Abstract
Dissemination of antibiotic resistance genes occurs mostly by conjugation, which mediates DNA transfer between cells in direct contact. Conjugative plasmids of the IncA/C incompatibility group have become a substantial threat due to their broad host-range, the extended spectrum of antimicrobial resistance they confer, their prevalence in enteric bacteria and their very efficient spread by conjugation. However, their biology remains largely unexplored. Using the IncA/C conjugative plasmid pVCR94ΔX as a prototype, we have investigated the regulatory circuitry that governs IncA/C plasmids dissemination and found that the transcriptional activator complex AcaCD is essential for the expression of plasmid transfer genes. Using chromatin immunoprecipitation coupled with exonuclease digestion (ChIP-exo) and RNA sequencing (RNA-seq) approaches, we have identified the sequences recognized by AcaCD and characterized the AcaCD regulon. Data mining using the DNA motif recognized by AcaCD revealed potential AcaCD-binding sites upstream of genes involved in the intracellular mobility functions (recombination directionality factor and mobilization genes) in two widespread classes of genomic islands (GIs) phylogenetically unrelated to IncA/C plasmids. The first class, SGI1, confers and propagates multidrug resistance in Salmonella enterica and Proteus mirabilis, whereas MGIVmi1 in Vibrio mimicus belongs to a previously uncharacterized class of GIs. We have demonstrated that through expression of AcaCD, IncA/C plasmids specifically trigger the excision and mobilization of the GIs at high frequencies. This study provides new evidence of the considerable impact of IncA/C plasmids on bacterial genome plasticity through their own mobility and the mobilization of genomic islands.
Author Summary
Multidrug resistance is a major health concern that complicates treatments of even the most common infections caused by bacteria. In recent years, IncA/C plasmids have emerged and spread in bacteria infecting humans, food-producing animals and food products, driving at the same time the dissemination of a broad spectrum of antibiotic resistance genes in environmental and in clinical settings. In this study, we have characterized the regulatory pathway that governs IncA/C plasmid dissemination. We have found that AcaCD, the master activator complex encoded by these plasmids, is not only essential for the dissemination of IncA/C plasmids but also activates unrelated mobile genetic elements in bacterial genomes, thereby further promoting the interspecies propagation of multidrug resistance and other adaptive traits at a very high frequency.
Introduction
Multidrug resistance (MDR) is steadily increasing in Gram-negative bacteria in both community and hospital settings, and represents a growing concern worldwide [1]. MDR usually results from the adaptation of microorganisms through various mutations or from the acquisition of foreign DNA by horizontal gene transfer. In recent years, conjugative plasmids of the IncA/C incompatibility group, which are prevalent in enteric bacteria, have become a substantial threat due to their broad host-range, their extended spectrum of antimicrobial resistance and their efficient spread by conjugation [2]. IncA/C plasmids were first identified more than 40 years ago from diseased fish infected by antibiotic-resistant Aeromonas hydrophila and Vibrio spp [3], [4]. For more than three decades IncA/C plasmids received relatively little attention, but the rapid dissemination of these MDR-carrying vectors among enteric pathogens recovered from food-producing animals, food products and humans have sprung intensive research at the epidemiological and genomic level. Several IncA/C plasmids are spreading the New Delhi metallo-lactamase bla NDM-1 gene and its variants, which confer resistance to all β-lactams except for monobactams and are widely distributed throughout all Gammaproteobacteria [5]–[8]. Resistance to β-lactams, aminoglycosides, chloramphenicol, folate pathway inhibitors (sulfonamides and trimethoprim), quinolones and tetracycline is also commonly conferred by these large plasmids (ca. 140 to 200 kb) [9]–[13]. IncA/C plasmids have also been shown to mobilize in trans the Salmonella genomic island 1 (SGI1), a 43-kb chromosomal mobile element carrying a class 1 integron that confers resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides and tetracycline (ACSSuT phenotype) [14]–[16]. SGI1 and related MDR-conferring genomic islands (GIs) have been found in a large variety of Salmonella enterica serovars and in Proteus mirabilis [17]. To date, the genetic regulatory network and the nature of the interactions allowing the specific mobilization of SGI1 by IncA/C helper plasmids remain largely unknown.
Comparative genomics studies of IncA/C plasmids isolated from Escherichia coli, S. enterica, Vibrio cholerae, Yersina pestis, Yersinia ruckeri, Klebsiella pneumoniae and Providencia stuartii have revealed their close relationship [9], [10], [18], [19]. IncA/C plasmids share a common core set of genes exhibiting more than 99% identity that is disrupted by antibiotic-resistance conferring cassettes. While several of these conserved genes are involved in conjugative transfer (tra genes) and replication (repA), most have unknown functions. IncA/C plasmids are distantly related to the integrative and conjugative elements (ICEs) of the SXT/R391 family, which are also broadly distributed among several species of Enterobacteriaceae and Vibrionaceae [10], [19], [20]. Although many IncA/C plasmids have been fully sequenced to date, little is known about their basic biology, and most importantly about the regulation of their dissemination by conjugation.
Previously, we have identified and characterized the IncA/C conjugative plasmid pVCR94 from the epidemic isolate V. cholerae O1 El Tor F1939 [19]. pVCR94 transfers at very high frequency (10−2 to 10−1) across species and genera, and mediates resistance to co-trimoxazole, chloramphenicol, streptomycin, ampicillin and tetracycline [19]. Here, we have characterized the regulatory mechanisms that control the conjugative transfer function of IncA/C plasmids. Two transcriptional repressors, acr1 and acr2, were shown to repress the expression of two plasmid-encoded conserved genes, acaC and acaD, that are essential for the activation of IncA/C tra gene expression. Chromatin immunoprecipitation coupled to exonuclease digestion (ChIP-exo) and RNA sequencing (RNA-seq) assays were used to characterize the AcaCD regulon. Finally, bioinformatics analyses and experimental evidence revealed that the AcaCD regulon expands beyond IncA/C plasmid-borne genes to include GIs such as SGI1 and another unrelated GI of Vibrio mimicus. Altogether these results reveal a mechanism by which IncA/C conjugative plasmids play a role more important than previously estimated in bacterial genome dynamics.
Results and Discussion
Identification of repressors of IncA/C plasmids transfer
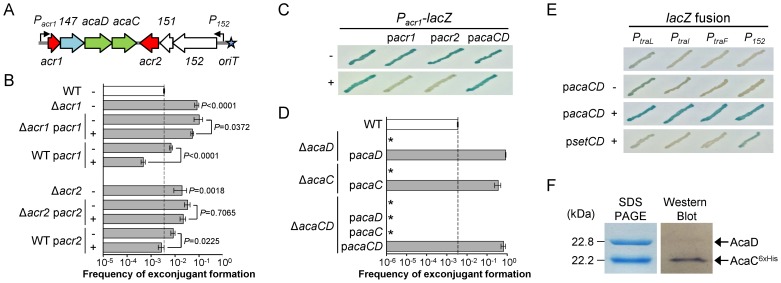
Comparative genomics previously revealed a set of six genes coding for putative transcriptional regulators in pVCR94 that are also conserved in nearly all IncA/C plasmids [19] (Table S1). To determine whether these genes are involved in the regulation of pVCR94 transfer, we constructed in-frame deletions and tested the ability of the resulting mutants to transfer by conjugation. For convenience, we carried out all of our assays using the 120,572-bp pVCR94ΔX mutant (referred to as pVCR94 in the rest of the paper), in which a single cluster containing all antibiotic resistance genes except for sul2 conferring resistance to sulfamethoxazole has been deleted [19]. Deletion of vcrx025, which codes for a putative HUβ-like DNA-binding protein, had no effect on plasmid transfer (3.94×10−3 exconjugant/donor for pVCR94Δvcrx025 compared to 3.51×10−3 exconjugant/donor for wild-type, P = 0.4400, two-tailed Student's t-test). Despite several attempts, we were unable to delete vcrx027, which codes for a Cro-like transcriptional regulator, suggesting that its absence is lethal. The adjacent gene vcrx028 is predicted to code for a toxin protein (addiction module killer protein, IPR009241), suggesting that the co-transcribed gene vcrx027 encodes its cognate antitoxin (see Dataset S1).
The remaining four putative regulator genes cluster near the origin of transfer (oriT) (Figure 1A). Deletion of either vcrx146 (acr1), which codes for a putative Ner-like DNA-binding protein, or vcrx150 (acr2), which codes for a predicted H-NS-like DNA-binding protein, resulted in a 23- and 5.6-fold increase of the frequency of transfer, respectively, thereby suggesting that both genes code for repressors of pVCR94 transfer (Figure 1B). For this reason, vcrx146 and vcrx150 were renamed acr1 and acr2, for IncA/C repressors 1 and 2, respectively. Complementation of the Δacr1 and Δacr2 mutations by ectopic expression of the corresponding genes under control of an arabinose-inducible promoter (P BAD) did not restore the wild-type transfer frequency (Figure 1B). However, overexpression of either acr1 or acr2 decreased transfer of wild-type pVCR94 by 14 and 3 fold, respectively. Since acr1 is the first gene of an operon structure (see Dataset S1) containing the putative transcriptional activator genes vcrx148 (acaD) and vcrx149 (acaC) (see below), we tested whether acr1 or acr2 are able to repress expression from Pacr1, the promoter driving expression of the operon containing acr1 (Figure 1A). To do this, we cloned Pacr1 upstream of a promoterless lacZ gene and monitored the β-galactosidase activity upon expression of acr1 or acr2 driven by P BAD. While Pacr1 was transcriptionally active in the absence of arabinose, no β-galactosidase activity was detected upon expression of either acr1 or acr2, confirming that both proteins are capable of directly repressing expression from Pacr1 (Figure 1C).
Figure 1. Regulation of IncA/C plasmids.
