Abstract
Cytogenetic chromosome maps offer molecular tools for genome analysis and clinical cytogenetics and are of particular importance for species with difficult karyotypes, such as camelids (2n = 74). Building on the available human–camel zoo-fluorescence in situ hybridization (FISH) data, we developed the first cytogenetic map for the alpaca (Lama pacos, LPA) genome by isolating and identifying 151 alpaca bacterial artificial chromosome (BAC) clones corresponding to 44 specific genes. The genes were mapped by FISH to 31 alpaca autosomes and the sex chromosomes; 11 chromosomes had 2 markers, which were ordered by dual-color FISH. The STS gene mapped to Xpter/Ypter, demarcating the pseudoautosomal region, whereas no markers were assigned to chromosomes 14, 21, 22, 28, and 36. The chromosome-specific markers were applied in clinical cytogenetics to identify LPA20, the major histocompatibility complex (MHC)-carrying chromosome, as a part of an autosomal translocation in a sterile male llama (Lama glama, LGL; 2n = 73,XY). FISH with LPAX BACs and LPA36 paints, as well as comparative genomic hybridization, were also used to investigate the origin of the minute chromosome, an abnormally small LPA36 in infertile female alpacas. This collection of cytogenetically mapped markers represents a new tool for camelid clinical cytogenetics and has applications for the improvement of the alpaca genome map and sequence assembly.
Key words: alpaca, BAC library, cytogenetics, FISH, minute chromosome, translocation
The development of cytogenetic maps for mammalian species constitutes a key feature for understanding the architecture and comparative evolution of chromosomes and karyotypes. Most domestic species have received considerable attention over the years due to their importance as production, model, or companion animals. Detailed cytogenetic maps are available for individual cattle (Goldammer et al. 2009; Di Meo et al. 2011) and pig (see Raudsepp and Chowdhary 2011) chromosomes and for the whole genome in horses (Raudsepp et al. 2008), dogs (Breen et al. 2004; Breen 2008), cats (Davis et al. 2009), river buffalo (Di Meo et al. 2008), and sheep (Di Meo et al. 2007). These maps have been critical for anchoring genetic linkage and radiation hybrid maps, as well as genome sequence draft assemblies of these species to physical chromosomes. Also, cytogenetically assigned markers are important in clinical studies for precise demarcation of chromosome abnormalities and aberration breakpoints (reviewed by Ducos et al. 2008; Lear and Bailey 2008; Rubes et al. 2009; Raudsepp and Chowdhary 2011).
Even though the domestication of camelid species dates back to approximately 7000 years ago (Kadwell et al. 2001), as long back as that of cattle (Taberlet et al. 2011), horses (Groeneveld et al. 2010), and dogs (Galibert et al. 2011), and considering that alpacas and llamas are gaining popularity as production and companion animals, camelid cytogenetics and physical chromosome mapping lag far behind those of other domesticated species. Reports about the karyotypes of camelid species date back to the 1960s, when first an erroneous diploid number of 2n = 72 was proposed (Capanna and Civitelli 1965; Hungerford and Snyder 1966), which was quickly corrected to 2n = 74 (Hsu and Benirschke 1967; Taylor et al. 1968; Koulischer et al. 1971; Hsu and Benirschke 1974). These studies from 50 years ago have been followed by only about 20 published reports describing normal or aberrant chromosomes in these species (e.g., Fowler 1990; Wilker et al. 1994; Hinrichs et al. 1997; Drew et al. 1999; Hinrichs et al. 1999; Tibary 2008), and only 1 effort has been made to develop molecular cytogenetic tools for camelids (Balmus et al. 2007).
One of the main complications in camelid cytogenetics is their particularly difficult karyotype. Despite distinct anatomical and physiological differences and the specialized adaptations of the 6 extant species, namely, the Bactrian (Camelus bactrianus, CBA) and dromedary (Camelus dromedarius, CDR) camels, alpaca (Lama pacos, LPA), llama (Lama glama, LGL), vicugna (Vicugna vicugna, VVI), and guanaco (Lama guanicoe, LGU; Stanley et al. 1994), their karyotypes are extremely conserved, with the same diploid numbers and almost identical chromosome morphology and banding patterns (Bunch et al. 1985; Bianchi et al. 1986; Di Berardino et al. 2006; Balmus et al. 2007). Morphological similarities and the relatively small size of some of the autosomes present serious challenges for identifying individual chromosomes within a species. The development of banding methods has helped resolve chromosome identification in several mammalian karyotypes, but not in camelids. Similarities in G-banding patterns between different chromosome pairs have resulted in discrepant karyotype arrangements in different studies (Bunch et al. 1985; Bianchi et al. 1986; Vidal-Rioja et al. 1989; Zhang et al. 2005; Di Berardino et al. 2006; Balmus et al. 2007).
Likewise, the 2 recent remarkable attempts to generate chromosome band nomenclature for the alpaca (Di Berardino et al. 2006) and the dromedary camel (Balmus et al. 2007) provide no common platform for chromosome identification. As a result, and in contrast to other domestic species, camelids still lack an internationally accepted chromosome nomenclature, which sets serious limitations for the advance of physical gene mapping and clinical cytogenetics, as well as for efficient cross talk between laboratories.
Lessons from other mammalian species with difficult karyotypes show that clinical cytogenetics can benefit from the development of physical maps that provide molecular markers for the identification of individual chromosomes, chromosome regions, or bands. An outstanding example is the domestic dog, a mammalian species with a high diploid number (2n = 78) and a set of morphologically similar (acrocentric) autosomes that gradually decrease in size (Breen et al. 1999; Breen 2008). The need for unambiguous identification of individual canine chromosomes led to the generation of a collection of molecular markers for chromosome identification by fluorescence in situ hybridization (FISH; Breen et al. 1999; Breen et al. 2004; Breen 2008) and, subsequently, to a standardized chromosome nomenclature.
Building on these experiences, we developed a genome-wide set of molecular markers for the alpaca, assigned the markers to individual chromosomes by FISH, and applied the new tool in alpaca and llama clinical cytogenetics.
Materials and Methods
Animals
A depository of fixed cell suspensions and chromosome slides of alpacas and llamas of the Molecular Cytogenetics and Genomics Laboratory at Texas A&M University was used for molecular cytogenetic analyses in this study. The depository was established in 2005 and currently contains samples from 56 alpacas and 4 llamas. The samples have been cytogenetically characterized, cataloged, and stored at –20 °C.
Cell Cultures, Chromosome Preparations, and Karyotyping
Metaphase and interphase chromosome spreads were prepared from peripheral blood lymphocytes according to standard protocols (Raudsepp and Chowdhary 2008a). The cells were dropped on clean, wet glass slides and checked under phase contrast microscope (×300) for quality. Chromosomes were stained with Giemsa, counted, and arranged into karyotypes using the Ikaros (MetaSystems GmbH) software. A minimum of 20 cells were analyzed per individual. Aberrant chromosomes were further analyzed by G- (Seabright 1971) and C-banding (Arrighi and Hsu 1971). The remaining cell suspensions were stored at –20 °C until needed.
