Abstract
Dendritic cells (DCs) are potent professional antigen presenting cells (APC) that activate naïve T cells. Interaction of ICAM-1 and LFA-1 molecules on each cell is required for T cell conjugation to DCs which leads to naïve CD4+ T cell activation and proliferation. Nanoparticles capable of blocking LFA-1/ICAM-1 interaction were studied as inhibitors of T cell conjugation to DCs. Primary DCs were primed with ovalbumin, then treated with a peptide that binds ICAM-1 (LABL), a peptide that binds LFA-1 (cIBR) or the same peptides covalently linked to the surface of poly(dl-lactic-co-glycolic acid) nanoparticles (NPs). LABL-NPs and cIBR-NPs rapidly bound to DCs and inhibited T cell conjugation to DCs to a greater extent than the free peptides, unconjugated nanoparticles (NPs), anti-ICAM-1 antibodies and anti-LFA-1 antibodies. In addition, DCs treated with NPs or with cIBR-NPs stimulated the proliferation of T cells, but DCs treated with LABL-NPs did not stimulate T cell proliferation. Nanoparticles targeting ICAM-1 or LFA-1 also altered cytokine production by DC cocultured with T cells when compared to free ligands suggesting these NPs may offer a unique tool for shaping T cell response.
Keywords: peptides, nanoparticles, targeted delivery, dendritic cells, T cell
Professional antigen presenting cells (APC) such as dendritic cells (DCs) help orchestrate immune responses to foreign antigens by capturing antigen and loading it onto major histocompatibilty complex (MHC) class II. The antigen-primed APC then present the antigen to naïve CD4+T cells which express cognate T cell receptors.1 The resulting immunological synapse formed between T cells and APCs often initiates signaling events for T cell proliferation and effector function such as cytokine production.2 Leukocyte function-associated antigen-1 (LFA-1; primarily on T cells) binding to intercellular adhesion molecule 1 (ICAM-1; primarily on DCs) can prolong the immunological synapse and support T cell activation.3-5 Targeting of these molecules is known to modify T cell activation.6
Molecules that mediate cell adhesion or signaling are typically present in large numbers at the cell-cell interface. Several studies have shown that multivalency can enhance the binding of ligands to these types of receptors and shift the response of targeted cells.7-10 For example, multivalent presentation of anti-CD20 monoclonal antibody fragments can enhance the targeting of B cell antigen CD20.10 Similarly, multiple copies of peptide ligands on a polymer backbone can improve binding to CD21 on B cells when compared to free ligand.11 Arrays of RGD peptides on micelles have been consistently found to dramatically enhance binding to αvβ3 receptors in contrast to free RGD peptides.12
The response of cells targeted by multivalent ligands is often unexpected. Antigen valency has been found to be a key parameter affecting binding to B cells and cellular response.13 High valency antigen arrays induced antibody production by B cells, while low valency antigen arrays did not.13 Multivalent ligands induced calcium influx in a dose dependent manner, whereas the same molar concentration of free ligand did not.13 Ligand valency also affects the response of leukocytes undergoing rolling adhesion. For example, multivalent L-selectin ligands clustered L-selectin and induced L-selectin shedding, but the corresponding monovalent ligands did not.14-15 Thus, multivalent ligands can affect cell response in addition to enhancing ligand binding.
The binding of LFA-1 and ICAM-1 is also controlled by changes in avidity resulting from receptor clustering.16-17 Cell adhesion mediated by ICAM-1 and LFA-1 involves multivalent interaction between these two molecules on opposing cells (APC and T cell). LABL (ITDGEATDSG) is a peptide modeled after the I domain of LFA-1 which is the binding site of ICAM-1. cIBR (cyclo 1,12 Pen-PRGGSVLVTGC) is a cyclic peptide derived from domain 1 of ICAM-1 which binds to the I domain of LFA-1.18-19 LABL and cIBR peptides inhibit homotypic and heterotypic T cell adhesion as well as mixed lymphocyte reactions.20-22
Nanoparticles modified with these peptides (cLABL-NPs and cIBR-NPs) were previously used to specifically target ICAM-1 and LFA-1 expressing cells, respectively.23-25 NPs targeting these receptors were found to bind cells and rapidly internalize via receptor-mediated endocytosis. In addition, cIBR-NP blocked the adhesion of T cells to lung epithelial cells expressing a high level of ICAM-1.23 Since the oligomeric states of LFA-1 and ICAM-1 molecules contribute to their ability to regulate T cell responses, it was hypothesized that these nanoparticles may bind with high avidity to ICAM-1 or LFA-1 on DCs. Such targeted NPs were suspected to be better inhibitors of T cell conjugation to DCs compared to free ligands and a potential tool to alter cell response.
Results
Characterization of nanoparticles and peptide-conjugated nanoparticles
A variety of targeted and control NPs were synthesized. The size of all PLGA NP formulations was less than 200 nm and NPs possessed a negative zeta potential (Table 1). Low polydispersity values (<0.1) suggested a relatively narrow particle size distribution. LABL and cIBR peptides were successfully conjugated to PLGA-NPs. The zeta potential of LABL-NPs was more negative than unconjugated NPs, presumably due to the LABL peptide having a net charge of −3. cIBR-NPs had a less negative charge than unconjugated NPs. Pluronic-COOH groups were probably masked by the cIBR peptide, which has a net charge of +1. The amounts of LABL and cIBR peptides attached to NPs were determined by quantifying the unconjugated peptide remaining in the medium after the conjugation reaction.23 LABL was conjugated with a high density, while cIBR density closely matched a previous publication (Table 2).
Table 1. Nanoparticle properties of specified formulations*.
| NP | LABL-NP | cIBR-NP | |
|---|---|---|---|
| Effective diameter (nm) | 154.6 ± 10.0 | 172.6 ± 2.5 | 171.9 ± 4.3 |
| Polydispersity | 0.026 ± 0.030 | 0.070 ± 0.039 | 0.055 ± 0.005 |
| Zeta potential (mV) | −21.4 ± 0.4 | −34.7± 2.0 | −13.9 ± 0.8 |
Values are representative of three experiments (mean ± S.D.).
Table 2. Density of peptides on the surface of nanoparticles*.
| Size (nm) |
Total surface area (m2/g of PLGA) |
peptide density (pmol/cm2) |
|
|---|---|---|---|
| LABL-NP | 172.6 | 25.9 | 265.5 ± 78.5 |
| cIBR-NP | 171.9 | 26.1 | 39.5 ± 3.0 |
Values are representative of three experiments (mean ± S.D.)
23 Relative expression of ICAM-1 and LFA-1 on DCs and T cells
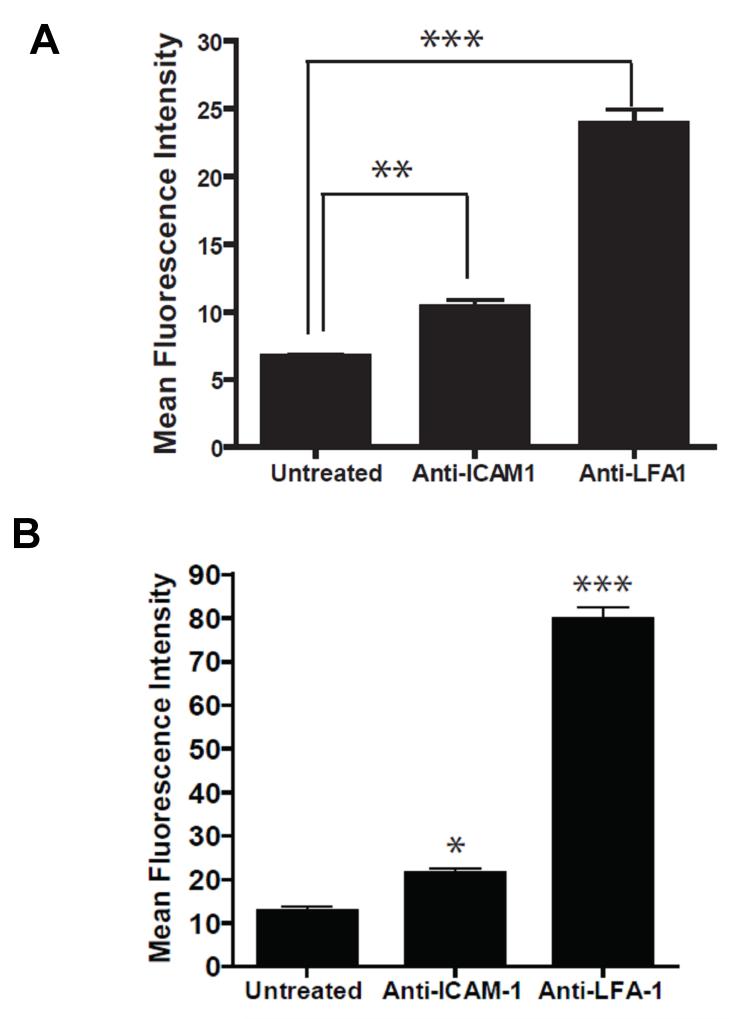
Expression levels of ICAM-1 and LFA-1 on activated bone marrow derived dendritic cells (DCs) matured with TNF-α and primed with OVA were determined using fluorescent anti-ICAM-1 and anti-LFA-1. DCs expressed both ICAM-1 and LFA-1 indicating that either LABL or cIBR could be used to target these adhesion molecules on DCs (Figure 2A). For these studies we used primary CD4+ T cells, which express an ovalbumin specific T cell receptor (OT-II). Flow cytometry data showed minimal expression of ICAM-1 on primary OT-II T cells but high expression of LFA-1 as expected (Figure 2B). Although NPs were not directly incubated with OT II T cells, the relative expression of these two receptors was provided for reference.
