A germline mutation in the 3′-untranslated region of KRAS (rs61764370, KRAS-variant: TG/GG) which disrupts microRNA regulation has previously been associated with altered patient outcome and drug sensitivity in various cancers. Our study suggests this KRAS-variant is a potential predictive biomarker for poor platinum response in recurrent/metastatic head and neck squamous cell carcinoma patients.
Keywords: KRAS-variant, cisplatin, cetuximab, p16 expression, head and neck squamous cell carcinoma
Abstract
Background
A germline mutation in the 3′-untranslated region of KRAS (rs61764370, KRAS-variant: TG/GG) has previously been associated with altered patient outcome and drug resistance/sensitivity in various cancers. We examined the prognostic and predictive significance of this variant in recurrent/metastatic (R/M) head and neck squamous cell carcinoma (HNSCC).
Patients and methods
We conducted a retrospective study of 103 HNSCCs collected from three completed clinical trials. KRAS-variant genotyping was conducted for these samples and 8 HNSCC cell lines. p16 expression was determined in a subset of 26 oropharynx tumors by immunohistochemistry. Microarray analysis was also utilized to elucidate differentially expressed genes between KRAS-variant and non-variant tumors. Drug sensitivity in cell lines was evaluated to confirm clinical findings.
Results
KRAS-variant status was determined in 95/103 (92%) of the HNSCC tumor samples and the allelic frequency of TG/GG was 32% (30/95). Three of the HNSCC cell lines (3/8) studied had the KRAS-variant. No association between KRAS-variant status and p16 expression was observed in the oropharynx subset (Fisher's exact test, P = 1.0). With respect to patient outcome, patients with the KRAS-variant had poor progression-free survival when treated with cisplatin (log-rank P = 0.002). Conversely, KRAS-variant patients appeared to experience some improvement in disease control when cetuximab was added to their platinum-based regimen (log-rank P = 0.04).
Conclusions
The TG/GG rs61764370 KRAS-variant is a potential predictive biomarker for poor platinum response in R/M HNSCC patients.
Clinical trial registration numbers
introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most frequent cancer worldwide [1]. Risk factors for HNSCC include tobacco and alcohol consumption, as well as human papillomavirus (HPV) infection [2, 3]. Recent studies have concluded that HPV-positive (+) and -negative (−) HNSCC represent distinct diseases [4]. Despite recent advances in multimodality treatment, ∼20% of HPV(+) and ∼50% of HPV(−) HNSCC patients experience treatment failure and subsequent disease-related death at 5 years. The current standard of care for first-line recurrent and/or metastatic (R/M) disease is a platinum-based combination regimen such as cisplatin, 5-fluorouracil and cetuximab [5]. However, treatment efficacy is still limited in these patients with overall survival (OS) <1 year. Clearly, novel predictive biomarkers for cisplatin resistance in R/M HNSCC patients are greatly needed.
While KRAS-activating mutations are well-established biomarkers for poor prognosis in various cancers, these mutations are extremely rare in HNSCC (<5%), limiting their prognostic value [6]. However, KRAS activity can be altered by means other than somatic mutations. Recent work has demonstrated that a germline, functional single-nucleotide polymorphism in the KRAS 3′-untranslated region (rs61764370, KRAS-variant) alters the expression and activity of otherwise ‘wild-type’ KRAS. Current evidence suggests this variant disrupts let-7 microRNA binding, thus decreasing KRAS-negative regulation and initiating gain of function [7]. Not surprisingly, the presence of this KRAS-variant has been shown to predict poor outcome in many cancer types. In head and neck cancer, after controlling for confounders of survival, this variant also predicts reduced survival [8].
In this study, we evaluated the association of this KRAS-variant with clinical outcome in R/M HNSCC patients treated with cisplatin, cisplatin + cetuximab or docetaxel + bortezomib. Additionally, we established the sensitivity of non-variant and KRAS-variant HNSCC cell lines to cisplatin as well as altered gene expression in HNSCC tumors. We found that patients with KRAS-variant tumors had worse outcome when given cisplatin, yet potentially improved response when cetuximab was added in combination.
