Abstract
Purpose
Recurrently mutated genes in myelodysplastic syndrome (MDS) are pathogenic drivers and powerfully associated with clinical phenotype and prognosis. Whether these types of mutations predict outcome after allogeneic hematopoietic stem-cell transplantation (HSCT) in patients with MDS is not known.
Patients and Methods
We used massively parallel sequencing to examine tumor samples collected from 87 patients with MDS before HSCT for coding mutations in 40 recurrently mutated MDS genes.
Results
Mutations were identified in 92% of patients, most frequently in the ASXL1 (29%), TP53 (21%), DNMT3A (18%), and RUNX1 (16%) genes. In univariable analyses, only TP53 mutations were associated with shorter overall (OS; hazard ratio [HR], 3.74; P < .001) and progression-free survival (HR, 3.97; P < .001). After adjustment for clinical variables associated with these end points, mutations in TP53 (HR, 2.30; P = .027), TET2 (HR, 2.40; P = .033), and DNMT3A (HR, 2.08; P = .049) were associated with decreased OS. In multivariable analysis including clinical variables, complex karyotype status, and candidate genes, mutations in TP53 (HR, 4.22; P ≤ .001) and TET2 (HR, 1.68; P = .037) were each independently associated with shorter OS. Nearly one half of patients (46%) carried a mutation in TP53, DNMT3A, or TET2 and accounted for 64% of deaths. Three-year OS in patients without these mutations was 59% (95% CI, 43% to 72%), versus 19% (95% CI, 9% to 33%) in patients with these mutations.
Conclusion
Mutations in TP53, TET2, or DNMT3A identify patients with MDS with shorter OS after HSCT.
INTRODUCTION
Diagnosis and predicted prognosis of patients with myelodysplastic syndrome (MDS) are largely determined by morphologic and clinical measures.1,2 Recurrent somatic mutations, which are drivers of MDS pathogenesis and can be powerfully associated with clinical phenotype, are not currently incorporated into the routine clinical care of patients with this disorder.3,4 Somatic mutations are common in MDS, with > 75% of patients carrying ≥ one abnormality in the 30 most frequently mutated genes.5–7 Abnormalities in specific genes, such as NRAS, RUNX1, and TP53, have been associated with prognostically important variables, including elevated bone marrow blast proportion and severe thrombocytopenia.3 Therefore, it is likely that acquired mutations could also predict response to specific interventions, such as treatment with hypomethylating agents or survival after hematopoietic stem-cell transplantation (HSCT).8
Calculation of risks, benefits, and timing of HSCT is often difficult in MDS.9–11 Older age and comorbidities typical of patients with MDS are frequently associated with unacceptable risk of early death after transplantation. Even in younger and generally healthier patients, deciding when HSCT is appropriate can be challenging. In particular, patients with poor prognostic features may be directed to transplantation because they have few treatment options available or because standard therapeutics are not expected to provide durable responses.
Clinical features can help identify patients with MDS likely to benefit from HSCT. In addition to age, these variables include International Prognostic Scoring System risk group, ferritin level, monosomal karyotype, and disease burden before transplantation.10,12,13 Donor features, such as degree of HLA matching, age, and sex, as well as preparative regimen also influence outcome. However, molecular genetic information is not routinely used to predict outcome for patients with MDS undergoing HSCT. The current survival rate for HSCT using well-matched donors in patients with MDS is only 40% at 5 years, even in this highly selected population, with relapse and disease-specific mortality responsible for the bulk of deaths.14 Better risk stratification before HSCT would allow for more accurate evaluation of potential benefit and could improve outcome in patients with MDS selected for transplantation and identify those for whom novel transplantation approaches are appropriate while preventing unnecessary transplantation in patients unlikely to benefit from the procedure.
Whether somatic mutations are important markers of response to HSCT is not known. To address this question, we examined samples from patients with MDS undergoing HSCT to determine whether somatic changes in frequently mutated genes are associated with long-term outcome after transplantation.
PATIENTS AND METHODS
Patient Samples
A total of 125 patients with MDS who underwent a first allogeneic bone marrow or peripheral-blood HSCT at the Dana-Farber Cancer Institute from 2004 to 2009 were considered for inclusion in this study. Patients were excluded if a tumor sample collected before transplantation was not available (34 patients) or if the sequencing assay could not be completed for their sample.4 After these exclusions, 87 patients were included in the analysis cohort. No difference in overall survival (OS) was noted between included and excluded patients (P = .44). All samples were collected with patient consent under an institutional review board–approved protocol in accordance with the Declaration of Helsinki. Patient characteristics are listed in Table 1, and details of their conditioning regimens are listed in the Data Supplement.
Table 1.