(A) Schematic representation of the regulatory region of IncA/C plasmids. Genes and promoters are represented by arrowed boxes and angled arrows, respectively. Repressors and activators are shown in red and green, respectively. Genes of unknown function are shown in white and the gene coding for a putative lytic transglycosylase is shown in light blue. Locus names vcrxXXX are abbreviated as XXX. The oriT locus is depicted by a blue star. (B) Effect of the deletion and the overexpression of the negative transcriptional regulator-encoding genes acr1 and acr2. Conjugation assays were carried out using as donors E. coli BW25113 Nx containing pVCR94ΔX2 (WT) or its Δacr1 and Δacr2 mutants. Complementation and overexpression assays were performed with (+) or without (−) arabinose for the expression of acr1 (pacr1) or acr2 (pacr2) from the inducible P BAD promoter. E. coli MG1655 Rf was used as the recipient. Transfer frequencies are expressed as a number of exconjugant per recipient CFUs. The bars represent the mean and standard deviation values obtained from at least three independent experiments. Statistical analyses were performed on the logarithm value of the means using one-way ANOVA with Tukey's multiple comparison test. P-values are indicated next to the bars when comparison referred to WT or next to the brackets when comparing two bars. (C) The constitutive promoter of acr1 (Pacr1) is repressed by Acr1 and Acr2. Activity of Pacr1 was monitored from a single-copy, chromosomally integrated lacZ transcriptional fusion (Pacr1-lacZ). Colorimetric assays were carried out on LB medium supplemented with 40 µg/ml of X-Gal and induction with (+) or without (−) arabinose to express acr1, acr2 or acaCD from P BAD on pacr1, pacr2 or pacaCD, respectively. (D) AcaC and AcaD are essential for conjugative transfer. Transfer assays were carried out using E. coli BW25113 Nx containing pVCR94ΔX2 (WT) or the mutants ΔacaC, ΔacaD or ΔacaCD. Complementation assays were performed by expressing acaC, acaD or acaCD from P BAD on pacaC, pacaD and pacaCD, respectively. Recipient strains and statistical analyses were as described for panel B. All P-values are below 0.0001 when compared to the WT. The asterisk indicates that frequency of exconjugant formation was below the detection limit (<10−8). (E) AcaCD is the direct activator of tra gene promoters. Activity of the PtraL, PtraI, PtraF and P152 was monitored from single-copy, chromosomally integrated lacZ transcriptional fusions. Colorimetric assays were performed as described in panel C with expression of acaCD or setCD from P BAD on pacaCD or psetCD (pGG2B), respectively. (F) AcaD co-purifies with AcaC. Coomassie blue-stained SDS-PAGE and Western blot analysis of AcaC purified using a Ni-NTA affinity chromatography. AcaD and 6×His-tagged AcaC were co-expressed in E. coli BL21(DE3) from pacaDC 6×His. Western blot analysis was performed using a monoclonal antibody against the 6×His-tag.
AcaC and AcaD are key activators of IncA/C plasmids transfer
The cluster of genes acr1-vcrx147-acaDC-acr2 is extremely well conserved and remain syntenic among all IncA/C plasmids except for XCN1_p from Xenorhabdus nematophila in which it is completely absent (Figures 2 and S1). Interestingly, the IncA/C plasmids pAM04528 and pSN254 from S. enterica, which were reported to be non-self-transferable [10], [21], [22], code for a truncated AcaC protein resulting from a frameshift mutation (acaC263) (Figure 2), suggesting a key role of acaC in the activation of IncA/C-plasmid transfer. Unfortunately, no data is currently available about the intercellular mobility of five other plasmids coding for truncated AcaC or AcaD proteins (Figure 2) [23], [24]. However, pRA1 was reported to transfer at a frequency of 10−3 [9] despite a GTG insertion at the 3′ end of acaC (acaC523ΩGTG) that slightly alters the primary sequence of AcaC C-terminus (Figure 2).
Figure 2. Molecular phylogenetic analysis of the acr1-vcr147-acaDC-acr2 locus by Maximum Likelihood method.
The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model [61]. The tree with the highest log likelihood (−5461.6977) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.6567)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 45 nucleotide sequences (Table S2). Codon positions included were 1st+2nd+3rd+Noncoding. There were a total of 2469 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [58]. The background color of each leaf indicates the original host species from which each plasmid was isolated. Insertions (Ω), deletion (Δ) and frameshift (Ψ) mutations are indicated where appropriate. *, Although pYT3 shares sequence identity with the IncA/C plasmids, it lacks an IncA/C-specific replication initiation gene (repA) and contains an IncFIB replicon instead (Figure S1) [62].
Pfam analyses (database v27.0) revealed that the conserved gene products AcaC and AcaD exhibit weak homology with the FlhC (Pfam PF05280) and FlhD (PF05247) domains, respectively (Table S1) [19]. FlhC and FlhD form a heteromeric complex playing a key role in the transcriptional activation of flagellar genes in Gram-negative bacteria such as E. coli [25]. We thus hypothesized that AcaC and AcaD (IncA/C activator subunits C and D) could acts as the master activator of tra genes in IncA/C plasmids. We constructed three null mutants, ΔacaC, ΔacaD and ΔacaCD, of pVCR94 and tested their ability to transfer by conjugation. All three mutations abolished conjugative transfer, thereby confirming that acaCD plays an essential role for IncA/C plasmid transmission (Figure 1D). Trans-complementation of these mutations under control of P BAD restored and even outperformed transfer of pVCR94 (Figure 1D). Importantly, the ΔacaCD mutant could not be complemented by expression of either acaC or acaD. Only the simultaneous expression of both acaC and acaD restored the transfer of pVCR94ΔacaCD, strongly suggesting that these two genes code for an FlhCD-like activating complex (Figure 1D). AcaC and AcaD share 34% and 23% identity with the master activator subunits SetC and SetD encoded by SXT/R391 ICEs. As expected from the very poor percentage of identity, the ΔacaCD mutation was not complemented by setCD expressed from the IPTG-inducible promoter Ptac (Figure S2).
To test whether AcaCD is necessary and sufficient to drive the expression of tra genes of pVCR94, we cloned the promoter regions of traL (PtraL), traI (PtraI) and traF (PtraF) upstream of a promoterless lacZ gene. The genes traL, traI and traF are likely candidate for AcaCD activation as traI codes for the predicted relaxase that would initiate conjugative transfer of pVCR94 at oriT while traL and traF code for two predicted sex pilus assembly proteins. We also tested the promoters Pacr1 and P152 that drive the expression of acaCD and vcrx152, respectively (Figure 1A). All but Pacr1 were inactive in the absence of acaCD, without arabinose induction or upon expression of setCD (Figure 1C, E). The constitutive expression from Pacr1 seemed to remain unaffected by AcaCD overexpression. In contrast, expression of AcaCD alone in cells lacking pVCR94 was sufficient to trigger the expression from PtraL, PtraI, PtraF and P152 (Figure 1E).
Collectively, these results are consistent with a model in which acr1 represses acaCD expression and its own, thereby preventing expression of the tra genes. Accordingly, under the appropriate conditions, repression by Acr1 would be alleviated, allowing expression of acaCD that in turn would enable expression of the tra genes and other genes such as vcrx152. Although acr2 seems to be part of the same operon-like structure as vcrx152, its expression is not up-regulated by AcaCD and likely not driven by P152 (Dataset S1). Instead acr2 seems to be constitutively expressed. As a mild H-NS-like general repressor, Acr2 could dampen acr1 and acaCD expression to prevent overexpression of the conjugative machinery, which is likely deleterious to the fitness of the host [26]. The H-NS-like protein Sfh encoded by the IncHI1 conjugative plasmid R27 has been shown to provide a stealth function helping the transmission of R27 into a naïve host by preventing titration of the cellular pool of H-NS by the A+T-rich sequences of the plasmid [27]. The locus occupied by acr2 in IncA/C plasmids contains the gene int in the SXT/R391 ICEs (Figure S3) [20]. int codes for the integrase which allows SXT/R391 ICEs to remain quiescent in the host chromosome [28].
Although the regulation loci of IncA/C conjugative plasmids and SXT/R391 ICEs encode similar transcriptional activators (AcaCD and SetCD, respectively), IncA/C plasmids carry acr1 and an extra acr2 gene, while lacking a homolog of setR (Figure S3). In SXT/R391 ICEs, setR codes for a λ CI-related transcriptional repressor that prevents the expression of setC and setD [29], [30]. Like λ CI, SetR responds to DNA damage by RecA*-dependent autoproteolysis, which alleviates the repression of setCD, thereby allowing excision and transfer of SXT/R391 ICEs [30]. Consistent with the absence of a setR homolog, transfer of IncA/C plasmids has been shown to be recA-independent [19].
AcaC and AcaD assemble as a heteromeric activator complex
Because deletion of either acaC or acaD abolished pVCR94 transfer, and both mutations could be individually complemented in trans, we sought to test whether AcaC and AcaD could form a heteromeric transcriptional activator complex. To investigate this possibility, we constructed a C-terminally 6×His-tagged version of acaC, acaC 6×His that was expressed together with acaD from Ptac. AcaC6×His was purified by Ni-NTA affinity chromatography and the sample was analyzed on a 12% SDS-polyacrylamide gel. Interestingly, two bands were detected after Coomassie blue staining with molecular weights consistent with AcaC and AcaD (Figure 1F). Western blot assays using anti-6×His-tag antibodies only revealed the smallest 22.2-kDa band which corresponds to AcaC6×His. The additional band that co-purified with AcaC6×His has a molecular weight of 22.8 kDa and was confirmed to be AcaD by mass spectrometry. Therefore, our results suggest that, like FlhC and FlhD, AcaC and AcaD assemble as a heteromeric complex.