Marker Selection and Primer Design
Human–camel zoo-FISH data (Balmus et al. 2007) were used to select regions in the human genome that are homologous to individual alpaca chromosomes. Based on this, 24 human orthologs in segments homologous to 18 alpaca chromosomes (16 autosomes and the sex chromosomes) were identified in the National Center for Biotechnology Information (NCBI) Human Genome Map Viewer (http://www.ncbi.nlm.nih.gov/projects/genome/guide/human/). Whenever possible, human genes were selected according to their likely involvement in reproduction or other economically important traits in alpacas. The alpaca genomic sequence for each gene was retrieved from the Ensembl Genome Browser (http://useast.ensembl.org/index.html), masked for repeats (Repeat)Masker: http://www.repeatmasker.org/), and used for the design of polymerase chain reaction (PCR) primers in Primer3 software (http://frodo.wi.mit.edu/primer3/), as well as overgo primers in or around the PCR amplicons (Gustafson et al. 2003). Additionally, PCR and overgo primers for 22 genes, expected to map to 22 different alpaca chromosomes, were designed from alpaca complementary DNA (cDNA) sequences (generated by L. Wachter and kindly provided by Pontius J, Johnson WE, unpublished data). Details of all selected genes and the PCR and overgo primers are presented in Table 1 and Supplementary Table 1, respectively.
Table 1.
List of gene-specific markers and their cytogenetic locations in alpaca and human chromosomes and in human sequence map
| Gene symbol | cDNA IDa | Gene name | Alpaca cytogenetic location | Human cytogenetic location | Human sequence map (chr:Mb) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| AGPAT2 | Lgnuc411 | 1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) | 4q35-36 | 9q34.3 | 11:19.5 | |||||
| ARHGDIG | Lgnuc612 | Rho GDP dissociation inhibitor (GDI) gamma | 18q12-q13 | 16p13.3 | 16:00.3 | |||||
| ASIP | — | Agouti signaling protein | 19q13-q14 | 20q11.2-q12 | 20:32.8 | |||||
| ATP6AP1 | Lgnuc610 | ATPase, H+-transporting, lysosomal accessory protein 1 | Xq25 | Xq28 | X:153.6 | |||||
| BAG4 | — | BCL2-associated athanogene 4 | 26q13 | 8p11.23 | 08:38.0 | |||||
| BRE | Lgnuc82 | Brain and reproductive organ-expressed (TNFRSF1A modulator) | 15q22-q23 | 2p23.2 | 02:28.1 | |||||
| C6orf211 | Lgnuc618 | Chromosome 6 open reading frame 211 | 8q24-q26 | 6q25.1 | 08:31.7 | |||||
| CAT56 | — | MHC class I region proline-rich protein CAT56 | 20q13 | 6p21.33 | 06:30.5 | |||||
| CDC42BPB | Lgnuc584 | CDC42 binding protein kinase beta (DMPK-like) | 6q33 | 14q32.3 | 15:43.3 | |||||
| CSTF2T | — | Cleavage stimulation factor, 3ʹ pre-RNA, subunit 2, 64kDa, tau variant | 11q21 | 10q11 | 10:53.4 | |||||
| DSCC1 | — | Defective in sister chromatid cohesion 1 homologue (S. cerevisiae) | 25q14 | 8q24.12 | 10:00.8 | |||||
| DYRK1A | Lgnuc737 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 1q26-q31 | 21q22.13 | 21:38.7 | |||||
| EDN3 | — | Endothelin 3 | 19q23 | 20q13.2-q13.3 | 20:57.8 | |||||
| FDFT1 | — | Farnesyl diphosphate farnesyltransferase 1 | 31q12-q13 | 8p23.1-p22 | 08:11.6 | |||||
| FGF5 | — | Fibroblast growth factor 5 | 2q21-q22 | 4q21 | 05:21.1 | |||||
| FGFR2 | — | Fibroblast growth factor receptor 2 | 11q22 | 10q26 | 12:03.2 | |||||
| GNB1L | Lgnuc743 | Guanine nucleotide binding protein (G protein), beta polypeptide 1-like | 32q13-q14 | 22q11.2 | 22:19.7 | |||||
| HEYL | — | Hairy/enhancer-of-split related with YRPW motif-like | 13q22-q23 | 1p34.3 | 01:40.0 | |||||
| HS3ST3A1 | — | Heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 | 16p13 | 17p12 | 17:13.3 | |||||
| HSD17B12 | Lgnuc524 | Hydroxysteroid (17-beta) dehydrogenase 12 | 33q12 | 11p11.2 | 11:43.7 | |||||
| KITLG | — | KIT ligand | 12q22-q23 | 12q22 | 13:28.8 | |||||
| LARP4B | Lgnuc417 | La ribonucleoprotein domain family, member 4B | 35q13-q14 | 10p15.3 | 10:00.8 | |||||
| LMO3 | Lgnuc510 | LIM domain only 3 (rhombotin-like 2) | 34q12-q13 | 12p12.3 | 12:16.7 | |||||
| LPGAT1 | Lgnuc63 | Lysophosphatidylglycerol acyltransferase 1 | 23q14-q15 | 1q32 | 04:31.9 | |||||
| MITF | — | Microphthalmia-associated transcription factor | 17q14 | 3p14.2-p14.1 | 04:09.7 | |||||
| NF1 | — | Neurofibromin 1 | 16q14-q15 | 17q11.2 | 17:29.4 | |||||
| NPTN | Lgnuc606 | Neuroplastin | 27q13 | 15q22 | 16:13.8 | |||||
| PAX3 | — | Paired box 3 | 5q33-q35 | 2q35 | 05:43.0 | |||||
| RAB38 | — | RAB38, member RAS oncogene family | 10q12-q14 | 11q14 | 12:27.8 | |||||
| RAG1 | Lgnuc460 | Recombination activating gene 1 | 10q25-q26 | 11p13 | 11:36.5 | |||||
| RALYL | — | RALY RNA binding protein-like | 29q13 | 8q21.2 | 09:25.0 | |||||
| RB1CC1 | — | RB1-inducible coiled-coil 1 | 29q15 | 8q11 | 08:53.5 | |||||
| SLC22A13 | — | Solute carrier family 22 (organic anion transporter), member 13 | 17q13 | 3p21.3 | 03:38.3 | |||||
| SLC36A1 | — | Solute carrier family 36 (proton/amino acid symporter), member 1 | 3q13-q16 | 5q33.1 | 07:30.8 | |||||
| SLC45A2 | — | Solute carrier family 45, member 2 | 3q33-q34 | 5p13.2 | 05:33.9 | |||||
| SOX2 | — | SRY (sex determining region Y)-box 2 | 1q21-q23 | 3q26.3-q27 | 06:01.4 | |||||
| STS-XY | — | Steroid sulfatase (microsomal), isozyme S | Xp16; Yq11 | Xp22.32 | X:0.7; Y:17.6 | |||||
| TGFBR3 | — | Transforming growth factor, beta receptor III | 9q25 | 1p33-p32 | 02:32.1 | |||||
| TRBV30 | Lgnuc355 | T cell receptor beta variable 30 | 7q24 | 7q34 | 09:22.5 | |||||
| TTR | Lgnuc409 | Transthyretin | 24q13-q14 | 18q12.1 | 18:29.1 | |||||
| TYRP1 | — | Tyrosinase-related protein 1 | 4q21 | 9p23 | 09:12.6 | |||||
| Unknown transcript | Lgnuc134 | Alpaca scaffold_48:270613:271380:1 | 2q33 | 4p15.3 | 4:00 | |||||