Patient Demographic and Clinical Characteristics (N = 87)
| Characteristic | No. | % |
|---|---|---|
| Age, years | ||
| Median | 58 | |
| Range | 19-73 | |
| ≥ 60 | 34 | 39 |
| Female sex | 27 | 31 |
| FAB at transplantation | ||
| RA | 24 | 28 |
| RARS | 7 | 8 |
| RAEB | 42 | 48 |
| RAEB-t | 1 | 1 |
| CMML | 5 | 6 |
| MDS-U | 1 | 1 |
| Other/unknown | 7 | 8 |
| Karyotype | ||
| −7/7q− or +1 (± other) | 15 | 17 |
| Normal | 28 | 32 |
| Complex | 28 | 32 |
| Other (any not listed above) | 8 | 9 |
| Unknown | 8 | 9 |
| Blasts, % | ||
| Median | 5 | |
| Range | 0-23 | |
| < 5 | 42 | 48 |
| 5-10 | 32 | 37 |
| 11-30 | 13 | 15 |
| Hemoglobin, g/dL | ||
| Median | 9.9 | |
| Range | 7.1-16.2 | |
| < 8.0 | 12 | 14 |
| 8.0-9.99 | 32 | 37 |
| 10.0-11.99 | 24 | 28 |
| ≥ 12.0 | 19 | 22 |
| ANC, cells/μL | ||
| Median | 1,233 | |
| Range | 14-57,288 | |
| < 500 | 22 | 25 |
| 500-1,499 | 28 | 32 |
| 1,500-9,999 | 32 | 37 |
| ≥ 10,000 | 4 | 5 |
| Unknown | 1 | 1 |
| Platelets | ||
| Median | 64,000 | |
| Range | 4,000-290,000 | |
| < 50,000 | 28 | 32 |
| 50,000-149,000 | 50 | 57 |
| ≥ 150,000 | 9 | 10 |
| Patient-donor sex matching | ||
| Male-male | 34 | 39 |
| Male-female | 26 | 30 |
| Female-male | 9 | 10 |
| Female-female | 18 | 21 |
| Type of conditioning regimen | ||
| Myeloablative | 25 | 29 |
| Nonmyeloablative | 62 | 71 |
| Conditioning regimen | ||
| Busulfan and fludarabine ± other | 53 | 61 |
| Cyclophosphamide/TBI | 30 | 34 |
| Other | 4 | 5 |
| Donor cell source | ||
| PBSCs | 80 | 92 |
| BM | 7 | 8 |
Abbreviations: ANC, absolute neutrophil count; BM, bone marrow; CMML, chronic myelomonocytic leukemia; FAB, French-American-British, MDS-U, myelodysplastic syndrome unclassified; PBSC, peripheral-blood stem cell; RA, refractory anemia; RAEB, refractory anemia with excess blasts; RAEB-t, refractory anemia with excess of blasts in transformation; RARS, refractory anemia with ringed sideroblasts; TBI, total-body irradiation.
Sample Processing, DNA Sequencing, and Mutation Analysis
DNA was extracted from bone marrow mononuclear cells or peripheral-blood samples collected before transplantation (median, 18 days; range, 9 to 119 days). Whole-genome amplification of DNA for each sample was performed using the REPLI-g kit from Qiagen (Venlo, the Netherlands). A genotype fingerprint of 22 common single-nucleotide polymorphisms (SNPs) for each sample was generated by matrix-assisted laser desorption/ionization time-of-flight genotyping (Sequenom, San Diego, CA). Target regions of 40 genes (Data Supplement) and genotype fingerprint regions were enriched using the HaloPlex polymerase chain reaction or Custom SureSelect hybrid capture system (Agilent Technologies, Santa Clara, CA) according to manufacturer instructions. Barcoded samples were pooled in equimolar amounts and subjected to 100-nucleotide paired-end sequencing on an Illumina Hi Seq 2000 (San Diego, CA). Sequence reads were aligned to the human genome (build 37) using the Burroughs-Wheeler algorithm.15 The Genome Analysis Toolkit (https://www.broadinstitute.org/gatk/) was used to clean and locally realign reads before calling missense and insertion/deletion variants using MuTect.16,17 Sample identity was confirmed by matching fingerprint genotype calls. Synonymous variants, noncoding variants more than 6 bases from splice junctions, or germline polymorphisms present in databases of normal genomes (dbSNP 132 or National Heart, Lung and Blood Institute Exome Sequencing Project) at a population frequency ≥ 1% were discarded. Remaining variants were considered candidate somatic mutations.
Statistical Methods
OS was calculated from date of transplantation to date of death, and surviving patients were censored at the date on which they were last known to be alive. Progression-free survival (PFS) was calculated from time of transplantation to date of relapse, progression, or death and was censored at the last date known to be alive and progression free. Curves were constructed for OS and PFS using the Kaplan-Meier method and compared using a log-rank test. Cox models were constructed to adjust for clinical and transplantation characteristics, and a backward elimination selection algorithm was used, with candidate variables having a univariable P value < .20. A time-dependent variable for interaction between mutation status and natural log of time was also included in these models to test the proportional hazards assumption. Cumulative incidence of nonrelapse death and relapse with or without death was calculated using competing risks from time of HSCT to relapse and nonrelapse death and compared using the Gray test. Associations of continuous measures between groups were assessed using a Wilcoxon rank sum test; for categorical variables, they were assessed using Fisher's exact test. P values are unadjusted, two sided, and considered significant at the .05 level.
