Abstract
Eukaryotes and bacteria can be infected with a wide variety of RNA viruses. On average, these pathogens share little sequence similarity and use different replication and transcription strategies. Nevertheless, the members of nearly all RNA virus families depend on the activity of a virally encoded RNA-dependent polymerase for the condensation of nucleotide triphosphates. This review provides an overview of our current understanding of the viral RNA-dependent polymerase structure and the biochemistry and biophysics that is involved in replicating and transcribing the genetic material of RNA viruses.
Keywords: RNA virus, Retrovirus, RdRp, Reverse transcriptase, Dynamics
Introduction
Infections with RNA viruses place a constant burden on our healthcare systems and economy [1, 2]. Over the past century, this has been particularly true for infections with the Human immunodeficiency virus 1 (HIV-1), Influenza A virus (IAV), Rotavirus (RotaV), West Nile virus (WNV), Dengue virus (DV), Measles virus (MV), and the Porcine reproductive and respiratory syndrome virus (PRRSV) [2–6]. But also emergent RNA viruses can have considerable consequences, such as the Severe acute respiratory syndrome-related coronavirus (SARS-CoV) in 2003 [7] and, more recently, the Middle East Respiratory Syndrome coronavirus (MERS-CoV) [8] and the Schmallenberg virus (SBV) [9]. One way to limit the impact of RNA viruses and retroviruses is to prevent their replication, and a thorough understanding of the replication and transcription of these pathogens is, therefore, essential.
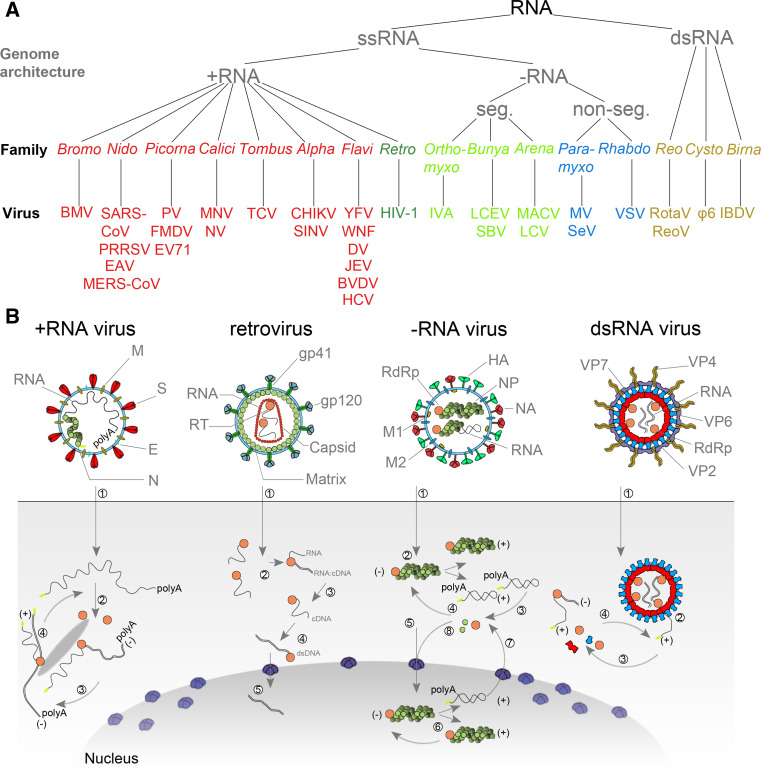
RNA virus genomes can consist of double-stranded RNA (dsRNA) or single-stranded (ssRNA) (Fig. 1a). In turn, the ssRNA viruses can be classified into positive sense (+RNA) and negative sense (−RNA) viruses (Fig. 1a), depending on the translatability of their genetic material. As summarised for four model RNA pathogens in Fig. 1b, all RNA viruses use dedicated replication and transcription strategies to amplify their genetic material. The common denominator of these strategies is a conserved RNA-dependent polymerase domain [10–12].
Fig. 1.
Taxonomy and replication strategies of RNA viruses. a Simplified taxonomy of the genome architecture of the RNA viruses described in this review. See main text for used abbreviations. b (+RNA virus) Infection with a +RNA virus—as exemplified here with a CoV-like virion—releases a single-stranded RNA genome into the cytoplasm (1) [81, 173, 174]. (2) Translation of the 5′-terminal open-reading frame of the genome produces the viral replicase. (3) This multi-enzyme complex includes RdRp activity (orange) and associates with intracellular membranes before −RNA synthesis commences. Newly synthesised −RNAs are subsequently used to produce new +RNAs (4), which are typically capped (yellow) and polyadenylated (polyA). (Retrovirus) HIV-1 genomes are packaged as ssRNA in virions. When the ssRNA is released (1) a cDNA copy is synthesised by the RT (2). The RNA is next degraded by the intrinsic RNase H activity in the RT (3) and the single stranded cDNA converted to dsDNA (4). The dsDNA is imported in the nucleus (5) for integration into the host’s genetic material. (−RNA virus) (1) As illustrated here with an IAV-like particle, infection with an −RNA virus releases a viral RNA genome that is associated with a viral polymerase (orange) and nucleoprotein (green). (2) In the case of non-segmented −RNA viruses, these complexes support transcription to produce viral mRNAs or cRNAs. (3) Viral mRNAs are next translated and new viral proteins complex with cRNAs to synthesise new vRNAs. (5) The vRNA-containing complexes of some segmented −RNA viruses are imported into the nucleus of the host cell, where (6) the RdRp produces mRNAs or cRNAs. (7) mRNAs are transported to the cytoplasm, while cRNAs are bound by new viral proteins to form cRNPs for −RNA synthesis. (dsRNA virus) Fully duplexed RNA genomes lack cap and polyA elements. (1) The RdRp (orange), therefore, transcribes the viral genome inside the capsid of the virion (blue and red), so viral mRNAs can be (2) released into the cytoplasm as illustrated here with a rotavirus-like virion. In the cytoplasm the mRNA is translated (3) or replicated by newly synthesised viral RdRps (4) [175, 176]
This review will start with a discussion of the structure of the polymerase domain and its key catalytic residues. Section “Beyond the conserved core” will subsequently focus on the additional domains that are present in polymerase proteins and how they are coordinated. Section “Template recognition, initiation, elongation and regulation” will concentrate on the interactions of the RNA-dependent polymerase with the viral promoter and the initiation of RNA synthesis. Finally, Sect. “Motion and fidelity” will discuss the dynamics and fidelity of RNA-dependent polymerases. A simplified taxonomy of the RNA viruses that will be discussed in these sections is shown in Fig. 1a.
The RNA-dependent polymerase domain
Nomenclature and organisation of the conserved elements in the RNA-dependent polymerase domain
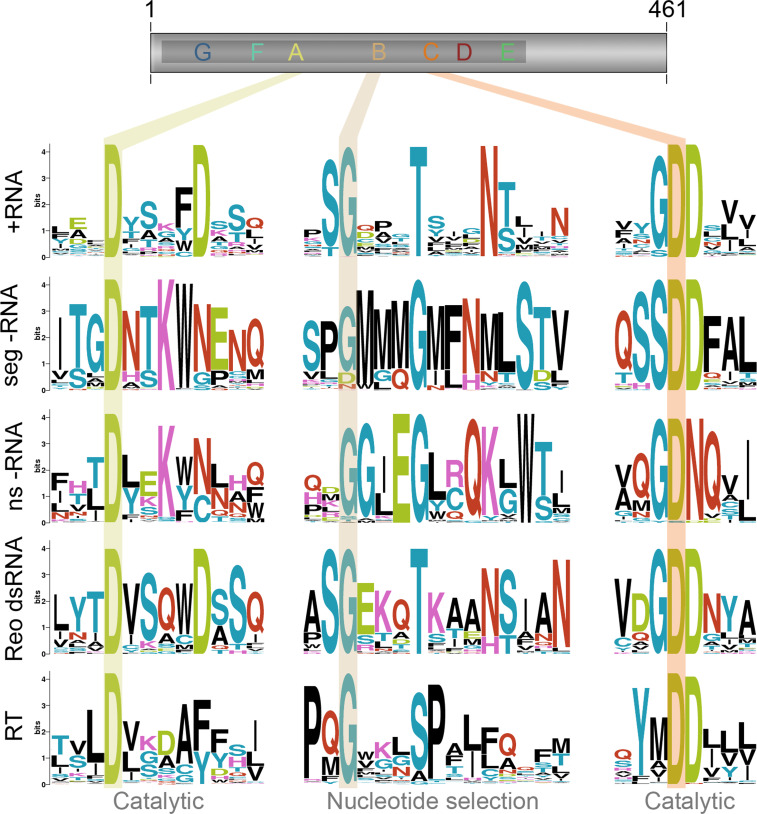
The RNA-dependent polymerase domain is a member of the superfamily of template-dependent nucleic acid polymerases and typically <400 amino acids in length [10, 11] (Fig. 2). Its sequence is highly variable on average, with some regions showing less than ~10 % conservation [11, 13]. Strong amino acid conservation can be observed, however, in regions that are directly involved in nucleotide selection or catalysis, such as the strictly conserved glycine and aspartates in the centre of the domain [11, 13] (Fig. 2; see Sects. “Function of the conserved aspartates and lysine in the polymerase catalytic site”, “Motifs and structural flexibility are important for nucleotide selection” for details). The prototypic RNA virus RNA polymerase domain harbours seven of such regions or motifs, which are arranged in the order G, F1–3, A, B, C, D and E from amino- to carboxy-terminus [10, 13] (Fig. 2). The only exception to this scheme can be found in the Infectious bursal disease virus (IBDV) and related viruses, where motif C is encoded upstream of motif A [14].
