues (Figure 5F–H) cleaved the high-tR SβS isomer (tR = 24.2 min). Because this result was in contrast to our findings with substrates GβG (Figure 2), GβS (Figure 3), and SβG (Figure 4), preparative chiral chromatography was used for the isolation of each isomer (for additional details, see Supporting Information and Figure S3) and the resulting enantiopure compounds were used to derive MTPA(R) esters that aided in the assignment of absolute configurations to the low-tR (Sβ(R)S) and high-tR (Sβ(S)S) isomers by 1H NMR spectroscopy (for additional details, see Supporting Information and Figure S4). As was the case with racemic substrates GβG, GβS, and SβG, we again conclude that each LigE/LigP homologue exhibits β(R)-stereospecificity whereas each LigF homologue catalyzes β(S)-ether cleavage of SβS enantiomers and that the isomers simply elute in reverse order in this case.
Figure 5.
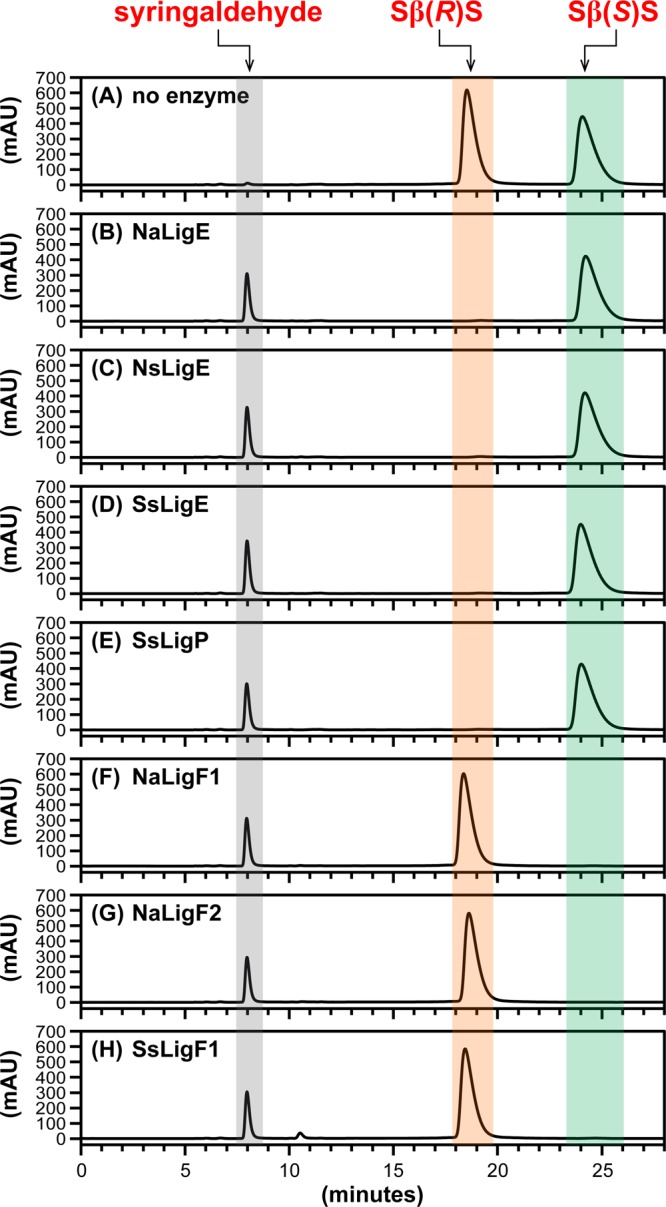
HPLC chromatographic traces (CHIRALPAK AY-H column, λ = 280 nm) of β-etherase enzyme assay samples from cosubstrates racem-SβS and glutathione. Chromatographic regions for syringaldehyde (gray), Sβ(R)S (orange), and Sβ(S)S (green). peak elution times are highlighted by shading. (A) No enzyme added, 0 h sample, where the ratio of peak area integrals of Sβ(R)S to Sβ(S)S was ∼1:1. After 1 h incubation with either enzymatic catalyst: (B) NaLigE, (C) NsLigE, (D) SsLigE, (E) SsLigP, (F) NaLigF1, (G) NaLigF2, or (H) SsLigF1. Structures of syringaldehyde, Sβ(R)S, and Sβ(S)S are shown in Figure 1D. Abbreviations: Na, N. aromaticivorans strain DSM12444; Ns, Novosphingobium sp. strain PP1Y; Ss, Sphingobium sp. strain SYK-6. See Experimental Section for details.