methods
patients
Tumor samples were collected from three previously published clinical trials for KRAS-variant testing; (i) 24 samples from HN0582 (NCT00503997), a phase II trial for efficacy and toxicity of induction pemetrexed and oxaliplatin in patients with locally advanced HNSCC [9], (ii) 22 samples from HN0501 (NCT00425750), phase II trial of combination weekly bortezomib and docetaxel in patients with R/M HNSCC [10], and (iii) 57 samples from E5397 (NCT00003809), a randomized, double-blind, placebo-controlled phase III evaluation of cisplatin + placebo versus cisplatin + C225 in patients with R/M HNSCC [11].
materials
KRAS-variant determination
Genomic DNA from cell lines and formalin-fixed paraffin-embedded tumors was isolated as previously described and 100 ng analyzed in a CLIA-certified laboratory for the KRAS-variant [7] (Mira Dx, New Haven, CT).
head and neck squamous cell carcinoma cell lines and MTS assay
Eight HNSCC cell lines (Cal27, UNC7, FaDu, SKN3, SCC6, SCC61, UPCI:SCC90, and UMSCC47) were obtained and maintained in cell culture as previously published [12]. To assess in vitro proliferation in the presence of cisplatin, MTS assays were used to estimate relative cell growth as previously described [12]. Cisplatin was purchased from Sigma-Aldrich (St Louis, MO).
RNA isolation and DNA microarray analysis
Total RNA was purified from frozen tumors (∼10–20 mg of wet tissue per sample) using the Qiagen RNeasy Mini Kit according to the manufacturer's recommendations (Qiagen, Valencia, CA). Gene expression data were generated as previously described using Affymetrix Human Genome U133 plus 2.0 GeneChip [13] and is available from GEO (GSE36110).
p16 status determination
p16 status was determined by immunohistochemistry using a p16 mouse monoclonal antibody (predilute, mtm-CINtech, E6H4). p16 positivity was determined by diffuse staining with >70% of the tumor staining positive as previously described [4].
statistics
statistical analysis of KRAS-variant incidence and clinical correlations
KRAS-variant incidence was reported by frequency and percentage of TG/GG samples with respect to total samples with known KRAS-variant status. Descriptive statistics were used to characterize patient demographics and disease characteristics. Associations between KRAS-variant status and patient characteristics/response were evaluated using Fisher's exact tests. Progression-free survival (PFS) was defined as the time between date of study entry and date of progression or death from any cause, censored at date of last disease assessment. OS was defined as the time between date of study entry and date of death, censored at last contact. Event-time distributions were estimated by the Kaplan–Meier method and compared using log-rank tests. Association of KRAS-variant genotype with OS and PFS was evaluated using univariate Cox proportional hazard regression modeling. If any association achieved statistical significance, multivariable Cox regression models were further fitted by controlling important patient/clinical variables. All P values are two-sided and a level of P < 0.05 was considered statistically significant.
statistical analysis of microarray data
Microarrays were normalized using frozen robust multiarray analysis [14]. Empirical Bayes moderated t-statistics were computed to compare gene expression from KRAS-variant (TG/GG) samples to non-variant (TT) samples using LIMMA [15]. Benjamini–Hotchberg multiple testing correction was applied to the corresponding P values [16].
results
tumor samples and KRAS genotypes
Detailed patient characteristics of the clinical trial cohorts have previously been published [9–11]. Of the combined 103 samples evaluated, KRAS-variant status could be determined in 95/103 (92%). More specifically, 19/22 samples (86%) from HN0501, 54/57 (95%) from E5397 and 22/24 (92%) from HN0582 yielded interpretable KRAS status information (Table 1). The overall allelic frequency of KRAS-variant (TG/GG) tumors was 32% (30/95). As a surrogate marker of HPV status, p16 expression was determined in 26 oropharynx tumors with available unstained tumor slides [KRAS TT/HPV(+): 9 (35%), KRAS TG/GG/HPV(+): 7 (27%), KRAS TT/HPV(−): 6 (23%), and KRAS TG/GG/HPV(−): 4 (15%)]. Although the numbers were small, there was no association between KRAS-variant status and p16 expression (Fisher's exact test, P = 1.0).
Table 1.