Fig. 2.
Key conserved residues of the RNA polymerase domain. Motifs A–C reside in the middle of the typical RNA-dependent polymerase domain as shown here in the schematic of the poliovirus 3Dpol subunit. They are involved in catalysis and nucleotide selection and the residues involved in these processes are highly conserved. The key residues of these motifs are shaded across the RNA polymerase domains of positive strand RNA viruses (+RNA), segmented negative strand RNA viruses (seg −RNA), non-segmented negative strand RNA viruses (ns −RNA), double strand RNA viruses of the reovirus family (Reo dsRNA), and reverse transcriptases (RT). Sequence logo images were created using prosite accession numbers PDSC50507 and PDOC50878
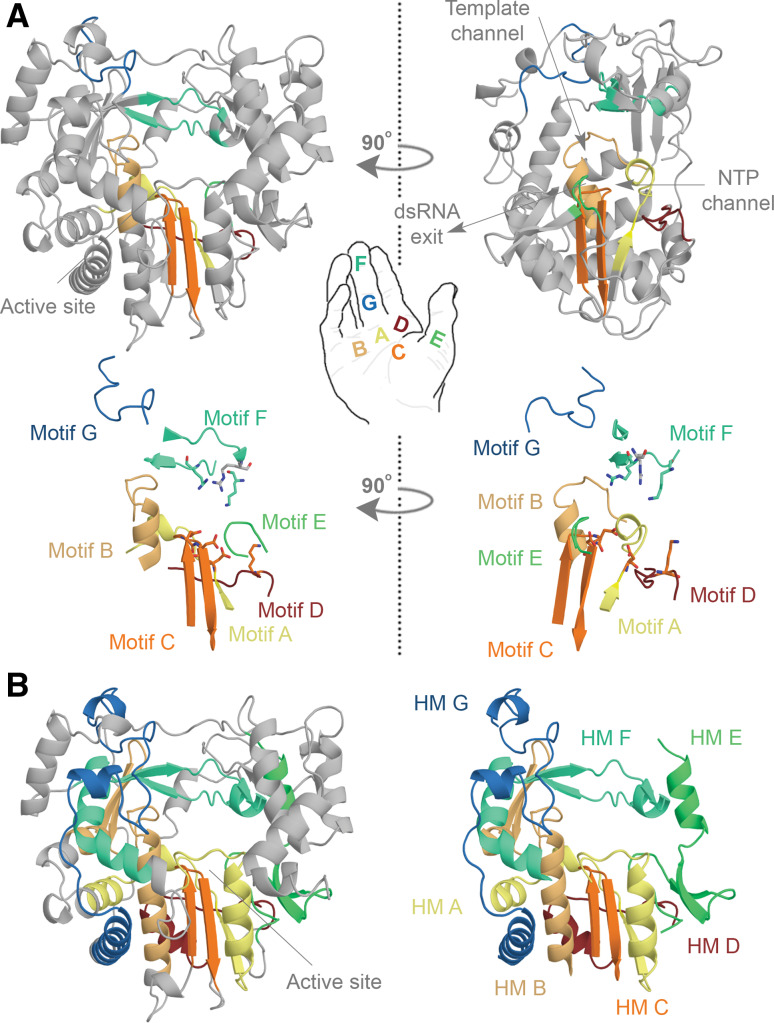
Each of the seven motifs in the RNA polymerase domain adopts a specific and conserved fold [10] (Fig. 3a). However, for most the conservation of the folds extends beyond the regions of sequence similarity into so-called homomorphs [15]. Together, these conserved structural elements make up approximately 75 % of the RNA-dependent polymerase domain [15] (Fig. 3b). In the RdRp structures that are currently available for +RNA, dsRNA, and Retroviridae (Fig. 4)—no structures are presently in the PDB for −RNA viruses—these elements define an RNA entry grove at the top of the polymerase, an RNA exit channel at the front, and a channel for the entry of nucleotides at the rear (Fig. 3a) [16–22].
Fig. 3.
Conserved structural elements in the RNA virus polymerase. a Structure of the FMDV RdRp. The motifs A, B, C, D, E, F, and G are colour coded yellow, gold, orange, red, light green, aquamarine, and blue. Overall the polymerase structure adopts a shape that resembles a cupped right hand. Herein, motifs A–E lie on the palm, while motif F and G are part of the fingers. In the side-view of the enzyme the location of the template and NTP channels is indicated. b Conserved structural elements of the FMDV RdRp. Homomorphs A–G were mapped according to Ref. [15] and colour coded yellow, gold, orange, red, light green, aquamarine, and blue, respectively. Images A and B are based on PDB accession 2E9R
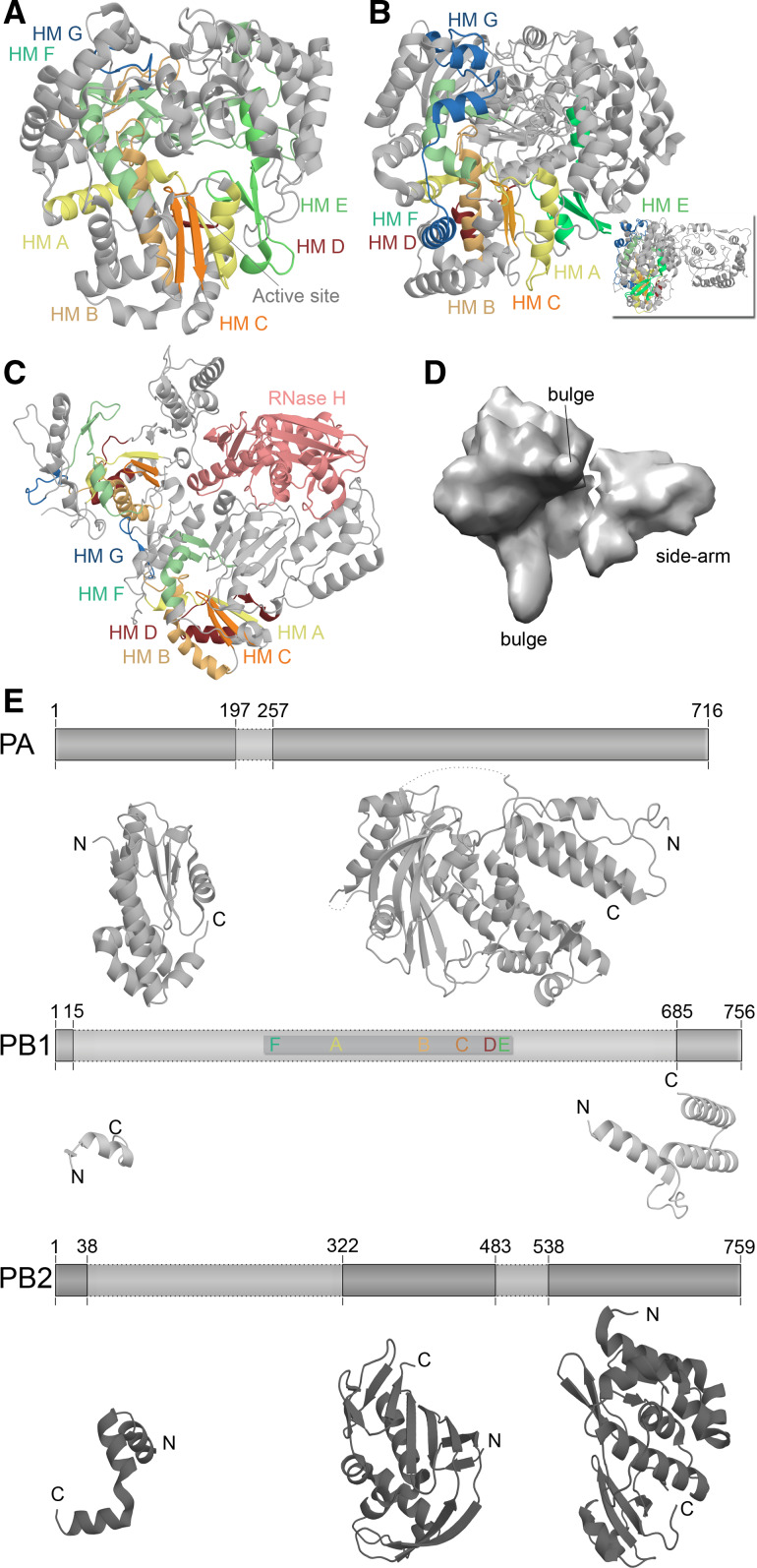
Fig. 4.