| Unknown transcript | Lgnuc681 | Alpaca scaffold_374:105849:106822:1 | 30q12-q14 | 18q21 | 18:00 |
a “Lgnuc” designates alpaca cDNA sequences (Perleman P, Pontius, J, unpublished data)
Alpaca CHORI-246 BAC Library Screening and BAC DNA Isolation
Overgo primers were radioactively labeled with [32P] 2ʹ-deoxyadenosine triphosphate (dATP) and [32P] deoxycytidine triphosphate (dCTP; Amersham Biosciences) as previously described (Gustafson et al. 2003). Equal amounts of 25 or less overgo probes were pooled and hybridized to high-density filters of the CHORI-246 alpaca bacterial artificial chromosome (BAC) library (http://bacpac.chori.org/library.php?id=448). The hybridization solution, containing the labeled probes, 20× SSPE, 10% sodium dodecyl sulfate, 5% dry milk, 100× Denhardt’s solution, and 50% formamide, was denatured by boiling for 10min, chilled, and hybridized to library filters at 42 °C for 16h. The filters were washed 3 times in 2× SSPE at 55 °C for 15min, exposed to autoradiography films over intensifying screens for 2–3 days at –80 °C, and the autoradiograms were developed. Positive BAC clones were identified and picked from the library. The BAC clones corresponding to individual genes (Supplementary Table 1) were identified by PCR using gene-specific primers and BAC cell lysates as templates. Isolation of DNA from individual BACs was carried out with the Plasmid Midi Kit (Qiagen) according to the manufacturer’s protocol. The quality and quantity of BAC DNA was evaluated by gel electrophoresis and nanodrop spectrophotometry.
BAC DNA Labeling and FISH
The physical location of the genes was determined by FISH to alpaca metaphase and/or interphase chromosomes according to our protocols (Raudsepp and Chowdhary 2008a). Briefly, DNA from individual BAC clones was labeled with biotin-16-deoxyuridine, 5ʹ-triphosphate (dUTP) or digoxigenin (DIG)-11-dUTP, using Biotin- or DIG-Nick Translation Mix (Roche), respectively. Differently labeled probes were hybridized in pairs to metaphase/interphase chromosomes. Biotin and DIG signals were detected with avidin–fluorescein isothiocyanate and anti-DIG-Rhodamine, respectively. Images for a minimum of 10 metaphase spreads and 10 interphase cells were captured for each experiment and analyzed with a Zeiss Axioplan2 fluorescence microscope equipped with Isis Version 5.2 (MetaSystems GmbH) software. Alpaca chromosomes were counterstained with 4ʹ-6-diamidino-2-phenylin-dole (DAPI) and identified according to the nomenclature proposed by Balmus and colleagues (2007) with our modifications for LPA12, 24, 26, 27, 29, 33, 36, and Y (see Results).
Generation of Probes for LPA36, the Minute Chromosome, and the Sex Chromosomes
Probes for LPA36, LPAX, and LPAY were amplified and biotin- or DIG-labeled by degenerate oligonucleotide–primed PCR (DOP-PCR; Telenius et al. 1992; Rens et al. 2006), and the sequences of the probes originated from the alpaca flow karyotype (Stanyon R, Perelman P, Stone G, unpublished data). A probe for the abnormally small homologue of LPA36, the minute chromosome, was generated by chromosome microdissection, as previously described (Kubickova et al. 2002). Briefly, chromosome spreads from 3 animals carrying the minute chromosome were prepared on glass-membrane slides. Ten copies of the minute per animal were microdissected using the PALM MicroLaser system (P.A.L.M. GmbH, Bernried, Germany) and collected into a PCR tube containing 20 µL of 10 mmol Tris–HCl (pH 8.8). Chromosomal DNA was amplified and labeled with Spectrum Orange-dUTP (Vysis) by DOP-PCR (Telenius et al. 1992; Rens et al. 2006). Additionally, repeat-enriched blocking DNA was prepared by microdissection and DOP-PCR amplification of all alpaca centromeres. The labeled minute DNA was mixed with unlabeled centromeric DNA, denatured, preannealed to block repetitive sequences, and hybridized to normal and minute-carrying alpaca metaphase spreads as described earlier.
Comparative Genomic Hybridization
Genomic DNA from a normal male alpaca (control) and from 2 minute carriers (case) was isolated and directly labeled by nick translation (Abbott, Inc.) with SpectrumGreen-dUTP (Vysis) and SpectrumOrange-dUTP (Vysis), respectively. Labeled control and case DNA (each ~500ng) were mixed with 20 µg of unlabeled alpaca repetitive DNA and 35 µg of salmon sperm DNA (Sigma) and cohybridized to metaphase spreads of a normal male alpaca. The comparative genomic hybridization (CGH) process and analysis of the results were carried out as described in detail by Hornak and colleagues (Hornak et al. 2009). For each CGH experiment, the red:green signal ratio was calculated for 10 metaphase spreads using the Isis-CGH software (MetaSystems, GmbH). A red:green ratio of >1.25:1 was indicative of chromosomal material gain, whereas a ratio of <0.75:1 indicated loss.
Results
A Map of Molecular Cytogenetic Markers for the Alpaca Genome
The alpaca CHORI-246 genomic BAC library was screened with primers corresponding to 44 alpaca genes and expressed sequence tags. Altogether, 151 BAC clones were isolated and identified for the gene content (Supplementary Table 1). Most of the genes were found in 2 or more clones, whereas each of the following 8 genes—BAG4, C6orf211, CDC42BPB, FGFR2, LMO3, NF1, PAX3, and SLC22A13—corresponded to only 1 BAC. One clone (which gave the strongest and cleanest PCR amplification) for each of the 44 genes was selected for labeling and FISH mapping (Supplementary Table 1). Each alpaca BAC clone produced a strong and clean FISH signal at 1 distinct location, and there were no chimeric clones or those that recognized multiple sites across the genome.