β-Etherase Assays with RpHypGST
In seeking to identify other potential β-etherases with the ability to cleave lignin model substrates, we constructed a phylogenetic tree from an alignment of closely related LigE/LigP and LigF homologues (Figure 6). R. palustris strain CGA009 is a bacterium previously shown to metabolize aromatic monomers likely to be derived from native lignin.41 Thus, we cloned an R. palustris gene that encodes a hypothetical Lig β-etherase (RpHypGST, having 36% amino acid sequence similarity to SsLigE) and purified recombinant protein to be tested for activity in the same β-etherase assays. Recombinant RpHypGST was expressed and purified as either N-terminally tagged (affording N-RpHypGST) and C-terminally tagged (affording C-RpHypGST) His8 fusions (see Experimental Section). N-RpHypGST and C-RpHypGST were each assayed using GSH and racem-GβG as cosubstrates. Under conditions identical to those where the sphingomonad Lig β-etherases exhibited β-etherase activity, neither substrate degradation nor release of the expected product (vanillin, Figure 1A) was detected (data not shown). We conclude that neither recombinant RpHypGST protein is a catalyst of β-etherase activity with these substrates.
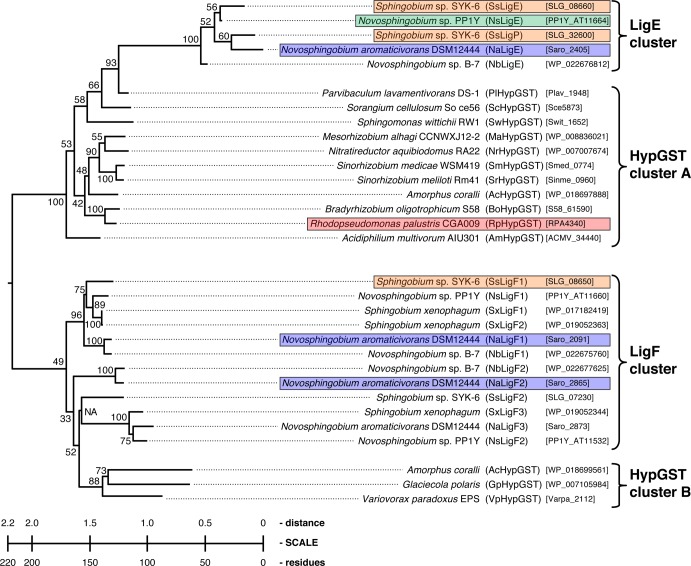
Figure 6.
Phylogenetic tree of aligned β-etherase amino acid sequences (10,000 bootstrap trials, 111 seeds). The 31 aligned sequences depicted were from the 15 most similar sequences to each SsLigE and SsLigF1 found in the BLASTP database, in addition to the sequence encoding RpHypGST. Gene symbols and locus tags are shown in parentheses and brackets. The LigE cluster shows the five closely related LigE-like sequences. HypGST cluster A shows the 11 divergent sequences from the SsLigE BLASTP search. The LigF cluster shows the 12 closely related LigF sequences (gene symbol numerals indicate relatedness to SsLigF1, where “1” indicates most similar). HypGST cluster B shows the three divergent sequences from the SsLigF1 BLASTP search. The LigE and LigF enzymes from selected strains that were tested for β-etherase activity in this study are highlighted by color: Sphingobium sp. strain SYK-6 (orange), Novosphingobium sp. strain PP1Y (green), N. aromaticivorans strain DSM12444 (blue), and R. palustris CGA009 (red).
Although the putative GSH binding domain was conserved across all sequences in both the LigE cluster and the HypGST cluster A (Figure 6), sequence analysis revealed several dissimilarities between the two clades. We found that each of the amino acid sequences in the HypGST cluster A, including RpHypGST, were significantly shorter (230 amino acids) than those in the LigE cluster (264–280 amino acids), which includes the four LigE homologues with confirmed β-etherase activity. Further, the putative GST superfamily substrate binding domains (residues 95–132) were conserved within a clade but dissimilar across the two clusters,42,43 suggesting that they bind different substrates. While this work was being reviewed, another member of HypGST cluster A encoded in Sorangium cellulosum (Figure 6) was also reported to be inactive with β-ether-linked substrates.23 Given these findings, we propose that the shorter sequences in HypGST cluster A from nonsphingomonad strains have an alternative glutathione-dependent function and do not encode active β-etherase enzymes.