Identification of AcaCD targets in pVCR94
To get a better understanding of the mechanisms governing transfer regulation of IncA/C conjugative plasmids, and identify genes of pVCR94 expressed under the direct control of AcaCD, we undertook an exhaustive characterization of the AcaCD regulon using transcriptome sequencing and ChIP-exo [31]. For these experiments, we used E. coli MG1655 Rf carrying pVCR94ΔacaCD and the same strain bearing a chromosomally integrated single copy of pacaDC 3×FLAG that expresses the native AcaD subunit along with a C-terminal 3×FLAG tagged AcaC subunit under control of Ptac. Based on the transfer frequency of the ΔacaCD mutant complemented with acaDC 3×FLAG, we concluded that the 3×FLAG did not significantly affect the function of the tagged AcaC subunit compared to its wild-type counterpart (Figure S2).
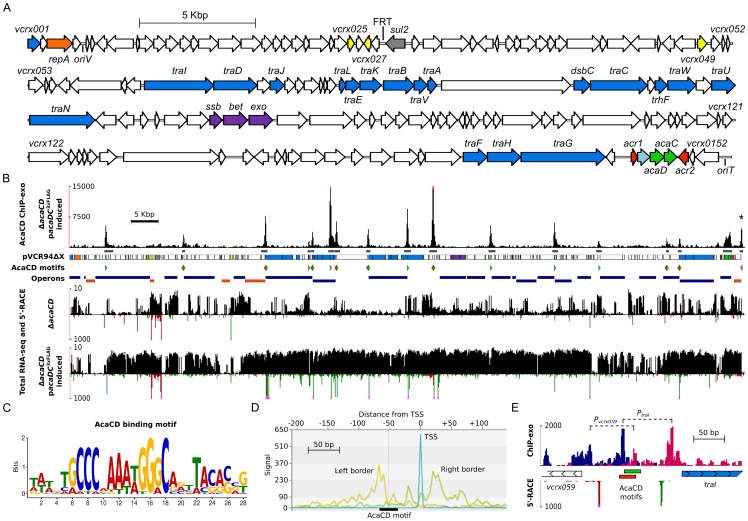
ChIP-exo data analyses revealed 17 major AcaCD enrichment peaks, all located within intergenic regions (Figure 3A, B and Table S3). Most of the genes or operons found downstream of these peaks play key roles in conjugative transfer (Figure 3, Figure S4 and Table S3). For instance, AcaCD-binding sites were found upstream of the traL, traV, dsbC, traN and traF genes that are predicted to be involved in the formation of the mating pore. One peak was also found upstream of the gene coding for the putative relaxase gene traI. The presence of an AcaCD-binding site is also well correlated with transcriptional activity since the expression of 86 out of 152 genes, including genes located downstream of AcaCD-binding sites, is clearly increased under the same conditions used for ChIP-exo when compared to a ΔacaCD mutant (Figure 3B and Dataset S1). In contrast, the vast majority of genes that are not significantly affected by the expression of AcaCD appeared to be either inactive or likely constitutively expressed. Their function is unknown or not directly tied to conjugative transfer. Examples of such genes in pVCR94 include repA (vcrx003) involved in plasmid replication, a putative toxin-antitoxin system (vcrx028 and vcrx027), the sul2 (vcrx029) gene conferring resistance to sulfamethoxazole, as well as acr1 and acr2 that negatively regulate the expression of the acr1-vcrx147-acaDC operon (Figure 1A, C and Dataset S1).
Figure 3. In-depth analysis of the AcaCD regulon in pVCR94.
(A) Genetic organization of pVCR94ΔX adapted from Carraro et al. [19]. The circular map of the plasmid was linearized at the start position of gene vcrx001. The locations and orientations of ORFs are indicated by arrowed boxes. Colors are coded by function as follow: white, unknown; blue, conjugative transfer; light blue, lytic transglycosylase; orange, replication; gray, antibiotic resistance; yellow, putative regulatory function; purple, recombination; green, activator; red, repressor. The origin of replication (oriV) and the origin of transfer (oriT) are also indicated. The position of the scar resulting from the deletion of the antibiotic resistance gene cluster is indicated by FRT. sul2, resistance to sulfamethoxazole. (B) Results of ChIP-exo and RNA-seq experiments on E. coli MG1655 Rf carrying pVCR94ΔacaCD with or without a single chromosomal copy of pacaDC 3×FLAG expressing the native AcaD subunit along with a C-terminal 3×FLAG-tagged AcaC subunit induced by IPTG. The first track plots the number of ChIP-exo reads mapped as a function of the position in pVCR94ΔX (black bars). Pink dots at the top of peaks indicate a signal beyond the represented y-axis maximal value. The second track shows the position of ChIP-exo enrichment peaks found by MACS [63] (dark gray). The asterisk at the top of the rightmost peak indicates a MACS false negative result manually incorporated in the figure. The third track is a representation of the pVCR94ΔX genes using the same color code as in panel A. The fourth track indicates the position of the AcaCD-binding motifs found by MAST [64] within ChIP-exo peaks using the AcaCD logo shown in panel C. Green arrows, motifs on positive DNA strand; red arrows, motifs on negative DNA strand. The fifth track represents Rockhopper's [65] predicted operons. Dark blue, operons transcribed from positive DNA strand; orange, operons transcribed from negative DNA strand. The remaining four tracks show the total RNA-seq read densities (black bars; log scale) and the genome-wide 5′-RACE signals (green and red bars respectively on the positive and negative DNA strands; linear scale) for cells harboring either pVCR94ΔacaCD or pVCR94ΔacaCD complemented with pacaDC 3×FLAG and induced by IPTG. Pink dots at the top of 5′-RACE signals indicate a signal beyond the represented y-axis maximal value. (C) Motif sequence recognized by AcaCD in pVCR94ΔX obtained by MEME [64] using the AcaCD-binding sequences generated from ChIP-exo experiments. (D) VAP aggregate profile [66] showing ChIP-exo and 5′-RACE density signals centered on the AcaCD-binding motif (black box). Yellow line, density of reads mapping on the positive DNA strand (Left border); green line, density of reads mapping on the negative DNA strand (Right border). X-axis displays the distance in nucleotides from the aggregated transcription start site shown in blue (TSS). (E) Organization of vcrx059 and traI divergent promoters revealed by ChIP-exo and 5′-RACE for pVCR94ΔacaCD complemented with IPTG-induced pacaDC 3×FLAG. The first track plots ChIP-exo read densities at single nucleotide resolution as in panel D. Dark blue, density of reads mapping on the positive DNA strand; magenta, density of reads mapping on the negative DNA strand. The second track shows the two AcaCD-binding motifs found by MAST within the ChIP-exo peak between vcrx059 (white arrows) and traI (blue arrows) genes. Motif corresponding to the positive DNA strand is represented in green and motif corresponding to the negative DNA strand is shown in red. The last track represents 5′-RACE signals as described in panel B. The exonuclease-protected regions of the vcrx059 and traI promoters are indicated by dashed lines.
De novo motif discovery of DNA sequences bound by AcaCD was carried out using MEME (Multiple Em for Motif Elicitation) [32] (Figure 3C). We next used MAST (Motif Alignment and Search Tool) [33] to determine the precise location of potential AcaCD-binding motifs on the entire sequence of pVCR94 (Figure 3B, Figure S4, and Table S3). Statistically significant motifs that were localized within ChIP-exo peaks were conserved. We next analyzed the localization of AcaCD-binding motifs relative to transcription start sites obtained using a genome-wide 5′-RACE (Rapid Amplification of cDNA Ends) methodology (Figure 3B, D, Figure S4 and Table S3). We observed a similar promoter profile, compatible with a class 2 activator [34], for all transcription start sites located between an AcaCD-binding motif and a gene in the same orientation (Figure S4). This interpretation is consistent with our high resolution ChIP-exo data that reveals a protected region containing a distal AcaCD motif (Figure 3D), and supported by previous observations of the promoter-bound RNA polymerase holoenzyme complex footprint between position −54 to +22 relative to the transcription start site [28]. An interesting example of this organization is the regulation of the vcrx059 and traI divergent promoter region, in which two partially overlapping transcription initiation complexes are detected (Figure 3E).
In addition to measuring transcriptome expression in pVCR94ΔacaCD and pVCR94ΔacaCD complemented with IPTG-induced pacaDC 3×FLAG, we also performed RNA-seq on wild-type pVCR94 (Dataset S1). Our results indicate little differences between the transcriptome expression levels of wild-type pVCR94 and pacaDC 3×FLAG-complemented pVCR94ΔacaCD (Pearson coefficient of 0.87). On the contrary, these two transcriptomes are clearly different from pVCR94ΔacaCD (Pearson coefficients of 0.12 and −0.04, respectively). In total, pVCR94 and pacaDC 3×FLAG-complemented pVCR94ΔacaCD share 76 differentially expressed genes out of 88 when compared to pVCR94ΔacaCD, which further supports the high similarity between their gene expression profiles (Dataset S1). These findings suggest that AcaCD in wild-type pVCR94 is at least partially active in E. coli under laboratory conditions because an appropriate activating signal is already being sensed and/or the expression of acaC and acaD is not efficiently repressed. This hypothesis is consistent with the fact that acr1 and acr2 negatively regulate conjugative transfer efficiency while not completely abolishing it (Figure 1B). Our results strikingly contrast with the conclusion drawn by others that most of the backbone of IncA/C plasmid pAR060302, including tra genes, is transcriptionally inactive in E. coli [35]. This interpretation is puzzling given the very high nucleotide identity between core regions of IncA/C plasmids, that reads per kilobase per million reads (RPKM) expression values reported for pAR060302 genes expressed at a “low level” such as repA reach several thousand units [35], and that pAR060302 is self-transferable at high frequencies by conjugation under laboratory conditions similar to ours [22].