Figure 2.
(A) ICAM-1 and LFA-1 expression on dendritic cells and on (B) T cells. OVA-primed DCs and T cells were incubated with anti-ICAM-1 and anti-LFA-1 for 45 min on ice. Cells were washed and analyzed by flow cytometry. ** indicates p<0.001 and * indicates p<0.05.
Both LABL-NPs and cIBR-NPs exhibited rapid binding to DCs
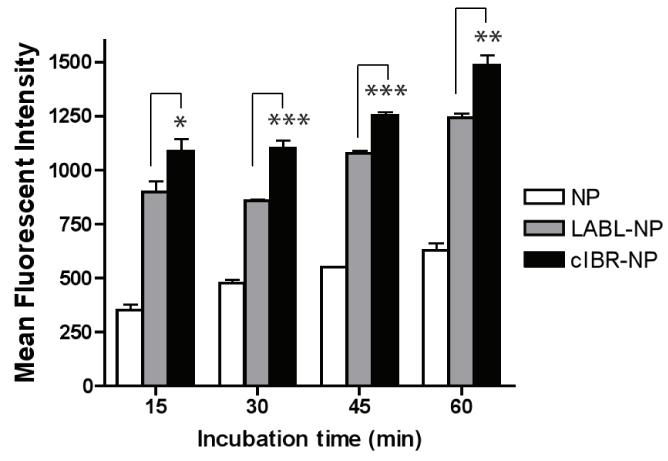
LABL peptide binds specifically to the domain 1 of ICAM-1, whereas cIBR peptide binds the I domain of LFA-1.18-19 LABL-NPs, cIBR-NPs and unconjugated NPs were incubated with DCs to investigate the binding by DCs. Immature DCs were stimulated with TNF-α and primed with OVA for 24 hrs prior to addition of NPs. In comparison to untargeted NPs, fluorescent intensities of DCs incubated with LABL-NPs or cIBR-NPs were much higher than cells incubated with untargeted NPs at all incubation times. Fluorescence intensities of DCs incubated 45 minutes with LABL-NPs or cIBR-NPs were ~2 and ~2.3 fold greater than NPs, respectively, indicating that the interactions of LABL-NPs and cIBR-NPs with DCs occurred more rapidly and to a greater extent compared to untargeted NPs (Figure 3).
Figure 3.
Binding of NPs, LABL-NPs or cIBR-NPs to dendritic cells. NPs, LABL-NPs and cIBR-NPs were incubated with OVA-primed and TNF-α activated DCs for 15, 30, 45 or 60 min at 37 °C and analyzed by flow cytometry. The interaction of cIBR-NPs with dendritic cells was significantly greater than LABL-NPs and untargeted NPs at all time points. The fluorescence of dendritic cells incubated with each of the NP types increased with time. * indicates p < 0.05, ** indicates p < 0.01 and *** indicates p < 0.001.
Fluorescence microscopy of DCs binding nanoparticles
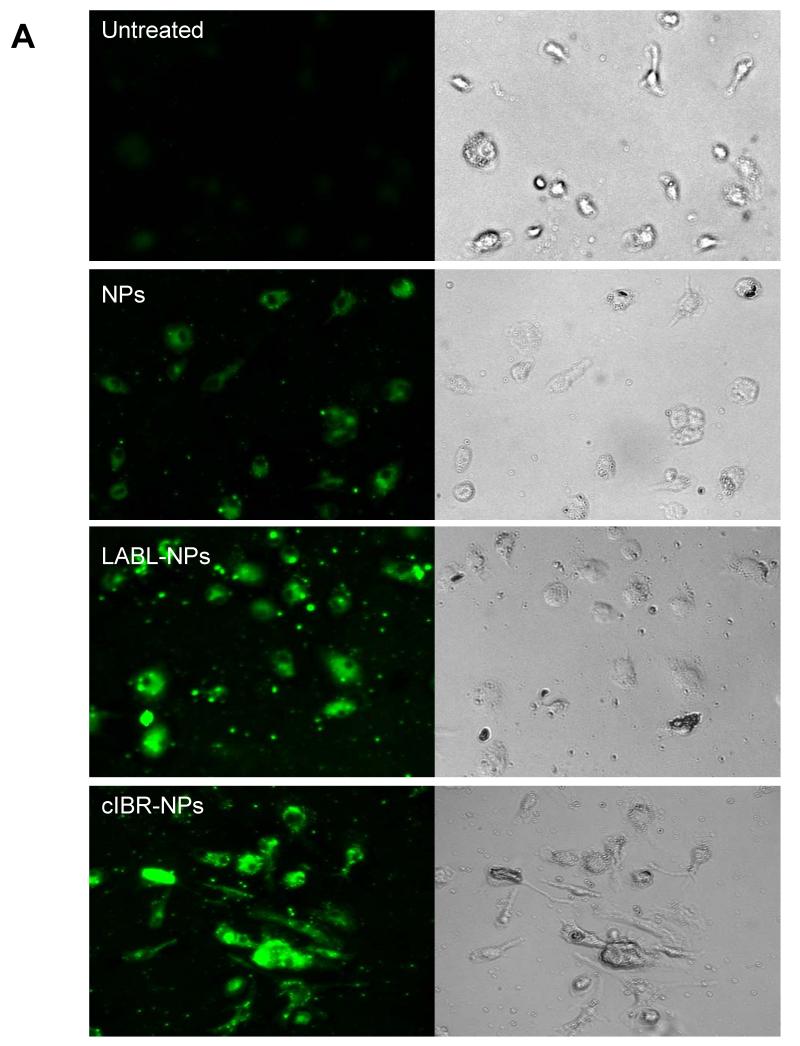
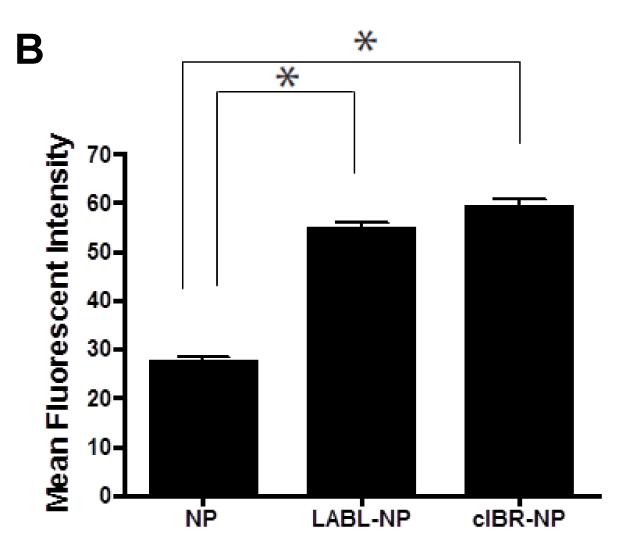
The binding of NPs, LABL-NPs or cIBR-NPs by DCs was also followed using fluorescence microscopy. Nanoparticle fluorescence rapidly localized to DCs (Figure 4A). Images collected using MetaMorph were analyzed using ImageJ software and showed ~2 times greater fluorescence intensity of DCs incubated with LABL-NPs compared to untargeted NPs at 40 min (Figure 4B), thus supporting flow cytometry results. DCs treated with cIBR-NPs demonstrated even higher fluorescent intensities. These results indicated that LABL-NPs and cIBR-NPs effectively targeted DCs.
Figure 4.
(A) Fluorescent micrographs of DCs in medium (untreated), DCs incubated with untargeted NPs, DCs incubated with LABL-NPs and DCs incubated with cIBR-NPs for 40 min. Punctate fluorescence patterns suggested the accumulation of NPs with the cells (B) Mean fluorescent intensities of DCs incubated with NP, LABL-NP and cIBR-NP were quantified from micrographs using ImageJ software. DCs were stimulated by TNF-α and primed with OVA for 24 hrs prior to the study. DCs were then incubated with NPs, LABL-NPs and cIBR-NPs for 40 min at 37 °C. * indicates p < 0.001.
The internalization of LABL-NPs and cIBR-NPs was supported by the punctate fluorescence pattern observed in the micrographs. Similar patterns were also reported for the uptake of untargeted NPs and cLABL-NPs in lung epithelial cells (A549 cells) and endothelial cells (HUVECs) expressing high levels of ICAM-1 in previous reports.24-25 The punctate pattern of cIBR-NPs appeared slightly more evident than LABL-NPs and untargeted NPs at 40 min of incubation (Figure 4A). These results supported the flow cytometry data.