RESULTS
Baseline clinical characteristics from our cohort of 87 patients who underwent HSCT for MDS at the Dana-Farber Cancer Institute are listed in Table 1. Among the 53 patients included in our cohort who died, 72% died as a result of disease, with or without other causes. Median follow-up for the cohort was 49 months (95% CI, 36 to 67).
Spectrum of Mutations and Chromosomal Abnormalities
We analyzed the coding sequence of 40 genes known to be recurrently mutated in MDS and related myeloid malignancies. We identified mutations in 35 of the 40 selected genes, ≥ one of which were present in 92% of samples (Fig 1; Data Supplement). Mutations in several genes associated with higher-risk MDS were more prevalent in this cohort than in previously reported populations of patients with MDS, including ASXL1 (29%), TP53 (21%), DNMT3A (18%), and RUNX1 (16%). In contrast, mutations in the TET2 (13%) and SF3B1 (9%) genes were relatively underrepresented relative to other cohorts.3,4,18–20
Fig 1.
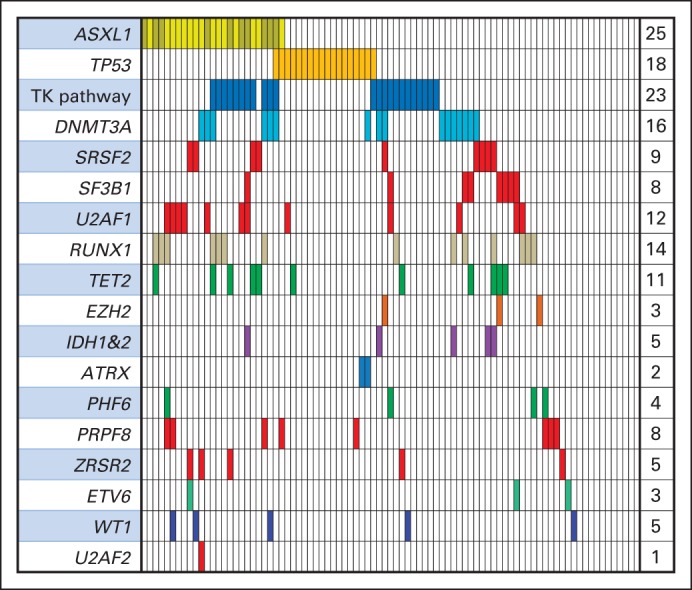
Spectrum of mutations in 87 patients in select myelodysplastic syndrome–associated genes. Each column represents an individual patient sample, and each colored cell represents mutation of gene or gene group listed to left of that row. No. of mutations for each row is indicated in column to right. Tyrosine kinase (TK) pathway genes include JAK2, NRAS, CBL, KRAS, PTPN11, BRAF, and CBLB.
Previously reported patterns of mutations were observed. Mutations in splicing factor genes were largely exclusive of one another, as were mutations in tyrosine kinase signaling genes.3,20 Mutations in specific genes have been associated with adverse prognostic features in patients with MDS, including elevated blast proportion and complex karyotype. In our transplantation cohort, samples with complex cytogenetics had a high frequency of TP53 mutations (57%), and TP53 mutations occurred almost exclusively in patients with complex karyotypes (89% v 17%; P < .001). TP53-mutant samples had a paucity of mutations in other genes (mean, 1.1 non-TP53 mutations per complex sample v 2.5 per noncomplex sample; P < .001).3,21
Clinical and Genetic Features and Transplantation Outcomes
First, we examined the hazard ratio (HR) of death associated with mutations in the 17 genes mutated in ≥ 5% of patients in this cohort (Table 2; Data Supplement). In this univariable analysis, only mutations in TP53 were significantly associated with shorter OS (HR, 3.74; 95% CI, 2.08 to 6.75; P < .001). TP53 mutations were also associated with shorter PFS (HR, 3.97; 95% CI, 2.22 to 7.10), and a trend toward shorter PFS for patients with DNMT3A mutations was observed (HR, 1.76; 95% CI, 0.98 to 3.19; P = .061; Data Supplement). No genetic mutations were associated with longer PFS or OS.
Table 2.