AcaCD drives the mobility of two unrelated classes of genomic islands
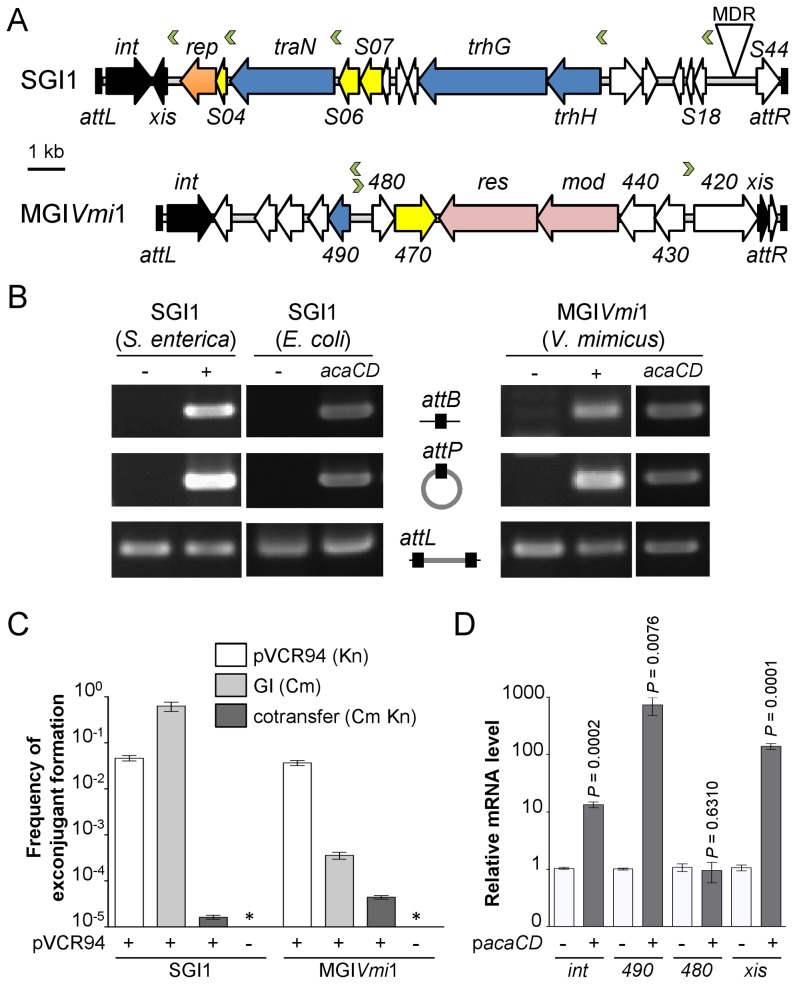
Determination of the AcaCD-bound DNA motif provides the opportunity to investigate the impact of IncA/C plasmids on genome dynamics. For instance, IncA/C plasmids are known to mobilize SGI1 in S. enterica by a yet uncharacterized mechanism [14], [15], [36]. SGI1 elements confer and propagate MDR in various pathogenic bacteria [16], [17], [37]. Data mining using the AcaCD-binding motif revealed putative sites upstream of the genes xis, S004/rep, traN and trhH/trhG likely involved in the mobility (recombination directionality factor and mobilization genes) of SGI1 (Figure 4A and Table 1). We carried out a similar analysis on the sequenced genome of V. mimicus VM573 and identified three AcaCD-binding motifs upstream of three genes, two of which coding for a predicted recombination directionality factor (VMD_06410, xis) and a distant homolog of Vcrx001 (VMD_06490, 29% of identity over 85 amino acid residues), which is a conserved key factor for conjugative transfer of IncA/C plasmids (Table 1) [19]. The third gene, VMD_06480, has no predicted function. The three genes are part of an unannotated 16 511-bp GI integrated into the 3′ end of yicC, that we named MGIVmi1 (Figure 4A and Table 1). MGIVmi1 could confer adaptive traits to its host, notably resistance to bacteriophages conferred by a putative type III restriction-modification (res-mod) (Figure 4A). SGI1 and MGIVmi1 are prototypes of two families of GIs that are phylogenetically unrelated to each other and to IncA/C plasmids.
Figure 4. IncA/C-dependent excision and transfer of GIs.
(A) Schematic representation of SGI1 from S. enterica Typhimurium DT104 and MGIVmi1 from V. mimicus VM573. The left and right junctions (attL and attR) within the host chromosome are indicated. SGI1 (42 596 bp, Genbank AF261825) and MGIVmi1 (16 511 bp, Genbank NZ_ACYV01000005) are integrated into the 3′ end of trmE and yicC (attL sides) in their respective hosts. ORFs with similar function are indicated by colors as follows: black, DNA recombination; orange, DNA replication; blue, conjugative transfer; yellow, regulatory function; pink, putative type III restriction-modification system; white, unknown functions; MDR, multidrug resistance locus. Green chevrons indicate the position and orientation of predicted AcaCD-binding sites (see Table 1). For clarity, ORF names S0XX were shortened as SXX for SGI1 and VMD_06XXX as XXX for MGIVmi1. (B) AcaCD induces SG1 and MGIVmi1 excision. Excision was detected by PCR on genomic DNA to specifically amplify the attB chromosomal site and the attP site resulting from the excision of the GIs in S. enterica Typhimurium or E. coli bearing SGI1 and V. mimicus bearing MGIVmi1. Integrated GIs were detected by amplification of the attL site. Assays were done in strains devoid of plasmid (−), bearing pVCR94ΔX3 (+) or only expressing acaCD (acaCD) from pacaDC 3×FLAG for assays in E. coli or pacaCD in V. mimicus. (C) Intraspecific mobilization of both GIs was assayed using E. coli MG1655 Rf bearing pVCR94ΔX3 and SGI1 or MGIVmi1 as a donor and the otherwise isogenic strain MG1655 Nx as a recipient. Exconjugants were selected for the acquisition of either GI, pVCR94ΔX3, and for cotransfer of both. Transfer frequencies are expressed as the number of exconjugant per donor CFUs. The bars represent the mean and standard deviation values obtained from three independent experiments. The asterisk indicates that the frequency of exconjugant formation was below the detection limit (<10−8). (D) AcaCD induces the expression of the putative excision and mobilization genes of MGIVmi1. Relative mRNA levels of int (VMD_06510), 490 (VMD_06490), 480 (VMD_06480) and xis (VMD_06410) were measured by RT-qPCR assays on cDNA of V. mimicus VM573 devoid of plasmid (−) or expressing acaCD from pacaCD (+). The bars represent the mean and standard deviation values obtained from three independent experiments. Comparison between the strain expressing or not AcaCD were done using two-tailed Student's t-tests and the P-values are indicated above to the bars.
Table 1. Prediction of putative AcaCD-binding sites in SGI1 and MGIVmi1.
| Element | Strand | Start | End | p-valuea | Matched Sequenceb | Downstream gene(s) |
| SGI1 | − | 2016 | 2043 | 3.2e-10 | TTCGCGCCCTAAAAGGGCAGATCCAGAG | xis |
| SGI1 | − | 3541 | 3568 | 1.2e-11 | TAGGTGCCCAAATAGGGCACTTCCAGAG | S004/rep |
| SGI1 | − | 6444 | 6471 | 6.1e-06 | CGTATGCCGCAAAAGGGCAAATAGCGAT | traN |
| SGI1 | − | 13484 | 13511 | 7.9e-09 | GGACTGCCCAAATTGGACAGTTTGGGAG | trhH/trhG |
| SGI1 | − | 16360 | 16387 | 2.6e-12 | AATGTGCCCAAAAAGGGCCAATACAGCG | S018 |
| MGIVmi1 | + | 6832 | 6859 | 1.7e-07 | AAAATGCCCAATCAGGACAGTTTGAGCG | VMD_06480/06470 |
| MGIVmi1 | − | 6827 | 6854 | 7.8e-06 | AAACTGTCCTGATTGGGCATTTTCGCAG | VMD_06490 |
| MGIVmi1 | + | 15770 | 15797 | 2.4e-06 | TTCAGTACCAAAATGGGCAATTACAGAT | VMD_06420/xis |
aOnly output matches with p-values below 1.0e-05 are shown and considered as significant hits.
bMatches were predicted by FIMO (MEME suite) based on the AcaCD logo presented in Figure 3C.
Based on these observations, we hypothesized that expression of acaCD either from pVCR94 or from pBAD30 would trigger the excision of both SGI1 and MGIVmi1 from their respective host chromosome. To verify this hypothesis, we monitored by PCR amplification the formation of a chromosomal attB site and of an attP site resulting from the circularization of the two GIs (Figure 4B). While no spontaneous excision of either GI was detected in the absence of pVCR94, both GIs excised from the chromosome not only in the presence of pVCR94 but also upon ectopic expression of acaCD in cells lacking pVCR94 (Figure 4B). Finally, to confirm that IncA/C plasmids can mobilize the novel GI MGIVmi1, we carried out inter- and intraspecific mating assays using pVCR94 as the helper plasmid and a modified MGIVmi1 carrying res::cat (Cm resistant) as a selectable marker. As a positive control, we used SGI1 and took advantage of its natural selectable markers. For both SGI1 and MGIVmi1, no exconjugant were observed in the absence of pVCR94 (Figure 4C). In contrast, both elements were specifically mobilized in its presence. On one hand, transfer of SGI1 occurred at very high frequency (almost all recipient cells received a copy of SGI1), but co-acquisition of pVCR94 was infrequent. This observation suggests that negative interactions may exist between these mobile genetic elements. On the other hand, MGIVmi1 transferred at a lower rate than pVCR94 (Figure 4C). We confirmed AcaCD-specific expression of the MGIVmi1-borne genes xis and VMD_06490 by real-time-quantitative PCR (RT-qPCR) (Figure 4D). int mRNA level was also specifically increased by AcaCD, despite the lack of a predicted AcaCD-binding motif in its promoter region. This observation suggests that its enhanced expression could be driven by the promoter upstream of xis on a large transcript containing attP on the circular excised form of MGIVmi1. For a reason that remains to be established and despite the presence of a predicted AcaCD-binding motif, VMD_06480 expression was not regulated by AcaCD.