In vitro cellular cytotoxicity of LABL-NPs or cIBR-NPs
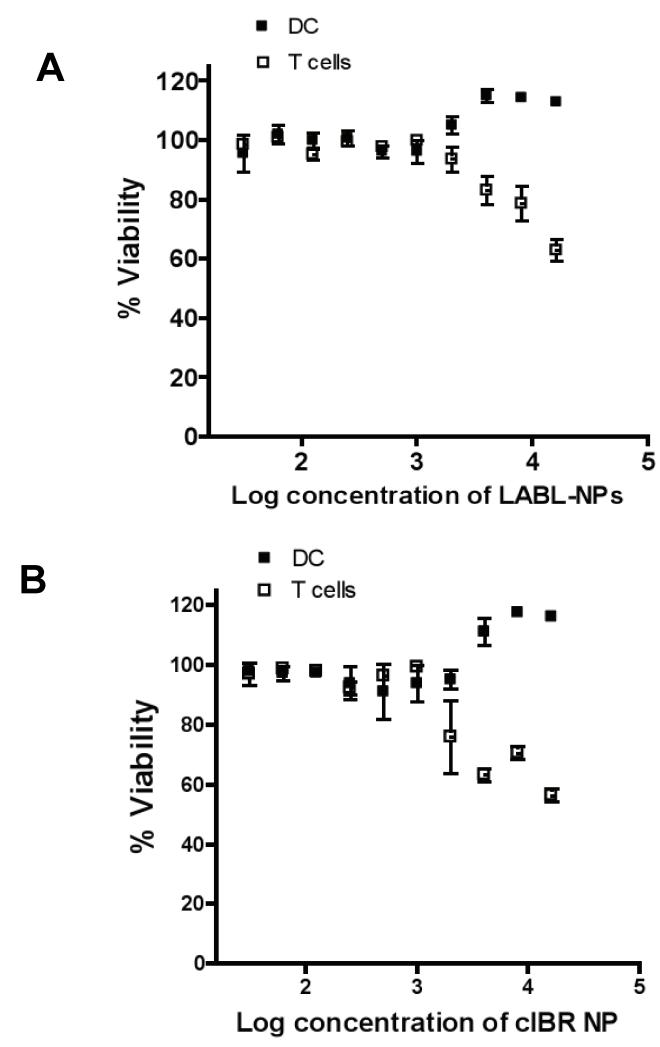
Cell viability studies indicated that LABL-NPs and cIBR-NPs were minimally cytotoxic to DCs and T cells. The average cell viability was greater than 95% at all concentrations tested when LABL-NPs and cIBR-NPs were incubated with DCs for 24 hrs (Figure 5A and 5B). For reference, the viability of T cells exposed to LABL-NPs and cIBR-NPs relative to untreated cells was also evaluated. After 24 hrs of treatment, a decrease in T cell viability was observed at LABL-NPs concentrations higher than 16 mg/ml (IC50 = 25.6 mg/ml). The IC50 of cIBR-NPs with T cells was 3.4 mg/ml. From our results, we inferred that DCs were highly viable after incubation with these particles for 30 min in the conjugation study. DC cultures were washed to remove any free NPs prior to addition of primary T cells; therefore, T cell cytotoxicity data is only offered for references.
Figure 5.
Dendritic cell and T cell viability in the presence of (A) LABL-NPs or (B) cIBR-NPs. LABL-NPs and cIBR-NPs were minimally cytotoxic to DCs and T cells at the concentrations used for ensuing studies.
Peptide-NPs significantly inhibited the conjugation of DCs and T cells
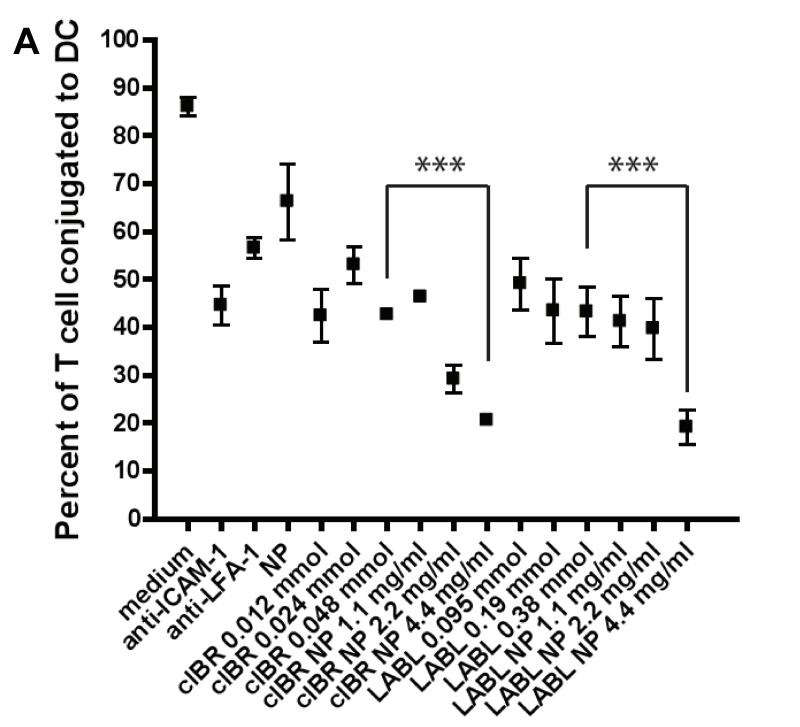
The induction of cell proliferation and cytokine production in resting T cells requires binding of LFA-1 on T cells and its receptor, ICAM-1 on DCs to allow prolonged signaling.3-5 The effect of LFA-1/ICAM-1 blockade on conjugate formation between T cells isolated from B6.129S7-Rag1tm1Mom Tg(TcraTcrb)425Cbn mice and mature DCs primed with OVA was investigated. LABL-NP blockade of ICAM-1 on DCs led to a substantial decrease in the number of T cells bound to DCs (Figure 6). Pretreatment of DCs with LABL-NPs resulted in up to a 76% decrease in T cell binding to DCs compared to T cells incubated with untreated DCs. From these results, we inferred that LABL-NPs bound ICAM-1 expressed on DCs and blocked the availability of ICAM-1 to interact with LFA-1 on T cells. The number of T cells interacting with DCs pretreated with cIBR-NPs was decreased up to 78% compared to T cells incubated with untreated DCs. Untargeted NPs decreased the T cell conjugation to DCs by only 23%.
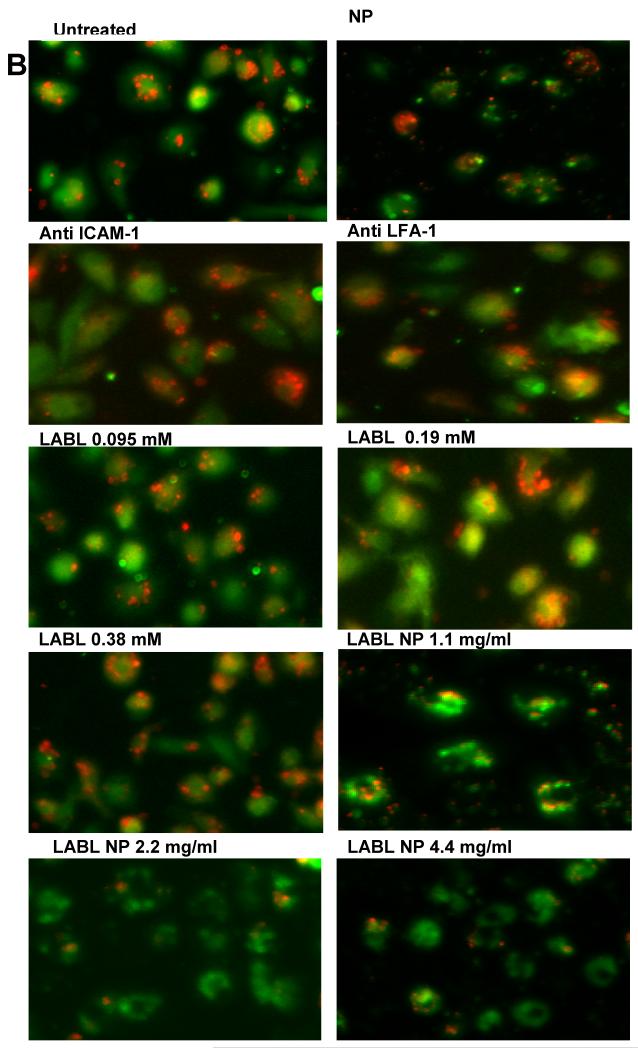
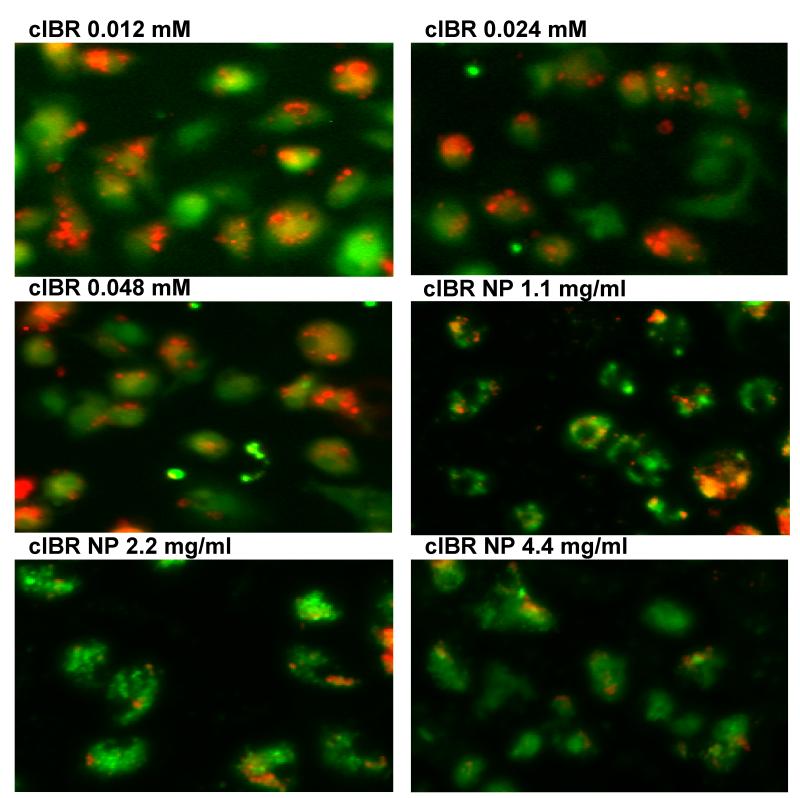
Figure 6.