Structural differences among RNA virus polymerases. a Structure of the ϕ6 RdRp P2 based on PDB accession 1HI0. b Structure of the JEV polymerase based on PBD entry 4K6 M. Inset depicts 90° rotation of polymerase to visualise the N-terminal methyltransferase domain. c Structure of the HIV-1 RT based on PDB accession 3V4I. The RT is comprised of the p66 (left) and p51 (right) protein subunits. Only the p66 subunit has an RNase domain (pink). Homomorphs A–G are colour coded yellow, gold, orange, red, light green, aquamarine, and blue, respectively in Fig. 4a–c. d EM model of the IAV RdRp based on PDBe entry EMD-2213. Structural features were identified by Moeller et al. [79]. e The IAV polymerase consists of the subunits PA, PB1, and PB2. Six of the seven canonical RNA-dependent polymerase domains motifs are found in PB1, which are colour coded as in Fig. 3. Presently only significant structural information is available for PA and PB2. Figure based on PBD entries 2VY6, 2W69, 2ZNL, 2ZTT, 3EBJ, and 4CB4
Following the convention for cellular DNA-dependent DNA polymerases (DdDp), the seven motifs and homomorphs are grouped into three subdomains. These subdomains are called fingers, palm and thumb in reference to the polymerase domain’s likeliness to a cupped right hand (Fig. 3a). The finger subdomain loops that interconnect the fingers with the thumb in the RNA-dependent RNA polymerases (RdRps) of +RNA and dsRNA viruses—for instance Foot and mouth disease virus (FMDV), Pseudomonas phage phi6 (ϕ6), and Japanese encephalitis virus (JEV) (Fig. 4)—are called the fingertips [23]. This connection creates an overall “closed-hand” conformation that is unique to RdRps and generally not seen in crystal structures of DdDps or reverse transcriptases (RTs) (Fig. 4c).
Function of the conserved main structural elements
The three subdomains work together to facilitate the binding of RNA and nucleotides (NTPs) [17–20]. The thumb subdomain contains various residues that are involved in RNA binding and these generally pack into the minor groove of the template RNA [20]. In some polymerases, the thumb also contains sequences that protrude into the template channel to help stabilise the initiating NTPs on the ssRNA template (see Sect. “Template recognition, initiation, elongation and regulation” for details on initiation) [17, 18, 24]. Crucially, these protrusions are also able to undergo relatively large conformational rearrangements to facilitate translocation of the template following the first condensation reaction [17, 25, 26]. The other residues of the thumb subdomain contribute to the formation of the NTP tunnel, which sits at an ~110° angle with the template channel (Fig. 3a). The cavity is lined with positively charged amino acids [17–20], though charge interactions are likely not sufficient to guide NTPs into the interior. Indeed, molecular dynamics (MD) simulations have shown that the residues of the NTP channel can also explore a relatively large amount of space [27], which may allow the RdRp to actively “pump” NTPs down the channel [27].
The fingers subdomain residues mainly pack into the major groove of the RNA template. Furthermore, upon entry of the template they are able to unstack the single strand at position +3 [20] (Fig. 5a). The non-conserved structural elements of the fingers subdomain play a role in RNA binding as well [17, 18, 20]. In particular, the fingertips of dsRNA and some +RNA virus RdRps allow the polymerase to “hold” the template without extensive conformational changes [18, 28]. The variations and extensions in the fingers subdomain have also been shown to play roles in protein–protein interactions, phosphorylation, oligomerisation, and nuclear import [24, 29–31]. In contrast, the HIV-1 RT lacks such extensions and adopts a more “open-hand” or U-shaped binding cleft [21] (Fig. 4c). As a consequence, the RT structure must rearrange its subdomains to coordinate the binding of the template, nascent strand, and incoming dNTPs.
Fig. 5.
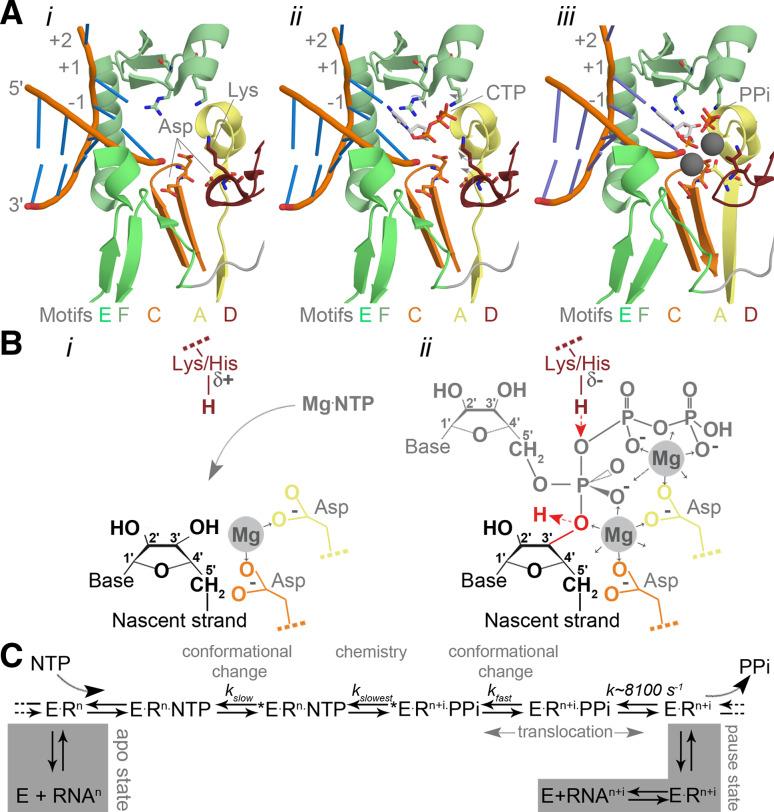
Catalysis in the RNA virus polymerase active site. a Structure of the PV active site as it moves from a native state or elongation complex (i) to an open complex (ii), and a closed complex (iii). The closed complex depicted here shows the active site after catalysis. Highlighted are the metal-binding aspartates of motifs A and C, and the lysine of motif D that acts as general acid. Colour coding by motif as in Fig. 3. Image based on PDB accessions 3Ol6, 3OLB, and 3OL7. b Schematic presentation of the RdRp active site. The aspartates (Asp) of motif A (yellow) and C (orange) bind divalent metal ions (marked Mg and shaded grey), which are used to coordinate the formation of a new phosphodiester bond at the 3′-OH (red in panel ii) of the nascent strand (yellow). The general acid (red Lys/His in panel ii) is positioned near the β-phosphate of the incoming NTP to protonate the PPi leaving group. c Simplified schematic of the kinetic steps of RNA polymerases. Asterisk indicates closed complex
The NTP and template entry channels meet at the palm subdomain (Fig. 3a). This is a structure that is comprised of a central, partially formed three-stranded β-sheet, which is also present in RNA-recognition motifs (RRMs). The relative movement of these strands within the RRM is vital to catalysis and dependent on NTP binding. Only when a correct NTP binds can motif A and motif C align and the RRM fold be completed [20] (Fig. 5a). This catalytically competent conformation of the active site is often referred to as the closed complex (not to be confused with the “closed-hand”, which refers to the overall structure of the RdRp) [20].