HR of Death Associated With Clinical Features and Mutations in Univariable Analyses
| Variable | Univariable |
Adjusted* |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Blasts (≥ 5% v < 5%) | 1.84 | 1.05 to 3.21 | .032 | |||
| Conditioning regimen (nonmyeloablative v ablative) | 2.03 | 1.04 to 3.94 | .037 | |||
| Karyotype (complex v other) | 2.16 | 1.24 to 3.77 | .007 | |||
| Donor type (unrelated v related) | 2.05 | 1.07 to 3.90 | .029 | |||
| Genetic mutation (present v absent) | ||||||
| TP53 | 3.74 | 2.08 to 6.75 | < .001 | 2.30 | 1.10 to 4.81 | .027 |
| TET2 | 1.68 | 0.79 to 3.57 | .18 | 2.40 | 1.07 to 5.38 | .033 |
| DNMT3A | 1.44 | 0.77 to 2.69 | .26 | 2.08 | 1.00 to 3.26 | .049 |
| PRPF8 | 1.23 | 0.53 to 2.89 | .63 | 1.06 | 0.44 to 2.55 | .89 |
| SF3B1 | 1.08 | 0.43 to 2.71 | .87 | 2.33 | 0.85 to 6.42 | .1 |
| CBL | 1.07 | 0.39 to 2.98 | .89 | 0.99 | 0.35 to 2.87 | .99 |
| ZRSR2 | 0.92 | 0.29 to 2.95 | .89 | 1.01 | 0.31 to 3.66 | .98 |
| SRSF2 | 0.90 | 0.36 to 2.77 | .83 | 1.20 | 0.46 to 3.12 | .71 |
| KDM6B | 0.80 | 0.25 to 2.56 | .7 | 1.09 | 0.32 to 3.67 | .89 |
| AXSL1 | 0.74 | 0.40 to 1.37 | .34 | 0.75 | 0.38 to 1.47 | .4 |
| NRAS | 0.74 | 0.27 to 2.06 | .57 | 0.63 | 0.22 to 1.78 | .38 |
| RUNX1 | 0.70 | 0.32 to 1.55 | .38 | 0.70 | 0.30 to 1.64 | .41 |
| U2AF1 | 0.54 | 0.22 to 1.37 | .2 | 0.76 | 0.29 to 2.02 | .58 |
| SUZ12 | 0.50 | 0.12 to 2.05 | .33 | 0.68 | 0.15 to 3.11 | .62 |
| WT1 | 0.44 | 0.11 to 1.82 | .26 | 0.54 | 0.13 to 2.30 | .41 |
| JAK2 | 0.23 | 0.03 to 1.66 | .14 | 0.31 | 0.04 to 2.28 | .25 |
| PRPF40B | 0.22 | 0.03 to 1.56 | .13 | 0.19 | 0.03 to 1.43 | .11 |
Abbreviation: HR, hazard ratio.
Adjusted for blast percentage, conditioning regimen, complex karyotype, and donor type.
Several patient and disease features are known to affect survival after HSCT for MDS, including patient age, sex, karyotype, and disease burden before transplantation. We next examined how these clinical and disease-related variables were associated with OS in our cohort. Patients with ≥ 5% blasts had significantly worse OS compared with those with < 5% (HR, 1.84; P = .032), as did patients with a complex karyotype compared with those without (HR, 2.16; P = .007). Differences in conditioning regimens (myeloablative v nonmyeloablative; HR, 2.03; P = .037) and donor type (unrelated v related; HR, 2.05; P = .029) had a similar impact on OS (Data Supplement). There were no significant differences in survival for patients stratified by age (≥ 60 v < 60 years), sex, or donor sex.
Because mutations could be associated with adverse clinical features associated with OS, we performed an adjusted multivariable analysis to assess the association of patient mutations in TP53 and other genes with OS (Table 2). Adjustment for presence of a complex karyotype, > 5% blasts, donor type, and conditioning regimen identified mutations in TP53, TET2, and DNMT3A as significantly associated with shorter OS. For TP53 mutations, which were strongly associated with complex karyotype, nonmyeloablative conditioning, and unrelated donor, the HR of death decreased (HR, 2.30; P = .027) yet remained significant. The OS curve for patients with TP53 mutations is shown in Figure 2A. Within the group of patients with complex cytogenetics, TP53 mutations highlighted a subset with significantly shorter OS. In contrast, patients with complex cytogenetics and no TP53 mutation had OS comparable to that of patients without complex cytogenetics (Fig 2B).
Fig 2.