The 44 BACs were assigned to 31 alpaca autosomes and the sex chromosomes (Figure 1). The clone containing the steroid sulfatase (STS) gene mapped to both the LPAXpter and Ypter and was considered pseudoautosomal (Figure 2). Thus, the gene-specific BACs were assigned to 33 chromosomes, of which 11 chromosomes were demarcated by 2 distinctly located markers, either on the same arm (acrocentrics) or on 2 different arms (submetacentrics; LPA16 and LPAX). The relative order of all syntenic markers was determined by dual-color FISH (Figure 2). No markers were assigned to 5 chromosomes, namely, LPA14, 21, 22, 28, and 36 (Figure 1).
Figure 1.

A cytogenetic gene map of the alpaca genome. Karyotype arrangement and ideograms are adapted from Balmus and colleagues (2007). The band nomenclature is corrected according to ISCN (1995). Chromosomes with ideograms adjusted for the alpaca are marked with a star.
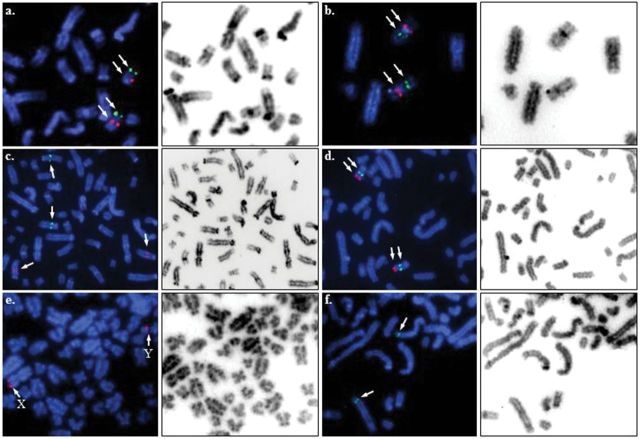
Figure 2.
Partial alpaca metaphase spreads showing FISH results (left, arrows) and corresponding inverted DAPI images (right) for selected markers mapped in this study: a. EDN3 (green) and ASIP (red) on LPA19; b. NF1 (green) and HS3ST3A1 (red) on LPA16; c. RAB38 (green) on LPA10 and TYRP1 (red) on LPA4; d. RALYL (green) and RB1CC1 (red) on LPA29; e. STS (red) on LPAX and LPAY; f. FGFR2 (green) on LPA11.
Precise cytogenetic locations of all BACs were determined by aligning the DAPI bands with the G-band nomenclature proposed by Balmus and colleagues (2007). However, we changed chromosome band numbering in compliance with the guidelines for human nomenclature (ISCN 1995) by designating centromeres as p11/q11 and starting band numbering on both arms from the centromere. New ideograms were generated for LPA12, 24, 26, 27, 29, 33, 36, and Y (Figure 1), because LPA12, 29, 33, and 36 are submetacentric and not acrocentric as their counterparts in the dromedary camel karyotype (Balmus et al. 2007); LPAY is a small acrocentric compared to the submetacentric CDRY, and the banding pattern of LPA24, 26, and 27 differed from their CDR counterparts (Figure 1; Supplementary Figure 1). Otherwise, the locations of all genes in the alpaca chromosomes were in agreement with the predictions of human–camel zoo-FISH data (Balmus et al. 2007).
Cytogenetic Findings
In the past 7 years (2005–2011), the Molecular Cytogenetics and Genomics Laboratory at Texas A&M University (http://vetmed.tamu.edu/labs/cytogenics-genomics), in close collaboration with the Department of Animal Sciences at the Oregon State University, has received samples from 51 alpacas (both Suri and Huacaya) and 1 llama. The animals were referred for chromosome analysis due to various reproductive and/or developmental disorders, including abnormal sexual development, gonadal dysgenesis, subfertility, and sterility. Also, control samples were procured from a number of normal alpacas and llamas.
Among the phenotypically abnormal animals, chromosome abnormalities were detected in 12 cases (23%). Abnormal karyotypes included XX/XY chimerism, XY sex reversal, an autosomal translocation, and the presence of an abnormally small LPA36, also known as a minute chromosome. Notably, the frequency of minute carriers was 17.7% of females with reproductive problems. A summary of the cytogenetic findings is presented in Table 2.
Table 2.
Summary of cytogenetic finding in 51 alpacas and 1 llama subjected to chromosome analysis due to reproductive problems and/or abnormal sexual development
| Species | Karyotype | Chromosomal abnormality | Phenotype | Number of cases |
|---|---|---|---|---|
| Alpaca | 74,XXm | Minute chromosome | Infertile female | 8 |
| 74,XX/74,XY | Blood chimerism | Co-twin to a male | 2 | |
| 74,XY | Sex reversal | Female | 1 | |
| Llama | 73,XY(t20;?) | Autosomal translocation | Infertile male | 1 |
Application of Molecular Tools in Camelid Clinical Cytogenetics
Autosomal Translocation in a Sterile Male Llama
A 10-year-old male llama was presented for chromosome analysis due of infertility. Clinical examination showed that ~75% of his sperm had abnormal morphology (midpiece defects, nuclear and acrosomal vacuoles), whereas the testes and accessory glands appeared normal on ultrasound checkup.
Cytogenetic analysis determined that the llama had an abnormal karyotype 73,XY carrying an autosomal translocation. The derivative chromosome, as determined by G-banding, was submetacentric with size and morphology similar to the X chromosome (Figure 3a). The G-banding pattern suggested the probable involvement of LGL11 and LGL17 (Figure 3b), although cytogenetic identification of the origin of the translocation remained ambiguous.
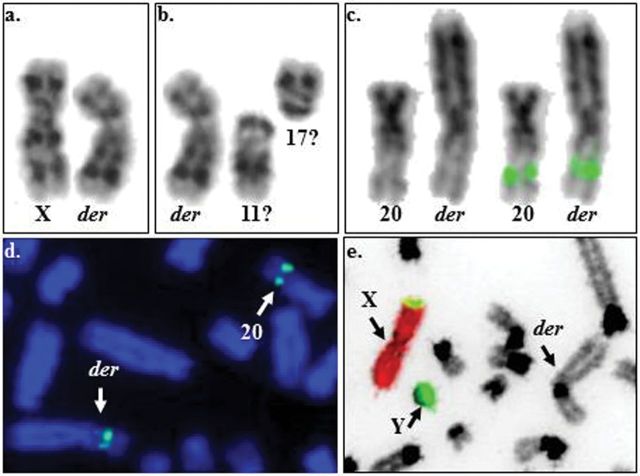
Figure 3.
Autosomal translocation in a male llama. a. G-banded LGLX (left) and the derivative chromosome (der; right); b. G-banded der (left) and LGL11 and 17 (right)—thought to be involved in the formation of the der; c. side-by-side presentation of LGL20 and the der as inverted DAPI images (left) and with CAT56 signal (right) d. partial metaphase showing FISH signals by CAT56 on LGL20 and the der (arrows); e. chromosome painting with LPAX (red) and Y (green) showing that der (arrow) is of autosomal origin.