Function of the conserved aspartates and lysine in the polymerase catalytic site
The palm subdomain motifs A, C and D are the most essential elements in the RNA polymerase domain. This is particularly evident from the strict conservation of the N-terminal aspartates in the Dx4–5D and xDD consensus sequences (here ‘x’ represents any residue) of motifs A and C (Fig. 2). Mutational analysis of these residues has shown that their absence abolishes or greatly alters RNA-dependent polymerase activity in vitro and in vivo [32–36]. This is in line with structural studies showing that the N-terminal aspartates coordinate two divalent metal ions that are crucial for polymerase function [17–20] (Fig. 5a, B). In contrast, motif D typically contains a sole lysine or histidine as signature amino acid [37, 38]. This residue is likely protonated in the environment of the active site and its mutation strongly reduces the activity of the RNA polymerase [37, 39–41]. Moreover, an arginine to lysine mutation at motif D’s position in the polymerase of a Hepatitis C virus (HCV) strain increases the activity of this RdRp [42].
The polymerase reaction creates new phosphodiester bonds between NTPs using RNA as template. The NTP substrates involved in this reaction are coordinated by two metal ions, which are bound by the conserved aspartates of motifs A and C (Fig. 5a, panel i). They also position the NTP’s triphosphate optimally for attack by the sugar moiety of the nascent strand once its 3′ carbon has lost a proton [17–20] (Fig. 5b). The N-terminal aspartate of motif C thus uses a metal ion to fix the α-phosphate of the incoming NTP and reduce the pKa of the nascent RNA’s 3′-OH to facilitate the attack (Fig. 5b, panel ii) [43]. The amino-terminal carboxylate of motif A, on the other hand, stabilises the β- and γ-phosphates with its divalent metal as well as the pentacovalent (phosphorane) intermediate that forms during catalysis (Fig. 5b, panel ii). Structural and biochemical analyses have shown that the formation of this transient structure is dependent on the attack of the NTP’s α-phosphate by the 3′-OH, which is often the rate-limiting catalytic step in NTP condensation [37, 43] (Fig. 5b).
Motif D’s lysine or histidine assists the N-terminal aspartate of motif A in coordinating the β-phosphate of the incoming NTP, analogous to the trigger-loop in DdDps [38, 44]. However, when the positively charged side chain of motif D approaches the β-phosphate, it can protonate the PPi leaving group as well (Fig. 5b, panel ii). This second protonation step in the active site is not essential for the polymerase reaction, since catalysis can still take place when motif D’s lysine has been mutated to a residue with a different pKa. The polymerase reaction rate will nevertheless be 50- to 1,000-fold higher when a lysine or histidine is present [38, 45]. Recent data even suggests that motif D can coordinate the export of the PPi group from the active site once catalysis has taken place [45], thereby triggering the translocation of the RNA.
Motifs and structural flexibility are important for nucleotide selection
Biochemical and structural analyses have shown that polymerase catalysis is a multi-step process that includes at least one NTP recognition event (Fig. 5C). This step must ensure that the catalytic residues and the substrate are not placed in a catalytically competent position before a correct NTP is bound. In part, this is achieved by motif D, which needs to bind the β-phosphate of the incoming NTP to trigger active site closure (Fig. 5a, b). Crucially, due to its highly flexible nature it can only adopt this position when a correct NTP is bound [44–46] and it is now believed that getting motif D near the β-phosphate is the second most rate-limiting step (k slow) in the RNA polymerase catalytic cycle after chemistry (Fig. 5c).
Motif D’s conformational change is thus the final checkpoint in the NTP selection process and other motifs facilitate the steps that lead up to this event. Subtle movements in motif B, for instance, help position the template and the incoming NTP via direct interactions between the base and conserved residues in the N-terminus of motif B [20, 47]. Motif F, on the other hand, contributes several evenly spaced, positively charged amino acids that coordinate the negatively charged triphosphate of the NTP (Fig. 5a) [17–20]. In the case of HIV-1 RT, the binding of a dNTP also triggers a rotation of the motif F-containing homomorph towards the polymerase active site [21]. As this motion creates a relatively tight fit between active site, dNTP, and the basic residues of motif F, only the binding of the correct dNTP can trigger it [21].
In +RNA RdRps, selection between NTPs and dNTPs is supported by motif B’s conserved asparagine and the C-terminal aspartate of motif A (Fig. 2). Both form a crucial hydrogen bond with the 2′-OH of the incoming NTP [48] and mutation of conserved motif B residues cripples polymerase activity [42, 49]. In dsRNA RdRps, residues with equivalent functions are an aspartate that is positioned just N-terminal of motif A and a conserved serine in motif B [17]. In the HIV-1 RT, on the other hand, these residues are a tyrosine just N-terminal of motif A and, most frequently, a phenylalanine at the same position as the +RNA virus asparagine, since their side chains are better suited to facilitate a hydrophobic interaction with the 2′-H of the incoming dNTP [21].
Beyond the conserved core
Additional enzymatic domains
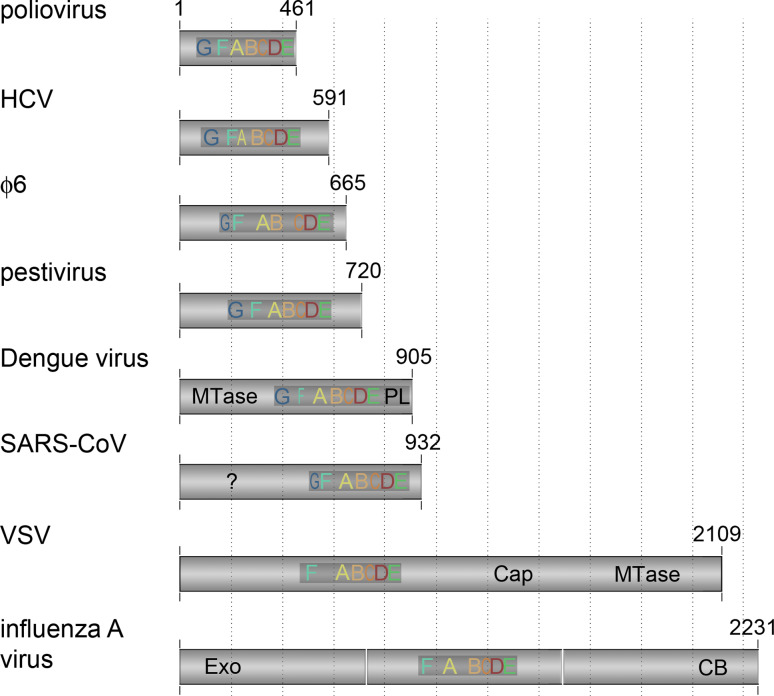
Additional domains frequently flank the polymerase domain (Fig. 6). Good examples are the polymerase subunits of the Bunya- and Arenaviridae, which contain both an N-terminal endonuclease domain as well as a C-terminal polymerase domain [50, 51]. An even more striking example is the Vesicular stomatitis virus (VSV) RdRp (Fig. 6). In electron microscopy (EM) images, this enzyme appears as a central density that is surrounded by globular appendages [52], which harbour enzymatic activities that are required for viral mRNA5’ cap synthesis: a 2′-O-methyltransferase (MTase), a guanine-N7-MTase, and a polyribonucleotidyl-transferase (PRNTase) [53, 54]. Such activities can also be found in the RdRps of Flavivirus genus members, albeit with a guanylyltransferase (GTase) instead of a PRNTase [55, 56]. As is shown in the inset of Fig. 4b, the MTase of NS5 is connected to the polymerase domain via a flexible linker. It is likely that the discrete domains of other RNA virus polymerases are connected via such linker sequences as well [57]. The benefit of such flexible attachments is that it allows domains to work in trans or even separately from the polymerase domain, as was demonstrated for the MTase and endonuclease domains of the Sendai virus (SeV) and Lymphocytic choriomeningitis virus (LCV) RdRps [50, 58]. Moreover, accessory domains can sometimes also be deleted from the polymerase subunit without disrupting the activity of the polymerase domain [24, 41].
Fig. 6.
