Abstract
Engineering resistance genes to gain effector recognition is emerging as an important step in attaining broad, durable resistance. We engineered potato resistance gene R3a to gain recognition of the virulent AVR3aEM effector form of Phytophthora infestans. Random mutagenesis, gene shuffling and site-directed mutagenesis of R3a were conducted to produce R3a* variants with gain of recognition towards AVR3aEM. Programmed cell death following gain of recognition was enhanced in iterative rounds of artificial evolution and neared levels observed for recognition of AVR3aKI by R3a. We demonstrated that R3a*-mediated recognition responses, like for R3a, are dependent on SGT1 and HSP90. In addition, this gain of response is associated with re-localisation of R3a* variants from the cytoplasm to late endosomes when co-expressed with either AVR3aKI or AVR3aEM a mechanism that was previously only seen for R3a upon co-infiltration with AVR3aKI. Similarly, AVR3aEM specifically re-localised to the same vesicles upon recognition by R3a* variants, but not with R3a. R3a and R3a* provide resistance to P. infestans isolates expressing AVR3aKI but not those homozygous for AVR3aEM.
Introduction
In a process known as effector triggered immunity (ETI), plant disease resistance (R) genes can facilitate immunity to phylogenetically diverse and unrelated pathogens that express cognate effector molecules [1]. Effectors that are recognised by R genes, either directly or indirectly, and provoke successful plant defences are genetically defined as avirulence (Avr) genes. The best described group of plant R gene products contains a nucleotide-binding (NB) domain and leucine-rich repeats (LRRs), collectively known as NB-LRRs [2]. NB-LRRs are strictly regulated by plants as, upon their activation, many elicit programmed cell death (PCD) as part of the hypersensitive response (HR) which prevents further spread of disease in plant tissues [3]. Together with effectors, R genes are at the forefront of host/pathogen co-evolution [4]. NB-LRRs are one of the largest gene families in plants and more than 750 members have recently been described in potato [5]–[6]. The organisation of many NB-LRRs into physically-linked clusters is providing insight into their evolution, which can involve duplication followed by diversification.
In agriculture, successful deployment of R genes to control important diseases in crop plants has so far been hampered by the ability of pathogens often to rapidly circumvent detection by the host plant's innate immune system. Advances in studying pathogen effector diversity coupled with the ability to engineer R genes offers the opportunity to develop more durable resistances that specifically target essential effectors and known variants [7]–[8].
The Phytophthora infestans effector AVR3a is an essential effector for this pathogen to cause late blight on potato. Stable silencing of Avr3a in the P. infestans isolate 88069 significantly reduces infection in susceptible Solanum tuberosum (potato) cv. Bintje and in the model solanaceous plant species Nicotiana benthamiana [9]–[10]. Two forms of AVR3a are prevalent in current P. infestans isolates and differ in only two amino acids within the mature protein; AVR3aE80M103 (AVR3aEM) and AVR3aK80I103 (AVR3aKI) [11]–[12]. AVR3aKI elicits ETI upon recognition by the potato resistance protein R3a, a member of the coiled-coil (CC) NB-LRR gene family [13]. This response is evaded by AVR3aEM which consequently is free to promote virulence [9], [11], [13]. However, a weak R3a-dependent response to AVR3aEM can be observed under UV light [14]. The mechanism of R3a-mediated recognition of AVR3a has been investigated recently [15]. Upon activation by AVR3aKI, both the effector protein and R3a rapidly re-localize from the host cytoplasm to late endosomes, components of the endocytic pathway, which is thought to be a prerequisite for subsequent HR development. The un-recognised AVR3aEM form of the effector does not cause this re-localisation. There is no evidence of direct interaction between AVR3aKI and R3a, but bimolecular fluorescence complementation (BiFC) assays reveal that the two proteins are in close proximity at late endosomes [15].
Artificial evolution has previously been used to alter the recognition specificity of the potato CC-NB-LRR resistance protein Rx to gain recognition of different strains of Potato virus X (PVX) and a distantly related virus, Poplar mosaic virus (PopMV) [16]–[17]. Random mutagenesis, screening for beneficial mutations and designed amalgamation of these mutations has been used to generate transgenic plants of the model species N. benthamiana that are resistant to previously virulent strains. DNA shuffling, also known as directed evolution, was first developed in the early 1990 s and has since been used to generate a wide variety of novel genes and proteins [18]. DNA shuffling has previously been used in the functional analysis of the resistance gene Pto [19]. In this study, the LRR of R3a has been subjected to error-prone PCR and iterative rounds of DNA shuffling to identify gain-of-recognition variants (R3a*) through functional screening in N. benthamiana. R3a* gene products with varying degrees of AVR3aEM recognition were generated in three rounds of DNA shuffling and a subsequent round of site-directed mutagenesis. The best-performing clones from each round were taken forward for further analysis and compared to wild-type R3a. R3a* variants demonstrated significantly improved gain-of-recognition towards AVR3aEM that manifested itself as a gain of re-localisation to late endosomes, but not yet as a gain of resistance.
Materials and Methods
Construction of plasmid vectors
The plasmid pBinPlus.R3a [13] was amplified with the primer pairs R3a-5-Asc/R3a-1564-Bam-M and R3a-1564-Bam-P/R3a-3-Not (Table S1) and the AscI/BamHI and BamHI/NotI digested amplification products cloned in a three way ligation in to AscI/NotI digested binary vector pGRAB [20] to produce pGRAB.35S::R3a. A derivative of the former plasmid, pGRAB.R3a::R3a, was produced in which the Cauliflower mosaic virus 35S (35S) promoter sequence was replaced with the R3a promoter sequence. This derivative was produced by digestion of pBinPlus.R3a with PmeI and XhoI, and insertion of the released fragment, containing 2358 bases upstream of the translational start site, in to pGRAB.35S::R3a that had been treated with BspEI, T4 DNA polymerase and XhoI in order.
Mutagenesis and DNA shuffling
Shuffling of 2283 bp of the R3a LRR region was performed as described by Stemmer [18]. First round PCRs were carried out with the primer pair R3a-1564-Bam-P/R3a-3-Not in the presence of 0.5 mM MnCl2 as described by Leung et al. [21] with Taq DNA polymerase (Roche, Mannheim, Germany). Following DNaseI treatment, fragments of circa 500 bp were reassembled through forty rounds of primer-less thermo-cycling. Gel-purified products of circa 2.3 kb were amplified through thirty cycles of PCR and, following gel-purification, digested with BamHI and NotI prior to cloning in to pGRAB.35S::R3a digested with the same enzymes. The ligated population was amplified in E. coli resulting in a population with a complexity of circa 125 K, a vector background of circa 14% and a base mutation rate of 0.43%. Aliquots of plasmid, gel-purified on account of plasmid instability, were transformed in to A. tumefaciens cells. In the second and third rounds PfuUltra II Fusion HS DNA polymerase (Stratagene, La Jolla, CA, USA) was used in thermo-cycling reactions to produce shuffled populations with lower mutation rates, less than 0.05%. The second and third round populations had complexities of circa 200 K and 150 K, respectively, and both had a vector background of circa 10%.
Site-directed mutagenesis
A mixture of two templates, the wild-type gene and clone Rd1-2 from the first round of shuffling containing the Q931R codon substitution, was used in PCR amplifications with the primer pairs R3a-1564-Bam-P/R3a-1740W-M, R3a-1740W-P/R3a-1841W-M, R3a-1841W-P/R3a-2743W-M, R3a-2743W-P/R3a-3028-M. The products of the primary PCRs were used in an overlap PCR reaction with the flanking primer pair R3a-1564-Bam-P/R3a-3028-M. The product of the secondary PCR reaction was digested with BamHI and AatII and cloned between the same unique sites of pGRAB.R3a::R3a. A population of circa 50K clones, with a vector background of 1% and random base mutation rate of 0.05% was produced.
Plant growth conditions
N. benthamiana plants were grown in a glasshouse with a 16 h day period at 22°C and an 8 h night period at 18°C. Supplementary lighting was provided below 200 W m−2 and screening above 450 W m−2.
Screening of mutated R3a clones
DNA populations prepared from E. coli were transformed in to Agrobacterium tumefaciens strain AGL1 [22] carrying the helper plasmids pSoup and pBBR1MCS1.VirGN54D [23] for screening. Agrobacterium cultures grown from single transformed colonies were co-infiltrated with cultures of Agrobacterium transformed with pGRAB.35S::AVR3aEM according to the method of Engelhardt et al. [15] in to N. benthamiana leaves with each of the components at the same final OD600 of 0.1–0.5. Reference mixtures were infiltrated in to opposing half-leaves and between two and seven days post infiltration leaves were inspected to assess visible symptoms and plant auto-fluorescence under day-light and 365 nm illumination from a Blak-Ray lamp (UVP, Upland, CA, USA), respectively.