(A) Blocking of DC-T cell conjugate formation. LABL-NPs and cIBR-NPs inhibited the binding of T cells to DCs to a much greater extent than free LABL and cIBR peptides. (B) Micrographs of T cells (red) binding DCs (green). LABL-NPs and cIBR-NPs exhibited greater inhibition oT cell conjugation to DCs than LABL peptides, anti-ICAM-1, anti-LFA-1, NPs and untreated DCs. DCs were incubated with TNFα and primed with OVA for 24 hrs prior to incubation with samples. DCs were washed and T cells were cocultured for 2 hours and T cells and DCs were imaged and counted. *** indicates p<0.001.
In addition, free peptides were incubated at molar concentrations corresponding to the molar amount of peptides presented on the surface of nanoparticles. Peptide-conjugated nanoparticles blocked the binding of T cells to DCs significantly better than free peptides. Furthermore, inhibition of the T cell conjugation to DCs was further enhanced by increasing LABL-NP or cIBR-NP concentration. These data were also analyzed by counting T cells conjugated to DCs (supplementary Figure 1) and the results were in agreement with the analysis reported here using Image J software.
Effects of LABL-NPs and cIBR-NPs on T cell proliferation
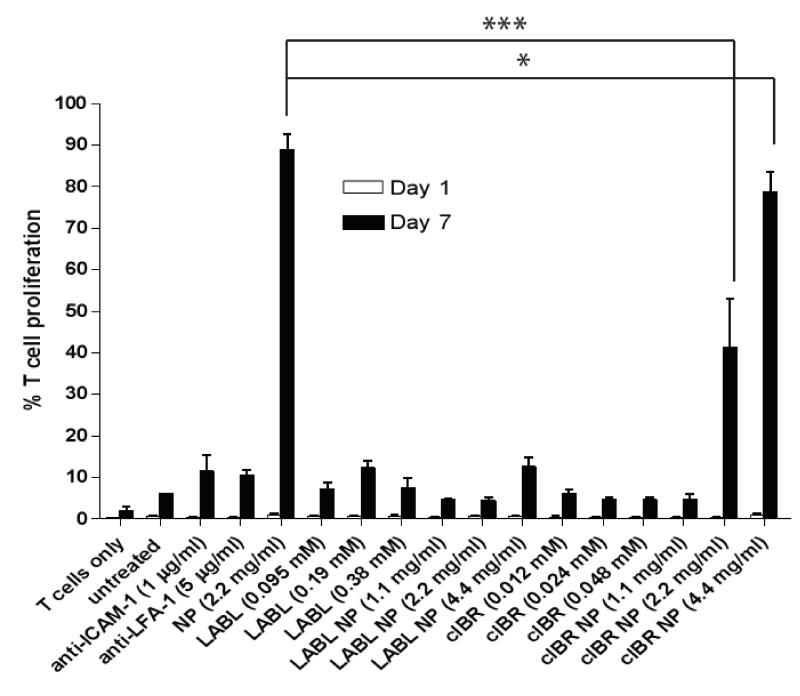
To examine the effect of NPs on T cell proliferation, carboxyfluorescein diacetate succimidyl ester (CFSE) fluorescence dilution in OVA-specific T cells was analyzed after co-incubation with DCs for 24 hrs (1 day) and 168 hrs (7 days). DCs were pretreated with untargeted nanoparticles (NPs), antibodies, free peptides or peptide-conjugated NPs. The three different molar concentrations of free peptides matched the molar concentration of peptide conjugated to NPs. Division of cells was calculated from the percentage of cells having diluted fluorescent intensity using Flowjo software. Untargeted NPs and cIBR-NPs incubated with DCs led to a substantial increase in the number of T cells undergoing division. We observed that 89%, 78% and 41% of T cells divided following co-incubation with DCs which had been pretreated with NPs (2.2 mg/ml) or cIBR-NPs (4.4 and 2.2 mg/ml) for 7 days, respectively. In contrast, T cells incubated with untreated DCs only proliferated ~6%.
The proliferation of T cells incubated with DCs pretreated with anti-ICAM-1, anti-LFA-1, LABL peptide, LABL-NPs (4.4, 2.2 and 1.1 mg/ml) or cIBR peptide were not substantially altered by the treatments (Figure 7). There were no significant differences in the levels of T cell proliferation between the groups of DCs treated with these samples when compared to untreated DCs. T cells alone without DCs were used as a negative control and yielded a T cell proliferation of only ~1.8%.
Figure 7.
The proliferation of T cells after coculture with DCs treated with samples. The percent of T cells dividing after incubation for 24 hrs (1 day) and 168 hrs (7 days) was determined by flow cytometry. DCs incubated with TNFα and primed with OVA were incubated with samples for 30 min at 37 °C. After removing samples, OVA specific T cells were added and incubated with DCs for 7 days. cIBR-NPs and untargeted NPs stimulated the proliferation of OTII T cells but other samples did not significantly alter T cell proliferation. *** indicates p<0.001 and * indicates p<0.05.
Cytokine production of DCs and OT-II T cells
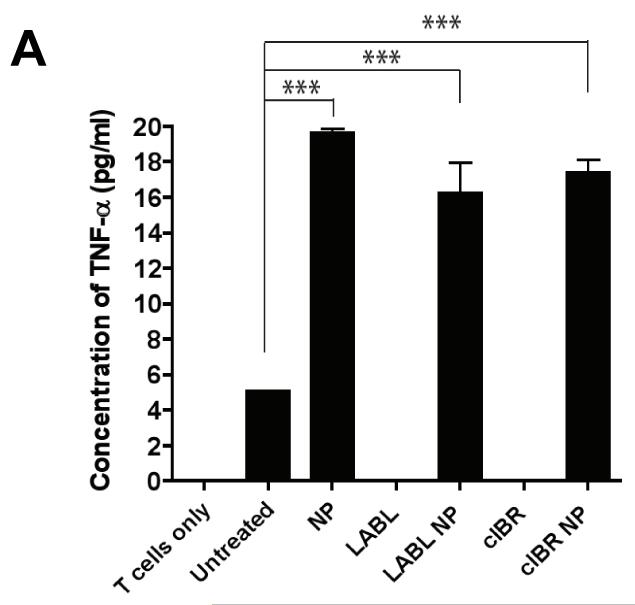
One marker for DC maturation and T cell stimulation is the production of cytokines. Cytokines are produced by both antigen presenting cells such as DCs and by T cells. DCs predominantly produce TNF-α, IL-1 and IL-6.26 IL-2 is produced by CD4+ T cells.27 To observe cytokine production of DCs after treatment with peptides or nanoparticles, DCs matured with TNF-α and primed with OVA were incubated with LABL peptide, cIBR peptide, LABL-NPs, cIBR-NPs, or NPs for 30 min. DC cultures were washed and then OT-II T cells were cocultured with DCs for 7 days and cytokine production was determined by enzyme-linked immunoassay (ELISA). DCs incubated with LABL-NPs, cIBR-NPs or NPs secreted significantly higher amounts of TNF-α (4-5 fold), relative to untreated DCs. In contrast, the amounts of TNF-α produced by DCs treated with LABL and cIBR peptide solutions were very low similar to untreated DCs (Figure 8A).
Figure 8.
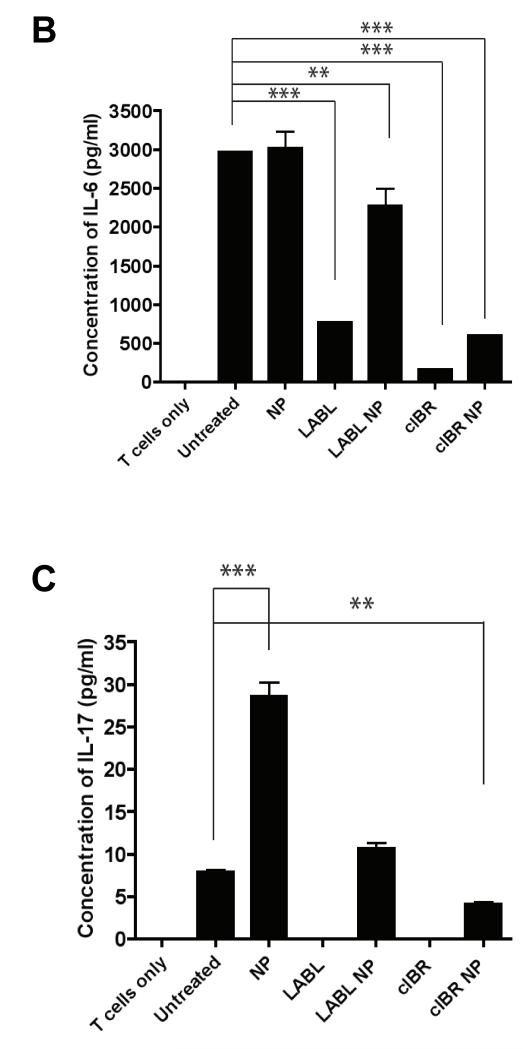
The production of cytokines in cocultures where DCs were pretreated with LABL-NPs, cIBR-NPs, untargeted NPs, LABL peptide or cIBR peptide. (A) All nanoparticles increased the secretion of TNF-α compared to untreated DCs. (B) LABL-NPs, cIBR-NPs, LABL and cIBR peptides decreased the amount of IL-6 detected in the coculture of DCs and T cells relative to untreated DCs. Untargeted NPs increased the level of IL-6 production in the coculture of DCs and T cells (C) Untargeted NPs induced the production of IL-17 from activated OT-II T cells. LABL-NPs, cIBR-NPs, LABL and cIBR peptides did not stimulate the production of IL-17. *** indicates p<0.001 and ** indicates p<0.01.