Sizes and additional domains of RNA virus polymerases. The RNA-dependent polymerase domain consists of approximately 400 amino acids. The size of the rest of the subunit varies significantly among RNA-dependent polymerases, as does the conservation of motifs F and G. The additional sequences of the polymerase subunit often contain functional domains, such as a methyltransferase (MTase), exonuclease (Exo), capping domain (Cap), or cap-binding domain (CB). The SAR-CoV RdRp has an additional N-terminal domain that is conserved, but to which no function has been ascribed yet (?). Figure based on alignments from references [15, 39, 83, 177, 178]
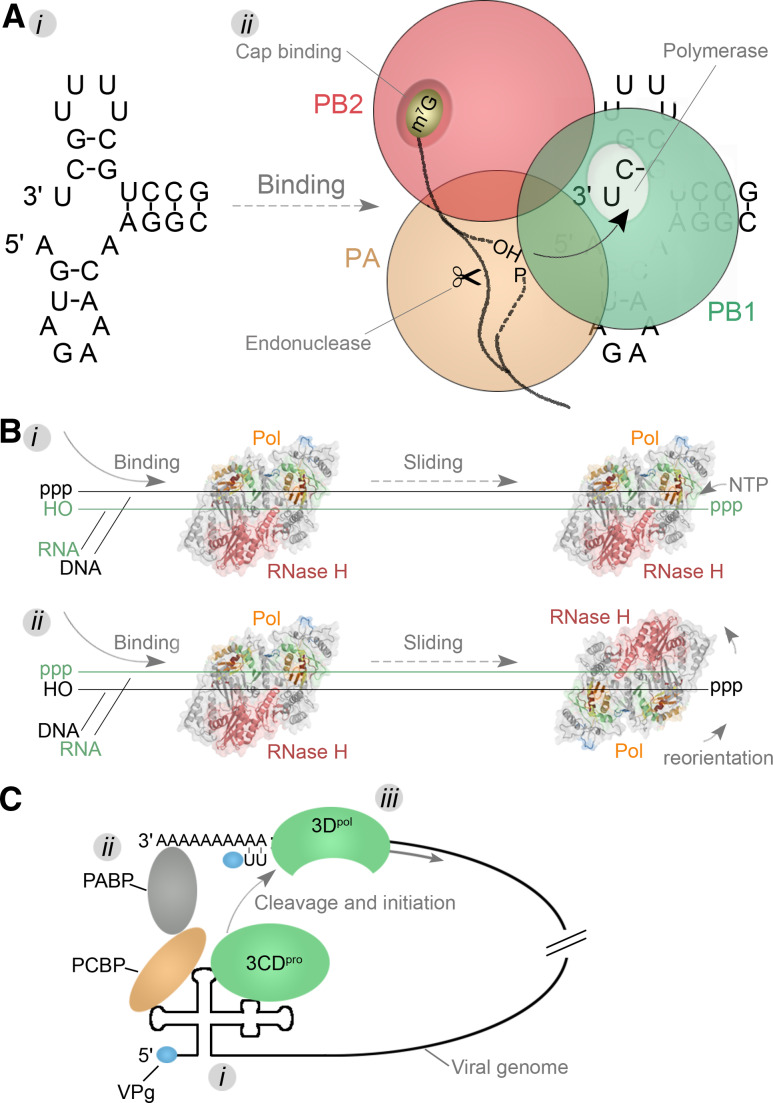
The IAV genome, in contrast, encodes an RdRp in which its multiple activities are separated out into three individual polypeptides, called PA, PB1, and PB2 (Figs. 4, 6). The three subunits are encoded by three negatively stranded genome segments (vRNAs), which are replicated and transcribed in the nucleus of the host in complexes (vRNPs) of heterotrimeric polymerase and a helical assembly of nucleoprotein (NP) monomers (Fig. 1). The RdRp motifs F–E are housed in PB1 [59, 60], while a metal-dependent endonuclease site is present in PA and a cap-binding domain resides in PB2 (Fig. 6). It is now widely accepted that IAV RNA synthesis initiates replication de novo, whereas transcription initiates in a primer-dependent manner (see Sect. “Template recognition, initiation, elongation and regulation” for details). In the latter scenario, a 5′ capped pre-mRNA molecule is bound by the PB2 subunit (Fig. 7a). The pre-mRNA is then cleaved by the PA subunit 9–15 nt downstream of the cap and moved into the PB1 active site for template-dependent extension [59, 60]. How the molecular hand-off is performed and the 3′-OH of the capped primer transferred to PB1 is presently unknown.
Fig. 7.
Template recognition mechanisms. a i The IAV RNA promoter forms through hybridization of the terminal ends of a viral genome segment. ii The structure is next bound by the heterotrimeric RdRp complex that consists of subunits PB1 (green), PB2 (red), and PA (orange). During transcription, the PB2 subunit binds the cap of pre-mRNAs. These pre-mRNAs are next cleaved by the PA subunit and transferred to the PB1 subunit. Here nucleotide condensation takes place on the 3′-OH of the capped primers. b The HIV-1 RT is comprised of the subunits p66 and p51, of which only the p66 subunit has an active polymerase (colour coded as Fig. 4) and RNase domain (pink). The enzyme can bind to the template in a random orientation and switch between the activities by tumbling around its axis once a polymerase (i) or RNase (ii) substrate has been encountered. c A model for the circularisation of the PV genome. First viral proteins and the 3CDpro cleavage intermediate bind to conserved 5′ structures on the genome (i). Next a protein bridge is formed that brings together the 5′ and 3′-ends of the genome (ii). This step subsequently ensures that the viral protease is activated and the viral polymerase 3Dpol is released in close proximity to the 3′-end of the viral genome. Here it can initiate -RNA synthesis using VPg as protein primer
Such a hand-off between enzymatic domains has been studied for the HIV-1 RT. This enzyme is effectively a dual-polymerase complex that consists of the viral p66 subunit, which contains an RNase H domain and a polymerase domain [61], and its cleavage product p51, which lacks the former domain [62] (Fig. 4c). During retrotranscription, the two active sites in the enzyme must catalyse a multi-step process, including (1) RNA-templated cDNA synthesis using a lys3 tRNA as primer, (2) dsDNA synthesis using the cDNA strand as template, and (3) hydrolysis of the incorporated lys3 RNA primer sequences [63] (Fig. 1b). Although the two enzymatic activities of RT can work in trans [64], fluorescence resonance energy transfer (FRET) assays demonstrated that the enzyme can flip along its axis and switch between the activities (Fig. 7b). This principle allows RNA hydrolysis and NTP condensation to effectively work in cis, irrespective of how RT initially bound [65, 66]. Interestingly, such an apparent dissociation-rebinding event is in contrast with the intramolecular translocation of the nascent strand between the polymerase and exonuclease sites in DdDps [67, 68]. It will, therefore, be interesting to see how other viral RNA-dependent polymerases orchestrate a hand-off between active sites.
Multimerisation
Oligomerisation of RdRps has been reported for Poliovirus (PV), FMDV, HCV, Norovirus (NV), and IBDV enzymes. It can result in cooperative template binding, lattice formation, and a stimulation of viral RNA polymerase activity in vitro [69–72], which in HCV appears to be specific for initiation [73]. Also inactive RdRps can stimulate activity and participate in array formation, which led to the hypothesis that multimerisation evolved to stabilise polymerases [69]. Mutagenesis studies suggest that the dimerisation of the HCV RdRp is mediated by the thumb domain [74], whereas in PV 3Dpol residues in N-terminal part of the polymerase domain and two aspartates of the so-called interface I—an interaction interface between the thumb subdomain of one RdRp and the palm of another—play a role [46, 69]. Mutation of the interacting residues also results in viral growth defects [75].
Polymerase multimerisation may also play a role in IAV infections [76]. In the replication cycle of this −RNA virus, the viral RdRp synthesises new vRNAs through a complementary +RNA (cRNA) intermediate, which is bound by newly produced viral RdRp and NP to form cRNPs (Fig. 1). Crucially, during the synthesis of vRNAs, the cRNP can be stimulated by both active as well as inactive secondary RdRps [77], which suggests that cooperation takes place. In contrast, experiments with vRNPs suggest that secondary −RNA virus polymerases are trans-acting and only able to complement the activity of the resident RdRp during the synthesis of cRNA copies [78, 79]. Curiously, no evidence has been found for either cooperation or trans-acting activity during transcription, which suggests that cap-snatching, mRNA synthesis, and polyadenylation all occur in cis [78] and putatively without multimerisation. It is presently not known how any of the two replication-specific interactions between the IAV polymerases are mediated.