Symptom scoring
Circa five week old plants were used for symptom scoring with two adjacent, expanded leaves, both circa 90 mm in length, being used for infiltrations. Symptoms were scored on an arbitrary scale for nine days after infiltration. Symptom scores were plotted; the areas under the curve determined and mean scores calculated by dividing by the duration. A linear mixed modelling approach was adopted using GenStat for Windows, 16th edition (VSN International Ltd., Hemel Hempstead, UK). The data were analysed in two stages: first, the stability of the phenotypes over repeated experiments was examined by fitting a model with experiment as a fixed effect. Infiltration mixture, leaf age, position of infiltration site on leaf, experiment and their interactions were set as fixed effects with plant and leaf within an individual plant as the random effects. This allowed for terms in the model which specifically tested for significant interactions with experiment. Lack of any significant interaction with the infiltration mixtures would provide evidence that the relative responses of the phenotypes were consistent. Having determined that the infiltration mixtures behaved consistently over the experiments, the second stage fitted a model with experiment as a random effect with plant and leaf within plant nested below this. Multiple comparison tests then examined the differences in response amongst the infiltration mixtures.
Virus-induced gene silencing (VIGS) of SGT1 and HSP90
Tobacco rattle virus (TRV)-induced gene silencing in N. benthamiana was performed as described previously [14]. Agrobacterium cultures transformed with the binary TRV RNA1 construct, pBINTRA6, or the TRV RNA2 vector constructs PTV00, PTV:eGFP, PTV:HSP90 or PTV:SGT1 were re-suspended to OD600 = 0.5 for the RNA1 construct and OD600 = 1.0 for the RNA2 constructs. Re-suspended RNA1 and RNA2 cultures were mixed in a 1∶1 ratio and infiltrated into non-cotyledonous leaves of N. benthamiana plants at the 5-leaf stage. For each of the biological replicates, six plants per treatment were used and six plants were used as non-TRV controls. Three weeks after treatment with the VIGS constructs, plants were infiltrated with culture mixtures (OD600 = 0.5) designed to express R3a, Rd2-1, Rd3-1 or Rd4-1 and AVR3aKI or AVR3aEM. HRs were scored at 6dpi.
Confocal laser scanning microscopy
Imaging was performed on a Leica TCS-SP2 AOBS microscope (Leica Microsystems) using HCX APO L, 40x/0.8, and 63x/0.9 water dipping lenses or a Zeiss 710 using a Plan APO 40x/1.0 water dipping lens. Images were collected using line by line sequential scanning. The optimal pinhole diameter and the same gain levels were used within experiments. YFP and CFP were imaged using 514 nm and 405 nm excitation, respectively, and emissions were collected between 520–563 nm and 455–490 nm, respectively. Photoshop CS5.1 software (Adobe Systems) was used for post-acquisition image processing.
Agrobacterium tumefaciens transient assays (ATTAs)
Functional Agrobacterium tumefaciens transient assays (ATTAs) were carried out in N. benthamiana. Cultures carrying pGRAB.R3a::R3a, pGRAB.R3a::Rd2-1, pGRAB.R3a::Rd3-1, pGRAB.R3a::Rd4-1 or pGRAB empty vector were re-suspended as described before to OD600 = 0.1 for each construct. Each of the five resuspensions was infiltrated into separate areas of leaves. Four leaves on each of sixteen plants were infiltrated in each replicate. Two days post infiltration, leaves were detached and infiltration sites inoculated with AVR3aKI homozygous P. infestans isolate 7804.b or AVR3aEM homozygous isolate 88069. Leaves were incubated in transparent sealed boxes at 100% humidity in a cool room and covered for the first 12 hours. Lesion sizes were measured up to 15dpi.
Production of transgenic potato plants
R3a wild-type gene and the three modified versions Rd2-1, Rd3-1 and Rd4-1 were cloned under R3a native regulatory elements in a pCambia-based binary vector with kanamycin resistance as a selectable marker, using standard restriction enzyme methods to create pSIM2093, pSIM3027, pSIM3028 and pSIM3029 respectively. Rpi-vnt1 under its native regulatory elements was cloned in to pSIM401 to generate pSIM1620. The binary vectors were then transformed into Agrobacterium strains AGL1 and LBA4404 for plant transformation. Ranger Russet was transformed as described in Duan et al. [24]. For each construct twenty to thirty lines were regenerated with kanamycin selection. These were tested for late blight resistance and assessed at 7dpi.
Results
Identification of R3a mutants with enhanced AVR3aEM recognition
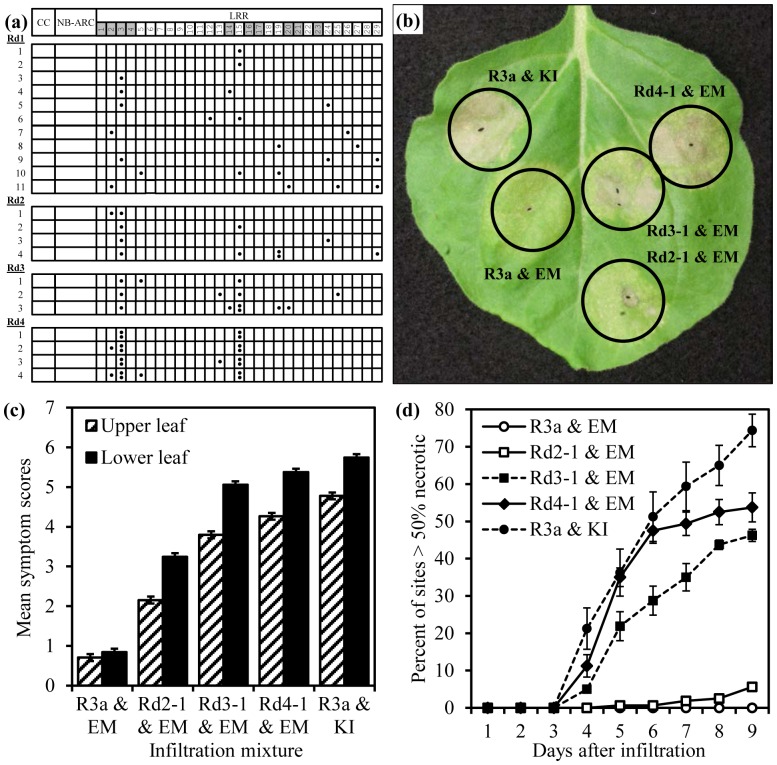
A binary vector, pGRAB.35S::R3a, containing the R3a open reading frame (Accession number AY849382.1) under the control of Cauliflower mosaic virus 35S promoter and terminator sequences was produced to allow specific mutation and shuffling of the R3a LRR domain. In this plasmid a unique BamHI site was introduced silently at nucleotide position 1567 in the ORF, ninety nucleotides upstream of the sequence encoding the LRR domain as denoted by Huang et al. [13]. Following mutagenesis and DNA shuffling of the LRR region, regenerated recombinant clones were screened for enhanced AVR3aEM recognition through Agrobacterium-mediated transient expression in co-infiltrations with binaries expressing the virulent elicitor, AVR3aEM. Clones were screened for enhanced recognition through comparison of visible symptoms of PCD and induced auto-fluorescence with reference to the responses produced by the wild-type R3a gene and AVR3aEM. Clones with putatively improved recognition isolated from primary screens were screened again to confirm the phenotypic improvement and to eliminate auto-activators. In the first cycle, screening of approximately three thousand clones identified eleven R3a* clones with improved phenotypes. The eleven clones from the first round are referred to as Rd1-1 to Rd1-11 and contained, in addition to synonymous changes, base changes that resulted in between one and four amino acid substitutions (Fig. 1a; Table S2). Three of the clones contained single amino acid substitutions (Rd1-1 [K920E], Rd1-2 [Q931R], Rd1-3 [R618Q]) identifying these changes as being responsible for enhanced recognition. Interestingly, the single amino acid substitution K920E in R3a was recently also identified in a complementary study by Segretin et al. [25] as a substitution that enhances recognition towards AVR3aEM. The largest numbers of amino acid substitutions were found in LRRs #3 and #15 and included those found in the three clones with single substitutions (Fig. 1a).
Figure 1. Four rounds of mutagenesis and shuffling identified R3a mutants with enhanced recognition of AVR3aEM and disease responses (R3a*).