Furthermore, the amount of IL-6 produced by coculture of T cells with DCs pretreated with untargeted NPs was not significantly different from the coculture of untreated DCs and T cells. Conversely, the amount of IL-6 produced by coculture of T cells with DCs pretreated with LABL peptide, cIBR peptide, LABL-NPs or cIBR-NPs were lower than coculture of untreated DCs and T cells (Figure 8B). These results suggested that LABL peptide, cIBR peptide, LABL NPs and cIBR-NPs, did not stimulate IL-6 production, which is predominantly produced by DCs.26 Untargeted NPs did not alter IL-6 production relative to untreated DCs suggesting that these particles neither bound specifically to cell adhesion molecules nor blocked the activation of the OTII T cells.
Proliferation of naïve CD4+ T cells into effector T cells is also regulated by mature DCs. Coculture of OT-II T cells with DCs that had been pre-incubated with untargeted NPs enhanced T cell proliferation and production of IL-17, when compared to other treatments or controls (Figure 8C). From these results we concluded the presence of untargeted NPs promotes activation of T cells and may increase production of TGFβ, which is normally considered immunosuppressive. Yet, in the presence of IL-6, TGF-β promotes the expression of IL-17 and the differentiation of Th17 CD4 cells.28 These findings are further supported by the low levels of IL-6 produced by LABL, cIBR, LABL-NPs and cIBR-NPs treatments which correlate with the production of IL-17 since IL-6 in combination with TGF-β induces Th17 differentiation from naïve CD4+ T cells.28 Finally, the amount of IL-2 was undetectable in all media of DCs cocultured with OT-II T cells.
Discussion
A critical step in the development of an immune response is the activation of T cells. This process transitions naïve CD4+ T cells into effector cells. T cell activation begins with antigen recognition through a highly specific interaction between T cell receptor (TCR) and antigenic peptide presented by major histocompatibility complex (MHC) molecules on the surface of antigen presenting cells.1 The activation of a T cell by an APC requires the reorganization of receptors and ligands to form an immunological synapse. This dynamic and highly organized structure maintains the cell-cell interaction.29 The immunological synapse is formed by a re-arrangement of receptors that form supramolecular activation clusters. The central cluster is comprised of TCR-peptide-MHC interaction which is surrounded by LFA-1 and ICAM-1 adhesion molecules, among others, in the mature immunological synapse.30 The binding of TCRs to peptide-MHC complexes further activates LFA-1 on T cells that recognize and bind ICAM-1 on APC.2 This interaction strengthens the conjugation of APC and T cells and completes the immunological synapse which can lead to efficient T cell activation.29-30
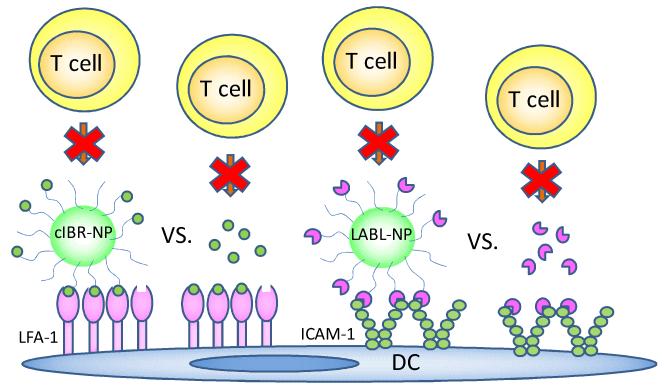
ICAM-1 belongs to the immunoglobulin superfamily and is expressed constitutively at low levels on APC, endothelial cells, and other cell types.16 The upregulation of ICAM-1 can be induced by inflammatory mediators such as IL-1.31 Generally, the binding of cell adhesion molecules is enhanced by oligomerization and clustering of the adhesion molecules.16 ICAM-1 rearranges into non-covalent homodimers on the cell surface via interactions between domain 4 (D4).16 Moreover, D4-D4 dimers can then be non-covalently linked by interaction of domain 1, resulting in a “W” shaped tetramer that forms linear arrays that are likely bent into circular arrays. This organization of ICAM-1 influences formation of the immunological synapse (Figure 1).16
Figure 1.
T cell conjugation to DCs maybe blocked by the binding of cIBR-NP to LFA-1 or LABL-NP to ICAM-1 on DCs.
The binding of LFA-1 to ICAM-1 is also tightly regulated. The interaction of LFA-1 to ICAM-1 is driven by cluster formation and a change in LFA-1 conformation leading to higher avidity and affinity, respectively.17 Binding of TCRs to peptide-MHC complexes generates intracellular signals leading to an increase in avidity of LFA-1 binding by forming a cluster.17, 32 The adhesion of DCs and T cells involves LFA-1 and ICAM-1 clusters, made up of thousands of molecules.16 The high densities of ICAM-1 on APCs and LFA-1 on T cells are necessary for firm, long-lasting conjugation. Thus, the nature of T cell conjugation to DCs suggests that multivalent inhibitors may pose an interesting intervention.
LABL and cIBR peptides have been shown to bind specifically to ICAM-1 and LFA-1, respectively.19-20 These peptides also specifically bind their respective ICAM-1 and LFA-1 receptors when conjugated to nanoparticles.23, 25 Presenting these as multivalent arrays on the nanoparticle surface was hypothesized to provide more potent intervention strategies due to enhanced binding avidity, triggered endocytosis of receptors, and changes in cell response (Figure 1).33-34
In this study, pretreatment of DCs with LABL-NPs yielded up to a ~76% decrease in T cell conjugate formation with DCs suggesting that LABL-NPs bound to ICAM-1 on DCs and blocked the availability of ICAM-1 to interact with LFA-1 on T cells. Similar results were obtained when DCs were pre-treated with cIBR-NPs. The quantity of T cells binding to DCs treated with cIBR-NPs was reduced by 78% when compared to untreated control. LABL-NPs and cIBR-NPs blocked T cell conjugation to DCs to a greater extent than LABL peptide, cIBR peptide, anti-ICAM-1 antibody, anti-LFA-1 antibody and unconjugated NPs.
Inhibition of T cell conjugation to DCs was most likely due to an increase in the binding avidity of multivalent peptides and perhaps internalization of cell adhesion molecules facilitated by the nanoparticles.33-34 Multiple copies of LABL or cIBR peptide conjugated to the surface of nanoparticles were hypothesized to induce the clustering of LFA-1 and ICAM-1, hence enhancing the binding avidity of peptides to activated, high-density receptors. Welder et al has shown that monovalent soluble ICAM-1 is unable to bind efficiently to LFA-1 expressing cells unless it is rendered multivalent by coupling to polystyrene microspheres.34 Furthermore, the peptide-NPs were suspected to sustain inhibition by inducing receptor internalization. Internalization of clustered ICAM-1 on endothelial cells is known to be triggered by multimeric anti-ICAM-1 conjugated on microspheres, whereas monomeric anti-ICAM-1 was not efficiently internalized.33 LABL-NPs and cIBR-NPs have also been shown to be internalized into lung epithelial cells and HUVEC cells expressing ICAM-1 and T cells expressing LFA-1, respectively.23-25 Here, the punctate staining patterns observed in DCs treated with NPs indicated uptake, and presumably receptor internalization.
LABL-NPs, cIBR-NPs, and NPs induced TNF-α production which may result from endocytosis or processing of particles in DCs.35 Indeed, PLGA nanoparticles have been shown to induce proinflammatory cytokine production and increase T cell proliferation.36-38 Co-incubation of T cells with DC that had been pretreated with untargeted NPs led to high levels of TNF-α, IL-6 and IL-17 and augmented T cell proliferation relative to untreated DCs. A similar observation was previously reported in DCs treated with PLGA microspheres.39 LABL-NPs decreased the production of IL-6 from DCs and/or OT-II T cells and also resulted in a decrease the amount of IL-17, presumably by inhibiting T cell differentiation into Th17 cells.
DCs pretreated with cIBR-NPs also induced TNF-α production and proliferation of OT-II T cells in a dose dependent manner. Significantly lower levels of IL-6 and IL-17 were produced when comparing cIBR-NPs to other NPs or to untreated coculture. In addition, it was unclear whether the T cells that proliferate following stimulation of T cells with cIBR-NP treated DC were Th1, Th2 or Th17 cells. Based on previous reports, LFA-1 does not seem to deliver a costimulatory signal but improves engagement of TCR by promoting adhesion of T cells to APC.3 Blockade of LFA-1 on DCs would, therefore, not be expected to inhibit T cell activation by DCs. However it could change the T cell differentiation by changing the microenvironment of T cell during activation. Although cIBR-NPs blocked T cell conjugation to DCs initially, this effect was most likely transient, since substantial T cell proliferation was observed at day 7. Data also showed that increasing cIBR-NP dose ultimately increased T cell proliferation.
Conclusion
Chemical conjugation of LABL and cIBR peptides to PLGA NPs generated efficient targeting to ICAM-1 and LFA-1 receptors on DCs, respectively. LABL-NPs and cIBR-NPs were found to be effective inhibitors of T cell conjugation to DCs. LABL-NPs and cIBR-NPs blocked T cell conjugation to DCs more efficiently than untargeted NPs, free peptides and antibodies. Compared to controls, T cell proliferation was arrested when T cells were incubated in the presence of DCs treated with LABL-NPs, whereas DCs treated with cIBR-NPs or untargeted NPs dramatically stimulated the division of T cells. These findings suggested the potential of LABL-NPs to change the character of mature DCs, yet block immunological synapse formation without stimulating T cell proliferation and point to cIBR-NPs as inducers of T cell expansion. All NPs increased the production of TNF-α compared to free ligands or untreated controls, but the amount of IL-6 or IL-7 depended on NP type. Collectively, results suggested that LABL-NPs and cIBR-NPs function as distinct immune modulators that dramatically differ from soluble peptide inhibitors of ICAM-1 or LFA-1.