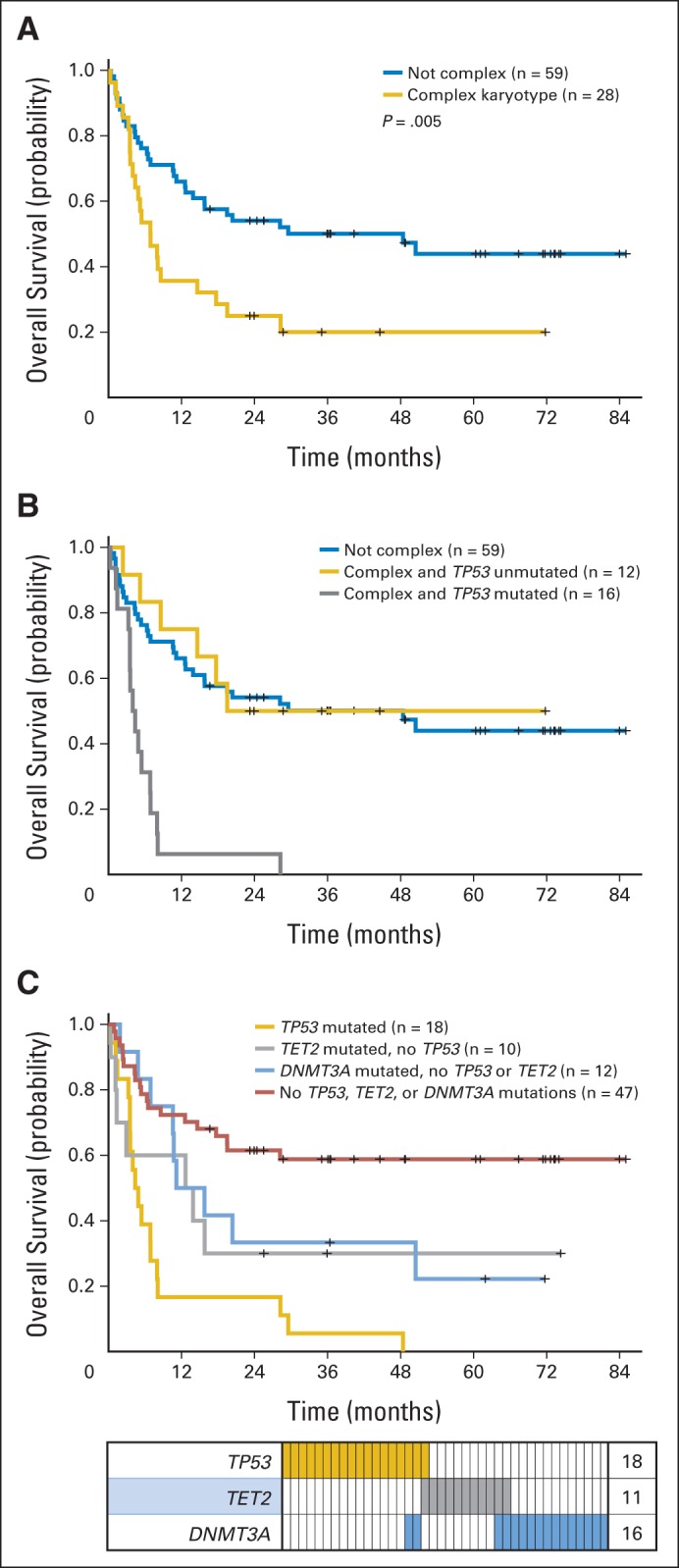
Overall survival (OS) by TP53 and DNMT3A mutation status. OS of patients (A) with and without complex karyotype and (B) with complex karyotype stratified by TP53 mutation status and compared with survival of patients with noncomplex karyotype; (C) OS and mutation distribution showing overlap between patients with TP53, TET2, and DNMT3A mutations. Each column indicates individual patient; colored bars represent mutations of genes in that row.
For patients with a TET2 or DNMT3A mutation, the adjusted HR for OS was statistically significant and comparable to that for patients with a TP53 mutation (Table 2). However, most patients with a DNMT3A or TET2 mutation did not have a complex karyotype and were not more likely to have an elevated bone marrow blast percentage before transplantation, yet these patients had shorter OS compared with patients without such mutations (Fig 2C). TP53, TET2, and DNMT3A mutations occurred in largely nonoverlapping groups of patients, and nearly one half of the cohort (47%) had a mutation in ≥ one of these three genes. Together, these genes identified a population of patients with substantially shortened OS (median, 7.4 months; 95% CI, 4.4 to 12.6 v not reached [NR]; 95% CI, 19.5 to NR) and PFS (median, 4.6 months; 95% CI, 2.8 to 8.0 v NRl 95% CI, 6.3 to NR).
100-Day Landmark and Regression Analyses
Survival to day 100 after HSCT for MDS is an important clinical time point, after which the risk of relapse begins to outweigh the risk of transplantation-related mortality. In our cohort, 72 patients reached this milestone. We hypothesized that mutations, as disease-intrinsic abnormalities, would be more closely associated with risk of relapse after HSCT rather than other causes of transplantation-related mortality. Because the exact cause of death can be difficult to discern, we performed a planned landmark analysis with patients surviving to day 100. In univariable analyses, donor type and conditioning regimen were no longer predictive of OS in this group, but mutations in TP53 remained strongly associated with OS and PFS. Differences were observed in multivariable Cox models created by examining the entire cohort of patients and those in the day-100 landmark group (Table 3). The model for the entire cohort identified mutations in TP53 and TET2 as independent predictors of OS. In the day-100 analysis, TP53 and DNMT3A mutations emerged as independent predictors of OS, along with a complex karyotype. Patients with TET2 mutations had slightly more early deaths that patients with mutations in DNMT3A alone, but both groups had comparable long-term survival rates (Fig 2C). Thus, mutations in TP53, TET2, and DNMT3A are predictors of survival, and 60% of patients with MDS without these mutations were alive and disease free 3 years after HSCT.
Table 3.
Multivariable* Models Identifying Independent Significant Risk Factors for OS
| Variable | HR | 95% CI | P |
|---|---|---|---|
| Entire cohort (N = 87) | |||
| Genetic mutation (present v absent) | |||
| TP53 | 4.22 | 2.30 to 7.76 | < .001 |
| TET2 | 2.29 | 1.05 to 5.00 | .037 |
| Day-100 landmark analysis (n = 72) | |||
| Karyotype (complex v other) | 2.85 | 1.35 to 6.47 | .013 |
| Genetic mutation (present v absent) | |||
| TP53 | 3.78 | 1.81 to 7.89 | < .001 |
| DNMT3A | 2.62 | 1.15 to 5.96 | .022 |
Abbreviations: HR, hazard ratio; OS, overall survival.