(a) Schematic showing locations of non-synonymous mutations found in the LRRs of R3a* clones isolated from the four rounds of mutagenesis and shuffling (Rd1 to Rd4). LRRs containing amino acids under diversifying selection are shaded above. (b) Representative N. benthamiana leaf showing responses of best performing clones from second, third and fourth rounds (Rd2-1, Rd3-1 & Rd4-1) to AVR3aEM (EM), compared to responses of wild-type R3a to AVR3aKI (KI) and AVR3aEM five days after co-infiltration with resistance genes under transcriptional control of R3a promoter. (c) Mean disease scores from the four experiments, each of nine days duration, for different infiltration mixtures in upper (hatched) and lower (solid) paired leaves. Error bars show +/− standard error. (d) Time-course of percentage of sites showing necrosis development, greater than 50% necrosis of individual infiltrated sites, for the five infiltrated mixtures. Mean percentages of the four experiments. Each experiment includes data for 40 infiltration sites (upper and lower leaves combined) and error bars show +/− standard errors.
A second round of shuffling was carried out to combine beneficial changes and remove deleterious substitutions using the eleven isolated clones, Rd1-1 to Rd1-11, and the wild-type gene as starting material. Screening of approximately six hundred recombinant clones identified four which gave responses comparable to or greater than the best performing clone from the first round, Rd1-1 [K920E]. All of these R3a* clones from the second round, Rd2-1 to Rd2-4, contained the amino acid substitution E620D in LRR #3 in addition to at least one other coding change (Fig. 1a; Table S2).
As recognition of AVR3aEM improved and gave stronger responses it became more difficult to discriminate the differences in responses between modified clones with AVR3aEM and also in comparison to the response of the wild-type R3a gene with AVR3aKI. Therefore, a third round of shuffling, using the clones from the first and second rounds of shuffling and the wild-type gene as starting material, was performed. However, the shuffled LRR domains were cloned in to a binary vector, pGRAB.R3a::R3a, containing the R3a gene or R3a* variants under the wild-type R3a promoter, rather than the strong 35S promoter. The purpose of this was to protract the timing of the cell death responses and facilitate the discrimination of differences (Fig. 1b). Screening of approximately 300 clones from this third round population identified three R3a* clones, Rd3-1 to Rd3-3, that gave responses greater than the best performing clone from the second round of shuffling, Rd2-1 [T585A, E620D]. All of these clones contained the E620D change found in the second round clones and a number of other amino acid changes including at least one change in LRR #15. A previously conducted comparison of functional R3a with three paralogous, non-functional resistance gene analogues revealed 13 positions under diversification [13]. These positions, with the exception of one in the CC-domain, are located in LRRs 1 to 4 and 14 to 23. Intriguingly, the majority of mutations present in the clones from the second and third rounds are within these LRRs (Fig. 1a).
One limitation of shuffling is in recombining beneficial mutations that are in close proximity due to the limited frequency of crossing-over. Therefore, as multiple mutations in LRRs #3 and #15 had been found to enhance AVR3aEM recognition, an alternate approach was adopted. A population of site-directed mutants was produced using degenerate oligonucleotides that encoded different pairs of amino acids at positions 585 (T/A), 618 (R/Q), 620 (E/D), 918 (R/G), 920 (K/E), 923 (D/G) and 931 (Q/R) with the potential for 128 different permutations. Screening of approximately 400 clones from this population with the best performing clone from the third round of shuffling (Rd3-1 [E620D, L668P, Q931R]) as a reference, identified eight clones that produced responses with AVR3aEM comparable to or greater than the reference. Five of these clones were found to have identical sequences. Representative unique clones, Rd4-1 to Rd4-4, contain the amino acid substitutions R618Q, E620D, K920E and Q931R, but lacked either of the designed substitutions R918G or D923G (Table S2).
Iterative rounds of shuffling have progressively improved AVR3aEM recognition by R3a* variants, producing faster and stronger PCD responses upon co-infiltration
The enhanced recognition of AVR3aEM was assessed more accurately in single leaf comparisons. The R3a* constructs Rd2-1, Rd3-1 and Rd4-1 were transiently expressed under the control of the native R3a promoter with AVR3aEM and the responses compared with those produced by the wild-type gene with AVR3aEM and AVR3aKI. The best performing clone from the first round was not included in this analysis on account of the relatively poor response produced when it was under the control of the R3a promoter.
Symptom development on N. benthamiana was monitored for 9 days after infiltration in four independent experiments. In each experiment two adjacent, expanded leaves on each of twenty plants were infiltrated with the five infiltration mixtures in a circularly permuted arrangement to account for possible intra-leaf position effects. Symptoms were scored on an arbitrary scale ranging from 0 (no symptoms) to 10 (complete necrosis of the infiltrated area). A progressive increase in the recognition of AVR3aEM was observed for the three R3a* clones with the necrotic response produced by Rd4-1 being close to that produced by the wild-type gene with AVR3aKI (Fig. 1b).
A mixed model with experiment as a fixed effect was fitted to test for consistent responses of the infiltration mixtures over repeated experiments. The experiments showed significant differences in mean response (p = 0.001), but there was no significant interaction (p = 0.306) between experiment and infiltration mixture indicating that the relative responses of the phenotypes were stable over repeated experiments. In addition, there were no significant interactions between experiment and the other fixed effects.
Since the phenotypes were determined to be stable, a second analysis of the data with experiment as a random effect was now fitted. This showed highly significant effects (p<0.001) from infiltration mixture and leaf age (upper vs lower leaf). Further, there was a highly significant interaction (p<0.001) between infiltration mixture and leaf age because the combination of R3a and AVR3aEM did not show a difference in mean scores between younger and older leaves, in contrast to the other combinations which showed significant differences (Fig. 1c). Position of infiltration site on leaf was non-significant and all other interactions of fixed effect were also non-significant. Multiple comparisons using Bonferroni correction with an experiment-wise significance level of 5% showed significant differences between the mean symptom scores for all infiltration mixtures within either younger or older leaves with a comparison-wise significance level of 0.0011 (Table S3). Ordering of the responses was the same for both younger and older leaves with R3a and AVR3aKI> Rd4-1 & AVR3aEM> Rd3-1 & AVR3aEM> Rd2-1 & AVR3aEM> R3a and AVR3aEM. While the clone Rd2-1 from the second round gave symptoms when co-expressed with AVR3aEM, it rarely produced an HR phenotype as shown in figure 1d, which shows the proportion of sites with more than 50% of the infiltrated area necrotic. Despite improved recognition of AVR3aEM by the R3a* variants relative to wild-type R3a, their recognition of AVR3aKI was not impaired (data not shown), indicating that this specificity had not been significantly attenuated. Further, when expressed from the strong 35S promoter in the absence of AVR3aEM or AVR3aKI none of the clones produced necrosis, indicating that they maintained appropriate control mechanisms and were not auto-activators (Fig. S1).
R3a* recognition of Avr3aEM is dependent on HSP90 and SGT1
Previous studies by Bos et al. [14] demonstrated that R3a-dependent recognition of AVR3aKI involves both SGT1 (suppressor of the G2 allele of skp1) and HSP90 (heat shock protein 90) that are required for the activation of other R proteins [26]–[27]. Their involvement in the AVR3aEM-dependent responses was tested through Tobacco rattle virus (TRV)-based gene silencing of SGT1and HSP90 in N. benthamiana with TRV-based expression of truncated GFP (eGFP) as a control. Three biological replicates for R3a, Rd2-1, Rd3-1 and one for Rd4-1, with infiltrations in two leaves of each of six plants per TRV-based silencing construct, revealed that both SGT1 and HSP90 are required to mediate an HR upon R3a*-based recognition of AVR3aKI and AVR3aEM (Fig. 2, Fig. S2). HRs were abolished for all infiltrations on TRV:SGT1 inoculated plants and there were almost no HRs recorded on TRV:HSP90 inoculated plants (Fig. 2). The HRs were not affected on plants inoculated with TRV:eGFP.
Figure 2. HR responses resulting from R3a* recognition of AVR3aEM and AVR3aKI, like those caused by wild-type R3a recognition of AVR3aKI, are dependent on SGT1 and HSP90.
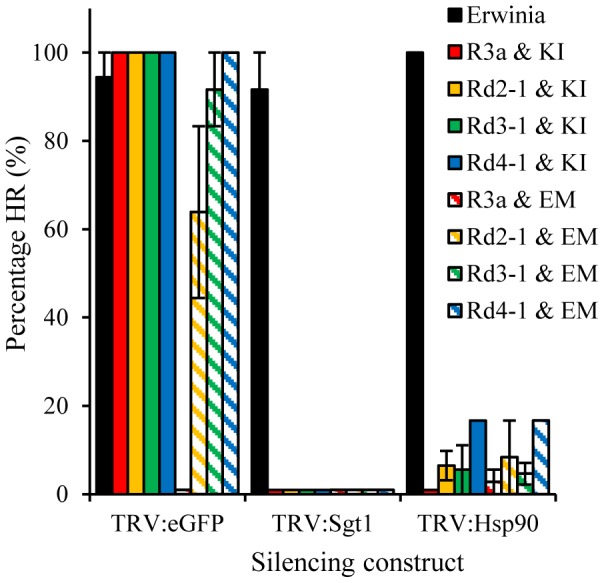
SGT1- and HSP90-silenced plants were produced using TRV-based vectors. These plants and control plants inoculated with TRV:eGFP were infiltrated with different combinations of Agrobacterium cultures designed to express R3a, R3a* variants, AVR3aKI (KI) or AVR3aEM (EM). The percentage of sites (N = 12) showing HR responses six days after infiltration was recorded. The graph shows the mean percentages from three independent experiments with the exception that the dependence on HSP90 of Rd4-1 responses was only tested in a single experiment. The non-host bacterial pathogen Erwinia amylovora was used as a control for an SGT1- and HSP90-independnet HR response. Error bars show +/− standard error. Zero values have been transformed to 1% to facilitate their observation.