Molecular cytogenetic analysis by FISH using LPAX and LPAY flow-sorted paints showed the presence of normal XY sex chromosomes and confirmed the autosomal origin of the derivative chromosome (Figure 3c). Dual-color FISH with all 41 autosomal BAC clones refuted the involvement of LGL11 and LGL17 in the translocation. Instead, FISH revealed that the short arm of the derivative chromosome corresponds to LGL20 (Figure 3d), the chromosome carrying the MHC (our unpublished data). The origin of the long arm of the aberrant chromosome remains as yet undetermined.
The Minute Chromosome in Infertile Alpacas
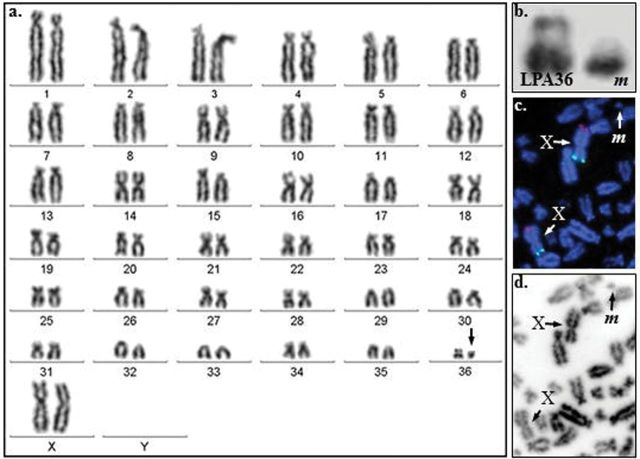
Among the 11 infertile females, 8 animals had karyotypes with an extremely small LPA36—the minute (Figure 4a). In all cases, the condition was heterozygous. Otherwise, chromosome number (74,XX) and gross morphology of other chromosomes in these animals were normal. Cytogenetic analysis determined that the minute is morphologically submetacentric, shows no distinct G-banding pattern, but stains positively by C-banding (Figure 4b), and is probably largely heterochromatic. However, it was not possible to identify the origin of the minute by conventional cytogenetic analysis.
Figure 4.
The minute chromosome. a. Karyotype of a female alpaca carrying the minute chromosome (arrow); b. G-banded LPA36 and the minute (m); c. FISH with STS (green) and ATP6AP1 (red) on LPAX, and d. the same image as inverted DAPI. The minute is shown as m (arrow).
Molecular hybridizations with flow-sorted LPA36 and microdissected minute probes to metaphase spreads of a minute carrier showed FISH signals not only on LPA36 and the minute but also on all centromeres and intercalary heterochromatic regions (Figure 5a,5b). In addition, the flow-sorted LPA36 also contained DNA from another small autosome, LPA34 (Figure 5a,5b). Although FISH results confirmed the largely heterochromatic nature of the normal and minute LPA36, they did not bring us closer to understanding the origin of the abnormality.
Figure 5.

The minute chromosome. a. FISH with a microdissected minute probe on a metaphase spread of a minute carrier: signals are seen on all centromeres and on the minute (m, arrow); b. FISH with a flow-sorted LPA36+LPA34 probe on a minute carrier: the minute, LPA36, and LPA34 are indicated by arrows (left: FISH signals; right: inverted DAPI); c. CGH results with the genomic DNA of a normal male (green) and a female minute carrier (red). Arrows show the gain on the X and the loss on the Y chromosome.
Next, in order to test a working hypothesis that the minute results from a deletion rather than a translocation, CGH experiments were carried out on normal male metaphase spreads using genomic DNA from a normal male and a minute-carrying female as hybridization probes. No regions of genomic imbalance between the control and minute-carrying animal were detected, providing no experimental proof to the deletion theory (Figure 5c).
Finally, FISH with 2 terminally located LPAX markers (STS and ATP6AP1) on metaphase spreads of minute carriers showed that the X chromosome in these animals is normal, thus challenging the hypothesis that the missing part of the minute has translocated to LPAX (Weber A, personal communication).
Discussion
This study reports the generation of a genome-wide collection of 151 gene-containing BAC clones and the construction of a 44-marker cytogenetic map for the alpaca. According to our best knowledge, this is the first cytogenetic gene map for the alpaca or any other camelid species and the first application of the CHORI-246 alpaca genomic BAC library (http://bacpac.chori.org/library.php?id=448). Until now, the only molecular probes for camelids were whole chromosome paints from the flow karyotype of the dromedary camel, which have been used for camel–human, camel–cattle, and camel–pig zoo-FISH studies (Balmus et al. 2007), for the study of chromosome evolution in Cetartiodactyla (Kulemzina et al. 2009) and ruminants (Kulemzina et al. 2011), as well as for the identification of the X and Y chromosomes in the alpaca karyotype (Di Berardino et al. 2006).
The BAC-based chromosome map, as presented in this study, confirms all and refines some of the known zoo-FISH homologies. For example, assignment of 2 genes from HSA9 (TYRP1, HSA9p23; AGPAT2, HSA9q34.2) to LPA4 improved the demarcation of homologous regions between the human sequence map and the alpaca chromosome. Likewise, zoo-FISH homologies were refined for 10 autosomes and the X chromosome by mapping 2 gene-specific markers on each (Figure 1, Table 1). In clinical cytogenetics, these markers will have a potential use for demarcating inversion and translocation breakpoints and determining the origin of complex rearrangements.
In some instances, particularly when 1 human chromosome shared evolutionary homology with 2 or more segments in the alpaca genome, the isolated BACs did not map to the expected alpaca chromosome. Instead, FISH signals were observed in another alpaca chromosome, which is homologous to the same human counterpart. This might be due to the relatively low resolution (~5Mb, Scherthan et al. 1994) and rather broad demarcation of evolutionary breakpoints by zoo-FISH. Therefore, no markers were assigned to LPA21, 22, and 28, which correspond to parts of HSA1, 5, and 2, respectively. In the case of LPA14, which corresponds one-to-one to HSA13 (Balmus et al. 2007), the BAC clone containing the mapping pseudogene (ATP5EP2) mapped to a different alpaca chromosome (data not shown).