In contrast to the above examples of multimerisation, which all involve multiple copies of essentially the same polypeptide, the positive-stranded CoVs have been hypothesised to use two distinct polymerase activities for their RNA synthesis [80]. The CoVs are well known for their uniquely large ~30-kb +RNA genomes, of which approximately two-thirds code for numerous viral replicase functions that associate with cellular membranes upon translation [81] (Fig. 1). Nearly three decades ago, bioinformatics analyses predicted that a canonical RNA polymerase domain resided among these replicase functions in a 932 amino acid-long mature protein [82] (Fig. 6). Since then, biochemical experiments of the CoV replicase protein have not only confirmed this prediction [33, 83], but also revealed a non-canonical, multimeric RdRp that is able to recognise the 3′ terminus of the viral genome [84, 85]. It has been proposed that these two enzymes may cooperate to improve or to achieve better control over initiation and elongation, putatively functioning as primase and primer-dependent polymerase [80].
Template recognition, initiation, elongation and regulation
Template recognition
The association between the heterotrimeric IAV polymerase and viral RNA can be solely achieved through interactions with a partially complementary RNA promoter, which consists of the conserved 5′ and 3′ terminal sequences that are present at each genome segment or complementary genome segment [86]. A similar sequence element and one that is also based on complementarity can be found in the genomic ends of the genomes of the Arena- and Bunyaviridae [87]. It is currently understood that the IAV polymerase first associates with the 5′ end of the promoter, before the 3′ terminus is bound [88–90]. Mutagenesis data suggest that the binding process creates structural rearrangements in the promoter, changing its conformation to a cork-screw-like structure (Fig. 7a). It is assumed that conformation directs the viral transcription and replication activity of the polymerase [91–93]. Other experiments have shown that the promoter is also vital for the packaging of viral genome segments into virions [94] and potentially in overcoming host restrictions [95].
In Flaviviridae, RdRp recruitment and binding are partially guided by circularisation of the genome. RNA structures, such as the stem-loop/promoter structure in the 5′ UTR of the genome, are strongly involved in the initiation of replication as well [96, 97]. The involvement of the 5′ UTR can be explained by the interaction it establishes with the conserved elements of the 3′ UTR, thereby circularising the genome akin to the segmented −RNA viruses. Such a long-range interaction can also effectively position the conserved 3′-terminal sequence 5′-CUOH opposite the 5′-terminal end [96, 98–100], which would facilitate recognition of the 3′-terminal end and minimise initiation on non-viral templates [101].
In contrast, the plant virus Brome mosaic virus (BMV) requires an additional viral protein to bring the viral 2a polymerase and genome together. The interaction is mediated by protein 1a, which can bind both RdRp and conserved so-called box B motifs in the viral RNA [102]. The helicase domain of 1a may also stimulate RdRp initiation [103], which is believed to take place on a conserved 3′-terminal cytosine during replication and an internal cytosine during subgenomic transcription [104]. The involvement of conserved (3′-terminal) cytosine residues appears to be a recurring theme in RNA virus replication or transcription, as they are also used for initiation by the Reovirus (ReoV), Machupo virus (MACV), and the Equine arteritis virus (EAV) [17, 105, 106].
In infections with members of the Picornaviridae, both circularisation and the use of viral protein factors can be observed (Fig. 7c). Here the 5′ structures in the genome present a binding platform for viral and cellular proteins. The most downstream of the these 5′ elements is currently believed to serve as promoter for both +RNA and −RNA synthesis [107] (Fig. 7c, marked i), while the upstream cloverleaf structure functions as binding site for the cellular poly(rC)-binding protein (PCBP) and the viral 3CDpro polyprotein cleavage intermediate [108–110]. To setup PV 3Dpol for RNA synthesis, the various proteins already associated with the viral genome must first bind the C-terminal part of the cellular poly(A) binding protein (PABP) [111], which effectively circularises the viral genome (Fig. 7c, marked ii). In turn, the circularisation stimulates the self-cleavage of the 3CDpro precursor and thereby the activation of the RdRp 3Dpol (Fig. 7c, marked iii), whose activity is inhibited as long as its N-terminus has not been processed [112, 113]. Typically the RdRp next initiates on the cis-replication element (CRE) in the viral genome [114].
De novo initiation
In spite of the wide range of replication strategies used by RNA viruses, their polymerases can use only two mechanisms to initiate the synthesis of RNA: de novo initiation or primer-dependent initiation (Fig. 8). The de novo initiation mechanism can be subdivided in 3′ terminal initiation or internal initiation. Some polymerases can also use a combination of both which is called prime-realign initiation [91, 115]. The advantage of the latter strategy is that no genetic information is lost, since internal residues are better protected from cellular exonucleases.
Fig. 8.
Modes of initiation. schematic presentation of a de novo initiation, b primer-dependent initiation, and c protein-dependent initiation. In the latter scenario, the tyrosine residue that is present at position 3 of VPg is used as primer
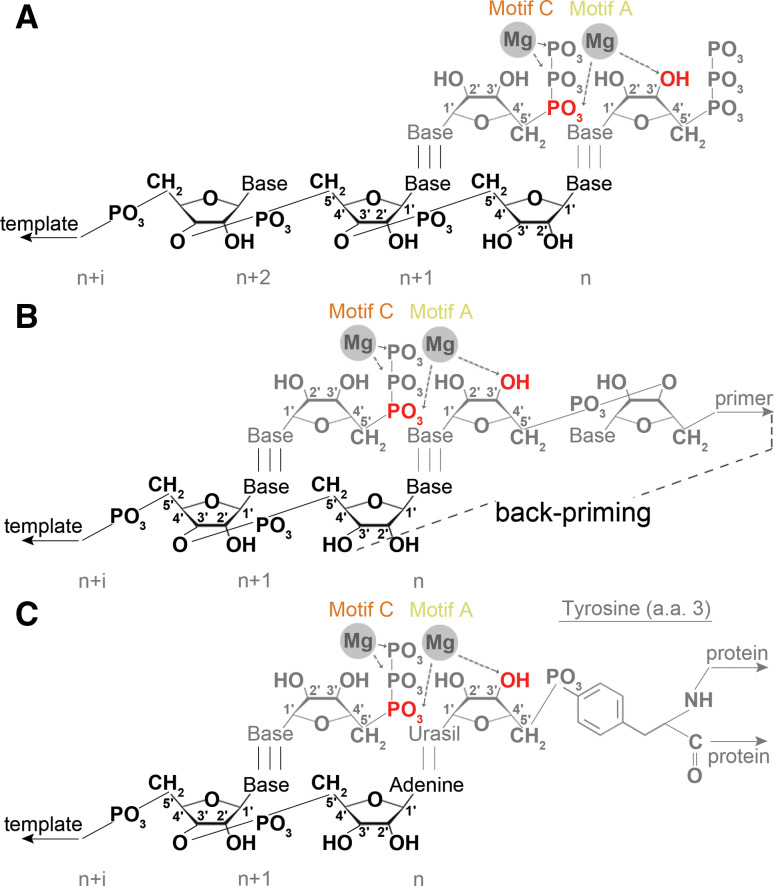
During de novo initiation, an initiating nucleotide serves as primer for a second nucleotide. These two NTPs are base paired with positions +1 and +2 of the template, respectively (Fig. 8a). Since base pairing with the template is not sufficient to hold both NTPs correctly positioned for catalysis, polymerases can also employ a number of techniques to stabilise the interaction. The ReoV RdRp λ3 uses a loop that protrudes from the palm domain to bind the negative triphosphate backbone of the NTP at position +1, which is typically GTP [17]. Other residues in the active site, most notably an arginine (R518) in motif F and a serine (S682) in motif B, form hydrogen bonds with the carbonyl and NH group of this GTP [17], while a conserved glutamine in motif C binds the 2′-OH of the GTP’s ribose [17]. Together these interactions not only stabilise the initiating GTP, but also provide nucleoside selection to keep the 3′-terminal cytosine present in both the plus and minus strand of ReoV RNAs preserved.