Compared to TRV:eGFP inoculated plants, SGT1 and HSP90 silenced plants were morphologically stunted, a phenotype that had been reported previously [14]. Nevertheless, upon infection with the bacterial pathogen Erwinia amylovora that produces a SGT1 and HSP90 independent non-host response in N. benthamiana [28], all plants were able to mount the expected HR response (Fig. 2, Fig. S2).
In a gain of mechanism, R3a* variants re-localize to late endosomes upon co-infiltration with Avr3aKI or Avr3aEM
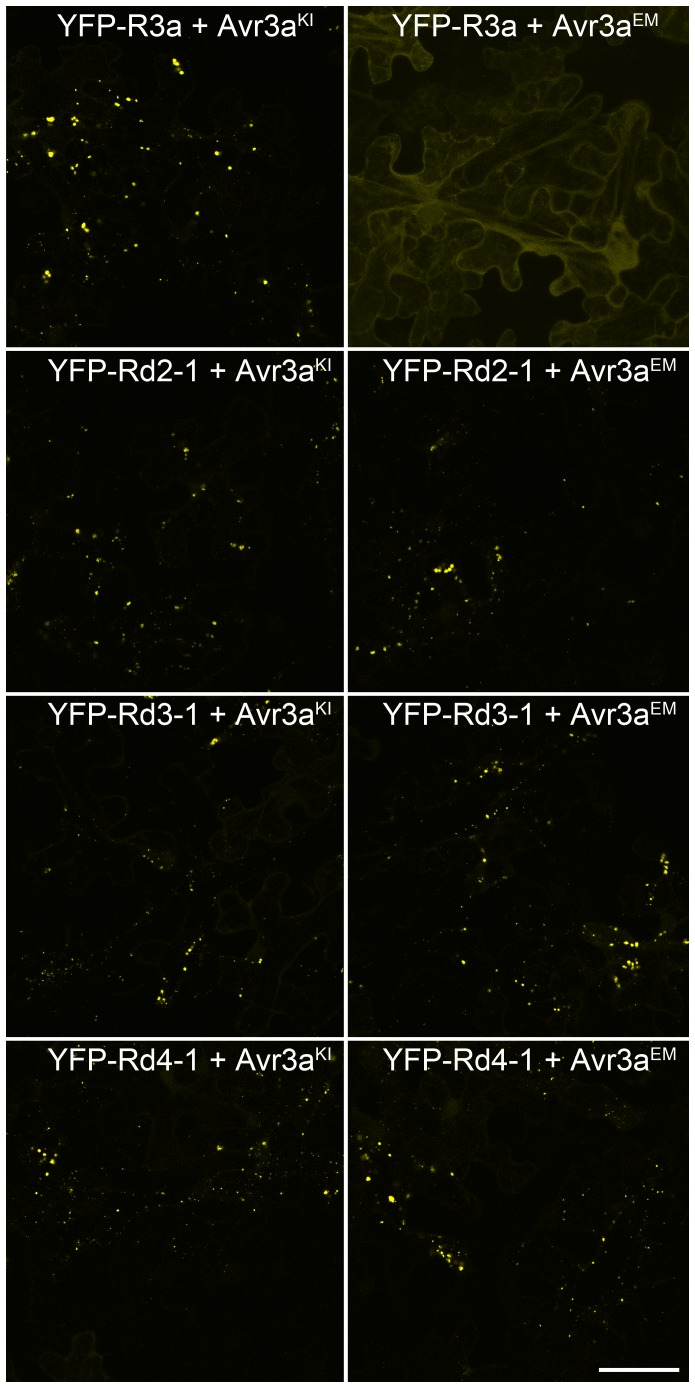
In a previous study, Engelhardt et al. [15] demonstrated that, upon recognition of AVR3aKI but not AVR3aEM, wild-type R3a re-localizes from the host cytoplasm to specific late endosomes that can be labelled with the cyan fluorescent protein marker PS1-CFP [29]. This re-localization was found to be a pre-requisite for subsequent HR development for untagged R3a co-expressed with AVR3aKI [15]. To study if R3a* variants with enhanced recognition of AVR3aEM had gained the capacity to re-localize upon detection of AVR3aEM and continued to exhibit this phenotype following detection of AVR3aKI, N-terminal fusions of R3a* variants Rd2-1, Rd3-1 and Rd4-1 with yellow fluorescent protein (YFP) were generated as described previously with expression of these constructs driven by a 35S promoter. Western-blot analysis of protein extracts from inoculated tissue demonstrated the integrity of the fusion proteins (Fig. S3). As demonstrated for YFP-R3a wild-type fusions by Engelhardt et al. [15], YFP-R3a* fusions did not elicit HRs alone or in the presence of AVR3aKI or AVR3aEM, probably due to steric hindrance of the signalling domains of R3a (data not shown).
As anticipated, following transient expression in N. benthamiana, all YFP-R3a/R3a* fusions when expressed by themselves displayed cytoplasmic localizations (Fig. S4). In accord with the observations described by Engelhardt et al. [15], the localisation of YFP-R3a remained cytoplasmic upon co-infiltration with AVR3aEM (Fig. 3), but changed to fast moving, PS1-CFP labelled vesicles, following recognition of AVR3aKI. The YFP-R3a* fusions of Rd2-1, Rd3-1 and Rd4-1 proteins maintained this mechanistically characteristic re-localisation following co-expression with AVR3aKI (Fig. 3). However, in contrast to YFP-R3a, all selected YFP-R3a* variants also displayed highly reproducible re-localization to PS1-CFP labelled vesicles after the perception of AVR3aEM (Fig. 3; Fig S5).
Figure 3. YFP fusions to R3a* variants re-localize to vesicles after the perception of both of AVR3aKI and AVR3aEM, whereas YFP-R3a remains cytoplasmic in the presence of AVR3aEM.
Two days after infiltration of mixtures of Agrobacterium cultures designed to express AVR3aKI, AVR3aEM, YFP-R3a or YFP fusions to the R3a* variants, infiltrated N. benthamiana leaf tissue was examined under a confocal laser scanning microscope. Scale bar = 50 µm.
AVR3aKI and AVR3aEM re-localize to endosomes upon co-infiltration with R3a* variants but not, in the case of AVR3aEM, with wild-type R3a
As shown by Engelhardt et al. [15] AVR3aKI, but not AVR3aEM, also re-localizes from the cytoplasm to endosomes upon co-expression with R3a. This was demonstrated by N-terminal fusions of AVR3aKI and AVR3aEM to green fluorescent protein as well as by BiFC, also known as split-YFP, assays [9], [15], [30]. The latter revealed that wild-type R3a and AVR3aKI are found in close proximity at PS1-CFP labelled vesicles [15].
To investigate if the vesicular co-association of R3a and AVR3aKI was extended to the R3a* variants, BiFC was used to analyse and localize protein–protein interactions in planta. As described previously for wild-type R3a, the N-terminal portion of YFP, YN, was fused to the N-terminal end of the R3a* variants Rd2-1, Rd3-1 and Rd4-1. The constructs used to express the C-terminal portion of YFP, YC, fused to AVR3aKI and AVR3aEM were as described previously [15] with all constructs being transiently expressed in N. benthamiana from the 35S promoter.
In accord with previous findings [15], co-expression of YN-R3a with YC-AVR3aKI gave strong YFP fluorescence, whereas co-expression with YC-AVR3aEM did not give detectable YFP fluorescence (Fig. 4). Like the YN fusion to wild-type R3a, all the YN-R3a* fusions when co-expressed with AVR3aKI gave strong, punctate, YFP signals (Fig. 4), but unlike the wild-type fusion also gave YFP fluorescence signals at PS1-CFP labelled vesicles when co-expressed with AVR3aEM (Fig. 4; Fig. S6). This indicates that AVR3aEM is also within close proximity of the re-localized R3a* gene products. Thus, in line with the gain of recognition of AVR3aEM by the R3a* variants and subsequent necrosis responses, the R3a* variants and AVR3aEM show the same mechanistic re-localization as observed for R3a and AVR3aKI.