Final model obtained from backward-elimination selection algorithm candidates included variables with univariable P < .20.
DISCUSSION
To explore the value of somatic mutations in predicting outcome in the setting of HSCT for MDS, we used deep, massively parallel sequencing to examine 40 genes in samples from 87 patients with MDS before allogeneic transplantation. We identified mutations in three genes—TP53, TET2, and DNMT3A—which were each associated with shorter OS after adjustment for clinical factors associated with poor outcome after stem-cell transplantation. Mutations of ≥ one of these genes were found in nearly one half of patients in this cohort. Mutations of other genes associated with poor prognosis in prior studies, such as RUNX1, ASXL1, SRSF2, and U2AF1, were not associated with differences in OS in our cohort of patients who underwent HSCT (Data Supplement).3,22–24 This may have been the result of disease-modifying effects of conditioning and transplantation or because of the fact that the prognostic significance of these gene mutations is more pronounced in lower-risk patients, of whom there were few in this study. In contrast, TP53 mutations have independent prognostic value, even in higher-risk patients with MDS, in whom they are most commonly found.3,21
The DNMT3A and TET2 genes encode epigenetic modifiers that regulate DNA methylation, and both are recurrently mutated in MDS, acute myeloid leukemia, and other hematologic malignancies. In acute myeloid leukemia, mutations of both genes are enriched in patients with intermediate-risk karyotypes and are associated with poor prognosis.25,26 In MDS, the clinical significance of DNMT3A mutations is less clear but also seems to be unfavorable, whereas TET2 mutations are not associated with survival.5,7,18,19,27,28 Both TET2 and DNMT3A mutations are relatively promiscuous and often co-occur with other mutated genes that can predict outcomes. For example, in a study of lower-risk patients with MDS, DNMT3A mutations were not associated with OS in univariable analysis. However, the DNMT3A-mutant/SF3B1-wild-type subgroup did have shorter OS.19 In our transplantation cohort of largely higher-risk patients, SF3B1 mutations were rare, and most DNMT3A-mutant samples were SF3B1 wild type (88%).
DNMT3A and TET2 mutations identified in pretransplantation samples were largely from patients without adverse clinical features known to predict poor outcome. Most of these patients did not have a complex karyotype and were not more likely to have an elevated bone marrow blast percentage before transplantation. Nevertheless, we found that patients with a TET2 or DNMT3A mutation were at increased risk of relapse and death after transplantation, particularly when other predictive variables were considered. We conclude that consideration of TET2 and DNMT3A mutation status can help predict the risk of mortality in patients with MDS.
In MDS, TP53 mutations have long been known to be associated with karyotype, elevated bone marrow blast percentage, and severe thrombocytopenia.3,29–31 Despite these links with prognostically adverse clinical features, TP53 mutations have strong and independent prognostic significance for patients with MDS.3,21,30–32 In our study, TP53 mutation status was the most significant predictor of mortality after transplantation. All 18 TP53-mutant patients died before 5 years after transplantation, and 83% had MDS present at time of death (Table 4). Their short median survival of 4.6 months is striking and suggests no HSCT benefit in this group. If TP53 mutations are associated with resistance to specific conditioning regimens or indicative of rapidly progressive disease, standard transplantation may not be a viable therapeutic option for this patient population. In the case of reduced-intensity transplantation, the pace of disease may not allow enough time for adequate graft-versus-tumor effect to take place. Clinical strategies to reinforce graft-versus-MDS effect or intensify conditioning or pretransplantation therapy or strategies that include post-HSCT therapy may be particularly beneficial in these patients.33 Although TP53 mutation status was not associated with patient age (P = .60), 16 of the 18 patients with TP53-mutant disease in this study received reduced-intensity conditioning, and all but one had an unrelated donor (Data Supplement). It remains possible that myeloablative conditioning might mitigate the impact of these adverse TP53 mutations (Data Supplement). For most patients with MDS, this would not be a viable option, because of advanced age or comorbid diseases. We conclude that TP53 mutation status is an important factor to consider when assessing the potential risks and benefits of HSCT for MDS.
Table 4.
Causes of Death and Cumulative Incidence of Relapse or NRM
| Incidence/Cause | All Patients (N = 87) |
TP53 Mutated (n = 18) |
TP53 Nonmutated (n = 69) |
P* | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||
| Cumulative incidence of relapse | < .001 | ||||||
| 100 days | 16 | 50 | 7 | ||||
| 95% CI | 9 to 24 | 25 to 71 | 5 to 20 | ||||
| 2 years | 41 | 83 | 30 | ||||
| 95% CI | 31 to 51 | 52 to 95 | 20 to 42 | ||||
| Cumulative incidence of NRM | .048 | ||||||
| 100 days | 11 | 11 | 12 | ||||
| 95% CI | 6 to 19 | 1 to 30 | 3 to 15 | ||||
| 2 years | 22 | 11 | 25 | ||||
| 95% CI | 14 to 31 | 2 to 30 | 15 to 35 | ||||
| Total No. of deaths | 53 | 18 | 35 | ||||
| Cause of death | |||||||
| Disease (± other)† | 38 | 72 | 15 | 83 | 23 | 66 | |
| Transplantation related/other‡ | 15 | 28 | 3 | 17 | 12 | 34 | |
Abbreviation: NRM, nonrelapse mortality.