Discussion
Recently, it has been shown that GST superfamily enzymes from Sphingobium sp. strain SYK-6 have the ability to act as stereospecific β-etherases using lignin model compounds as substrates.15,18 These so-called Lig β-etherases have been shown to cleave lignin dimers composed of guaiacyl monomers. In this study, we investigated whether Lig β-etherases from Sphingobium sp. strain SYK-6 also exhibit enzyme activity with substrates that contain syringyl units, the other major monomeric constituent of lignin. Further, we investigated whether other bacteria possess sequence-related proteins with similar or different substrate or stereospecificities as those reported for the Sphingobium sp. strain SYK-6 enzymes.
This study reveals for the first time that (a) several species of sphingomonads encode glutathione-dependent enzymes that catalyze cleavage of β-ether linkages that are found in lignin, (b) each Lig homologue cleaves guaiacyl-β-guaiacyl, guaiacyl-β-syringyl, syringyl-β-guaiacyl, and syringyl-β-syringyl β-ether-linked substrates, and (c) with each substrate, LigE/LigP and their homologues exhibit β(R)-stereospecificity whereas LigF and its homologues have β(S)-ether stereospecificity. These results show that methoxy group ring substitutions on the aromatic monomeric units are not inhibitory to the function of these β-etherase enzymes. Rather, sphingomonads use enzymes with active sites that are receptive to variably methoxylated rings. Also, these findings give insight into how a set of β-etherase pathway enzymes from different species accommodate substrates containing the multiple chiral centers (i.e., at carbons α and β) that exist in the β-ether-linked structures found in lignin.21,22 The NAD-dependent dehydrogenases oxidize and eliminate the chiral center at carbon α, forming α-keto-β(R)- and α-keto-β(S)-enantiomers. Further, the existence of both β(R)- and β(S)-ether enantiomers in nature is overcome by the evolution of separate glutathione-dependent enzymes with either β(R)- or β(S)-ether-cleaving reaction mechanisms.
Our results predict that a single organism may contain multiple β(R)-etherases (e.g., SsLigE and SsLigP) or numerous β(S)-etherases (e.g., NsLigF1 and NsLigF2), each of which is capable of catalyzing cleavage of GβG, GβS, SβG, and SβS enantiomers. In sphingomonads Sphingobium sp. SYK-6, Novosphingobium sp. strain PP1Y, N. aromaticivorans strain DSM12444, and another Novosphingobium strain with sequence-related homologues to Lig enzymes, strain B-7 (Figure 6), it appears that metabolism of α-keto-β-ether-linked compounds is achieved via catalysis by multiple Lig β-etherases with overlapping function. However, it is possible that variations of the pathway may exist in closely related bacteria. For example, a phylogenetic tree constructed from an alignment of LigE/LigP and LigF homologues (Figure 6) reveals that five LigE/LigP homologues belonging to four sphingomonad strains (Figure 6, LigE cluster) were more closely related to each other than the next 11 sequences identified in the SsLigE BLASTP search (Figure 6, HypGST cluster A). The genome of each sphingomonad encodes multiple LigF homologues. Five such sphingomonad strains encode closely related putative LigF enzymes (Figure 6, LigF cluster) that exhibited sequence dissimilarity with the three nonsphingomonad LigF homologues (Figure 6, HypGST cluster B), perhaps because the HypGST sequences encode different functions.
Overall, BLASTP analysis predicts that six sphingomonads (α-Proteobacteria of the order Sphingomonadales) encoded Lig homologues that were aligned in the phylogenetic tree (Figure 6). Additional BLASTP searches within the genomes of Sphingobium sp. strain SYK-6 and Novosphingobium strains B-7, PP1Y, and DSM12444, each of which had multiple sequences in the phylogenetic tree, revealed that each organism encoded both the LigE homologue needed for β(R)-enantiomer degradation, and the LigF homologue required for catabolism of β(S)-enantiomers. Of these, only Sphingobium sp. strain SYK-6 encoded multiple β(R)-specific (SsLigE and SsLigP) and multiple β(S)-specific enzymes (SsLigF1 and SsLigF2). However, Novosphingobium sp. strain PP1Y, Novosphingobium sp. strain B-7, and N. aromaticivorans strain DSM12444 were each found to encode a single LigE homologue and multiple sequences with LigF homology. Also, all four sphingomonad strains additionally encode multiple NAD-dependent dehydrogenases that catalyze the formation of the α-ketones required for β-ether cleavage activity.