Figure 4. Both YC-AVR3aKI and YC-AVR3aEM when co-expressed with YN-R3a* fusions give vesicle associated YFP fluorescence like YC-AVR3aKI and YN-R3a, whereas YC-AVR3aEM and YN-R3a do not.
Two days after infiltration of mixtures of Agrobacterium cultures designed to express YC-AVR3aKI, YC-AVR3EM, YN-R3a or YN fusions to the R3a* variants, infiltrated N. benthamiana leaf tissue was examined under a confocal laser scanning microscope. Representative images from two experiments. Scale bar = 50 µm.
R3a* variants maintain resistance towards AVR3aKI-expressing P. infestans isolates but have not gained resistance towards AVR3aEM homozygous isolates
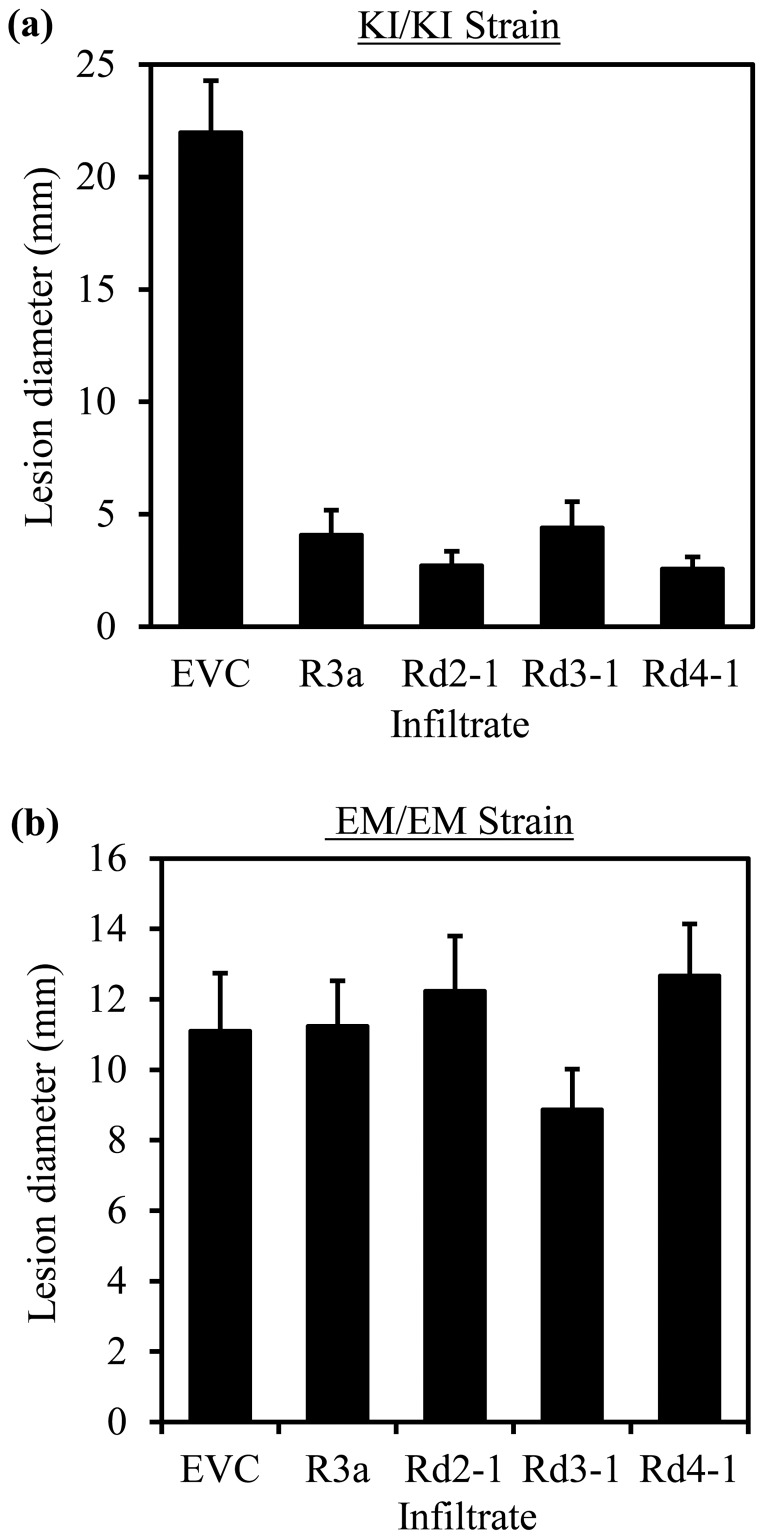
To evaluate if R3a* variants with gain of AVR3aEM recognition and re-localisation mechanism yield effective disease resistance, transient and stable expression systems were utilised. Agrobacterium tumefaciens transient assays (ATTAs) in N. benthamiana have successfully been used to demonstrate function for late blight resistance gene products such as R2, Rpi_STO1 [31] and R3b [32]. Selected R3a* clones Rd2-1, Rd3-1 and Rd4-1 were transiently expressed in N. benthamiana using the R3a promoter in ATTAs alongside wild-type R3a and an empty vector control. Infiltrated leaf areas were challenged two days after infiltration with AVR3aKI or AVR3aEM homozygous P. infestans isolates via drop inoculation. Disease progression was monitored by measuring visible lesion diameters in multiple independent experiments and analysis of variance was carried out using GenStat on the data from individual experiments to test for significant differences. Multiple comparisons were performed using Bonferroni correction with a significance level of 5% and a comparison-wise error rate of 0.005.
In three experiments ATTA sites were inoculated with the AVR3aKI homozygous P. infestans isolate 7804.b. In all three experiments the wild-type R3a and the R3a* variants significantly reduced spread of P. infestans relative to the empty vector control and there were no significant differences between the different R3a forms (Fig. 5a). This result indicates that the selected mutations in the LRR do not impair the resistance induced by AVR3aKI. Likewise ATTA sites were inoculated with the AVR3aEM homozygous isolate 88069 [9] in five experiments. In four of the five experiments there were no significant differences in P. infestans spread between any of the R3a forms and the empty vector control (Fig. 5b). In the fifth experiment there was significantly increased spread with the empty vector control, but the R3a* variants showed no significant differences from the wild-type gene which does not provide resistance against AVR3aEM homozygous isolates. Co-infiltrations of R3a, R3a*, AVR3aKI and AVR3aEM constructs were carried out contemporaneously in all experiments to confirm that the conditions were conducive to HR development (data not shown).
Figure 5. R3a and R3a* variants expressed from Agrobacterium reduce the spread of a P. infestans strain expressing AVR3aKI, but not the spread of a strain expressing only AVR3aEM.
(a) Means of lesion diameters measured 12 days after drop inoculation of agro-infiltrated areas with strain 7804.b (KI/KI). (b) Means of lesion diameters measured 8 days after drop inoculation of agro-infiltrated areas with strain 88069 (EM/EM). (a) and (b) show representative experiments from sets of three and five repeated experiments, respectively. For both (a) and (b), error bars show +/− standard errors, N = 30. EVC indicates empty vector control.
To confirm these results and to rule out potential adverse effects and limitations of the ATTA system in N. benthamiana, transgenic potato plants were generated. The wild-type R3a gene and the R3a* genes for Rd2-1, Rd3-1 and Rd4-1 were transformed into the potato cultivar Ranger Russet using the R3a promoter and terminator to regulate gene expression and stability. Transgenic Ranger Russet lines expressing R3a and the three R3a* variants were compared to Ranger Russet lines containing the Rpi_vnt1 gene [33] and non-transgenic Ranger Russet plants as positive and negative controls, respectively. Transgenic lines were challenged with the Mexican isolate P6752, which is heterozygous for AVR3aKI and AVR3aEM, and the US isolate US-8 BF-6, which is homozygous for AVR3aEM. The transgenic potato plants expressing R3a, Rpi_vnt1 and the R3a* variants, but not the non-transgenic Ranger Russet, demonstrated high levels of resistance towards the heterozygous isolate P6752 (Fig. 6). Thus, the transient ATTA data and the transgenic plants corroborate the conclusion that the selected mutations in the LRR do not negatively impact on resistance towards P. infestans isolates expressing Avr3aKI. The transgenic Ranger Russet lines expressing Rpi_vnt1 provided resistance towards the AVR3aEM homozygous isolate US-8 BF-6. However, none of the transgenic lines expressing the R3a* or R3a genes, which had been shown to be resistant to isolate P6752, were able to control disease development of isolate US-8 BF-6 (Fig. 6).
Figure 6. R3a and R3a* variants expressed via the R3a promoter in transgenic plants protect the susceptible cultivar Ranger Russet from P. infestans strain P6752, which is heterozygous for AVR3aKI and AVR3aEM, but not from strain US-8 BF-6, which is homozygous for AVR3aEM.