Unlike SGI1, which seem to encode a large set of proteins likely dedicated to mobilization (TraN, TrhG and TrhH), MGIVmi1 only encodes a distant homolog of Vcrx001 (VMD_06490), which is strongly induced by AcaCD (Figure 4D). Based on these differences, we speculate that MGIVmi1 is not as efficient as SGI1 to hijack the conjugative apparatus encoded by IncA/C plasmids. Characterization of the genetic and molecular mechanisms of SGI1 and MGIVmi1 mobilization by IncA/C helper plasmids are ongoing.
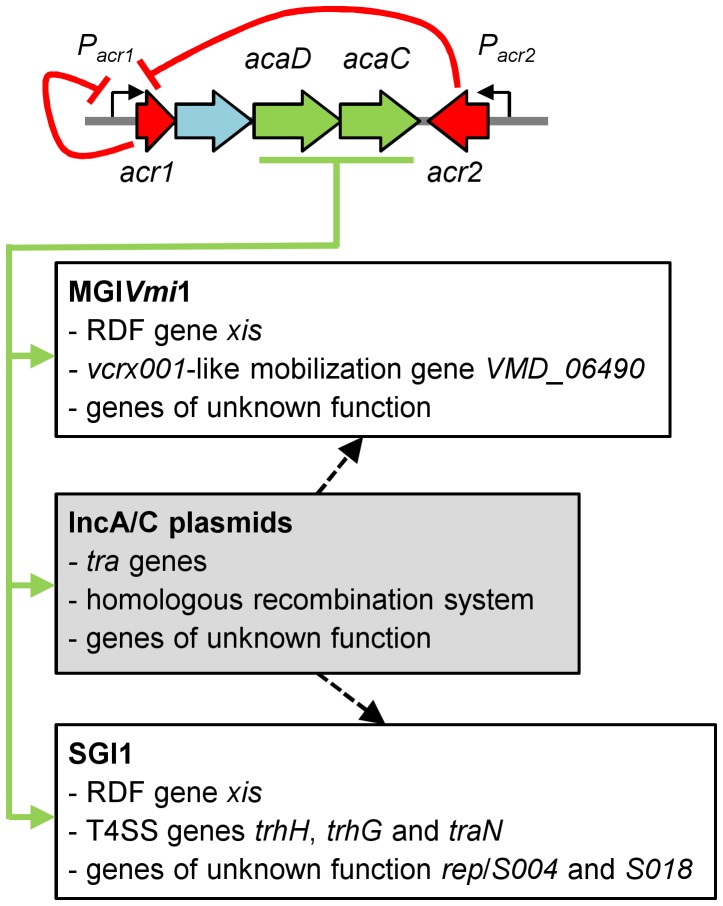
Altogether, these results unraveled a novel example of the multiple and intricate interactions linking phylogenetically unrelated mobile genetic elements. Moreover, our study provides new evidence that many GIs are not defective for their propagation but rather mobilizable GIs, whose biology is just linked and perfectly adapted to other self-transmissible helper elements [15], [38], [39], [40] (Figure 5).
Figure 5. Model of regulation of IncA/C plasmids and interaction with genomic islands.
Expression of the master activator complex AcaCD from the promoter of acr1 (Pacr1) is directly repressed by Acr1 and Acr2 (red arrows). AcaCD directly activates the expression of the transfer genes of pVCR94, as well as the expression of the bet/exo homologous recombination system and numerous genes of unknown function (green arrows). AcaCD triggered the excision of SGI1 and MGIVmi1 by directly activating the expression of the RDF genes xis. AcaCD activates the expression of the vcrx001-like mobilization gene and other genes of unknown function in MGIVmi1. AcaCD also activates the expression of three genes coding for putative component of type IV secretion system (T4SS), as well as genes of unknown function such as S004, rep and S018 in SGI1. IncA/C plasmids likely provide additional functions for genomic islands dissemination such as oriT recognition and processing or formation of the mating pore (black hatched arrows). Genes are color-coded as in Figure 1A.
Concluding remarks
IncA/C conjugative plasmids and SXT/R391 ICEs both rely on transcriptional activator complexes that are reminiscent of the FlhDC master activator of flagellar genes to enable expression of their tra genes. Nevertheless, AcaCD recognizes DNA motifs that are unrelated to the recognition motif of FlhDC [25]. Moreover, AcaCD and SetCD, the master activator of SXT/R391 ICEs, are not exchangeable and are thus expected to recognize completely unrelated DNA motifs. Our study revealed that the AcaCD regulon extends beyond the sole genes involved in the dissemination of IncA/C plasmids and that AcaCD acts as a beacon signaling the presence of a helper plasmid, allowing the propagation of two unrelated classes of GIs. Similarly, SetCD has been shown to trigger the excision of MGIs originating from various pathogenic Vibrio species [38], [39], [41], thereby suggesting that sequences bound by AcaCD- and SetCD-like activators can be easily mimicked by unrelated GIs to regulate their own gene expression. Clearly, our results and observations from others indicate that GIs are not necessarily defective or decaying mobile genetic elements, unable to propagate. Instead, most are likely quiescent parasites awaiting opportunities to hijack helper self-transmissible elements using diverse strategies [14], [38], [42]. Such intricate connections between various genetic elements support their major impact on the evolution of genomes and on the adaptation of bacteria to their environment, particularly in the current context of massive emergence of multidrug resistant pathogens worldwide. Ultimately, future research investigating the regulation of other mobile genetic elements relying on similar transcriptional activator complexes to regulate their own dissemination will unravel unforeseen regulatory networks linking self-transmissible and mobilizable elements.
Materials and Methods
Bacterial strains and bacterial conjugation assays
The bacterial strains used in this study are described in Table 2. The strains were routinely grown in Luria-Bertani (LB) broth at 37°C in an orbital shaker/incubator and were maintained at −80°C in LB broth containing 15% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 µg/ml; chloramphenicol (Cm), 20 µg/ml for E. coli, 30 µg/ml for S. enterica and 10 µg/ml for V. mimicus; erythromycin (Em), 10 µg/ml; kanamycin (Kn), 50 µg/ml or 10 µg/ml for single copy integrants of pOPlacZ; nalidixic acid (Nx), 40 µg/ml; rifampicin (Rf) 50 µg/ml; spectinomycin (Sp), 50 µg/ml; streptomycin (Sm), 200 µg/ml; sulfamethoxazole (Su), 160 µg/ml; tetracycline (Tc), 12 µg/ml; trimethoprim (Tm), 32 µg/ml. Conjugation assays were performed as described elsewhere [19]. To induce expression from pBAD30 and from pAH56 in complementation assays, mating experiments were carried on LB agar plates supplemented with 0.02% L-arabinose or 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), respectively.
Table 2. Strains and plasmids used in this study.
| Strains or plasmids | Relevant genotype or phenotype | References |
| Escherichia coli | ||
| BW25113 | F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ–, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [50] |
| BW25113 Nx | Nxr-derivative of BW25113 | [19] |
| MG1655 Nx | Nxr-derivative of MG1655 | [51] |
| MG1655 Rf | Rfr-derivative of MG1655 | [51] |
| BL21(DE3) | F−, ompT ΔhsdS gal dcm (DE3) | Novagen |
| β2163 | (F−) RP4-2-Tc::Mu ΔdapA::(erm-pir) (Kn Em) | [49] |
| Salmonella enterica | ||
| St038 | serovar Typhimurium; SGI1+ (Ap Cm Tc Su Sp Sm) | F. Malouin |
| Vibrio mimicus | ||
| VM573 | Patient with diarrhea; USA, 1990s; CT+ TCP+ MGIVmi1+ (Ap Sp Su) | [68] |
| Plasmids | ||
| pVCR94 | IncA/C conjugative plasmid; V. cholerae O1 F1939; Rwanda, 1994 | [19] |
| pVCR94ΔX | Sur-derivative of pVCR94 (Su) | [19] |
| pVCR94ΔX2 | Spr-derivative of pVCR94ΔX (Sp Su) | This study |
| pVCR94ΔX3 | Knr-derivative of pVCR94ΔX (Kn Su) | This study |
| pVCR94Δ25 | pVCR94 ΔX2 Δvcrx025 (Sp Su) | This study |
| pVCR94ΔacaC | pVCR94 ΔX2 ΔacaC (Sp Su) | This study |
| pVCR94ΔacaD | pVCR94 ΔX2 ΔacaD (Sp Su) | This study |
| pVCR94ΔacaCD | pVCR94 ΔX2 ΔacaCD (Sp Su) | This study |
| pVCR94Δacr1 | pVCR94 ΔX2 Δacr1 (Sp Su) | This study |
| pVCR94Δacr2 | pVCR94 ΔX2 Δacr2 (Sp Su) | This study |
| pBAD30 | ori p15A araC P BAD (Ap) | [45] |
| pacaC | pBAD30::acaC (Ap) | This study |
| pacaD | pBAD30::acaD (Ap) | This study |
| pacaCD | pBAD30::acaCD (Ap) | This study |
| pacr1 | pBAD30::acr1 (Ap) | This study |
| pacr2 | pBAD30::acr2 (Ap) | This study |
| pGG2B | pBAD30::setDC (Ap) | [69] |
| pSW23T | oriT RP4; oriV R6Kγ (Cm) | [49] |
| pAH56 | oriV R6Kγ; attP λ; lacI; P tac (Kn) | [70] |
| pJL148 | SPA-tag (CBP-TEV site-3×FLAG) (Kn) | [47] |
| pacaDC 3×FLAG | pAH56::acaDC 3×FLAG (Kn) | This study |
| psetDC 3×FLAG | pAH56::setDC 3×FLAG (Kn) | This study |
| pOPlacZ | pAH56 lacZ (Kn) | This study |
| pPromtraL | pOPlacZ PtraL-lacZ (Kn) | This study |
| pPromtraI | pOPlacZ PtraI-lacZ (Kn) | This study |
| pPromtraF | pOPlacZ PtraF-lacZ (Kn) | This study |
| pPromacr1 | pOPlacZ Pacr1-lacZ (Kn) | This study |
| pProm152 | pOPlacZ P152-lacZ (Kn) | This study |
| pINT-ts | oriR101; cI857; λp R-int λ (Ap Ts) | [70] |
| pET-24b(+) | oriV pBR322; lacI; P T7; 6×His; expression vector (Kn) | Novagen |
| pacaDC 6×His | pET-24b(+)::acaDC 6×His (Kn) | This study |
| pSIM6 | λRed recombination thermo-inducible encoding plasmid (Ts Ap) | [52] |
| pKD3 | PCR template for one-step chromosomal gene inactivation (Cm) | [50] |
| pKD13 | PCR template for one-step chromosomal gene inactivation (Kn) | [50] |
| pVI36 | PCR template for one-step chromosomal gene inactivation (Sp) | [51] |
| pCP20 | Flp recombinase thermo-inducible encoding plasmid (Ts Ap Cm) | [53] |
Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Kn, kanamycin; Nx, nalidixic acid; Sp, spectinomycine; Rf, rifampicin; Sm, streptomycine; Su, sulfamethoxazole; Tc, tetracycline; Tm, trimethoprim; Ts, thermosensitive; CT, cholera toxin; TCP, toxin co-regulated pilus.