The fifth sphingomonad that encodes putative β-etherases, Sphingomonas wittichii (Sm. wittichii) strain RW1, had a single LigE homologue (Figure 6, HypGST cluster A) that, based on sequence analysis, is more similar to RpHypGST (which had a shorter sequence and did not exhibit β-etherase activity) than to the confirmed β-etherases in the LigE cluster. Further, the Sm. wittichii genome did not encode a protein related to those that have β(S)-etherase activity or putative NAD-dependent Lig dehydrogenase activity, suggesting that the LigE homologue in Sm. wittichii does not encode a function related to β-ether catabolism. Another sphingomonad, Sphingobium xenophagum (Sb. xenophagum), encoded three homologues with potential β(S)-specific activity (SxLigF1, SxLigF2, and SxLigF3), but did not encode a LigE homologue. Given the high sequence similarity to enzymes with demonstrated β(S)-etherase activity, it is possible that Sb. xenophagum carries out β(S)-enantiomer catabolism with its various LigF homologues but uses alternative metabolic pathways for the degradation of β(R)-enantiomers.
Thirteen of the thirty-one sequences in the phylogenetic tree (Figure 6) are derived from nonsphingomonads, one from each of α- (of the order Rhodospirillales), β-, γ-, and δ-Proteobacteria, and nine from α-Proteobacteria (of the order Rhizobiales). Amorphus coralii (A. coralii) was the only nonsphingomonad that encoded both a LigE- and a LigF-like protein. However, unlike in Novosphingobium sp. strains B-7 and PP1Y, N. aromaticivorans strain DSM12444, and Sphingobium sp. strain SYK-6, the A. coralii genome encoded no sequences with homology to the NAD-dependent Lig dehydrogenases, suggesting that homologues from A. coralii are HypGSTs with alternative functions to those of the Lig β-etherases. Further, the A. coralii LigE homologue clustered with the other homologues with shorter sequences that we predict not to have β-etherase activity (Figure 6, HypGST cluster A). The genomes of Glaciecola polaris and Variovorax paradoxus EPS, each encode a single LigF-like sequence (Figure 6, HypGST cluster B), but did not encode homologues of any of the other essential β-etherase pathway enzymes. We therefore propose that the HypGST proteins in clusters A and B do not have activity as β-etherases with the lignin compounds used in this study.
Given that each of the LigE/LigP enzymes that we tested catalyzed β(R)-ether cleavage, whereas each LigF enzyme exhibited β(S)-stereospecificity, we propose that the β-etherase pathway functions similarly in Novosphingobium sp. strains B-7 and PP1Y, N. aromaticivorans strain DSM12444, and Sphingobium sp. strain SYK-6. These organisms appear to have adapted to the racemic nature of lignin by evolving multiple glutathione-dependent enzymes with complementary β-etherase stereospecificities. It will be intriguing to learn if the functions of the β-etherase pathway are unique to the sphingomonads as the availability of additional genome sequences pave the way for future studies of lignin catabolism in other bacteria.
Acknowledgments
This work was supported by the Department of Energy Office of Science’s Great Lakes Bioenergy Research Center, Grant DE-FC02-07ER64494. D.L.G. was supported by a NIGMS Biotechnology Training grant (Grant T32 GM08349). We thank Sally Ralph at the U.S. Forest Product Laboratory and members of the Ralph laboratory for aiding in synthesis, analysis, and characterization of model compounds.