Patient characteristics of three completed clinical trials in head and neck squamous cell carcinoma (HNSCC) patients; HN0501 (phase II docetaxel and bortezomib as first-line therapy for recurrent/metastatic HNSCC), E5397 (phase III cisplatin with/without cetuximab as first-line therapy for recurrent/metastatic HNSCC), and HN0582 (phase II pemetrexed and oxaliplatin induction followed by chemoradiation in newly diagnosed HNSCC)
| HN0501 |
E5397 |
HN0582 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TG/GG (N = 8) |
TT (N = 11) |
TG/GG (N = 12) |
TT (N = 42) |
TG/GG (N = 10) |
TT (N = 12) |
|||||||
| n | % | n | % | n | % | n | % | n | % | n | % | |
| Age (median, range) | 54 (35–62) | 54 (41–76) | 56 (42–83) | 58 (36–85) | 56 (36–66) | 57 (41–74) | ||||||
| Sex | ||||||||||||
| Male | 7 | 88 | 6 | 75 | 11 | 92 | 32 | 76 | 8 | 80 | 11 | 92 |
| Female | 1 | 12 | 2 | 25 | 1 | 8 | 10 | 24 | 2 | 20 | 1 | 8 |
| Unknown | 0 | – | 3 | – | 0 | – | 0 | – | 0 | – | 0 | – |
| Race | ||||||||||||
| White | 7 | 88 | 7 | 64 | 11 | 92 | 37 | 88 | 10 | 100 | 11 | 92 |
| Black | 1 | 12 | 1 | 9 | 1 | 8 | 4 | 10 | 0 | 0 | 1 | 8 |
| Others/unknown | 0 | 0 | 3 | 27 | 0 | 0 | 1 | 2 | 0 | – | 0 | – |
| Path differentiation | ||||||||||||
| Well differentiated | 0 | 0 | 0 | 0 | 2 | 20 | 5 | 12 | 0 | 0 | 0 | 0 |
| Moderately differentiated | 3 | 43 | 5 | 83 | 4 | 40 | 20 | 49 | 5 | 56 | 9 | 82 |
| Poorly differentiated | 4 | 57 | 1 | 17 | 4 | 40 | 16 | 39 | 4 | 44 | 2 | 18 |
| Unknown | 1 | – | 5 | – | 2 | – | 1 | – | 1 | – | 1 | – |
| Disease subsites | ||||||||||||
| Oral cavity | 2 | 25 | 3 | 30 | 1 | 8 | 10 | 24 | 0 | 0 | 1 | 8 |
| Oropharynx | 4 | 50 | 7 | 70 | 4 | 33 | 13 | 31 | 7 | 70 | 8 | 60 |
| Hypopharynx | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 10 | 0 | 0 | 1 | 8 |
| Larynx | 1 | 12 | 0 | 0 | 3 | 25 | 12 | 29 | 2 | 20 | 1 | 8 |
| Others | 1 | 12 | 0 | 0 | 4 | 33 | 3 | 7 | 1 | 10 | 1 | 8 |
| Unknown | 0 | – | 1 | – | 0 | – | 0 | – | 0 | – | 0 | – |
KRAS-variant status is associated with cisplatin resistance in R/M HNSCC patients
To determine whether KRAS-variant status was associated with cisplatin resistance in R/M HNSCC patients, survival analyses were carried out for patients in HN0501 and E5397 (Table 2). Both of these studies were powered to determine survival benefit and also enrolled similar first-line R/M patients [10, 11]. No prior cisplatin exposure as an R/M treatment was allowed in E5397 patients. HN0582 was not included in these analyses because the primary end point of this study was chemotherapy response rate rather than survival outcomes in newly diagnosed patients. The median follow-up time among patients with KRAS data was 8 months for E5397, 10 months for HN0501, and 20 months for HN0582. The median survival data and hazard ratios (HRs) with respect to KRAS-variant status are summarized in Table 2. The disease control and objective response rates did not differ by KRAS-variant status in both studies (supplementary Table S1, available at Annals of Oncology online).
Table 2.