Gray test.
Causes that include term “disease” indicate that myelodysplastic syndrome was present at time of death.
Including respiratory failure, sepsis, infection, cardiac, bleeding, second malignancy, graft-versus-host disease, veno-occlusive disease, and other.
The decision to perform HSCT in patients with MDS can be challenging, and our findings suggest that analysis of mutations can aid such decisions. Selecting patients for transplantation requires careful determination of the predicted risks and potential benefits of the procedure in comparison with other therapeutic approaches, including supportive care and clinical trials. Consideration of several host factors, disease features, and donor qualities can help in this determination, but uncertainty persists. To date, the presence of molecular genetic abnormalities has not been routinely included in the evaluation of patients before transplantation.
The results of this study identify a clinical setting in which genetic profiling of tumor samples can inform care decisions for patients with MDS. Although replication of these findings will be required, our study demonstrates how mutations in TET2, DNMT3A, and TP53 identify a significant fraction of HSCT recipients with poor long-term survival, for whom alternatives to standard transplantation options should be considered. Patients without mutations in these genes have relatively good OS and PFS and could potentially be prioritized for HSCT.
Supplementary Material
Acknowledgment
We thank Peter Grauman for his assistance on this project.
Glossary Terms
- Germline polymorphism:
a difference in DNA sequence among individuals in the germ cells. Unlike somatic cell genetic mutations, these polymorphisms can be transmitted to an organism's offspring. Genetic polymorphisms may be the result of a chance process or may have been induced by external agents (such as viruses or radiation). Changes in DNA sequence that have been confirmed to be caused by external agents are generally called “mutations” rather than “polymorphisms”.
- Karyotype:
an organized chromosomal profile defining chromosomal arrangement and number. In a karyotype, chromosomes are photographically arranged and displayed in pairs, ordered by size. Chromosomal size, banding pattern, and centromere position are typically used as guides to determine chromosomal abnormalities, but improved resolution may be obtained by combining traditional banding techniques with genome-wide molecular cytogenetics such as multicolor fluorescent in situ hybridization (FISH) and locus-specific FISH.
- Somatic mutation:
a change in the genotype of a cancer cell. This is distinguished from a germline mutation, which is a change in the genotype of all the normal cells in a patient's body. Germline mutations may be passed to offspring, but somatic mutations may not.
- TP53:
gene encoding p53, a nuclear protein that plays an essential role in the regulation of cell cycle. Mutations in p53, resulting in proteins that fail to bind DNA, frequently occur in several human cancers, resulting in a loss of tumor-suppressor activity. Alterations of the TP53 gene occur as somatic mutations in human malignancies and as germline mutations in some cancer-prone families with Li-Fraumeni syndrome.
Footnotes
Listen to the podcast by Dr Estey at www.jco.org/podcasts
Processed as a Rapid Communication manuscript
Supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant No. 5K08DK091360 and an American Society of Hematology scholar award (R.B.) and by National Heart, Lung, and Blood Institute Grant No. R01HL082945, a Leukemia and Lymphoma Society scholar award, and the Yellow Diamond Foundation Fund (B.L.E.).