Glossary
Abbreviations and Nomenclature
- NAD
nicotinamide adenine dinucleotide
- GSH
glutathione
- GSSG
glutathione disulfide
- OMe
methoxyl
- GβG
α-(4-O-Me)-guaiacylglycerone-β-(1′-formyl)-guaiacyl ether
- GβS
α-(4-O-Me)-guaiacylglycerone-β-(1′-formyl)-syringyl ether
- SβG
α-(4-O-Me)-syringylglycerone-β-(1′-formyl)-guaiacyl ether
- SβS
α-(4-O-Me)-syringylglycerone-β-(1′-formyl)-syringyl ether
- Gβ-SG
β-glutathionyl-α-(4-O-Me)-guaiacylglycerone
- Sβ-SG
β-glutathionyl-α-(4-O-Me)-syringylglycerone
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- GST
glutathione-S-transferase
- Na
Novosphingobium aromaticivorans strain DSM12444
- Ns
Novosphingobium sp. strain PP1Y
- Rp
Rhodopseudomonas palustris strain CGA009
- Ss
Sphingobium sp. strain SYK-6
- HypGST
hypothetical glutathione-S-transferase
- His8
octa-histidine affinity tag
- Ni-NTA
nickel-nitrilotriacetic acid resin
- HSQC
(1H–13C) heteronuclear single quantum coherence (NMR spectroscopy)
- HMBC
(1H–13C) heteronuclear multiple-bond correlation (NMR spectroscopy)
- COSY
(1H–1H) correlation spectroscopy
- tR
retention time
- MTPA(R)
α(R)-methoxy-trifluoromethyl-phenylacetate
- MTPACl(S)
α(S)-methoxy-trifluoromethyl-phenylacetyl chloride
- GβG-MTPA(R)
β-(1′-formyl)-guaiacyl-α-(4-O-Me)-guaiacylglyceryl α(R)-methoxy-trifluoromethyl-phenyl-acetate
- SβS-MTPA(R)
β-(1′-formyl)-syringyl-α-(4-O-Me)-syringylglyceryl α(R)-methoxy-trifluoromethyl-phenyl-acetate
- GβG-propenone
α-(4-O-Me)-guaiacyl-β,γ-propenone-β-(1′-formyl)-guaiacyl ether
Supporting Information Available
Text describing gene cloning, synthetic details and NMR data, and accompaning references and figures showing SDS-12% PAGE gel images from studied enzyme preparations, synthetic schemes for preparation of β-etherase substrates, preparative chiral HPLC chromatographic separations, aligned 1H NMR spectra of studied esters, and 1H and 13C NMR spectra of model compounds and reaction products. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Lewis N. G.; Yamamoto E. Lignin—Occurrence, biogenesis and biodegradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1990, 41, 455–496. [DOI] [PubMed] [Google Scholar]
- Higuchi T., Lignin structure and morphological distribution in plant cell walls. In Lignin biodegradation: microbiology, chemistry and potential applications, Kirk T. K., Higuchi T., Chang H., Eds.; CRC Press: Boca Raton, FL, USA, 1980; Vol. I, pp 1–20. [Google Scholar]
- Freudenberg K. Biosynthesis and constitution of lignin. Nature 1959, 18346691152–5. [DOI] [PubMed] [Google Scholar]
- Sarkanen K. V.; Ludwig C. H. In Lignins: Occurrence, formation, structure and reactions; Sarkanen K. V., Ludwig C. H., Eds.; John Wiley & Sons: New York, 1971; pp 1–916. [Google Scholar]
- Dixon R. A.; Paiva N. L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 771085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler E. Lignin chemistry—Past, present and future. Wood Sci. Technol. 1977, 113169–218. [Google Scholar]
- Simmons B. A.; Logue D.; Ralph J. Advances in modifying lignin for enhanced biofuel production. Curr. Opin. Plant Biol. 2010, 133313–320. [DOI] [PubMed] [Google Scholar]
- Chen F.; Dixon R. A. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007, 257759–761. [DOI] [PubMed] [Google Scholar]
- Bugg T. D. H.; Ahmad M.; Hardiman E. M.; Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 2011, 223394–400. [DOI] [PubMed] [Google Scholar]
- Zakzeski J.; Bruijnincx P. C. A.; Jongerius A. L.; Weckhuysen B. M. The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 2010, 11063552–3599. [DOI] [PubMed] [Google Scholar]
- Masai E.; Katayama Y.; Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci., Biotechnol., Biochem. 2007, 7111–15. [DOI] [PubMed] [Google Scholar]