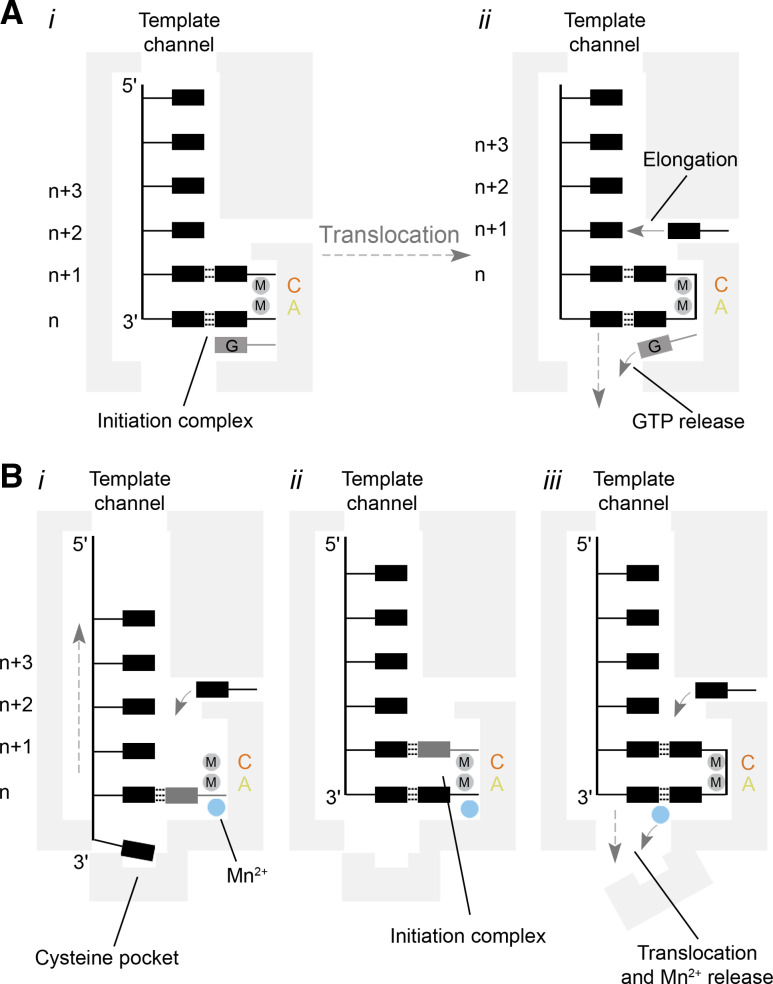
Analogously, in some +RNA virus RdRps a short polypeptide loop can protrude from the thumb domain into the active site to perform these functions [24, 25, 116]. In the Bovine viral diarrhoea virus (BVDV) RdRp, this so-called primer-loop is assisted with a non-templated GTP molecule (Fig. 9A). Positioned in a −1 position ~6 Å away from the catalytic site, the additional GTP creates a stacking platform for NTPs that bind at positions +1 and +2 [24] (Fig. 9a). In BVDV, the N1 and N2 of the guanine are bound by the carbonyl of a motif A threonine, while the hydrophobic proline and leucine residues in motif A and the thumb subdomain stabilise the guanine base. In addition, positively charged residues in motif E stabilise the triphosphate backbone of the GTP [24]. GTP also plays a role in de novo initiation of the JEV polymerase, as demonstrated by Surana et al. [117]. However, in contrast to its role in BVDV, the GTP here binds in an orientation that can prevent template binding. Addition of manganese can resolve this blocked pre-initiation state and it is possible that this mechanism evolved to limit non-templated RNA synthesis [117].
Fig. 9.
Models for template recognition and de novo initiation by the BVDV and ϕ6 polymerases. a Model of de novo RNA synthesis by a BVDV RdRp. The RdRp binds to the 3′-end of the viral genome (i). Then the template, initiating NTPs, and an additional GTP form an initiation complex that is stabilised by the closed C-terminal loop. After the first polymerase reaction, the GTP is released by opening of the C-terminal loop and translocation of the template–nascent strand duplex (ii). A new NTP can enter the active site via the NTP channel to start elongation. b Simplified model of de novo RNA synthesis by the ϕ6 RdRp. Template recognition occurs through the binding of the 3′-cytosine of the genome to a pyrimidine specificity pocket (i). At the same time, a nucleotide base-pairs with the second base of the template. Its triphosphate group is stabilised by a manganese ion that is coordinated by the palm domain. The 3′-cytosine next translocates to the n-position to base-pair with a second NTP (ii). After nucleotide condensation, the manganese ion triggers a rearrangement of the C-terminal and is subsequently released (iii). In contrast to the GTP in the NS5 RdRp, the manganese ion must be rebound for subsequent elongation
The RdRp of ϕ6 stabilises the initiating nucleotides with an additional, small C-terminal domain in at least three ways. First, it blocks the RNA exit channel, which fixes the template strand, not unlike the priming loop in other RdRps (Fig. 9b) [18]. A Mn2+ that is bound ~6 Å away from the catalytic site by residues in the palm subdomain also plays a role in template binding [18]. Second, it provides a small, pyrimidine-sized specificity pocket in which a glutamate hydrogen bonds with the NH2 of the 3′-terminal cytosine that is present in all ϕ6 genome segments (Fig. 9b, panel i). This also means that an incoming NTP first base pairs with position +2 of the template and that position +1 can be occupied when the template ratchets back from the cytosine pocket (Fig. 9b, panel ii) [18]. Third and last, the C-terminal domain contains a tyrosine (or tryptophan) on which the initiating GTP can stack and stably base pair with the conserved 3′-terminal C at the +1 site [18]. Crucially, after the formation of the dinucleotide, the C-terminal domain has to be displaced to allow RNA translocation and current evidence suggests that the non-catalytic Mn2+ is also involved in this step [26]. A non-catalytic Mn2+ has also been observed in other RdRps [118].
The RdRps encoded by some -RNA viruses, including the heterotrimeric RdRp of IAV, can perform both de novo initiation as well as primer-dependent initiation. The de novo mechanism is used to start the synthesis of cRNAs from vRNAs (Fig. 1). In turn, these cRNAs can be used as template for the production of new vRNAs. This latter process utilises a prime-realign mechanism that depends on high concentrations of initiating NTPs [91]. These initiating NTPs base pair with positions +4 and +5 of the cRNA 3′ terminus [91] and then translocate to bases +1 and +2 after the first nucleotide condensation as dinucleotide pppApG. Although it has been shown that the structure of the viral promoter and its sequence play an important role in this mechanism [91], it is presently unknown which residues of the −RNA virus RdRp are involved in stabilising the first nucleotide bond. It is also unknown how the pppApG is transferred to the 3′ terminus of the cRNA template.
Primer-dependent initiation
Primer-dependent initiation can be categorised in oligonucleotide-primed, protein-primed, and back-primed initiation, in which the 3′ terminus of the template forms a small hairpin (Fig. 8b, c). Since the use of a primed template requires a template channel that can accommodate several base pairs of A-form dsRNA, the Corona- and Picornaviridae polymerases such as PV 3Dpol and SARS-CoV nsp12 lack the palm or thumb subdomain protrusions found in most Flaviviridae and dsRNA viruses [33, 83, 119, 120]. Consistent with this observation, it was found that the HCV and ϕ6 polymerases can be changed into primer-dependent polymerases by mutating or deleting their de novo initiation platforms [121–123].
Although the PV polymerase 3Dpol can initiate on partial dsRNA, it uses a protein primer in vivo. Following its activation through the self-cleavage of 3CD (Fig. 7c), 3Dpol uridylates the PV protein VPg on the tyrosine 3 (Fig. 8c) using a back-slide mechanism on adenosine residues [124]. This creates a VPg(pUpUOH) molecule that can hybridise to the viral 3′ polyA tail and prime the synthesis of a complementary strand (Fig. 7c). A study of the related FMDV RdRp has shown that the terminal uridylate of this product can enter the active site and prime a duplication of the genomic 3′ end [125]. Furthermore, VPg(pUpUOH) can anneal to the 3′-terminal adenosine residues of the negative strand and thereby prime +RNA synthesis as well [107, 126, 127].