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) and/or an author's immediate family member(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Rafael Bejar, Genoptix (C), Celgene (C); David P. Steensma, Celgene (C), Genoptix (C), Amgen (C), Novartis (C); John Koreth, Spectrum (C), Millennium Pharmaceuticals (C), Eleven Biotherapeutics (C), Takeda Pharmaceuticals (C); Benjamin L. Ebert, Genoptix (C), Celgene (C) Stock Ownership: None Honoraria: Rafael Bejar, Genoptix, Celgene; David P. Steensma, Genoptix; John Koreth, OptiumHealth Education Research Funding: John Koreth, Millennium Pharmaceuticals, Otsuka, Prometheus Laboratories; Benjamin L. Ebert, Celgene Expert Testimony: None Patents, Royalties, and Licenses: Rafael Bejar, signatures for predicting survivability of myelodysplastic syndrome subjects (WO 2012/174419 A2); Kristen E. Stevenson, signatures for predicting survivability of myelodysplastic syndrome subjects (WO 2012/174419 A2); Donna Neuberg, signatures for predicting the survivability of myelodysplastic syndrome subjects (WO 2012/174419 A2), methods for determining response to hypomethylating agent; Benjamin L. Ebert, signatures for predicting survivability of myelodysplastic syndrome subjects (WO 2012/174419 A2) Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Rafael Bejar, Kristen E. Stevenson, R. Coleman Lindsley, Brenton G. Mar, David P. Steensma, Jerome Ritz, Donna Neuberg, Corey S. Cutler, Benjamin L. Ebert
Financial support: David P. Steensma, Benjamin L. Ebert
Provision of study materials or patients: Jerome Ritz, Robert Soiffer, Joseph H. Antin, Edwin Alyea, Philippe Armand, Vincent Ho, John Koreth, Corey S. Cutler
Collection and assembly of data: Rafael Bejar, Kristen E. Stevenson, Bennett Caughey, R. Coleman Lindsley, Jerome Ritz, Robert Soiffer, Joseph H. Antin, Edwin Alyea, Philippe Armand, Vincent Ho, John Koreth, Donna Neuberg, Corey S. Cutler, Benjamin L. Ebert
Data analysis and interpretation: Rafael Bejar, Kristen E. Stevenson, R. Coleman Lindsley, Petar Stojanov, Gad Getz, Jerome Ritz, Robert Soiffer, Donna Neuberg, Corey S. Cutler, Benjamin L. Ebert
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter MJ, Shen D, Shao J, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013;27:1275–1282. doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itzykson R, Kosmider O, Cluzeau T, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 9.Cutler CS, Lee SJ, Greenberg P, et al. A decision analysis of allogeneic bone marrow transplantation for the myelodysplastic syndromes: Delayed transplantation for low-risk myelodysplasia is associated with improved outcome. Blood. 2004;104:579–585. doi: 10.1182/blood-2004-01-0338. [DOI] [PubMed] [Google Scholar]
- 10.Deeg HJ, Scott BL, Fang M, et al. Five-group cytogenetic risk classification, monosomal karyotype, and outcome after hematopoietic cell transplantation for MDS or acute leukemia evolving from MDS. Blood. 2012;120:1398–1408. doi: 10.1182/blood-2012-04-423046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koreth J, Pidala J, Perez WS, et al. Role of reduced-intensity conditioning allogeneic hematopoietic stem-cell transplantation in older patients with de novo myelodysplastic syndromes: An international collaborative decision analysis. J Clin Oncol. 2013;31:2662–2670. doi: 10.1200/JCO.2012.46.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armand P, Kim HT, Cutler CS, et al. Prognostic impact of elevated pretransplantation serum ferritin in patients undergoing myeloablative stem cell transplantation. Blood. 2007;109:4586–4588. doi: 10.1182/blood-2006-10-054924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim ZY, Fiaccadori V, Gandhi S, et al. Impact of pre-transplant serum ferritin on outcomes of patients with myelodysplastic syndromes or secondary acute myeloid leukaemia receiving reduced intensity conditioning allogeneic haematopoietic stem cell transplantation. Leuk Res. 2010;34:723–727. doi: 10.1016/j.leukres.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 14.Saber W, Cutler CS, Nakamura R, et al. Impact of donor source on hematopoietic cell transplantation outcomes for patients with myelodysplastic syndromes (MDS) Blood. 2013;122:1974–1982. doi: 10.1182/blood-2013-04-496778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith AE, Mohamedali AM, Kulasekararaj A, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 19.Bejar R, Stevenson KE, Caughey BA, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 21.Kulasekararaj AG, Smith AE, Mian SA, et al. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br J Haematol. 2013;160:660–672. doi: 10.1111/bjh.12203. [DOI] [PubMed] [Google Scholar]
- 22.Thol F, Friesen I, Damm F, et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J Clin Oncol. 2011;29:2499–2506. doi: 10.1200/JCO.2010.33.4938. [DOI] [PubMed] [Google Scholar]
- 23.Graubert TA, Shen D, Ding L, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 25.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2011;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thol F, Winschel C, Lüdeking A, et al. Rare occurrence of DNMT3A mutations in myelodysplastic syndromes. Haematologica. 2011;96:1870–1873. doi: 10.3324/haematol.2011.045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter MJ, Ding L, Shen D, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko H, Misawa S, Horiike S, et al. TP53 mutations emerge at early phase of myelodysplastic syndrome and are associated with complex chromosomal abnormalities. Blood. 1995;85:2189–2193. [PubMed] [Google Scholar]
- 30.Kita-Sasai Y, Horiike S, Misawa S, et al. International prognostic scoring system and TP53 mutations are independent prognostic indicators for patients with myelodysplastic syndrome. Br J Haematol. 2001;115:309–312. doi: 10.1046/j.1365-2141.2001.03073.x. [DOI] [PubMed] [Google Scholar]
- 31.Horiike S, Kita-Sasai Y, Nakao M, et al. Configuration of the TP53 gene as an independent prognostic parameter of myelodysplastic syndrome. Leuk Lymphoma. 2003;44:915–922. doi: 10.1080/1042819031000067620. [DOI] [PubMed] [Google Scholar]
- 32.Jädersten M, Saft L, Smith A, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29:1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- 33.Ho VT, Vanneman M, Kim H, et al. Biologic activity of irradiated, autologous, GM-CSF-secreting leukemia cell vaccines early after allogeneic stem cell transplantation. Proc Natl Acad Sci U S A. 2009;106:15825–15830. doi: 10.1073/pnas.0908358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