Event-time distribution by KRAS-variant genotype and first-line recurrent/metastatic HNSCC treatment regimen in HN0501 (phase II docetaxel and bortezomib) and E5397 (phase III cisplatin with/without cetuximab)
| Survival outcome | Treatment regimens | KRAS genotype | # of events/N | Median survival (95% CI) | Log-rank P | Univariate | Wald P | Multivariablea | Wald P |
|---|---|---|---|---|---|---|---|---|---|
| HR (TG/GG versus TT) (95% CI) | HR (TG/GG versus TT) (95% CI) | ||||||||
| Progression-free survival | HN0501: docetaxel + bortezomib | TG/GG | 8/8 | 1.6 (1.1–4.7) | 0.89 | 0.93 (0.33–2.62 | 0.90 | – | – |
| TT | 8/11 | 1.7 (1.4–4.1) | |||||||
| E5397: overall cisplatin + placebo or cetuximab | TG/GG | 12/12 | 2.2 (1.0–4.5) | 0.002 | 2.82 (1.40–5.67) | 0.004 | 3.68 (1.50–9.03) | 0.004 | |
| TT | 40/42 | 4.7 (3.7–5.9) | |||||||
| E5397: cisplatin + placebo | TG/GG | 5/5 | 1.9 (0.0–3.4) | 0.002 | 4.94 (1.58–15.40) | 0.006 | 3.75 (0.78–18.12) | 0.10 | |
| TT | 22/23 | 3.9 (2.3–5.5) | |||||||
| E5397: cisplatin + cetuximab | TG/GG | 7/7 | 3.9 (1.8–4.6) | 0.04 | 2.65 (1.01–6.98) | 0.049 | 3.59 (0.96–13.41) | 0.057 | |
| TT | 18/19 | 5.8 (3.7–6.3) | |||||||
| Overall survival | HN0501: docetaxel + bortezomib | TG/GG | 8/8 | 6.7 (1.6–9.8) | 0.60 | 1.30 (0.49–3.50) | 0.60 | – | – |
| TT | 11/11 | 5.1 (1.3–12.6) | |||||||
| E5397: overall cisplatin + placebo or cetuximab | TG/GG | 12/12 | 7.3 (1.0–12.1) | 0.11 | 1.71 (0.89–3.31) | 0.11 | – | – | |
| TT | 38/42 | 8.2 (7.0–12.2) | |||||||
| E5397: cisplatin + placebo | TG/GG | 5/5 | 5.4 (0.0–12.1) | 0.09 | 2.38 (0.85–6.62) | 0.10 | – | – | |
| TT | 21/23 | 8.1 (6.1–13.5) | |||||||
| E5397: cisplatin + cetuximab | TG/GG | 7/7 | 8.0 (1.8–12.2) | 0.52 | 1.34 (0.55–3.25) | 0.52 | – | – | |
| TT | 17/19 | 8.2 (6.3–12.3) |
P-values in bold indicate P-values which are statistically significant.
aCovariates included PS (0 versus 1), disease status (previously untreated versus recurrent), cell differentiation (well/moderate versus poorly differentiation), primary site (oropharynx versus non-oropharynx), smoking history (≤40 versus >40 packs-years), alcohol consumption (<10 oz whiskey/week versus ≥10 oz whiskey/week) and treatment (when appropriate). N = 21 in the cisplatin + placebo arm and in the cisplatin + cetuximab arm.
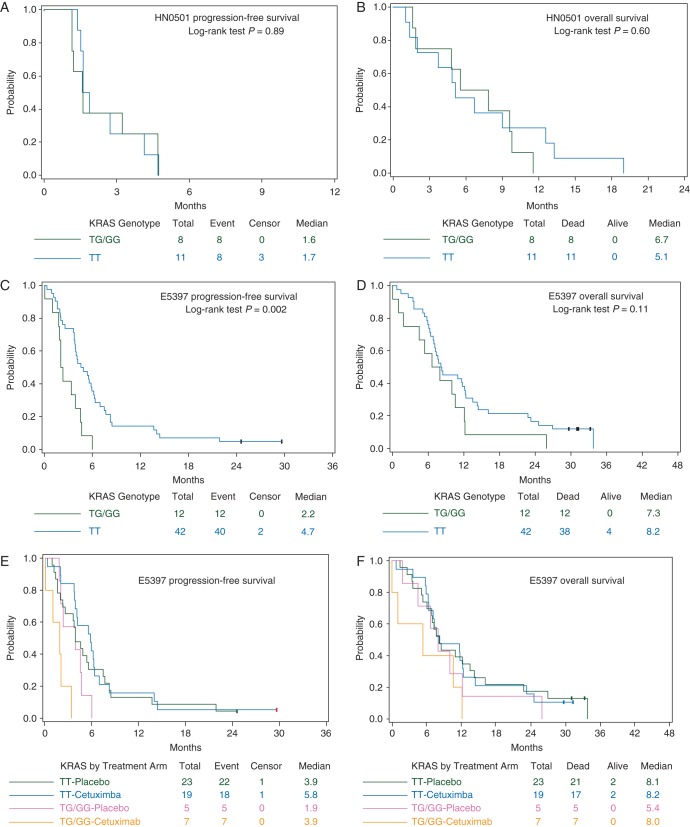
We further compared KRAS-variant status with PFS and OS in these studies separately, as patients were treated differently. In HN0501 (bortezomib + docetaxel), KRAS-variant status was not associated with PFS (median: 1.6 months in TG/GG versus 1.7 months in TT; log-rank P = 0.89; Figure 1A) or OS (median: 6.7 months in TG/GG versus 5.1 months in TT; log-rank P = 0.60; Figure 1B). However, in E5397, when the two treatment arms were combined (cisplatin + placebo and cisplatin + cetuximab), KRAS-variant status was a statistically significant predictor of poor PFS (median: 2.2 months in TG/GG versus 4.7 months in TT; log-rank P = 0.002; Figure 1C). Furthermore, in multivariable analyses, the HR for TG/GG versus TT with respect to PFS was 3.85 (95% confidence interval 1.59–9.35; Wald P = 0.003). While this association was significant for PFS, KRAS-variant status did not significantly associate with OS (median: 7.3 months in TG/GG versus 8.2 months in TT, log-rank P = 0.11; Figure 1D).