Non-transgenic plants were used as a control for susceptibility. Transgenic plants expressing R3a or Rpi-vnt1 were used as positive controls for resistance to P6752 or US-8 BF-6, respectively. Representative plants were photographed at 11dpi.
Discussion
The relatively narrow genetic basis of clonal potato cultivars in agriculture provides pathogens such as P. infestans with sufficient opportunity to adapt and overcome inducible host resistant responses and to thus cause disease on a global scale. Resistance responses rely on the direct or indirect recognition of modified-self or pathogen-derived molecules [1]. For example, in the first layer of inducible resistance, also referred to as PAMP triggered immunity (PTI), conserved pathogen-associated molecular patterns (PAMPs) and/or damage-associated molecular patterns (DAMPs) are recognised [1], [34], [35]. Successful pathogens circumvent this recognition with the help of effectors that perturb host resistance responses and promote effector-triggered susceptibility (ETS). However, by being in close proximity to the host, pathogen effectors provide the innate plant immune system with another opportunity for detection that is dependent on the presence of cognate plant R proteins that subsequently yield ETI [1], [36]. In nature, this closely entwined co-evolution between hosts and pathogens is evident in the diversification observed for effectors and R genes [4].
Indeed, P. infestans is known as a pathogen with ‘high evolutionary potential’ [37] and more than 560 RXLR-type candidate effectors have been described within the late blight pathogen genome [38]. The genomic organisation of RXLR effector genes, which are often in gene-poor and repeat-rich regions, is thought to facilitate their enhanced diversification by enabling non-homologous recombination. Oomycete RXLR-type effectors have been shown to evade detection by R gene products via transcriptional regulation [39], utilising functionally equivalent effectors that allow loss of recognised effectors [40], suppressor activity of unrelated effectors [41] and/or sequence diversity [11], [42]. Sequence diversity underpins virulence or avirulence behaviour for the essential P. infestans effector AVR3a. AVR3aKI determines avirulence on plants carrying R3a whereas AVR3aEM promotes virulence. It is thought that only AVR3aKI was present in the P. infestans strain responsible for the outbreak of late blight disease leading to the Irish Potato Famine in the 1840 s [43]. The AVR3aEM allele may have come to dominate in P. infestans populations once the resistance gene R3a was deployed in potato crops, quickly usurping the AVR3aKI allele. By using clonal potato varieties in current agriculture, the diversification of R genes is undermined and novel, naturally occurring resistances can only slowly be introgressed via breeding.
Functional R genes are often found in clusters that show evidence of duplication followed by diversification [44]. The phylogenetic NB-LRR gene grouping that contains homologs of the functional R3a gene in the potato genome has previously been described as CNL-8 [5]. This group is, after the R2 cluster (CNL-5), the second largest R gene cluster in potato where more than 750 NB-LRR-like genes have been identified [5]–[6]. Functional R3a, a homolog of the tomato I2 resistance gene that controls races of the fungus Fusarium oxysporum [45], was cloned alongside three paralogous sequences that provided insight into amino acid positions under diversification [13]. With one exception, positions under diversification reside in the LRR and cluster around two regions spanning LRRs 1 to 4 and 14 to 23 that were also identified as being important in this study.
Here we used, in addition to random mutagenesis and screening, DNA shuffling and targeted mutagenesis to enhance AVR3aEM recognition by R3a*. DNA shuffling, which emulates the natural evolutionary processes of mutation, recombination and selection, has proven a highly effective method for evolving new specificities/properties for a wide range of proteins that cannot be rationally designed and is of particular use in identifying mutations that are beneficial in combination [18]. Artificial evolution of a resistance gene to broaden its specificity has previously been performed on the gene Rx that protects potato plants from strains of PVX. In an initial Rx study, mutagenesis of the LRR region was performed on the basis that this region is the primary determinant of recognition specificity. Screening identified four mutations in the LRR region that affected elicitor recognition and activation functions [16]. Introduction of the mutated Rx genes in to the model host N. benthamiana as transgenes extended resistance to a normally resistance-breaking strain of PVX, HB, and a distantly related carlavirus, PopMV [16].
Our primary screen identified eleven mutants with enhanced AVR3aEM recognition containing in total 23 amino acid substitutions. However, only three of these (R618Q, K920E, Q931R), found in the clones with single amino acid substitutions, are known to be causative to the improved phenotype. The previous study performed by Segretin et al. [25] identified six amino acid substitutions in the LRR domain that enhanced AVR3aEM recognition: two of these were also found in our primary screen (L668P, K920E). The fact that we did not identify all the LRR mutations identified by Segretin et al. [25] and that they did not identify more mutations in the LRR region, demonstrates that neither screen was exhaustive.
The initial screen identified mutants with enhanced recognition of AVR3aEM when expressed from the strong 35S promoter. Amino acid changes in LRRs #3 and #15, and thus within regions known to be under diversifying selection [13], were prevalent in the clones from the first round of screening, suggesting these regions might be of particular importance. This suggestion was supported by the fact that all the clones from the second round of shuffling contained an amino acid change, E620D, in LRR #3 and sometimes additional changes in LRR #15, while all of the clones from the third round contained the E620D change and one or two amino acid changes in LRR #15. Using DNA shuffling it is more difficult to bring together combinatorially beneficial mutations that are in close proximity. Furthermore, random mutagenesis as a source of diversity has limitations in that single nucleotide changes can only convert a codon for one amino acid to a limited set of codons for other amino acids rather than all twenty possible amino acids. To circumvent the former problem a more directed approach was adopted in which a library that contained all permutations of the amino acid changes thought important (one in LRR #2, two in LRR #3 and four in LRR #15) was produced for screening. The aptness of this approach was shown by the fact that clones were obtained with enhanced recognition responses to AVR3aEM compared to the best performing clone from the third round of shuffling, Rd3-1, and that one of the possible 128 forms was prevalent. This form contained two amino acid changes in close proximity in each of LRRs #3 and #15; a combination that would have been difficult to obtain through DNA shuffling. Interestingly, an amino acid change at one of these positions, Q931, was also found in one of the clones recovered by Segretin et al. [25], though their “de-convolution” did not show their substitution at this position, proline instead of arginine, to improve AVR3aEM recognition. As shown by Segretin et al. [25] for the R3a mutants with enhanced AVR3aEM recognition, we did not note any reduction in the AVR3aKI recognition responses of the clones we isolated.
Our studies show that the recognition of AVR3aEM by the R3a* mutants we have isolated recapitulates the mechanistic processes of recognition of AVR3aKI by the wild-type R3a gene. It has previously been reported [14] that the HR triggered by R3a recognition of AVR3aKI is dependent on the ubiquitin ligase-associated protein SGT1 and HSP90. VIGS of SGT1 and, to a lesser degree, of HSP90 in our experiments inhibited the cell death responses induced by recognition of AVR3aEM by our R3a* mutants. Similarly, it has been shown that wild-type R3a re-localises from the cytoplasm to late endosomal compartments when co-expressed with AVR3aKI, but not when co-expressed with AVR3aEM [15]. We found that the mutants from the three later rounds still re-localised to endosomal compartments when co-expressed with AVR3aKI and, importantly, also re-localised to the same vesicles when co-expressed with AVR3aEM. The earlier study by Engelhardt et al. [15] also showed that the effector AVR3aKI itself relocalises from the cytoplasm to endosomes when co-expressed with wild-type R3a and is in close physical proximity to R3a, whereas AVR3aEM remains distributed through the cytoplasm. Our BiFC experiments show that AVR3aKI and the normally unrecognized form, AVR3aEM both traffic from the cytoplasm to vesicles when co-expressed with the R3a* forms. This re-localisation of R3a and AVR3KI was shown to be a prerequisite for the development of the HR [15]. Thus, the gain of AVR3aEM recognition R3a* variants have gained many aspects of the mechanism of the wild-type R3a response to AVR3aKI.
Although the R3a* variants produced in this study responded to AVR3aEM and produced HR responses when the elicitor was transiently expressed via Agrobacterium, critically they only provided resistance to P. infestans isolates that express AVR3aKI and not to isolates that express only AVR3aEM. Both transient expression via Agrobacterium in N. benthamiana and stable transgenic expression in potato corroborated this finding. Failure to protect from the pathogen itself was also reported for the R3a mutants identified by Segretin et al. [25]. That this was the case for mutants with single amino acid changes is perhaps not surprising given the large differences from the wild-type R3a/AVR3aKI response we observed when first and second round clones were expressed from the R3a promoter with AVR3aEM. For some pathogen/R gene combinations, e.g. PVX and Rx, the resistance responses can be separated from the HR [46] though the induction of necrotic responses by transient expression of the elicitor protein has been used to identify Rx mutations that, when expressed transgenically in the model host N. benthamiana, provide resistance to the pathogen itself. However, for P. infestans it has been suggested that the strength of the HR correlates with resistance levels [47].