Materials and Methods
Materials
LABL peptide (ITDGEATDSG, Mw 964.95) and cIBR peptide (cyclo 1,12 Pen-PRGGSVLVTGC, Mw 1174.50) were synthesized on a Pioneer peptide synthesizer (PerSeptive Biosystems, CA). Poly(dl-lactic-co-glycolic acid) (50:50) with terminal carboxyl group (PLGA, inherent viscosity 0.67dL/g, Mw ~90 kDa) was purchased from Lakeshore Biomaterials (Birmingham, AL, USA). Pluronic®F-127 was obtained from BASF Corporation. (Mount Olive, NJ, USA). 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide (sulfo-NHS) and 2-β mercaptoethanol were purchased from Thermo Fisher Scientific Inc. (Rockford, IL, USA). Coumarin-6 was purchased from Polysciences, Inc. (Warrington, PA, USA). Dialysis membrane (MwCO 100,000) was purchased from Spectrum laboratory Products Inc. (Rancho Dominguez, CA, USA). RPMI-1640 medium was obtained from Cellgro (Manassas, VA, USA). Tumor Necrosis Factor-α (TNF-α) was purchased from Promega (Madison, WI, USA). Granulocyte Macrophage-Colony Stimulating Factor (GM-CSF) was purchased from Peprotech Inc. (Rocky Hill, NJ, USA). Carboxyfluorescein diacetate succimidyl ester (CFSE) and 5-(and-6)-(((4cloromethyl) enzoyl)amino)tetramethylrhodamine (Orange CMTMR) were purchased from Invitrogen Corporation, (Carlsbad, CA). Monoclonal anti-human CD54 (ICAM-1) Domain 1 and Monoclonal anti-LFA-1 were purchased from Ancell (Bayport, MN, USA). CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (MTS) was purchased from Promega (Madison, WI, USA). Ovalbumin was purchased from Sigma (St. Louis, MO). Penicillin, streptomycin and L-glutamine were purchased from Cellgro (Manassas, VA, USA). IL-2 was generously provided by Dr. Christophe Nicot at the University of Kansas Medical Center. B6.129S7-Rag1tm1MomTg(TcraTcrb)425Cbn mice were purchased from Taconic Farms (Hudson, NY). C57BL/6 wildtype mice were purchased from Jax labs (Bar Harbor, ME).
Methods
Cell culture and isolation
Bone marrow derived dendritic cells were generated from C57BL/6 wildtype mice as described.40 Briefly, 2 × 106 cells were isolated from bone marrow, plated on bacterial Petri dishes and cultured in 10 ml DC media (RPMI-1640, 10% heat inactivated fetal bovine serum, 100 μg/ml penicillin-streptomycin, 50 μM β-mercaptoethanol, 20 ng/ml murine granulocyte macrophage colony stimulating factor (GM-CSF) (R&D), and 2 nM L-glutamine. At 72 hours (Day 3), 10 ml of fresh DC media was added to each dish. On days 6 and 8, 10 ml of supernatant and cells were removed, cells recovered by centrifugation (90 × g) and added back to the dish with fresh DC medium. On day 9 non-adherent cells were collected, labeled with (5 μM) 5-(and-6)-carboxyfluorescein diacetate, succinimidyl ester (CFSE) for 10 minutes at 37°C, washed, primed with 50 μg/ml ovalbumin, and matured overnight with 100 ng/ml TNF-α.
On day 10, T cells were isolated from B6.129S7-Rag1tm1Mom Tg(TcraTcrb)425Cbn mice spleens by passing spleens through a wire mesh. T cells were purified using a negative selection, mouse T cell enrichment kit according to manufacturers’ directions (EasySep).
PLGA nanoparticle preparation and characterization
PLGA nanoparticles and nanoparticles loaded with coumarin-6 were prepared by a solvent displacement method. In brief, PLGA was dissolved in acetone (18 mg/ml) containing coumarin-6 (50 μg/ml). The solution was gently infused into 0.1% Pluronic®F-127-COOH (25 ml) under mild stirring (300 RPM). Terminal hydroxyl groups on Pluronic®F-127 were converted to carboxyl groups according to a reported procedure.25, 41 The resultant nanoparticle suspension was dialyzed (100,000 MWCO) against a 0.2% mannitol solution for 48 hrs to remove excess surfactant. Particle size and zeta potential of nanoparticles were characterized using dynamic light scattering (ZetaPALS, Brookhaven instrument Inc.).
Conjugation of LABL and cIBR peptides to PLGA-nanoparticles
The N-terminus of peptide was covalently linked with the carboxyl groups of Pluronic®F-127-COOH coated on PLGA nanoparticles by carbodiimide chemistry.23 Specific binding of the LABL and cIBR peptides suggests they are radially pointing away from the particles surface (Figure 1). Nanoparticles (2.2 mg/ml) were buffered using 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.5) and incubated with 100 mM 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) and 50 mM N-hydroxysulfosuccinimide (sulfo-NHS) for 15 min. EDC was used to react with a carboxyl group on PLGA NPs and formed an amine-reactive O-acylisourea intermediate. Sulfo-NHS was added to stabilize this intermediate, hence increasing the efficiency of coupling reaction. Excess EDC and sulfo-NHS were removed by centrifugation (16,089 g, 10 min). Then, cIBR or LABL peptides (0.4 mg) were added and allowed to react with Pluronic®F-127-COOH on nanoparticles for 12 hrs at room temperature. Peptide conjugated NPs were collected by centrifugation (16,089 g, 10 min) and washed three times with purified water.
The conjugation efficiency was determined by quantifying the unconjugated ligand remaining in the reaction medium after nanoparticle separation. The peptide density on the surface of nanoparticles after reaction was calculated assuming a normal Guassian particle size distribution.23, 25 The amount of free peptides in reaction medium was analyzed by gradient reversed phase HPLC (SHIMADZU) using a C18 column. The HPLC consisted of SCL-10A SHIMADZU system controller, LC-10AT VP SHIMADZU liquid chromatograph, SIL-10A XL SHIMADZU autoinjecter set at 30 μl injection volume, DGU-14A SHIMADZU degasser, sample cooler, and SPD-10A SHIMADZU UV-Vis detector (220 nm). The HPLC-UV system was controlled by a personal computer equipped with SHIMADZU class VP Software. All separations were carried out using a Vydac® HPLC Protein and Peptide C18 column. Gradient elution was carried out to determine the amount of LABL peptide at constant flow of 1 ml/min, from 0% B to 8% B for 5 min, followed by 14.3% B at 17.5 min, 50% at 23 min and 70% B at 24-35 min. HPLC gradient system was programmed to separate cIBR peptide at constant flow of 1 ml/min, from 25% B for 5 min, 55% B at 25 min and 100%B at 25.1-30 min. Mobile phase compositions were (A) acetronitrile-water (5:95) with 0.1% TFA and (B) 100% acetonitrile with 0.1% trifluoroacetic acid (TFA).
LFA-1 and ICAM-1 expression on dendritic cells and T cells
The relative expression of LFA-1 and ICAM-1 on T cells and DCs were qualitatively assessed using flow cytometry (FACScan). DCs (4 × 105 cells/ml) were matured with TNF-α (1,000 U/ml) and primed with ovalbumin (OVA) (50 μg/ml) for 24 hrs. DCs were isolated by centrifugation, and incubated 45 minutes on ice with 80 μl of anti-ICAM-1 (0.05 mg/ml) or anti-LFA-1 (0.25 mg/ml) conjugated with FITC at 1:50 dilution. Cells were washed three times and analyzed by flow cytometry.
Splenic C57BL/6-TgN(OT-II.2a)-Rag1 T cells (2.2 × 106 cells/ml) were incubated with 80 μl of anti-ICAM-1-FITC (0.05 mg/ml) or anti-LFA-1-FITC (0.25 mg/ml) on ice for 45 min. Unbound antibodies were removed by rinsing three times with PBS after centrifugation (16,089 g, 2.5 min). The fluorescent intensity of cells was measured by a FACscan flow cytometer. Data analysis was performed using Cell Quest software (BD).
Binding and uptake of LABL-NPs and cIBR-NPs into DCs
The binding and uptake of LABL-NPs and cIBR-NPs encapsulated fluorescent dye was monitored using flow cytometry. DCs (1 × 105 cells/ml) were added and allowed to adhere on a 96 well-plate (200 μl/well) for 24 hr in the presence of TNF-α (1,000 U/ml) and OVA (50 μg/ml). DCs were washed three times with PBS and incubated with NPs, LABL-NPs or cIBR-NPs (2.2 mg/ml, 100 μl) at 37 °C for 15, 30, 45 and 60 min. Cells were washed three times with PBS and trypsinized for 3 min at 37°C. Then DCs were transferred to microcentrifuge tube and washed once with PBS by centrifugation (600 g, 2 min). The fluorescent intensity due to DC uptake of fluorescent NPs was measured using the FACscan flow cytometer. Data analysis was performed using Cell Quest software (BD).