- Sato Y.; Moriuchi H.; Hishiyama S.; Otsuka Y.; Oshima K.; Kasai D.; Nakamura M.; Ohara S.; Katayama Y.; Fukuda M.; Masai E. Identification of three alcohol dehydrogenase genes involved in the stereospecific catabolism of arylglycerol-β-aryl ether by Sphingobium sp. strain SYK-6. Appl. Environ. Microbiol. 2009, 75165195–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E.; Kubota S.; Katayama Y.; Kawai S.; Yamasaki M.; Morohoshi N. Characterization of the Cα-dehydrogenase gene involved in the cleavage of β-aryl ether by Pseudomonas paucimobilis. Biosci., Biotechnol., Biochem. 1993, 57101655–1659. [DOI] [PubMed] [Google Scholar]
- Masai E.; Katayama Y.; Kubota S.; Kawai S.; Yamasaki M.; Morohoshi N. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993, 3231–2135–140. [DOI] [PubMed] [Google Scholar]
- Masai E.; Ichimura A.; Sato Y.; Miyauchi K.; Katayama Y.; Fukuda M. Roles of the enantioselective glutathione S-transferases in cleavage of β-aryl ether. J. Bacteriol. 2003, 18561768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E.; Katayama Y.; Nishikawa S.; Yamasaki M.; Morohoshi N.; Haraguchi T. Detection and localization of a new enzyme catalyzing the β-aryl ether cleavage in the soil bacterium (Pseudomonas-paucimobilis SYK-6). FEBS Lett. 1989, 2492348–352. [DOI] [PubMed] [Google Scholar]
- Masai E.; Katayama Y.; Kawai S.; Nishikawa S.; Yamasaki M.; Morohoshi N. Cloning and sequencing of the gene a Pseudomonas-paucimobilis enzyme that cleaves β-aryl ether. J. Bacteriol. 1991, 173247950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall D. L.; Kim H.; Lu F.; Donohue T. J.; Noguera D. R.; Ralph J. Stereochemical features of glutathione-dependent enzymes in the Sphingobium sp. strain SYK-6 β-aryl etherase pathway. J. Biol. Chem. 2014, 289128656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T.; Sugimoto T.; Matsumoto Y.; Meshitsuka G. Erythro/threo ratio of β-O-4 structures as an important structural characteristic of lignin. I: Improvement of ozonation method for the quantitative analysis of lignin side-chain structure. J. Wood Sci. 2002, 483210–215. [Google Scholar]
- Sugimoto T.; Akiyama T.; Matsumoto Y.; Meshitsuka G. The erythro/threo ratio of β-O-4 structures as an important structural characteristic of lignin—Part 2. Changes in erythro/threo (E/T) ratio of β-O-4 structures during delignification reactions. Holzforschung 2002, 564416–421. [Google Scholar]
- Akiyama T.; Magara K.; Matsumoto Y.; Meshitsuka G.; Ishizu A.; Lundquist K. Proof of the presence of racemic forms of arylglycerol-β-aryl ether structure in lignin: studies on the stereo structure of lignin by ozonation. J. Wood Sci. 2000, 465414–415. [Google Scholar]
- Ralph J.; Peng J. P.; Lu F. C.; Hatfield R. D.; Helm R. F. Are lignins optically active?. J. Agric. Food Chem. 1999, 4782991–2996. [DOI] [PubMed] [Google Scholar]
- Picart P.; Müller C.; Mottweiler J.; Wiermans L.; Bolm C.; Domínguez de María P.; Schallmey A.. From gene towards selective biomass valorization: Bacterial β-etherases with catalytic activity on lignin-like polymers. ChemSusChem 2014, in press [DOI] [PubMed]
- Sonoki T.; Iimura Y.; Masai E.; Kajita S.; Katayama Y. Specific degradation of β-aryl ether linkage in synthetic lignin (dehydrogenative polymerizate) by bacterial enzymes of Sphingomonas paucimobilis SYK-6 produced in recombinant Escherichia coli. J. Wood Sci. 2002, 485429–433. [Google Scholar]
- Tanamura K.; Abe T.; Kamimura N.; Kasai D.; Hishiyama S.; Otsuka Y.; Nakamura M.; Kajita S.; Katayama Y.; Fukuda M.; Masai E. Characterization of the third glutathione S-transferase gene involved in enantioselective cleavage of the β-aryl ether by Sphingobium sp. Strain SYK-6. Biosci., Biotechnol., Biochem. 2011, 75122404–7. [DOI] [PubMed] [Google Scholar]
- Moore D. D. In Current protocols in molecular biology; Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., Eds.; John Wiley & Sons: New York, 2003. [Google Scholar]