Figure 1.
Kaplan–Meier survival plots by KRAS-variant status (variant TG/GG versus non-variant TT). (A) Progression-free survival from HN0501 (phase II trial of docetaxel + bortezomib in R/M HNSCC patients); (B) overall survival from HN0501; (C) progression-free survival from E5397 (randomized phase III trial of cisplatin + placebo versus cisplatin + cetuximab in R/M HNSCC patients); (D) overall survival from E5397; (E) progression-free survival by KRAS-variant status (variant TG/GG versus non-variant TT) and E5397 treatment arms (cisplatin + placebo versus cisplatin + cetuximab); and (F) overall survival by KRAS-variant status (variant TG/GG versus non-variant TT) and E5397 treatment arms (cisplatin + placebo versus cisplatin + cetuximab).
KRAS-variant HNSCC cell lines are more resistant to cisplatin in vitro
To confirm that KRAS-variant status affects cisplatin sensitivity in HNSCC, eight HNSCC cell lines were evaluated by MTS assay with increasing doses of cisplatin (1 nM–10 µM). Six of these cell lines were HPV-negative (CAL27, UNC7, FaDu, SKN3, SCC6, and SCC61), while two were HPV-positive (UPCI:SCC90 and UMSCC47). Three cell lines were positive for the KRAS-variant: CAL27, UNC7, and UMSCC47. After in vitro exposure to increasing doses of cisplatin, it was determined that CAL27 and UNC7 were two of the most cisplatin-resistant cell lines evaluated (supplementary Figure S1A, available at Annals of Oncology online). In fact, UNC7 demonstrated complete resistance to cisplatin at the highest dosage utilized (10 μM) (supplementary Figure S1B, available at Annals of Oncology online). Additionally, two of the three non-variant cell lines (SCC61 and SCC6) began demonstrating cisplatin sensitivity at the lowest doses in the study (1 and 10 nM). FaDu, a highly metastatic HNSCC cell line developed from a hypopharyngeal tumor [17], was the only non-variant cell line that demonstrated significant cisplatin resistance (supplementary Figure S1A, available at Annals of Oncology online). While the KRAS-variant appears to confer resistance to cisplatin in HPV(−) cells, this effect was not found in HPV(+) cell lines (supplementary Figure S1C, available at Annals of Oncology online).
addition of cetuximab to cisplatin may benefit R/M HNSCC patients with the KRAS-variant
Outcome with respect to treatment and KRAS-variant status was further examined comparing cisplatin + placebo versus cisplatin + cetuximab treatment arms in E5397 (Table 2 and Figure 1E–F). Although the sample size was very small, PFS was significantly improved in KRAS-variant (TG/GG) patients who received cetuximab versus placebo in univariate analysis (median: 1.9 months in cisplatin + placebo versus 3.9 months in cisplatin + cetuximab; log-rank P = 0.03), while this effect was not observed in the non-variant (TT) group (median: 3.9 months in cisplatin + placebo versus 5.8 months in cisplatin + cetuximab; log-rank P = 0.57). While KRAS-variant patients experienced enhanced disease control from the addition of cetuximab, this effect was not statistically significant for OS (TG/GG median: 5.4 months in cisplatin + placebo versus 8.0 months in cisplatin + cetuximab, log-rank P = 0.37; TT median: 8.1 months in cisplatin + placebo versus 8.2 months in cisplatin + cetuximab, log-rank P = 0.96). This preliminary investigation suggests that the KRAS-variant may provide potentially meaningful information regarding clinical benefit of cetuximab given with cisplatin in R/M HNSCC patients.