Molecular biology methods
Genomic and plasmid DNA were prepared using the Wizard Genomic DNA Purification Kit (Promega) and EZ-10 Spin Column Plasmid DNA Minipreps Kit (Biobasic), respectively, according to manufacturer's instructions. All the enzymes used in this study were purchased from New England BioLabs or Enzymatics. PCR assays were performed with the primers described in Table S4. The PCR conditions were as follows: (i) 3 min at 94°C; (ii) 30 cycles of 30 sec at 94°C, 30 sec at the appropriate annealing temperature, and 1 minute/kb at 68°C; and (iii) 5 min at 68°C. When necessary, PCR products were purified using a EZ-10 Spin Column PCR Products Purification Kit (Biobasic) according to manufacturer's instructions. E. coli was transformed by electroporation according to Dower et al. [43]. V. mimicus was transformed by electroporation according to Occhino et al. [44] with modified G buffer (200 mM sucrose, 1 mM Hepes, pH 7.5). Electroporation was carried out in a BioRad GenePulser Xcell apparatus set at 25 µF, 200 V and 1.8 kV using 1-mm gap electroporation cuvettes. Sequencing reactions were performed by the Plateforme de Séquençage et de Génotypage du Centre de Recherche du CHUL (Québec, QC, Canada).
Plasmids and strains construction
Plasmids and oligonucleotides used in this study are listed in Table 2 and S4. Complementation vectors derived from the pBAD30 vector [45]. The ORFs acr1, acr2, acaC, acaD and acaCD were amplified using primers pairs vcrx146.for/vcrx146.rev, vcrx150.for/vcrx150.rev, vcrx149(acaC).for/vcrx149(acaC).rev, vcrx148(acaD).for/vcrx148(acaD).rev, vcrx148(acaD).for/vcrx149(acaC).rev and cloned into the EcoRI restriction site of pBAD30 to generate pacr1, pacr2, pacaC, pacaD and pacaCD, respectively. The transcriptional fusion vector pOPlacZ contains the promoterless lacZ gene from E. coli K12 MG1655 with its native Shine-Dalgarno sequence. This plasmid was constructed by amplifying the lacZ gene using primer pair Op-lacZ-F/Op-lacZ-R and subsequent cloning into the 2 462-bp fragment of PstI-digested pAH56 using the Gibson assembly method [46]. PCR fragments containing the promoter region upstream of acr1, traL, traI, traF and vcrx152 were cloned into the PstI restriction site of pOPlacZ to produce pPromacr1, pPromtraL, pPromtraI, pPromtraF and pProm152, respectively. The vectors pacaDC 3×FLAG and psetDC 3×FLAG used for complementation assays and ChIP-exo experiments derived from pAH56. Briefly, the acaDC and setDC loci were amplified using primer pairs acaDF-NdeI/acaCR-HindIII and setDF-NdeI/setCR-HindIII, respectively, and cloned into pCR2.1 (Invitrogen) to generate pCR2.1::acaDC and pCR2.1::setDC. 3×FLAG was amplified from pJL148 [47] using the primer pair FlagF/FlagR and subsequently cloned into the HindIII site of pCR2.1::acaDC and pCR2.1::setDC to generate pCR2.1::acaDC 3×FLAG and pCR2.1::setDC 3×FLAG. The acaDC 3×FLAG and setDC 3×FLAG inserts were recovered by NdeI/SalI digestion and subsequently cloned into the NdeI/SalI-digested pAH56 to generate pacaDC 3×FLAG and psetDC 3×FLAG [48]. pacaDC 6×His was obtained by cloning of acaDC amplified with acaDF-NdeI/acaCR-HindIII into pET-24b(+) (Novagen). pRes for insertion of a Cm marker into MGIVmi1 was obtained cloning into EcoRI/BamHI-digested pSW23T [49] the 732-bp PCR fragment amplified using the primer pair GIVmi-res2F/GIVmi-res2R on genomic DNA of V. mimicus VM573.
Deletion mutants of pVCR94ΔX were constructed using the one-step chromosomal gene inactivation technique and are listed in the Table 1 [19], [50]. Primers used are listed in Table S4. The pVCR94ΔX derivatives pVCR94ΔX2 (Sp) and pVCR94ΔX3 (Kn) were constructed using primer pair 94DelXnoFRT.for/94DelXnoFRT.rev and pVI36 and pKD13 as templates, respectively [50], [51]. Subsequent deletions of vcrx025, acr1, acr2, acaC, acaD and acaCD were done on pVCR94ΔX2 using primer pairs 94Delvcrx025.for/94Delvcrx025.rev, 94Delvcrx146.for/94Delvcrx146.rev, 94Delvcrx150.for/94Delvcrx150.rev, 94DelacaC.for/94DelacaC.rev, 94DelacaD.for/94DelacaD.rev and 94DelacaD.for/94DelacaC.rev, respectively, and pKD3 as the template. The λRed recombination system was expressed using pSIM6 as described by Datta et al. [52]. When possible, the antibiotic resistance cassette was removed from the resulting construction by Flp-catalyzed excision using the pCP20 vector [53]. All deletions were designed to be non-polar and verified by PCR and antibiotic resistance profiling.
MGIVmi1 was labelled with a Cm resistance marker in V. mimicus VM573 by inserting the pSW23T-derivative suicide plasmid pRes into the putative type III restriction gene res. Briefly, pRes was mobilized from E. coli β2163 to V. mimicus VM573 and exconjugants were selected on LB agar plates supplemented with 10 µg/ml chloramphenicol in the absence of DL-α,ε-diaminopimelic acid according to Demarre et al. [49].
Proteins purification and analysis
E coli BL21(DE3) carrying pacaDC6×His was grown to OD600 nm = 0.5 and induced for 2 hours with 1 mM of IPTG. Purification of AcaC tagged with a 6×His C-terminal epitope was done using a Ni-NTA affinity chromatography following the manufacturer's instructions (Qiagen). Migration of the purified proteins on 12% SDS-PAGE gel and Western blotting experiments using anti-6×His tag antibody were done as described by the manufacturer (Life Technologies/Invitrogen). Protein identification was performed at the proteomics platform of the Université de Sherbrooke on an excised acrylamide gel slice digested with trypsin and subjected to LC/MS/MS analysis as previously described [54].
ChIP-exo experiments and RNA sequencing
A thorough description of the chromatin immunoprecipitation coupled with exonuclease digestion and RNA sequencing procedures is given in Text S1 and Table S5.
RNA isolation and qRT-PCR
Cells from V. mimicus VM573 that contain no vector or pacaCD were recovered after 2.5 hours of induction with 0.02% of arabinose. Total RNA extraction was done using an RNeasy minikit (Qiagen) following the manufacturer's instructions. Purified RNA samples were subsequently subjected to gDNA digestion using Turbo DNase (Ambion) following the manufacturer's instructions. RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). qRT-PCR assays were performed on the RNomics platform of the Laboratoire de Génomique Fonctionnelle de l'Université de Sherbrooke (http://lgfus.ca). Reverse transcription was performed on 2.2 µg total RNA with Transcriptor reverse transcriptase, random hexamers, dNTPs (Roche Diagnostics), and 10 units of RNAseOUT (Invitrogen Life Technologies) following the manufacturer's protocol in a total volume of 20 µl. All forward and reverse primers were individually resuspended to 20–100 µM stock solutions in Tris-EDTA buffer (IDT) and diluted as a primer pair to 1 µM in RNase DNase-free water (IDT). Quantitative PCR (qPCR) reactions were performed in 10-µl volumes in 96 well plates on a CFX-96 thermocycler (Bio-Rad) with 5 µL of 2× iTaq Universal SYBR Green Supermix (Bio-Rad), 10 ng (3 µl) cDNA, and 200 nM final (2 µl) primer pair solutions. The following cycling conditions were used: 3 min at 95°C; 50 cycles: 15 s at 95°C, 30 s at 60°C, 30 s at 72°C. Relative expression levels of int (VMD_06510), VMD_06490, VMD_06480 and xis (VMD_06410) were calculated using a model taking into account multiple stably expressed reference genes [55] and housekeeping genes rpoZ and gyrA evaluated by geNorm [56]. Primer design (see Table S4) and validation were evaluated as described elsewhere [57]. In every qPCR run, a no-template control was performed for each primer pair and a no-reverse transcriptase control was performed for each cDNA preparation. Experiments were carried out three times on three biological replicates and combined.