A recent, secondary mutation study of Rx provides some evidence that stepwise artificial evolution could be required to obtain an optimum combination between effector recognition and subsequent R gene activation and signal transduction [17]. In the Rx study, the resistance provided by one of the mutations in the LRR domain to PopMV was improved by random mutagenesis of the CC-NB-ARC1-ARC2 domains [17]. In addition to constitutively active mutants that by themselves gave necrotic responses, four mutants with enhanced responses to PopMV were isolated. For three of these mutants the improved phenotype was conferred by a single amino acid change, while for the fourth a pair of amino acid substitutions was required. The mutations, which affect activation sensitivity, were found to be located around the nucleotide-binding pocket of Rx. As mentioned previously, we have evidence that neither this screening nor the efforts from Segretin et al. [25] were exhaustive as both approaches yielded novel beneficial mutations. Whereas our study of R3a focused solely on the LRRs, in Segretin et al. [25] the entire R3a gene was subjected to mutagenesis. The latter study identified eight single amino acid changes that enhanced responses to AVR3aEM. Out of these, six occurred in the LRR domain and one in the CC domain. This substitution enhanced the response to AVR3aEM but also showed some auto-activation. The final substitution, found in the NB-ARC domain, sensitised the AVR3aEM response and broadened specificity to include an elicitor from another Phytophthora species [25]. Interestingly, this change occurred in the nucleotide-binding pocket and is adjacent to one of the sensitizing mutations found in Rx [17]. Broadening resistance gene specificity merely by introducing sensitizing mutations without improving recognition may have detrimental consequences in the field. However, a natural precedent for this has been found in PM3 resistance protein alleles in which the substitution of two amino acids in the NB domain enhances the HR and broadens the spectrum of resistance to wheat powdery mildew isolates [48]. Thus, additional efforts to combine novel mutations in R3a domains responsible for AVR3aEM recognition (LRR) and response (CC-NB-ARC1/ARC2) could further improve R3a* variants that already display gain of recognition and mechanistic re-localisation to ultimately yield genes that provide effective resistance in potato against isolates expressing AVR3aEM. Considering the importance of AVR3a to P. infestans, such a resistance, combined with other, mechanistically distinct R genes could provide a step towards more durable late blight control.
Supporting Information
R3a* variants are not auto-activators. N. benthamiana leaves were infiltrated with Agrobacterium cultures designed to express R3a or the R3a* variants from the strong 35S promoter. Mixtures of cultures designed to co-express AVR3aKI (KI) were used as positive controls for the induction of cell death. Leaves were examined under white-light and UV-B illumination. Photograph of representative leaf was taken five days after infiltration. In the absence of elicitor the R3a* variants, like R3a, do not produce visible cell death.
(TIF)
HR responses resulting from R3a* recognition of AVR3aEM and AVR3aKI, like those caused by wild-type R3a recognition of AVR3aKI, are dependent on SGT1 and HSP90. SGT1- and HSP90-silenced plants were produced using TRV-based vectors. These plants and control plants inoculated with TRV:eGFP were infiltrated with different combinations of Agrobacterium cultures designed to express R3a, R3a* variants, AVR3aKI (KI) or AVR3aEM (EM). Photographs show representative HR responses induced by each of the different mixtures on control TRV:eGFP inoculated plants, SGT1-silenced plants and HSP90-silenced plants. The non-host bacterial pathogen Erwinia amylovora was used as a control for an SGT1- and HSP90-independnet HR response.
(TIF)
Western blot analysis showing integrity of YFP fusion proteins. Soluble protein extracts were prepared from N. benthamiana leaf tissue two days after infiltration with cultures designed to express YFP fusions to R3a, Rd2-1, Rd3-1 or Rd4-1. The blot was probed with anti-GFP antibodies as described by Engelhardt et al. (2012). Protein sizes are indicated in kilodaltons (kD) and protein loading is shown by Ponceau S (PS) staining.
(TIF)
YFP fusions to R3a and the R3a* variants localize to the cytoplasm in the absence of AVR3a. N. benthamiana leaves were infiltrated with cultures designed to express YFP fusions to R3a, Rd2-1, Rd3-1 or Rd4-1. Leaf tissue was examined two days after infiltration under a confocal laser scanning microscope. Representative images are from five independent experiments. Scale bar = 20 µm.
(TIF)
In the presence of AVR3aEM YFP fusions to R3a* variants, but not YFP-R3a, re-localize to vesicles labelled by the prevacuolar compartment marker PS1-CFP. N. benthamiana leaves were infiltrated with mixtures of cultures designed to express PS1-CFP, AVR3aEM and YFP fusions to R3a, Rd2-1, Rd3-1 or Rd4-1. Tissue was examined two days after infiltration under a confocal laser scanning microscope. The left-hand panel shows YFP signal, the right-hand panel CFP signal and the central panel displays the merged signals. Representative images are from three independent experiments. Scale bar = 10 µm.
(TIF)
YC-AVR3aEM reconstitutes YFP fluorescence with YN fusions to the R3a* variants at vesicles labelled by the prevacuolar compartment marker PS1-CFP. Generation of the YFP signal indicates that AVR3aEM and the R3a* variants are in close proximity at the vesicles. N. benthamiana leaves were infiltrated with mixtures of cultures designed to express PS1-CFP, YC-AVR3aEM and YN fusions to Rd2-1, Rd3-1 or Rd4-1. Tissue was examined 2 d after infiltration under a confocal laser scanning microscope. Left-hand panel, YFP signal; right-hand panel, CFP signal; central panel, merged signals. Representative images from three experiments. Scale bar = 20 µm.
(TIF)
Primers used in this study.
(DOCX)
Nucleotide and amino acid changes found in R3a* clones.
(DOCX)
Mean symptom scores from four nine-day experiments for upper and lower leaves.
(DOCX)
Acknowledgments
The authors acknowledge Dr Sanwen Huang for kindly providing sequences of wild type R3a and the native regulatory elements.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
LS is supported by The James Hutton Institute and the University of Dundee through a joint PhD student bursary. PSMVW is supported by the U.S. Department of Agriculture through National Institute of Food and Agriculture (NIFA) project 2011-68004-30154 (http://www.csrees.usda.gov/). NC is employed by J.R. Simplot Company, Simplot Plant Sciences, 5369 West Irving Street, Boise, ID 83706, USA. This work was funded by the Rural & Environment Science & Analytical Services (RESAS) Division of the Scottish Government (http://www.scotland.gov.uk/Topics/Research/About/EBAR/research-providers) and the Biotechnology and Biological Sciences Research Council (BBSRC) (http://www.bbsrc.ac.uk/home/home.aspx) through the joint projects CRF/2009/SCRI/SOP & BB/H018441/1 (IH), BB/K018299/1 and RESAS funded work package 6.4. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- 2. van der Biezen EA, Jones JDG (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456. [DOI] [PubMed] [Google Scholar]
- 3. Heath MC (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334. [DOI] [PubMed] [Google Scholar]
- 4. Hein I, Gilroy EM, Armstrong MR, Birch PRJ (2009) The zig-zag-zig in oomycete–plant interactions. Mol Plant Pathol 10: 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jupe F, Pritchard L, Etherington GJ, MacKenzie K, Cock PJA, et al. (2012) Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics 13: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jupe F, Witek K, Verweij W, Śliwka J, Pritchard L, et al. (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J 76: 530–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Birch PRJ, Boevink PC, Gilroy EM, Hein I, Pritchard L, et al. (2008) Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr Opin Plant Biol 11: 373–379. [DOI] [PubMed] [Google Scholar]
- 8. Vleeshouwers VG, Raffaele S, Vossen JH, Champouret N, Oliva R, et al. (2011) Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol 49: 507–531. [DOI] [PubMed] [Google Scholar]
- 9. Bos JIB, Armstrong MR, Gilroy EM, Boevink PC, Hein I, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA 107: 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vetukuri RR, Tian Z, Avrova AO, Savenkov EI, Dixelius C, et al. (2011) Silencing of the PiAvr3a effector-encoding gene from Phytophthora infestans by transcriptional fusion to a short interspersed element. Fungal Biol 115: 1225–1233. [DOI] [PubMed] [Google Scholar]
- 11. Armstrong MR, Whisson SC, Pritchard L, Bos JIB, Venter E, et al. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA 102: 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cárdenas M, Grajales A, Sierra R, Rojas A, González-Almario A, et al. (2011) Genetic diversity of Phytophthora infestans in the Northern Andean region. BMC Genet 12: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGAA, Zhang N, et al. (2005) Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J 42: 251–261. [DOI] [PubMed] [Google Scholar]