Fluorescence microscopy of DCs binding/uptake with LABL-NPs or cIBR-NPs
Dendritic cells (1 × 106 cells/ml, 300 μl) were added into an 8-well plate and stimulated by TNF-α (1,000 U/ml) and OVA (50 μg/ml) for 24 hrs. Cells were washed with PBS and then incubated with NPs, LABL-NPs or cIBR-NPs (2.2 mg/ml, 300 μl) for 40 min at 37°C. Unbound nanoparticles were removed by washing three times with PBS and cells were fixed with 4% paraformaldehyde. Fluorescence micrographs were acquired using the FITC filter set of a Nikon Eclipse 80i microscope equipped for epifluorescence. Micrographs were captured using an Orca ER camera (Hamamatsu, Inc., Bridgewater, NJ) and analyzed by Metamorph, version 6.2 (Universal Imaging Corp., West Chester, PA). All images were corrected for variations in excitation light intensity.
In vitro cellular cytotoxicity of LABL-NPs and cIBR-NPs
MTS cell viability assays were performed to provide an assessment of the toxicity of LABL-NPs and cIBR-NPs on T cells and TNF-α stimulated and OVA primed DCs. Briefly, T cells (3.2 × 106 cells/ml) and DCs (1 × 105 cells/ml) were seeded on 96-well plates and incubated with various concentrations of LABL-NPs and cIBR-NPs for 24 hr. A tetrazolium salt MTS was applied and the incubation was continued for additional 4 hrs. MTS was converted by mitocondrial dehydrogenase enzyme in living cells to form a colored formazan product. The absorbance of the formazan product was recorded at 490 nm using a 96-well plate reader (Spectramax M5).
DCs and T cell conjugate formation
DCs (4 × 105 cells/ml) were stained by incubating with 5 ml of CFSE in PBS (10 μM) for 10 min at 37 °C in PBS. The staining was quenched by the addition of 25 ml of complete culture medium and incubation at 4°C for 10 min. The remaining dye was washed away by three washes with complete culture medium. T cells were incubated with 10 μl of CMTMR orange fluorescent dye (5 μM) in 10 ml of PBS for 30 min at 37°C. The reaction of the dye was quenched by incubating with complete culture medium for 30 min at 37°C. T cells were washed three times with complete culture medium. Untreated DCs were used as a positive control. Dendritic cells (4 × 105 cells/ml) primed with OVA (50 μg/ml) and matured in the presence of TNF-α (1,000 U/ml) were incubated with anti-ICAM-1 (1 μg/ml), anti-LFA-1 (5 μg/ml), NPs (2.2 mg/ml), LABL peptide (0.095, 0.19 and 0.38 mM), LABL-NPs (1.1, 2.2 and 4.4 mg/ml), cIBR peptide (0.012, 0.024 and 0.048 mM) or cIBR-NPs (1.1, 2.2 and 4.4 mg/ml) for 30 min at 37 °C. DCs were washed three times with PBS and incubated with T cells (2 × 106 cells/ml) for 2 hr at 37 °C. After incubation, DCs were washed three times with PBS and fixed with 4% paraformaldehyde. Cells were imaged using an Orca ER camera (Hamamatsu, Inc., Bridgewater, NJ) and imaged using Metamorph, version 6.2 (Universal Imaging Corp., West Chester, PA). Dendritic cells and T-cells attached to dendritic cells were measured by using Image J software per condition from all images. The analysis was performed by using color segmentation (RGB channel separation), which was made binary and followed by measurement of the area of R and G channels. Similar threshold limiting was applied to all images and colors. The results were reported by the percent of ratio of R channel area (corresponds to T cells) to G channel area (corresponds to DC). The percentage of T cell-conjugates was calculated.
T cell proliferation assay
Primary T cells isolated from C57BL/6-TgN(OT-II.2a)-Rag1 mice were labeled with carboxyfluorescein succinimidyl ester (CFSE) (5 μM) for 10 min at 37 °C, 5% CO2 to observe the dye dilution by cell division. The staining was quenched by the addition of 5 volumes of culture medium into T cells and incubated 10 min at 4°C. DCs (4 × 105 cells/ml) were matured in the presence of TNF-α (1,000 U/ml) and primed with OVA (50 μg/ml) for 24 hrs in 24-well plate. DCs were treated with anti-ICAM-1 (1 μg/ml), anti-LFA-1 (5 μg/ml), unconjugated NPs (2.2 mg/ml), LABL peptide (0.095, 0.19 and 0.38 mM), LABL-NPs (1.1, 2.2 and 4.4 mg/ml), cIBR peptide (0.012, 0.024 and 0.048 mM) or cIBR-NPs (1.1, 2.2 and 4.4 mg/ml) for 30 min, at 37 °C, 5% CO2 and washed three times with PBS. T cells (2 × 106 cells/ml) in serum free RPMI-1640, IL-2 and 1% penicillin-streptomycin were incubated with DCs 7 days at 37°C, 5% CO2. T cells collected after 24 hrs (1 day) and 168 hrs (7 days) were centrifuged at 16,089 g for 2 minutes and fixed with 4% paraformaldehyde. The CFSE dilution was measured by using FACScan flow cytometer. The percent of T cell proliferation was analyzed by calculating the percent of cells with diluted CFSE using FlowJo software.
Quantification of cytokines in cell culture supernatants by ELISA
DCs were matured in the presence of TNF-α (1,000 U/ml) and primed with OVA (50 μg/ml) for 24 hrs in 24-well plate. DCs were treated with anti-ICAM-1 (1 μg/ml), anti-LFA-1 (5 μg/ml), unconjugated NPs (2.2 mg/ml), LABL peptide (0.095, 0.19 and 0.38 mM), LABL-NPs (1.1, 2.2 and 4.4 mg/ml), cIBR peptide (0.012, 0.024 and 0.048 mM) or cIBR-NPs (1.1, 2.2 and 4.4 mg/ml) for 30 min, at 37 °C, 5% CO2 and washed three times with PBS. T cells (2 × 106 cells/ml) in serum free RPMI-1640, IL-2 and 1% penicillin-streptomycin were incubated with DCs 7 days at 37°C, 5% CO2. Supernatants of cell cultures were collected for cytokine detection. Secreted TNF-α, IL-2, IL6 and IL-17 were measured by ELISA assay (Cytokine Core Lab, Baltimore, Maryland). ELISA was performed in Nunc Maxisorb ELISA strips freshly coated with capture antibody for 16 hours before the assay was performed. Detecting antibody and the streptavidin-peroxidase conjugate were added into standard, all samples and controls. Premixed substrate solution (Neogen) was then added. The plate was read on a Molecular Divices ELISA plate reader. Curve fitting was selected among linear, quadratic and 4-point based on the best regression coefficient using the SoftPro software package.
Statistical analysis
Statistical evaluation of data was performed using an analysis of variance (one-way ANOVA). Newman-Keuls was used as a post-hoc test to assess the significance of differences. To compare the significance of the difference between the means of two groups, a t-test was performed; in all cases, a value of p < 0.05 was accepted as significant.
Supplementary Material
Supplementary figure 1. Blocking of the T cell conjugation to DCs. LABLNPs and cIBR-NPs inhibited the binding of T cells to DCs to a much greater extent than free LABL and cIBR peptides. DCs were incubated with samples for 30 min, washed and cocultured with T cells for 2 hours. T cells and DCs were imaged and counted. *** indicates p<0.001.
Acknowledgements
The authors acknowledge J. Krise for providing the fluorescence microscope. We would like to acknowledge support from the Royal Thai Government, the Coulter Foundation, and the Higuchi Biosciences Center as well as additional lab funding from the American Heart Association, the NIH (R03 AR054035, P20 RR016443 and T32 GM08359-11) and the Department of Defense. In addition, we acknowledge the support of National Institutes of Health (R01-AI063002 and R56-AI063002), COBRE (RO16443-11) and the Biomedical Research and Training Program.