Detection of SGI1 and MGIVmi1 excision
Excision of the GIs was detected by PCR on genomic DNA of the strains containing either SGI1 (S. enterica or E. coli) or MGIVmi1 (V. mimicus) using the primers listed in the Table S4. For SGI1, the attL site was amplified using primer pair SGI-1attL.for/SGI-1attL.rev in S. enterica and EcU7-L12.for/SGI-1attL.rev in E. coli [15]. The chromosomal site attB was detected using SGI-1attL.for/SGI-1attR.rev in S. enterica and EcU7-L12.for/Ec104D.rev in E. coli. The attP site carried by the extrachromosomal circular form of the element was amplified using the primer pair SGI-1attL.rev/SGI-1attR.for. Based on the same methodology for MGIVmi1 in V. mimicus, primer pairs GIVmi-A/GIVmi-B, GIVmi-A/GIVmi-D and GIVmi-B/GIVmi-C were used to detect attL, attB and attP, respectively.
Phylogenetic analyses
The molecular phylogenetic analysis of the acr1-vcr147-acaDC-acr2 locus was conducted in MEGA6 [58]. The nucleotide sequence of the 2452-bp sequence of pVCR94 starting at the initiation codon of acr1 and ending at the initiation codon of acr2 was used to search for homologous sequences in the Genbank Nucleotide collection (nr/nt) database using Megablast [59]. Phylogenetic analyses were computed using a nucleotide alignment generated by MUSCLE [60]. The evolutionary history was inferred by using the Maximum Likelihood method. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. Identical procedures were used for the molecular phylogenetic analysis of the 1101-bp repA gene of pVCR94 (Figure S1).
Data availability
Fastq files for each experiment were deposited at the NCBI Sequence Read Archive (SRA) under accession numbers SRX675564 and SRR1544064 for ChIP-exo, SRX675582 and SRR1544143 for total RNAseq as well as SRX675814 and SRR1544479 for 5′-RACE. Complete data from aligned reads for ChIP-exo and RNA-seq can also be visualized using the UCSC genome browser at http://bioinfo.ccs.usherbrooke.ca/pVCR94.html.
Supporting Information
Molecular phylogenetic analysis of the repA replication initiation gene by Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Hasegawa-Kishino-Yano model [67]. The tree with the highest log likelihood (−2386.8588) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.2014)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 45 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. There were a total of 1101 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [58]. The background color of each leaf indicates the original host species from which each plasmid was isolated. *, XNC1_p lacks the acr1-vcrx147-acaDC-acr2 regulation cluster.
(TIF)
The 3×FLAG epitope does not alter the function of AcaCD. The donor strain E. coli MG1655 Rf was used to transfer pVCR94ΔX or its ΔacaCD mutant (Su Sp) to the recipient strain MG1655 Nx. AcaCD and SetCD complementation assays were carried out with the native D subunit and the C subunit C-terminally fused to the 3×FLAG epitope expressed from an IPTG-inducible promoter (Ptac) provided by pAH56, i.e. pacaDC 3×FLAG (acaDC 3×FLAG), psetDC 3×FLAG (setDC 3×FLAG) or empty pAH56 (0) integrated into the attB λ locus. Exconjugants were selected as Nx Su Sp colonies. The bars represent the mean and standard deviation values obtained from at least 3 biological replicates. The asterisks indicate that the frequency of transfer was below the limit of detection (<10−8).
(TIF)
Comparison of the genetic context of genes coding for AcaCD and SetCD orthologs in IncA/C plasmids and SXT/R391 ICEs. Schematic representation of regulatory regions of pVCR94 from V. cholerae O1 El Tor (NC_023291.1) and SXT from V. cholerae O139 (AY055428.1). Arrows of similar color represent genes predicted to have similar functions: green, transcriptional activator; yellow, putative transcriptional regulator; red, transcriptional repressor; blue, conjugative transfer; light blue, putative lytic transglycosylase; orange, replication; black, site-specific recombination; white, unknown function. Blue stars indicate the position of origins of transfer (oriT). The orange star indicates the position of the origin of replication (oriV) of pVCR94 based on identity with pRA1 from Aeromonas hydrophila (NC_012885). The black star indicates the position of the attP site for chromosomal integration of SXT by site-specific recombination. The percent of identity of orthologous proteins are indicated on dashed lines.
(TIF)
Alignment of AcaCD-dependent promoters in pVCR94. The AcaCD motif is as represented in Figure 3C. AcaCD boxes obtained by MAST analysis are shown in bold green capital letters with their respective p-value and downstream regulated gene. The positions of the transcription start sites obtained from 5′-RACE data are indicated in bold blue capital letters and underlined (TSS). Shine-Dalgarno sequences (SD) are underlined while start codons are in capital letters. The approximate positions of the −35 and −10 regions are highlighted in gray. The length of spacers between the represented transcription start sites and the Shine-Dalgarno regions is indicated in base pairs. Since no clear 5′-RACE signal was observed for vcrx035 and vcrx098, the approximate positions of expected transcription start sites are underlined and shown in bold black capital letters.
(TIF)
Open reading frames (ORFs) of pVCR94 coding for putative transcriptional regulators.
(DOCX)
acr1-vcrx147-acaDC-acr2 orthologous clusters in IncA/C plasmids.
(DOCX)
AcaCD-regulated promoters identified by ChIP-exo and 5′-RACE.
(DOCX)
Primers used in this study.
(DOCX)
Illumina libraries sequenced in this study.
(DOCX)
RNAseq transcriptome profiling of wild-type pVCR94, pVCR94ΔacaCD and complemented pVCR94ΔacaCD.
(XLS)
Additional experimental procedures.
(DOCX)
Acknowledgments
We are grateful to François Malouin and Fabiano Thompson for the kind gift of S. enterica Typhimurium St038 and V. mimicus VM573, respectively. We thank Guillaume Lauzon, Dominic Poulin-Laprade, Pierre-Etienne Jacques and the Centre de calcul scientifique of the Université de Sherbrooke for technical assistance, as well as Eric Bordeleau and Alain Lavigueur for their insightful comments on the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Fastq files for each experiment were deposited at the NCBI Sequence Read Archive (SRA) under accession numbers SRX675564 and SRR1544064 for ChIP-exo, SRX675582 and SRR1544143 for total RNAseq as well as SRX675814 and SRR1544479 for 5′-RACE. Complete data from aligned reads for ChIP-exo and RNA-seq can also be visualized using the UCSC genome browser at http://bioinfo.ccs.usherbrooke.ca/pVCR94.html.
Funding Statement
This work was supported by the Fonds Québécois de la recherche sur la nature et les technologies (DM, SR and VB), a Discovery Grant and Discovery Acceleration Supplement from the Natural Sciences and Engineering Council of Canada (VB) and the Natural Science Foundation of China (31370149) (PL). VB holds a Canada Research Chair in molecular bacterial genetics. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2014) Antimicrobial resistance: global report on surveillance. World Health Organization. pp. 257. [Google Scholar]
- 2. Johnson TJ, Lang KS (2012) IncA/C plasmids: An emerging threat to human and animal health? Mob Genet Elements 2: 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aoki T, Egusa S, Kimura T, Watanabe T (1971) Detection of R factors in naturally occurring Aeromonas salmonicida strains. Appl Microbiol 22: 716–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wantanabe T, Aoki T, Ogata Y, Egusa S (1971) Anbtibiotics and drug resistance in animals. R factors related to fish culturing. Ann N Y Acad Sci 182: 383–410. [DOI] [PubMed] [Google Scholar]
- 5. Walsh TR, Weeks J, Livermore DM, Toleman MA (2011) Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11: 355–362. [DOI] [PubMed] [Google Scholar]
- 6. Peirano G, Ahmed-Bentley J, Fuller J, Rubin JE, Pitout JD (2014) Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: the first 3 years. J Clin Microbiol 52: 1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, et al. (2014) Prevalence and molecular characterisation of New Delhi metallo-beta-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents 30–7. [DOI] [PubMed] [Google Scholar]
- 8. Dortet L, Poirel L, Nordmann P (2014) Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, et al. (2009) Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol 191: 4750–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, et al. (2007) Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lindsey RL, Frye JG, Fedorka-Cray PJ, Meinersmann RJ (2011) Microarray-based analysis of IncA/C plasmid-associated genes from multidrug-resistant Salmonella enterica . Appl Environ Microbiol 77: 6991–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Folster JP, Pecic G, McCullough A, Rickert R, Whichard JM (2011) Characterization of bla(CMY)-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog Dis 8: 1289–1294. [DOI] [PubMed] [Google Scholar]
- 13. Arpin C, Thabet L, Yassine H, Messadi AA, Boukadida J, et al. (2012) Evolution of an incompatibility group IncA/C plasmid harboring bla CMY-16 and qnrA6 genes and its transfer through three clones of Providencia stuartii during a two-year outbreak in a Tunisian burn unit. Antimicrob Agents Chemother 56: 1342–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Douard G, Praud K, Cloeckaert A, Doublet B (2010) The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One 5: e15302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doublet B, Boyd D, Mulvey MR, Cloeckaert A (2005) The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55: 1911–1924. [DOI] [PubMed] [Google Scholar]
- 16. Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A (2006) The genetics of Salmonella genomic island 1. Microbes Infect 8: 1915–1922. [DOI] [PubMed] [Google Scholar]
- 17. Hall RM (2010) Salmonella genomic islands and antibiotic resistance in Salmonella enterica . Future Microbiol 5: 1525–1538. [DOI] [PubMed] [Google Scholar]
- 18. Fernandez-Alarcon C, Singer RS, Johnson TJ (2011) Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6: e23415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carraro N, Sauve M, Matteau D, Lauzon G, Rodrigue S, et al. (2014) Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front Microbiol 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, et al. (2009) Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5: e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allen KJ, Poppe C (2002) Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by beta-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can J Vet Res 66: 137–144. [PMC free article] [PubMed] [Google Scholar]
- 22. Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, et al. (2010) bla CMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob Agents Chemother 54: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han J, Lynne AM, David DE, Nayak R, Foley SL (2012) Sequencing of plasmids from a multi-antimicrobial resistant Salmonella enterica serovar Dublin strain. Food Research International 45: 931–934. [Google Scholar]