- 14. Bos JIB, Kanneganti T-D, Young C, Cakir C, Huitema E, et al. (2006) The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana . Plant J 48: 165–176. [DOI] [PubMed] [Google Scholar]
- 15. Engelhardt S, Boevink PC, Armstrong MR, Ramos MB, Hein I, et al. (2012) Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell 24: 5142–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farnham G, Baulcombe DC (2006) Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc Natl Acad Sci USA 103: 18828–18833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci USA 110: 21189–21194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stemmer WP (1994) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA 91: 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernal A, Pan Q, Pollack J, Rose L, Willets N, et al. (2005) Functional dissection of the Pto resistance gene using DNA shuffling. J Biol Chem 280: 23073–23083. [DOI] [PubMed] [Google Scholar]
- 20. Simpson CG, Lewandowska D, Liney M, Davidson D, Chapman S, et al. (2014) Arabidopsis PTB1 and PTB2 proteins negatively regulate splicing of a mini-exon splicing reporter and affect alternative splicing of endogenous genes differentially. New Phytol 203: 424–436. [DOI] [PubMed] [Google Scholar]
- 21. Leung DW, Chen E, Goeddel DV (1989) A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1: 11–15. [Google Scholar]
- 22. Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9: 963–967. [DOI] [PubMed] [Google Scholar]
- 23. van der Fits L, Deakin EA, Hoge JH, Memelink J (2000) The ternary transformation system: constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol Biol 43: 495–502. [DOI] [PubMed] [Google Scholar]
- 24. Duan H, Richael C, Rommens CM (2012) Overexpression of the wild potato eIF4E-1 variant Eva1 elicits Potato virus Y resistance in plants silenced for native eIF4E-1. Transgenic Res 21: 929–938. [DOI] [PubMed] [Google Scholar]
- 25. Segretin ME, Pais M, Franceschetti M, Chaparro-Garcia A, Bos JIB, et al. (2014) Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol Plant Microbe Interact 27: 624–637. [DOI] [PubMed] [Google Scholar]
- 26. Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem 279: 2101–2108. [DOI] [PubMed] [Google Scholar]
- 27. Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, et al. (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25: 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilroy EM, Hein I, van der Hoorn R, Boevink PC, Venter E, et al. (2007) Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J 52: 1–13. [DOI] [PubMed] [Google Scholar]
- 29. Saint-Jean B, Seveno-Carpentier E, Alcon C, Neuhaus JM, Paris N (2010) The cytosolic tail dipeptide Ile-Met of the pea receptor BP80 is required for recycling from the prevacuole and for endocytosis. Plant Cell 22: 2825–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438. [DOI] [PubMed] [Google Scholar]
- 31. Saunders DG, Breen S, Win J, Schornack S, Hein I, et al. (2012) Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum Immune receptor R2 to mediate disease resistance. Plant Cell 24: 3420–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li G, Huang S, Guo X, Li Y, Yang Y, et al. (2011) Cloning and characterization of R3b; members of the R3 superfamily of late blight resistance genes show sequence and functional divergence. Mol Plant Microbe Interact 24: 1132–1142. [DOI] [PubMed] [Google Scholar]
- 33. Foster SJ, Park TH, Pel M, Brigneti G, Sliwka J, et al. (2009) Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Interact 22: 589–600. [DOI] [PubMed] [Google Scholar]
- 34. Maffei ME, Arimura G, Mithöfer A (2012) Natural elicitors, effectors and modulators of plant responses. Nat Prod Rep 29: 1288–1303. [DOI] [PubMed] [Google Scholar]
- 35. Newman MA, Sundelin T, Nielsen JT, Erbs GI (2013) MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 16 4: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17: 644–655. [DOI] [PubMed] [Google Scholar]
- 37. Raffaele S, Win J, Cano LM, Kamoun S (2010) Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics 16: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- 39. Rietman H, Bijsterbosch G, Cano LM, Lee HR, Vossen JH, et al. (2012) Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Mol Plant Microbe Interact 25: 910–919. [DOI] [PubMed] [Google Scholar]
- 40. Van Poppel PM, Guo J, van de Vondervoort PJ, Jung MW, Birch PR, et al. (2008) The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol Plant Microbe Interact 21: 1460–1470. [DOI] [PubMed] [Google Scholar]
- 41. Wang Q, Han C, Ferreira AO, Yu X, Ye W, et al. (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23: 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gilroy EM, Tayor RM, Hein I, Boevink P, Sadanandom A, et al. (2011) CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol 190: 653–666. [DOI] [PubMed] [Google Scholar]
- 43. Yoshida K, Schuenemann VJ, Cano LM, Pais M, Mishra B, et al. (2013) The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2: e00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDowell JM, Simon SA (2006) Recent insights into R gene evolution. Mol Plant Pathol 7: , 437–448. [DOI] [PubMed] [Google Scholar]
- 45. Ori N1, Eshed Y, Paran I, Presting G, Aviv D, et al (1997) The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell 9: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vleeshouwers VGAA, van Dooijeweert W, Govers F, Kamoun S, Colon LT (2000) The hypersensitive response is associated with host and nonhost resistance to Phytophthora infestans . Planta 210: 853–864. [DOI] [PubMed] [Google Scholar]
- 48. Stirnweiss D, Milani SD, Jordan T, Keller B, Brunner S (2014) Substitutions of two amino acids in the nucleotide-binding site domain of a resistance protein enhance the hypersensitive response and enlarge the PM3F resistance spectrum in wheat. Mol Plant Microbe Interact 27: 265–276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
R3a* variants are not auto-activators. N. benthamiana leaves were infiltrated with Agrobacterium cultures designed to express R3a or the R3a* variants from the strong 35S promoter. Mixtures of cultures designed to co-express AVR3aKI (KI) were used as positive controls for the induction of cell death. Leaves were examined under white-light and UV-B illumination. Photograph of representative leaf was taken five days after infiltration. In the absence of elicitor the R3a* variants, like R3a, do not produce visible cell death.
(TIF)
HR responses resulting from R3a* recognition of AVR3aEM and AVR3aKI, like those caused by wild-type R3a recognition of AVR3aKI, are dependent on SGT1 and HSP90. SGT1- and HSP90-silenced plants were produced using TRV-based vectors. These plants and control plants inoculated with TRV:eGFP were infiltrated with different combinations of Agrobacterium cultures designed to express R3a, R3a* variants, AVR3aKI (KI) or AVR3aEM (EM). Photographs show representative HR responses induced by each of the different mixtures on control TRV:eGFP inoculated plants, SGT1-silenced plants and HSP90-silenced plants. The non-host bacterial pathogen Erwinia amylovora was used as a control for an SGT1- and HSP90-independnet HR response.
(TIF)
Western blot analysis showing integrity of YFP fusion proteins. Soluble protein extracts were prepared from N. benthamiana leaf tissue two days after infiltration with cultures designed to express YFP fusions to R3a, Rd2-1, Rd3-1 or Rd4-1. The blot was probed with anti-GFP antibodies as described by Engelhardt et al. (2012). Protein sizes are indicated in kilodaltons (kD) and protein loading is shown by Ponceau S (PS) staining.
(TIF)
YFP fusions to R3a and the R3a* variants localize to the cytoplasm in the absence of AVR3a. N. benthamiana leaves were infiltrated with cultures designed to express YFP fusions to R3a, Rd2-1, Rd3-1 or Rd4-1. Leaf tissue was examined two days after infiltration under a confocal laser scanning microscope. Representative images are from five independent experiments. Scale bar = 20 µm.
(TIF)
In the presence of AVR3aEM YFP fusions to R3a* variants, but not YFP-R3a, re-localize to vesicles labelled by the prevacuolar compartment marker PS1-CFP. N. benthamiana leaves were infiltrated with mixtures of cultures designed to express PS1-CFP, AVR3aEM and YFP fusions to R3a, Rd2-1, Rd3-1 or Rd4-1. Tissue was examined two days after infiltration under a confocal laser scanning microscope. The left-hand panel shows YFP signal, the right-hand panel CFP signal and the central panel displays the merged signals. Representative images are from three independent experiments. Scale bar = 10 µm.
(TIF)
YC-AVR3aEM reconstitutes YFP fluorescence with YN fusions to the R3a* variants at vesicles labelled by the prevacuolar compartment marker PS1-CFP. Generation of the YFP signal indicates that AVR3aEM and the R3a* variants are in close proximity at the vesicles. N. benthamiana leaves were infiltrated with mixtures of cultures designed to express PS1-CFP, YC-AVR3aEM and YN fusions to Rd2-1, Rd3-1 or Rd4-1. Tissue was examined 2 d after infiltration under a confocal laser scanning microscope. Left-hand panel, YFP signal; right-hand panel, CFP signal; central panel, merged signals. Representative images from three experiments. Scale bar = 20 µm.
(TIF)
Primers used in this study.
(DOCX)
Nucleotide and amino acid changes found in R3a* clones.
(DOCX)
Mean symptom scores from four nine-day experiments for upper and lower leaves.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.