Because the CHORI-246 BAC library was constructed from a female alpaca (http://bacpac.chori.org/library.php?id=448), we did not expect markers to be assigned to the Y chromosome. Nevertheless, a BAC clone for the STS gene produced FISH signals on both sex chromosomes, providing the first pseudoautosomal (PAR) marker for the alpaca genome. Interestingly, STS is an X-specific gene in humans (Skaletsky et al. 2003; Ross et al. 2005), and a non-PAR gene on horse sex chromosomes (Raudsepp and Chowdhary 2008b), whereas in other nonrodent mammals studied so far, STS belongs to the PAR (Raudsepp and Chowdhary 2008b; Das et al. 2009; Raudsepp et al. 2011). Thus, our results demarcate the location of the PAR in the alpaca sex chromosomes and provide the first gene-specific molecular marker for LPAY. Given that sex chromosome abnormalities are the most common viable cytogenetic defects associated with disorders of sexual development and reproduction in domestic animals (Villagomez and Pinton 2008; Villagomez et al. 2009), including camelids (Fowler 1990; Hinrichs et al. 1997; Drew et al. 1999; Hinrichs et al. 1999; Wilker et al. 1994; Tibary 2008), the BACs containing the STS gene will be of value for the identification of Y chromosome abnormalities in clinical studies.
Cytogenetic assignment of alpaca BAC clones in this study was carried out following the Giemsa (GTG)-banded chromosome nomenclature for the dromedary camel (Balmus et al. 2007) and not the one recently proposed for the alpaca (Di Berardino et al. 2006). Our primary argument was that the camel nomenclature is aligned with the human (Balmus et al. 2007) and other mammalian genomes (Kulemzina et al. 2009; Kulemzina et al. 2011), thus facilitating the development of gene-specific markers in the present and future studies. Also, Balmus and colleagues (2007) ordered chromosomes by size and not by morphological types as in the alpaca nomenclature (Di Berardino et al. 2006). The former seems to be the most logical approach in camelids, because heterochromatin and/or nucleolus organizer region (NOR) polymorphism in the short arms of some chromosomes (Bunch et al. 1985), (Bianchi et al. 1986), combined with either ambiguous or too similar banding patterns in others, make morphological classification arbitrary. Furthermore, inverted-DAPI-banding patterns of alpaca chromosomes in this study corresponded well to the GTG-banded camel chromosomes and ideograms (Balmus et al. 2007), further justifying our approach. The few minor differences between the alpaca and the dromedary camel homologues, namely, chromosomes 12, 24, 26, 27, 29, 33, 36, and Y, were adjusted in the resulting FISH map (Figure 1). However, despite the well-known evolutionary conservation of camelid karyotypes (Bianchi et al. 1986; Di Berardino et al. 2006; Balmus et al. 2007), it is anticipated that, with the expansion of the alpaca cytogenetic map, more differences between alpaca, dromedary camel, and other camelid chromosomes will be revealed.
Successful identification of one of the chromosomes involved in an autosomal translocation in an infertile male llama (Figure 3d) demonstrated the immediate utility of the markers in camelid cytogenetics. Also, erroneous calling of the aberrant chromosomes by G-banding (Figure 3b) highlighted the limitations of conventional cytogenetic methods. This is in line with experiences from other domestic species, in which the development of molecular cytogenetic markers has considerably improved the quality and depth of clinical cytogenetic studies (Breen 2008; Ducos et al. 2008; Lear and Bailey 2008; Rubes et al. 2009; Raudsepp and Chowdhary 2011). Efforts will be made to identify the other counterpart of the aberration; likely candidates could be LGL21 and 22. Interestingly, the translocation did not seriously affect meiosis because the animal produces sperm, though with morphological defects. The involvement of LGL20, the chromosome harboring the MHC (our unpublished data) in the translocation is noteworthy, though studies are needed to elucidate the possible genetic consequences of this rearrangement.
As expected, no markers were assigned to LPA36 because, to date, there is no knowledge about mammalian homology to the smallest autosome present in the karyotypes of all 6 extant camelid species (Bianchi et al. 1986; Balmus et al. 2007). Zoo-FISH studies with flow-sorted CDR36 in humans, pigs, cattle (Balmus et al. 2007), ruminants (Kulemzina et al. 2011), and other Cetartiodactyls (Kulemzina et al. 2009) concluded that the chromosome does not contain enough euchromatin to produce detectable FISH signals. Indeed, our cytogenetic studies and FISH results with normal and minute LPA36 paints support the idea that the chromosome is largely heterochromatic (Figure 5a–c).
The lack of LPA36-specific markers hinders the understanding of the origin of the minute. The minute might be either the result of a deletion or a translocation. Attempts to test the deletion theory by CGH were inconclusive because of the limited resolution of chromosome CGH. Similarly, the lack of specific markers for LPA36 did not allow testing the theory of a translocation. The only exception was the X chromosome, where FISH with markers from Xpter (STS) and Xqter (ATP6AP1) showed that both terminal segments were the same in minute carriers and controls and did not support LPA36/X translocation.
Because the minute is largely heterochromatic, we have considered the possibility that it is an accessory or a B chromosome. However, except for the heterochromatin, the minute in alpacas does not qualify as a typical B chromosome. In mammals, B chromosomes are found in some species, for example, canids; they are supernumerary to the standard karyotype, are completely heterochromatic or might contain amplified oncogenes, but are dispensable to the carrier (Vujosevic and Blagojevic 2004; Becker et al. 2011). In contrast, the minute in alpacas is not completely heterochromatic (Figure 4), there is no variation in its numbers between individuals, and most importantly, it has been detected in infertile individuals. Furthermore, in all our cases, the minute was heterozygous; suggesting that homozygosity for the aberration might not be viable.
Despite these arguments, one cannot exclude the possibility that the minute is a normal size polymorphism of LPA36, which can be found at a certain frequency in the alpaca population, and the association of the minute with infertility is accidental. Testing this hypothesis needs large cohort karyotyping in alpacas with confirmed records of fertility. Yet, the minute is a unique feature of the alpaca genome, and further molecular studies, including direct sequencing of LPA36, are needed to determine the origin and molecular nature of this chromosome.
In summary, this collection of cytogenetically mapped markers forms a foundation for molecular and clinical cytogenetics in camelids. These and additional FISH-mapped markers will help the improvement and standardization of chromosome nomenclature for the alpaca and other camelids, as well as for anchoring and validating radiation hybrid maps and the genome sequence assembly (Breen 2008; Raudsepp et al. 2008; Lewin et al. 2009). This is of particular importance in alpacas, a species in which many large sequence scaffolds have not yet been assigned to physical chromosomes (Ensembl: http://useast.ensembl.org/index.html). Finally, the 151 BAC clones containing specific alpaca genes can be used as baits for target-enrichment capture and next-generation sequencing (Mamanova et al. 2010; Horn 2012) to identify sequence variants and mutations associated with important health and disease phenotypes in these valued animals.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
Alpaca Research Foundation; Morris Animal Foundation (D09LA-004), GA (CR P506/10/0421); CEITEC (CZ.1.05/1.1.00/02.0068).
Supplementary Material
Acknowledgments
We are grateful to Leslie Wachter and Joan Pontius for making the alpaca cDNA sequences available for primer design and to Roscoe Stanyon and the late Gary Stone for providing flow-sorted probes for LPA36, X, and Y.