differentially expressed genes based on the KRAS-variant status in HNSCC
To gain further insight into the observed differences in clinical responses, we compared the gene expression profiles of 17 non-variant and 5 KRAS-variant tumors with available microarray data. After normalizing the datasets and employing empirical Bayes moderated t-statistics, we examined 25 probes with the smallest FDR-adjusted P values (supplementary Table 2 and Figure S2, available at Annals of Oncology online). Among the genes with known molecular functions, a few interesting trends emerged. Most notably, epidermal growth factor (EGF) was upregulated (log FC: 0.77; P = 6.00 × 10−5) in KRAS-variant tumors, suggesting that increased KRAS activity in these cells may still be dependent on upstream EGFR growth signals. This may explain the enhanced disease control observed in these patients with cetuximab treatment. Another interesting trend observed in KRAS-variant tumor gene expression is the upregulation of various cytoskeleton and microtubule associated proteins. DST (dystonin or bullous pemphigoid antigen 1, log FC: 1.88; P = 3.98 × 10−6) and MACF1 (log FC: 0.90; P = 4.22 × 10−5) are both members of the plakin family, responsible for junctional complex and cytoskeleton function reviewed in [18]. SYNJ2 (synaptojanin 2, log FC: 1.08; P = 2.58 × 10−4) is a regulatory lipid phosphatase and upregulation has previously been associated with metastatic spread [19]. Taken together, this gene expression profile suggests KRAS-variant HNSCC cells have a prometastatic gene signature due to enhanced expression of migratory machinery in this exploratory and hypothesis-generating study. Larger sample sizes will be required in future studies to further evaluate these putative genes for functional validation.
discussion
Our current study suggests that R/M HNSCC patients with KRAS-variant have worse prognosis and exhibit cisplatin resistance. We also found these patients may benefit from the addition of cetuximab to their treatment regimen, suggesting this mutation may be a predictive biomarker of treatment response, and not just a prognostic biomarker. Our gene expression studies supported these clinical results, demonstrating an upregulation of EGF and prometastasis genes.
Previous studies have evaluated the prevalence of this KRAS-variant in HNSCC; however, our study is the first to examine the frequency in a patient population comprised of mostly R/M disease, with known treatment, enabling evaluation of this mutation as a predictive biomarker. The worldwide allelic frequency of the TG/GG KRAS-variant is ∼6% [20]. Yet, incidence rates as high as 23% are observed in newly diagnosed non-small-cell lung cancer [20], ovarian cancer [21], and triple-negative breast cancer [22]. In a previous case-control study, Christensen et al. reported the frequency of TG/GG to be ∼15% in newly diagnosed HNSCC patients [8]. An association between incidence of HNSCC and specific primary sites (oral, pharyngeal, or laryngeal cancer) was not observed after stratification by potential confounders. However, in agreement with our study, the presence of this variant was significantly associated with poor survival, and these effects were mostly observed in oral cancer, but not pharyngeal or laryngeal tumors. Given the patient population in our study, the higher incidence of this mutation suggests two possibilities: (i) the presence of a KRAS-variant is associated with particularly aggressive de novo disease, or (ii) this variant confers increased resistance to the therapy for newly diagnosed HNSCC, enriching this variant within the R/M patients. Current evidence suggests these mechanisms may not be mutually exclusive.
Identifying biomarkers of cisplatin resistance is critically important, as cisplatin is the most commonly used chemotherapy in both newly diagnosed and R/M HNSCC. Thus, elucidation of such biomarkers would allow optimization and personalization of chemotherapeutic strategies, or personalized medicine. The association between the KRAS-variant and cisplatin resistance has been previously established in ovarian cancer [21]. Ratner et al. genotyped 536 epithelial ovarian cancers for the same variant and subset analysis determined variant patients exhibited significant platinum resistance. The clinical and in vitro data reported within our current study are consistent with these prior results, and support the hypothesis that cisplatin therapy should be reconsidered in KRAS-variant R/M HNSCC, and perhaps all cancer patients harboring this mutation.