- 24. Cooper KK, Mandrell RE, Louie JW, Korlach J, Clark TA, et al. (2014) Complete Genome Sequences of Two Escherichia coli O145:H28 Outbreak Strains of Food Origin. Genome Announc 2 10.1128/genomeA.00482-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu X, Matsumura P (1994) The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol 176: 7345–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernandez-Lopez R, Del Campo I, Revilla C, Cuevas A, de la Cruz F (2014) Negative feedback and transcriptional overshooting in a regulatory network for horizontal gene transfer. PLoS Genet 10: e1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, et al. (2007) An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315: 251–252. [DOI] [PubMed] [Google Scholar]
- 28. Hochhut B, Waldor MK (1999) Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC . Mol Microbiol 32: 99–110. [DOI] [PubMed] [Google Scholar]
- 29. Beaber JW, Hochhut B, Waldor MK (2002) Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae . J Bacteriol 184: 4259–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beaber JW, Hochhut B, Waldor MK (2004) SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427: 72–74. [DOI] [PubMed] [Google Scholar]
- 31. Rhee HS, Pugh BF (2012) ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol 10.1002/0471142727.mb2124s100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36. [PubMed] [Google Scholar]
- 33. Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54. [DOI] [PubMed] [Google Scholar]
- 34. Browning DF, Busby SJ (2004) The regulation of bacterial transcription initiation. Nat Rev Microbiol 2: 57–65. [DOI] [PubMed] [Google Scholar]
- 35. Lang KS, Danzeisen JL, Xu W, Johnson TJ (2012) Transcriptome mapping of pAR060302, a bla CMY-2-positive broad-host-range IncA/C plasmid. Appl Environ Microbiol 78: 3379–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kiss J, Nagy B, Olasz F (2012) Stability, entrapment and variant formation of Salmonella genomic island 1. PLoS One 7: e32497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boyd DA, Shi X, Hu QH, Ng LK, Doublet B, et al. (2008) Salmonella genomic island 1 (SGI1), variant SGI1-I, and new variant SGI1-O in Proteus mirabilis clinical and food isolates from China. Antimicrob Agents Chemother 52: 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Daccord A, Ceccarelli D, Burrus V (2010) Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol 78: 576–588. [DOI] [PubMed] [Google Scholar]
- 39. Daccord A, Mursell M, Poulin-Laprade D, Burrus V (2012) Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J Bacteriol 194: 5794–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bellanger X, Payot S, Leblond-Bourget N, Guedon G (2014) Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38: 720–760. [DOI] [PubMed] [Google Scholar]
- 41. Daccord A, Ceccarelli D, Rodrigue S, Burrus V (2013) Comparative analysis of mobilizable genomic islands. J Bacteriol 195: 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quiles-Puchalt N, Carpena N, Alonso JC, Novick RP, Marina A, et al. (2014) Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases. Proc Natl Acad Sci U S A 111: 6016–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dower WJ, Miller JF, Ragsdale CW (1988) High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16: 6127–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM (1998) Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol 29: 1493–1507. [DOI] [PubMed] [Google Scholar]
- 45. Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gibson DG (2011) Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, et al. (2004) Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res 3: 463–468. [DOI] [PubMed] [Google Scholar]
- 48. Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183: 6384–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, et al. (2005) A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156: 245–255. [DOI] [PubMed] [Google Scholar]
- 50. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ceccarelli D, Daccord A, Rene M, Burrus V (2008) Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 190: 5328–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Datta S, Costantino N, Court DL (2006) A set of recombineering plasmids for gram-negative bacteria. Gene 379: 109–115. [DOI] [PubMed] [Google Scholar]
- 53. Cherepanov PP, Wackernagel W (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14. [DOI] [PubMed] [Google Scholar]
- 54. Boisvert FM, Ahmad Y, Gierlinski M, Charriere F, Lamont D, et al. (2012) A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics 11: M111 011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brosseau JP, Lucier JF, Lapointe E, Durand M, Gendron D, et al. (2010) High-throughput quantification of splicing isoforms. RNA 16: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214. [DOI] [PubMed] [Google Scholar]
- 60. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 62. Tamamura Y, Tanaka K, Akiba M, Kanno T, Hatama S, et al. (2013) Complete nucleotide sequences of virulence-resistance plasmids carried by emerging multidrug-resistant Salmonella enterica Serovar Typhimurium isolated from cattle in Hokkaido, Japan. PLoS One 8: e77644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, et al. (2013) Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coulombe C, Poitras C, Nordell-Markovits A, Brunelle M, Lavoie MA, et al. (2014) VAP: a versatile aggregate profiler for efficient genome-wide data representation and discovery. Nucleic Acids Res 42: W485–W493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174. [DOI] [PubMed] [Google Scholar]
- 68. Thompson CC, Vicente AC, Souza RC, Vasconcelos AT, Vesth T, et al. (2009) Genomic taxonomy of Vibrios. BMC Evol Biol 9: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Garriss G, Poulin-Laprade D, Burrus V (2013) DNA-damaging agents induce the RecA-independent homologous recombination functions of integrating conjugative elements of the SXT/R391 family. J Bacteriol 195: 1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183: 6384–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular phylogenetic analysis of the repA replication initiation gene by Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Hasegawa-Kishino-Yano model [67]. The tree with the highest log likelihood (−2386.8588) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.2014)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 45 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. There were a total of 1101 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [58]. The background color of each leaf indicates the original host species from which each plasmid was isolated. *, XNC1_p lacks the acr1-vcrx147-acaDC-acr2 regulation cluster.
(TIF)
The 3×FLAG epitope does not alter the function of AcaCD. The donor strain E. coli MG1655 Rf was used to transfer pVCR94ΔX or its ΔacaCD mutant (Su Sp) to the recipient strain MG1655 Nx. AcaCD and SetCD complementation assays were carried out with the native D subunit and the C subunit C-terminally fused to the 3×FLAG epitope expressed from an IPTG-inducible promoter (Ptac) provided by pAH56, i.e. pacaDC 3×FLAG (acaDC 3×FLAG), psetDC 3×FLAG (setDC 3×FLAG) or empty pAH56 (0) integrated into the attB λ locus. Exconjugants were selected as Nx Su Sp colonies. The bars represent the mean and standard deviation values obtained from at least 3 biological replicates. The asterisks indicate that the frequency of transfer was below the limit of detection (<10−8).
(TIF)
Comparison of the genetic context of genes coding for AcaCD and SetCD orthologs in IncA/C plasmids and SXT/R391 ICEs. Schematic representation of regulatory regions of pVCR94 from V. cholerae O1 El Tor (NC_023291.1) and SXT from V. cholerae O139 (AY055428.1). Arrows of similar color represent genes predicted to have similar functions: green, transcriptional activator; yellow, putative transcriptional regulator; red, transcriptional repressor; blue, conjugative transfer; light blue, putative lytic transglycosylase; orange, replication; black, site-specific recombination; white, unknown function. Blue stars indicate the position of origins of transfer (oriT). The orange star indicates the position of the origin of replication (oriV) of pVCR94 based on identity with pRA1 from Aeromonas hydrophila (NC_012885). The black star indicates the position of the attP site for chromosomal integration of SXT by site-specific recombination. The percent of identity of orthologous proteins are indicated on dashed lines.
(TIF)
Alignment of AcaCD-dependent promoters in pVCR94. The AcaCD motif is as represented in Figure 3C. AcaCD boxes obtained by MAST analysis are shown in bold green capital letters with their respective p-value and downstream regulated gene. The positions of the transcription start sites obtained from 5′-RACE data are indicated in bold blue capital letters and underlined (TSS). Shine-Dalgarno sequences (SD) are underlined while start codons are in capital letters. The approximate positions of the −35 and −10 regions are highlighted in gray. The length of spacers between the represented transcription start sites and the Shine-Dalgarno regions is indicated in base pairs. Since no clear 5′-RACE signal was observed for vcrx035 and vcrx098, the approximate positions of expected transcription start sites are underlined and shown in bold black capital letters.
(TIF)
Open reading frames (ORFs) of pVCR94 coding for putative transcriptional regulators.
(DOCX)
acr1-vcrx147-acaDC-acr2 orthologous clusters in IncA/C plasmids.
(DOCX)
AcaCD-regulated promoters identified by ChIP-exo and 5′-RACE.
(DOCX)
Primers used in this study.
(DOCX)
Illumina libraries sequenced in this study.
(DOCX)
RNAseq transcriptome profiling of wild-type pVCR94, pVCR94ΔacaCD and complemented pVCR94ΔacaCD.
(XLS)
Additional experimental procedures.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Fastq files for each experiment were deposited at the NCBI Sequence Read Archive (SRA) under accession numbers SRX675564 and SRR1544064 for ChIP-exo, SRX675582 and SRR1544143 for total RNAseq as well as SRX675814 and SRR1544479 for 5′-RACE. Complete data from aligned reads for ChIP-exo and RNA-seq can also be visualized using the UCSC genome browser at http://bioinfo.ccs.usherbrooke.ca/pVCR94.html.
Fastq files for each experiment were deposited at the NCBI Sequence Read Archive (SRA) under accession numbers SRX675564 and SRR1544064 for ChIP-exo, SRX675582 and SRR1544143 for total RNAseq as well as SRX675814 and SRR1544479 for 5′-RACE. Complete data from aligned reads for ChIP-exo and RNA-seq can also be visualized using the UCSC genome browser at http://bioinfo.ccs.usherbrooke.ca/pVCR94.html.