References
- 1.Dustin ML, Shaw AS. Costimulation: Building An Immunological Synapse. Science. 1999;283:649–650. doi: 10.1126/science.283.5402.649. [DOI] [PubMed] [Google Scholar]
- 2.Dustin ML. T-cell Activation Through Immunological Synapses and Kinapses. Immunol Rev. 2008;221:77–89. doi: 10.1111/j.1600-065X.2008.00589.x. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann MF, et al. Distinct Roles for LFA-1 and CD28 During Activation of Naive T cells: Adhesion Versus Costimulation. Immunity. 1997;7:549–557. doi: 10.1016/s1074-7613(00)80376-3. [DOI] [PubMed] [Google Scholar]
- 4.Van Seventer GA, Shimizu Y, Horgan KJ, Shaw S. The LFA-1 Ligand ICAM-1 Provides An Important Costimulatory Signal for T cell Receptor-Mediated Activation of Resting T cells. J Immunol. 1990;144:4579–4586. [PubMed] [Google Scholar]
- 5.Springer TA, Dustin ML, Kishimoto TK, Marlin SD. The Lymphocyte Function-Associated LFA-1, CD2, and LFA-3 Molecules: Cell Adhesion Receptors of The Immune System. Annu Rev Immunol. 1987;5:223–252. doi: 10.1146/annurev.iy.05.040187.001255. [DOI] [PubMed] [Google Scholar]
- 6.Lunsford KE, Koester MA, Eiring AM, Horne PH, Gao D, Bumgardner GL. Targeting LFA-1 and CD154 Suppresses The In Vivo Activation And Development of Cytolytic (CD4-Independent) CD8+ T cells. J Immunol. 2005;175:7855–66. doi: 10.4049/jimmunol.175.12.7855. [DOI] [PubMed] [Google Scholar]
- 7.Simnick AJ, Valencia A, Liu R, Chilkoti A. Morphing Low-Affinity Ligands into High-Avidity Nanoparticles by Thermally Triggered SelfAssembly of a Genetically Enclosed Polymer. ACS Nano. 2010;4:2217–2227. doi: 10.1021/nn901732h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stukel JM, Li RC, Maynard HD, Caplan MR. Two-Step Synthesis of Multivalent Cancer-Targeting Constructs. Biomacromolecules. 2010;11:160–167. doi: 10.1021/bm9010276. [DOI] [PubMed] [Google Scholar]
- 9.Hong A, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr., Banaszak Holl MM. The Binding Avidity of a Nanoparticle-Based Multivalent Targeted Drug Delivery Platform. Chemistry and Biology. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Johnson RN, Kopeckova P, Kopecek J. Synthesis and Evaluation of Multivalent Branched HPMA Copolymer-Fab’ Conjugates Targeted to the B-Cell Antigen CD20. Bioconjugate Chem. 2009;20:129–137. doi: 10.1021/bc800351m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding H, Prodinger WM, Kopecek J. Identification of CD21-Binding Peptides with Phage Display and Investigation of Binding Properties of HPMA Copolymer-Peptide Conjugates. Bioconjugate chem. 2006;17:514–523. doi: 10.1021/bc0503162. [DOI] [PubMed] [Google Scholar]
- 12.Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. Selective Tumor Cell Targeting Using Low-Affinity, Multivalent Interactions. ACS Chem Biol. 2007;2:119–27. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 13.Puffer EB, Pontrello JK, Hollenbeck JJ, Kink JA, Kiessling LL. Activating B Cell Signaling with Defined Multivalent Ligands. ACS Chem Biol. 2007;2:252–62. doi: 10.1021/cb600489g. [DOI] [PubMed] [Google Scholar]
- 14.Gordon EJ, Sanders WJ, Keissling LL. Synthetic Ligands Point to Cell Surface Strategies. Nature. 1998;392:30–31. doi: 10.1038/32073. [DOI] [PubMed] [Google Scholar]
- 15.Mowery P, Yang ZQ, Gordon EJ, Dwir O, Spencer AG, Alon R, Kiessling LL. Synthetic Glycoprotein Mimics Inhibit L-Selectin Mediated Rolling and Promote L-Selectin Shedding. Chem Biol. 2004;11:725–32. doi: 10.1016/j.chembiol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Lebedeva T, Dustin ML, Sykulev Y. ICAM-1 Co-stimulates Target Cells to Facilitate Antigen Presentation. Curr Opin Immunol. 2005;17:251–258. doi: 10.1016/j.coi.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Hogg N, Laschinger M, Giles K, McDowall A. T-cell Integrins: More Than Just Sticking Points. J Cell Sci. 2003;116:4695–4705. doi: 10.1242/jcs.00876. [DOI] [PubMed] [Google Scholar]
- 18.Anderson ME, Siahaan TJ. Targeting ICAM-1/LFA-1 Interaction for Controlling Autoimmune Diseases: Designing Peptide and Small Molecule Inhibitors. Peptides. 2003;24:487–501. doi: 10.1016/s0196-9781(03)00083-4. [DOI] [PubMed] [Google Scholar]
- 19.Anderson ME, Siahaan TJ. Mechanism of Binding and Internalization of ICAM-1-Derived Cyclic Peptides by LFA-1 on The Surface of T cells: A Potential Method for Targeted Drug Delivery. Pharm Res. 2003;20:1523–1532. doi: 10.1023/a:1026188212126. [DOI] [PubMed] [Google Scholar]
- 20.Jois SDS, Siahaan TJ. A Peptide Derived from LFA-1 That Modulates T-cell Adhesion Binds to Soluble ICAM-1 Protein. J. Biomol. Struc. Dyn. 2003;20:635–644. doi: 10.1080/07391102.2003.10506880. [DOI] [PubMed] [Google Scholar]
- 21.Tibbetts SA, Seetharama JD, Siahaan TJ, Benedict SH, Chan MA. Linear and Cyclic LFA-1 and ICAM-1 Peptides Inhibit T cell Adhesion and Function. Peptides. 2000;21:1161–1167. doi: 10.1016/s0196-9781(00)00255-2. [DOI] [PubMed] [Google Scholar]
- 22.Tibbetts SA, et al. Peptides Derived from ICAM-1 and LFA-1 Modulate T cell Adhesion and Immune Function in A Mixed Lymphocyte Culture. Transplantation. 1999;68:685–692. doi: 10.1097/00007890-199909150-00015. [DOI] [PubMed] [Google Scholar]
- 23.Chittasupho C, Manikwar P, Krise JP, Siahaan TJ, Berkland C. cIBR Effectively Targets Nanoparticles to LFA-1 on Acute Lymphoblastic T cells. Mol Pharm. 2010;7:146–155. doi: 10.1021/mp900185u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang N, Chittasupho C, Duangrat C, Siahaan TJ, Berkland C. PLGA Nanoparticle-Peptide Conjugate Effectively Targets Intercellular Cell-Adhesion Molecule-1. Bioconjug Chem. 2008;19:145–152. doi: 10.1021/bc700227z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chittasupho C, Xie S, Baoum A, Yakovleva T, Siahaan TJ, Berkland C. ICAM-1 Targeting of Doxorubicin-Loaded PLGA Nanoparticles to Lung Epithelial Cells. European Journal of Pharmaceutical Sciences. 2009;37:141–150. doi: 10.1016/j.ejps.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Commins SP, Borish L, Steinke JW. Immunologic Messenger Molecules: Cytokines, Interferons, and Chemokines. J Allergy Clin Immunol. 2010;125:S53–72. doi: 10.1016/j.jaci.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Yamane H, Paul WE. Differentiation of Effector CD4 T cell Populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miossec P, Korn T, Kunchroo VK. Mechanisms of Disease Interleukin-17 and Type 17 Helper T Cells. The new england journal of medicine. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 29.Dustin ML. A Dynamic View of The Immunological Synapse. Semin Immunol. 2005;17:400–410. doi: 10.1016/j.smim.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Dustin ML. Modular Design of Immunological Synapses and Kinapses. Cold Spring Harbor Perspect Biol. 2009;1:a002873. doi: 10.1101/cshperspect.a002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothlein R, Dustin ML, Marlin SD, Springer TA. A Human Intercellular Adhesion Molecule (ICAM-1) Distinct from LFA-1. J Immunol. 1986;137:1270–1274. [PubMed] [Google Scholar]
- 32.Dustin ML, Springer TA. T-cell Receptor Cross-Linking Transiently Stimulates Adhesiveness Through LFA-1. Nature. 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 33.Muro S, Wiewrodt R, Thomas A, Koniaris L, Albelda SM, Muzykantov VR, Koval M. A Novel Endocytic Pathway Induced by Clustering Endothelial ICAM-1 or PECAM-1. J Cell Sci. 2003;116:1599–609. doi: 10.1242/jcs.00367. [DOI] [PubMed] [Google Scholar]
- 34.Welder CA, Lee DHS, Takei F. Inhibition of Cell Adhesion by Microspheres Coated with Recombinant Soluble ICAM-1. Journal of Immunology. 1993;150:2203–2210. [PubMed] [Google Scholar]
- 35.Yoshida M, Babensee JE. Molecular Aspects of Microparticle Phagocytosis by Dendritic Cells. J. Biomater. Sci. Polymer. 2006;17:893–907. doi: 10.1163/156856206777996844. [DOI] [PubMed] [Google Scholar]
- 36.Semete B, Booysen LIJ, Kalombo L, Venter JD, Katata B, Ramalapa JA, Verschoor JA, Swai H. In Vivo Uptake and Acute Immune Response to Orally Administered Chitosan and PEG Coated PLGA Nanoparticles. Toxicology and Applied Pharmacology. 2010 doi: 10.1016/j.taap.2010.09.002. article in press. [DOI] [PubMed] [Google Scholar]
- 37.Molavi O, Mahmud A, Hamdy S, Hung RW, Lai R, Samuel J, Lavasanifar A. Development of a Poly(D,L-lactic-co-glycolic acid) Nanoparticle Formulation of STAT3 Inhibitor JSI-124: Implication for Cancer Immunotherapy. Molecular Pharmaceutics. 2009;7:364–374. doi: 10.1021/mp900145g. [DOI] [PubMed] [Google Scholar]
- 38.Steenblock E, Fahmy TM. A Comprehensive Platform for Ex Vivo Tcell Expansion Based on Biodegradable Polymeric Artificial Antigenpresenting Cells. Molecular Therapy. 2008;16:765–772. doi: 10.1038/mt.2008.11. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida M, Babensee JE. Molecular Aspects of Microparticle Phagocytosis by Dendritic Cells. J. Biomater. Sci. Polymer. 2006;17:893–907. doi: 10.1163/156856206777996844. [DOI] [PubMed] [Google Scholar]
- 40.Lutz MB, et al. An Advanced Culture Method for Generating Large Quantities of Highly Pure Dendritic Cells from Mouse Bone Marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 41.Guerrouache M, Karakasyan C, Gaillet C, Canva M, Millot MC. Immobilization of A Functionalized Poly(ethylene glycol) onto β-Cyclodextrin-Coated Surfaces by Formation of Inclusion Complexes: Application toThe Coupling of Proteins. J Applied Polym Sci. 2006;100:2362–2370. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Blocking of the T cell conjugation to DCs. LABLNPs and cIBR-NPs inhibited the binding of T cells to DCs to a much greater extent than free LABL and cIBR peptides. DCs were incubated with samples for 30 min, washed and cocultured with T cells for 2 hours. T cells and DCs were imaged and counted. *** indicates p<0.001.