References
- Arrighi FE, Hsu TC. 1971. Localization of heterochromatin in human chromosomes. Cytogenetics. 10: 81–86 [DOI] [PubMed] [Google Scholar]
- Balmus G, Trifonov VA, Biltueva LS, O’Brien PC, Alkalaeva ES, Fu B, Skidmore JA, Allen T, Graphodatsky AS, Yang F, et al. 2007. Cross-species chromosome painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res. 15: 499–515 [DOI] [PubMed] [Google Scholar]
- Becker SE, Thomas R, Trifonov VA, Wayne RK, Graphodatsky AS, Breen M. 2011. Anchoring the dog to its relatives reveals new evolutionary breakpoints across 11 species of the Canidae and provides new clues for the role of B chromosomes. Chromosome Res. 19: 685–708 [DOI] [PubMed] [Google Scholar]
- Bianchi NO, Larramendy ML, Bianchi MS, Cortes L.. 1986. Karyological conservation in South American camelids. Experientia. 42: 622–624 [Google Scholar]
- Breen M. 2008. Canine cytogenetics–from band to basepair. Cytogenet Genome Res. 120: 50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M, Bullerdiek J, Langford CF. 1999. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res. 7: 401–406 [DOI] [PubMed] [Google Scholar]
- Breen M, Hitte C, Lorentzen TD, Thomas R, Cadieu E, Sabacan L, Scott A, Evanno G, Parker HG, Kirkness EF, et al. 2004. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 5: 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch TD, Foote WC, Maciulis A. 1985. Chromosome banding pattern homologies and NORs for the Bactrian camel, guanaco, and llama. J Hered. 76: 115–118 [Google Scholar]
- Capanna E, Civitelli MV. 1965. The chromosomes of three species of neotropical Camelidae. Mamm Chrom Newsl. 17: 75–79 [Google Scholar]
- Das PJ, Chowdhary BP, Raudsepp T. 2009. Characterization of the bovine pseudoautosomal region and comparison with sheep, goat, and other mammalian pseudoautosomal regions. Cytogenet Genome Res. 126: 139–147 [DOI] [PubMed] [Google Scholar]
- Davis BW, Raudsepp T, Pearks Wilkerson AJ, Agarwala R, Schäffer AA, Houck M, Chowdhary BP, Murphy WJ. 2009. A high-resolution cat radiation hybrid and integrated FISH mapping resource for phylogenomic studies across Felidae. Genomics. 93: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Berardino D, Nicodemo D, Coppola G, King AW, Ramunno L, Cosenza GF, Iannuzzi L, Di Meo GP, Balmus G, Rubes J. 2006. Cytogenetic characterization of alpaca (Lama pacos, fam. Camelidae) prometaphase chromosomes. Cytogenet Genome Res. 115: 138–144 [DOI] [PubMed] [Google Scholar]
- Di Meo GP, Goldammer T, Perucatti A, Genualdo V, Iannuzzi A, Incarnato D, Rebl A, Di Berardino D, Iannuzzi L. 2011. Extended cytogenetic maps of sheep chromosome 1 and their cattle and river buffalo homoeologues: comparison with the OAR1 RH map and human chromosomes 2, 3, 21 and 1q. Cytogenet Genome Res. 133: 16–24 [DOI] [PubMed] [Google Scholar]
- Di Meo GP, Perucatti A, Floriot S, Hayes H, Schibler L, Incarnato D, Di Berardino D, Williams J, Cribiu E, Eggen A, et al. 2008. An extended river buffalo (Bubalus bubalis, 2n = 50) cytogenetic map: assignment of 68 autosomal loci by FISH-mapping and R-banding and comparison with human chromosomes. Chromosome Res. 16: 827–837 [DOI] [PubMed] [Google Scholar]
- Di Meo GP, Perucatti A, Floriot S, Hayes H, Schibler L, Rullo R, Incarnato D, Ferretti L, Cockett N, Cribiu E, et al. 2007. An advanced sheep (Ovis aries, 2n = 54) cytogenetic map and assignment of 88 new autosomal loci by fluorescence in situ hybridization and R-banding. Anim Genet. 38: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew ML, Meyers-Wallen VN, Acland GM, Guyer CL, Steinheimer DN. 1999. Presumptive Sry-negative XX sex reversal in a llama with multiple congenital anomalies. J Am Vet Med Assoc. 215: 1134–1139 [PubMed] [Google Scholar]
- Ducos A, Revay T, Kovacs A, Hidas A, Pinton A, Bonnet-Garnier A, Molteni L, Slota E, Switonski M, Arruga MV, et al. 2008. Cytogenetic screening of livestock populations in Europe: an overview. Cytogenet Genome Res. 120: 26–41 [DOI] [PubMed] [Google Scholar]
- Fowler M. 1990. Twinning in llamas. Int Camelid J. 4: 35–38 [Google Scholar]
- Galibert F, Quignon P, Hitte C, André C. 2011. Toward understanding dog evolutionary and domestication history. C R Biol. 334: 190–196 [DOI] [PubMed] [Google Scholar]
- Goldammer T, Brunner RM, Rebl A, Wu CH, Nomura K, Hadfield T, Maddox JF, Cockett NE. 2009. Cytogenetic anchoring of radiation hybrid and virtual maps of sheep chromosome X and comparison of X chromosomes in sheep, cattle, and human. Chromosome Res. 17: 497–506 [DOI] [PubMed] [Google Scholar]
- Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, Pilling D, Negrini R, Finlay EK, Jianlin H, Groeneveld E, et al. ; GLOBALDIV Consortium 2010. Genetic diversity in farm animals–a review. Anim Genet. 41 Suppl 1: 6–31 [DOI] [PubMed] [Google Scholar]
- Gustafson AL, Tallmadge RL, Ramlachan N, Miller D, Bird H, Antczak DF, Raudsepp T, Chowdhary BP, Skow LC. 2003. An ordered BAC contig map of the equine major histocompatibility complex. Cytogenet Genome Res. 102: 189–195 [DOI] [PubMed] [Google Scholar]
- Hinrichs K, Buoen LC, Ruth GR. 1999. XX/XY chimerism and freemartinism in a female llama co-twin to a male. J Am Vet Med Assoc. 215: 1140–1141 [PubMed] [Google Scholar]
- Hinrichs K, Horin SE, Buoen LC, Zhang TQ, Ruth GR. 1997. X-chromosome monosomy in an infertile female llama. J Am Vet Med Assoc. 210: 1503–1504 [PubMed] [Google Scholar]
- Horn S. 2012. Target enrichment via DNA hybridization capture. Methods Mol Biol. 840: 177–188 [DOI] [PubMed] [Google Scholar]
- Hornak M, Hulinska P, Musilova P, Kubickova S, Rubes J. 2009. Investigation of chromosome aneuploidies in early porcine embryos using comparative genomic hybridization. Cytogenet Genome Res. 126: 210–216 [DOI] [PubMed] [Google Scholar]
- Hsu TC, Benirschke K. 1967. An atlas of mammalian chromosomes New York:Springer-Verlag; 1: folio40 [Google Scholar]