- Blommel P. G.; Fox B. G. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expression Purif. 2007, 55153–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A.; Lupardus P. J.; Albrow V. E.; Guzzetta A.; Powers J. C.; Garcia K. C.; Bogyo M. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat. Chem. Biol. 2009, 57469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazkova K.; Shuvalova L. A.; Minasov G.; Voburka Z.; Anderson W. F.; Satchell K. J. F. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. J. Biol. Chem. 2009, 2843926557–26568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan K.-L.; Cordero C. L.; Satchell K. J. F. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J. 2007, 26102552–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler E.; Eriksoo E. Guaiacylglycerol and its β-guaiacyl ether. Acta Chem. Scand. 1955, 9, 341–342. [Google Scholar]
- Landucci L. L.; Geddes S. A.; Kirk T. K. Synthesis of C-14-labeled 3-methoxy-4-hydroxy-α-(2-methoxyphenoxy)-β-hydroxypropiophenone, a lignin model-compound. Holzforschung 1981, 35267–70. [Google Scholar]
- Hishiyama S.; Otsuka Y.; Nakamura M.; Ohara S.; Kajita S.; Masai E.; Katayama Y. Convenient synthesis of chiral lignin model compounds via optical resolution: Four stereoisomers of guaiacylglycerol-β-guaiacyl ether and both enantiomers of 3-hydroxy-1-(4-hydroxy-3-methoxyphenyl)-2-(2-methoxy-phenoxy)-propan-1-one (erone). Tetrahedron Lett. 2012, 53, 842–845. [Google Scholar]
- Takeuchi M.; Hamana K.; Hiraishi A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2001, 51, 1405–1417. [DOI] [PubMed] [Google Scholar]
- Notomista E.; Pennacchio F.; Cafaro V.; Smaldone G.; Izzo V.; Troncone L.; Varcamonti M.; Di Donato A. The marine isolate Novosphingobium sp. PP1Y shows specific adaptation to use the aromatic fraction of fuels as the sole carbon and energy source. Microb. Ecol. 2011, 613582–594. [DOI] [PubMed] [Google Scholar]
- LaRoe S. L.; Wang B.; Han J.-I. Isolation and characterization of a novel polycyclic aromatic hydrocarbon-degrading bacterium, Sphingopyxis sp. strain M2R2, capable of passive spreading motility through soil. Environ. Eng. Sci. 2010, 276505–512. [Google Scholar]
- Xia Y.; Min H.; Rao G.; Lv Z. M.; Liu J.; Ye Y. F.; Duan X. J. Isolation and characterization of phenanthrene-degrading Sphingomonas paucimobilis strain ZX4. Biodegradation 2005, 165393–402. [DOI] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 2153403–410. [DOI] [PubMed] [Google Scholar]
- Masai E.; Kamimura N.; Kasai D.; Oguchi A.; Ankai A.; Fukui S.; Takahashi M.; Yashiro I.; Sasaki H.; Harada T.; Nakamura S.; Katano Y.; Narita-Yamada S.; Nakazawa H.; Hara H.; Katayama Y.; Fukuda M.; Yamazaki S.; Fujita N. Complete genome sequence of Sphingobium sp, strain SYK-6, a degrader of lignin-derived biaryls and monoaryls. J. Bacteriol. 2012, 1942534–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio V.; Petrillo M.; Cantiello P.; Naso B.; Cozzuto L.; Notomista E.; Paolella G.; Di Donato A.; Salvatore F. De novo sequencing and assembly of the whole genome of Novosphingobium sp. strain PP1Y. J. Bacteriol. 2011, 193164296–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S.; Gibson J. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl. Environ. Microbiol. 1988, 543712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Q.; Hall P. R.; Zhou X. Y. E.; Ranson H.; Hemingway J.; Meehan E. J. Structure of an insect δ-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003, 59, 2211–2217. [DOI] [PubMed] [Google Scholar]
- Ji X. H.; Johnson W. W.; Sesay M. A.; Dickert L.; Prasad S. M.; Ammon H. L.; Armstrong R. N.; Gilliland G. L. Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene. Biochemistry 1994, 3351043–1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.