We have also evaluated KRAS-variant patient outcome when treated with the only targeted agent approved for HNSCC patients: cetuximab. Our findings suggest that KRAS-variant patients experience improved disease control when cetuximab is added to cisplatin, albeit the results require additional confirmation due to our small sample size. In the gene expression profile associated with KRAS-variant tumors, it was noted that EGF, a potent ligand of EGFR, was upregulated in these lesions. It is attractive to hypothesize that cetuximab would be beneficial for these patients to block the progrowth signal provided by an upregulated EGF. Furthermore, a number of genes associated with microtubule and cytoskeleton function are also upregulated in KRAS-variant tumors. More specifically, MACF1 is upregulated in our current analysis and this protein has previously been reported to interact with ErbB2 and control microtubule capture during cell migration [23]. Taken together, the upregulation of an EGFR-specific growth stimulatory ligand (EGF), a promigratory phosphatase (SYNJ2), and various components of the microtubule/cytoskeletal architecture (DST and MACF1) provides intriguing evidence of an enhanced migratory or metastatic gene expression profile associated with KRAS-variant HNSCCs, and potentially explains inherent sensitivity to cetuximab treatment.
The results of our current study support the hypothesis that KRAS-variant is a biomarker of altered response to treatment in R/M HNSCC patients. Platinum-based regimens result in suboptimal disease control in these patients, and treatment with cetuximab in combination with platinum could be beneficial in this setting. This mutation appears to reflect a unique biology of various tumor subtypes, and further studies to delineate the clinical ramifications are warranted.
funding
The project was funded in part by NIH NIDCR R01 (DE017982) to CHC. The project was also funded in part by NIH R01 CA157749–01A1 to JBW. This study was coordinated in part by the Eastern Cooperative Oncology Group (Robert L. Comis) and supported in part by Public Health Service Grants CA23318, CA66636, CA21115, CA16116, CA27525, CA13650, CA9957 and from the National Cancer Institute, National Institutes of Health and the Department of Health and Human Services. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
disclosure
JBW has patented this mutation through Yale University and founded a company that has licensed this intellectual property. All remaining authors have declared no conflicts of interest.
Supplementary Material
references
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Maier H, Dietz A, Gewelke U, et al. Tobacco and alcohol and the risk of head and neck cancer. Clin Investig. 1992;70:320–327. doi: 10.1007/BF00184668. [DOI] [PubMed] [Google Scholar]
- 3.D'Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 6.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen BC, Moyer BJ, Avissar M, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–1007. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert J, Murphy B, Dietrich MS, et al. Phase 2 trial of oxaliplatin and pemetrexed as an induction regimen in locally advanced head and neck cancer. Cancer. 2012;118:1007–1013. doi: 10.1002/cncr.26364. [DOI] [PubMed] [Google Scholar]
- 10.Chung CH, Aulino J, Muldowney NJ, et al. Nuclear factor-kappa B pathway and response in a phase II trial of bortezomib and docetaxel in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2009;21:864–870. doi: 10.1093/annonc/mdp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burtness B, Goldwasser MA, Flood W, et al. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group Study. J Clin Oncol. 2005;23:8646–8654. doi: 10.1200/JCO.2005.02.4646. [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama H, Cheng H, Wirth P, et al. Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS One. 2010;5:e12702. doi: 10.1371/journal.pone.0012702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slebos RJ, Yi Y, Ely K, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 14.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–23. doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 17.Giard DJ, Aaronson SA, Todaro GJ, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 18.Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 19.Chuang YY, Tran NL, Rusk N, et al. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 2004;64:8271–8275. doi: 10.1158/0008-5472.CAN-04-2097. [DOI] [PubMed] [Google Scholar]
- 20.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratner ES, Keane FK, Lindner R, et al. A KRAS variant is a biomarker of poor outcome, platinum chemotherapy resistance and a potential target for therapy in ovarian cancer. Oncogene. 2012;31:4559–4566. doi: 10.1038/onc.2011.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paranjape T, Heneghan H, Lindner R, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12:377–386. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaoui K, Benseddik K, Daou P, et al. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc Natl Acad Sci USA. 2010;107:18517–18522. doi: 10.1073/pnas.1000975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.