- Hsu TC, Benirschke K. 1974. An atlas of mammalian chromosomes Berlin (Germany): Springer-Verlag; 1: folio 389 [Google Scholar]
- Hungerford DA, Snyder RI. 1966. Chromosomes of European wolf (Canis lupus) and of a Bactrian camel (Camelus bactrianus). Mamm Chrom Newsl. 20: 72 [Google Scholar]
- ISCN 1995. An international system for human cytogenetic nomenclature (1995). Basel (Switzerland): Karger; [Google Scholar]
- Kadwell M, Fernandez M, Stanley HF, Baldi R, Wheeler JC, Rosadio R, Bruford MW. 2001. Genetic analysis reveals the wild ancestors of the llama and the alpaca. Proc Biol Sci. 268: 2575–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulischer L, Tijskens J, Mortelmans J. 1971. Mammalian cytogenetics. IV. The chromosomes of two male Camelidae: Camelus bactrianus and Lama vicugna. Acta Zool Pathol Antverp. 52: 89–92 [PubMed] [Google Scholar]
- Kubickova S, Cernohorska H, Musilova P, Rubes J. 2002. The use of laser microdissection for the preparation of chromosome-specific painting probes in farm animals. Chromosome Res. 10: 571–577 [DOI] [PubMed] [Google Scholar]
- Kulemzina AI, Trifonov VA, Perelman PL, Rubtsova NV, Volobuev V, Ferguson-Smith MA, Stanyon R, Yang F, Graphodatsky AS. 2009. Cross-species chromosome painting in Cetartiodactyla: reconstructing the karyotype evolution in key phylogenetic lineages. Chromosome Res. 17: 419–436 [DOI] [PubMed] [Google Scholar]
- Kulemzina AI, Yang F, Trifonov VA, Ryder OA, Ferguson-Smith MA, Graphodatsky AS. 2011. Chromosome painting in Tragulidae facilitates the reconstruction of Ruminantia ancestral karyotype. Chromosome Res. 19: 531–539 [DOI] [PubMed] [Google Scholar]
- Lear TL, Bailey E. 2008. Equine clinical cytogenetics: the past and future. Cytogenet Genome Res. 120: 42–49 [DOI] [PubMed] [Google Scholar]
- Lewin HA, Larkin DM, Pontius J, O’Brien SJ. 2009. Every genome sequence needs a good map. Genome Res. 19: 1925–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. 2010. Target-enrichment strategies for next-generation sequencing. Nat Methods. 7: 111–118 [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP. 2008a. FISH for mapping single copy genes. Methods Mol Biol. 422: 31–49 [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP. 2008b. The horse pseudoautosomal region (PAR): characterization and comparison with the human, chimp and mouse PARs. Cytogenet Genome Res. 121: 102–109 [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP. 2011. Cytogenetics and chromosome maps. In: Rothschild MF, Ruvinsky A., editors. The genetics of the pig Oxfordshire (UK): CABI Press; p. 134–178 [Google Scholar]
- Raudsepp T, Das PJ, Avila F, Chowdhary BP. 2012. The pseudoautosomal region and sex chromosome aneuploidies in domestic species. Sex Dev. 6: 72–83 [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Gustafson-Seabury A, Durkin K, Wagner ML, Goh G, Seabury CM, Brinkmeyer-Langford C, Lee EJ, Agarwala R, Stallknecht-Rice E, et al. 2008. A 4,103 marker integrated physical and comparative map of the horse genome. Cytogenet Genome Res. 122: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W, Fu B, O’Brien PC, Ferguson-Smith M. 2006. Cross-species chromosome painting. Nat Protoc. 1: 783–790 [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature. 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubes J, Pinton A, Bonnet-Garnier A, Fillon V, Musilova P, Michalova K, Kubickova S, Ducos A, Yerle M. 2009. Fluorescence in situ hybridization applied to domestic animal cytogenetics. Cytogenet Genome Res. 126: 34–48 [DOI] [PubMed] [Google Scholar]
- Scherthan H, Cremer T, Arnason U, Weier HU, Lima-de-Faria A, Frönicke L. 1994. Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat Genet. 6: 342–347 [DOI] [PubMed] [Google Scholar]
- Seabright M. 1971. A rapid banding technique for human chromosomes. Lancet. 2: 971–972 [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 423: 825–837 [DOI] [PubMed] [Google Scholar]
- Stanley HF, Kadwell M, Wheeler JC. 1994. Molecular evolution of the family Camelidae: a mitochondrial DNA study. Proc Biol Sci. 256: 1–6 [DOI] [PubMed] [Google Scholar]
- Taberlet P, Coissac E, Pansu J, Pompanon F. 2011. Conservation genetics of cattle, sheep, and goats. C R Biol. 334: 247–254 [DOI] [PubMed] [Google Scholar]
- Taylor KM Hungerford DA Snyder RL Ulmer FAJr. 1968. Uniformity of kryotypes in the Camelidae. Cytogenetics. 7: 8–15 [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjöld M, Ponder BA, Tunnacliffe A. 1992. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 13: 718–725 [DOI] [PubMed] [Google Scholar]
- Tibary A. Reproductive disorders in alpacas and llamas. Proceedings of the 1st International Workshop on Camelid Genetics 2008. Feb 22–24Scottsdale, AZ: The Alpaca Research Foundation and The Alpaca Registry, Inc; [Google Scholar]
- Vidal-Rioja L, Larramendy ML, Semorile L. 1989. Ag-NOR staining and in situ hybridization of rDNA in the chromosomes of the South American camelids. Genetica. 79: 215–222 [DOI] [PubMed] [Google Scholar]
- Villagómez DA, Parma P, Radi O, Di Meo G, Pinton A, Iannuzzi L, King WA. 2009. Classical and molecular cytogenetics of disorders of sex development in domestic animals. Cytogenet Genome Res. 126: 110–131 [DOI] [PubMed] [Google Scholar]
- Villagómez DA, Pinton A. 2008. Chromosomal abnormalities, meiotic behavior and fertility in domestic animals. Cytogenet Genome Res. 120: 69–80 [DOI] [PubMed] [Google Scholar]
- Vujosević M, Blagojević J. 2004. B chromosomes in populations of mammals. Cytogenet Genome Res. 106: 247–256 [DOI] [PubMed] [Google Scholar]
- Wilker CE, Meyers-Wallen VN, Schlafer DH, Dykes NL, Kovacs A, Ball BA. 1994. XX sex reversal in a llama. J Am Vet Med Assoc. 204: 112–115 [PubMed] [Google Scholar]
- Zhang QL, Dong CS, He JP, He XY, Fan RW, Geng JJ, Ren YH. 2005. Study on the chromosomal karyotype and G-banding of Alpacas (Lama pacos). Yi Chuan. 27: 221–226 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.