The Orthomyxo-, Arena-, and Bunyaviridae use a primer-dependent initiation mechanism for transcription. In IAV, this process begins with the binding of a 5′ capped pre-mRNA molecule by the PB2 subunit (Fig. 7a, panel ii) [128, 129]. A crystal structure of the IAV PB2 subunit with cap analogue m7GTP demonstrated that its structure is similar though different from other known cap-binding proteins [128]. It coordinates binding via histidine and lysine residues that hydrogen bond with the 2′ OH of the ribose, a histidine and asparagine that coordinate the phosphates, and histidine, phenylalanine and lysine residues that sandwich the m7-methylated guanine base [128, 130]. After cap binding, the PA subunit cleaves the pre-mRNA 9–15 nt downstream of the cap with a strong preference for cleaving 3′ of guanine bases [59, 131] (Fig. 7a, panel ii). The endonuclease domain of the L-protein of Bunyaviridae shows a strong similarity to the IAV PA subunit [50, 51]. How the 3′-OH of the capped primer is transferred to the polymerase domain in the L-proteins of the Arena- and Bunyaviridae and the PB1 active site in IAV is presently unknown.
Motion and fidelity
Rates of motion within the polymerase structure
Several steps need to occur to allow the RNA-dependent polymerase to transition from an apo to a catalytically active state (Fig. 5c). The first takes place when the polymerase binds the template RNA. To facilitate RNA binding, the template channel has to undergo structural rearrangements. The thumb domain of the HIV-1 RT, for instance, has to move 20° relative to its position in the apo structure [132], whereas the HCV RdRp needs to contract its template channel from a width of 20.5 to 12.2 Å to interact with ssRNA [133]. In strong contrast, primer-dependent RdRps need to expand their template channel from a constricted apo form to one that can accommodate dsRNA [20, 27, 134]. In the PV RdRp this means a change from 15.7 to 18.7 Å [113]. Present evidence also suggests that the inherently flexible template channel of viral RNA-dependent polymerases can sample its conformational space rapidly [27, 133, 135], which thus makes it a very dynamic structure in apo form and able to respond quickly to an encounter with RNA.
The open complex that is formed after template binding is highly selective for base-pairing [20], in part due to motif D (see Sect. “The RNA-dependent polymerase domain”). In +RNA virus RdRps, correct nucleotide binding triggers the alignment of motif A to motif C and forces the N-terminal aspartate of motif A to bind metal ions [20]. This ostensibly subtle change completes the RRM-fold of the palm subdomain (the closed complex, Fig. 5a) and optimally positions the NTP for ribose binding by motifs A and B [20]. Interestingly, in some RNA polymerases the formation of the closed complex is fundamentally different. The HIV-1 RT, for instance, relies on large finger domain movements to close the active site and position the nascent RNA into the catalytic site, similar to DdDps [20, 132]. In the RdRps of dsRNA viruses, the RRM fold is already fully structured in the apo form, which means that only minor rearrangements of the side-chains are required to close their active site [17, 18].
The rate at which the residues in the active site sample their space is faster (k = 500−800 s−1) than the rate-limiting phosphoryl transfer step (k = 300 s−1) [136] (Fig. 5c, k slowest). Biochemical evidence suggests that the rate with which motif D takes the polymerase-RNA-NTP complex to a closed state is k = 520 s−1 [137], which is only somewhat faster than chemistry (Fig. 5c, k slow). So far the conformational change of motif D has only been observed with NMR [44], because it has proved too transient for crystallography. It is likely that the pre-catalytic conformational step is also dependent on the stabilisation of motifs A and F [27, 137].
After catalysis, the polymerase reverts to an open complex to allow translocation of the template-nascent strand duplex and pyrophosphate (PPi) release at k = 8,100 s−1 (Fig. 5c) [20, 136]. Viral polymerases that initiate de novo, however, must also transition from an initiation to an elongation conformation. As discussed in Sect. “Template recognition, initiation, elongation and regulation”, this involves rearrangements of several ångström in the thumb domain and the C-terminus of the ϕ6 RdRp [18] (Fig. 9b). Analogously, elements that stabilise the initiation of +RNA viruses such as the priming loop need to undergo a conformational change as well. Furthermore, such RdRps need to expand their template-nascent RNA exit channel again to facilitate the translocation of the now partially dsRNA template [133]. It is possible that such events, which only take place once during the synthesis of an RNA molecule, direct the polymerase briefly into a separate state or pause state (Fig. 5c), possibly akin to the pause states that can be observed during elongation [138]. Currently no experiments, which would likely require single-molecule techniques, have been performed to explore this.
Fidelity
As discussed in Sect. “The RNA-dependent polymerase domain”, polymerase fidelity is largely determined by the conserved motifs of the polymerase domain. Furthermore, in vitro experiments have shown that the incorporation of nucleotides is indeed relatively robust [139, 140]. However, the error rate of RNA virus polymerases is nevertheless several orders of magnitude higher than the mutation rate of DNA genomes [141–143]. Moreover, it is also easily influenced by many environmental factors. The fidelity of the HIV-1 RT, for instance, can decrease 9-fold when the pH of the in vitro reaction is raised from pH 6.5 to pH 8.0 [144]. Similarly, nucleotide analogues can have a dramatic impact on the mutation rate of RNA virus RNA synthesis as well [145], as do various divalent metals [136].
It has been argued that the relative low fidelity of RNA virus polymerases is responsible for the ability of RNA viruses to overcome bottlenecks imposed by host defences and antivirals. Indeed, RNA virus strains that contained high-fidelity polymerase were attenuated in vivo [46, 146]. Moreover, the growth of IAV, PV, FMDV, Chikungunya virus (CHIKV), and Human enterovirus 71 (EV71) in the presence of the nucleotide analogues such as ribavirin or 5-fluorouracil readily leads to the appearance resistance after several passages [147–151]. It is likely that the resistance mutations alter the positioning and dynamics of the motifs involved in nucleotide selection, which in turn optimises the RdRp’s ability to discriminate between NTPs [27].
In spite of the typically high error rate of viral RNA synthesis, dramatic mutations or deletions can be repaired. It was shown, for instance, that the RdRp of Turnip crinkle virus (TCV) is able to repair mutations in the 5′-CCUGCCCOH sequence of the 3′-end of its genome using a 3-step process. Counter intuitively, the first step in this process is the non-templated synthesis of an RNA primer, which can next be aligned to the 3′-end and extended in a primer-dependent fashion [152]. A variant of this repair mechanism has been observed for the DV RdRp, provided that deletions were smaller than 6 nt and did not impair circularisation of the genome [153]. Although the use of non-templated nucleotide condensation for repair seems paradoxical, since the reaction mostly creates a pool of quasispecies, it is possible that the wild-type sequence is eventually ‘filtered’ back from the quasispecies population [153]. As non-templated polymerisation has been observed for −RNA, dsRNA, and +RNA RdRps [154–157]—in some cases for polyA tailing of viral transcripts [155]—it will be interesting to see if the conserved mechanism is universally used for template repair.
Excision reaction
Although RNA virus polymerases do not contain a separate 3′-to-5′ exonuclease activity, they are able to perform a reversal of the polymerase reaction (Fig. 5c) that is typically referred to as pyrophosphorolysis. Hence, using PPi and the same active site residues as those involved in nucleotide condensation, 3′ nucleoside monophosphates can be removed from the nascent strand, including misincorporated nucleotides or nucleoside analogues such as lamivudine (3TC), zidovudine (AZT), and 3′-dNTPs [158–160]. Crucially, chain-terminated complexes are stable in the presence of heparin, suggesting that pyrophosphorolysis typically occurs in cis [160].
Some RNA virus polymerases can substitute PPi for an NTP molecule (typically ATP) [159]. These triphosphates are accommodated by an additional nucleoside binding pocket, which orients the γ-phosphate of the NTP towards the active site to accept the leaving monophosphate group [159]. In contrast to normal pyrophosphorolysis, this reaction results in the formation of dinucleoside tetraphosphate molecule. Through mutation of the NTP binding pocket, the NTP-dependent mechanism of excision can evolve into a mechanism that confers resistance to nucleoside analogues [132, 159, 161].
Unknowns and closing remarks
The RdRp ensures that the viral genetic code is replicated and transcribed, but only for some viruses it is understood how these two processes are separated. In infections with Alphaviridae, for instance, the switch between the two processes is largely conducted by the viral protease, since incompletely processed polyproteins only support replication [162]. In contrast, it is still unclear what dedicates the IAV RdRp to either replication or transcription [163]. Various co-factors have been discovered that bias the activity of the RdRp towards one or the other pathway, including other viral proteins [164] and small viral RNAs (svRNA) [165]. In Nidoviridae, the regulation of transcription and replication also has to account for the synthesis of both full-length and multiple subgenome-length mRNAs [166]. Again, additional viral proteins have been proposed to guide this process, such as nsp1 of EAV [167], but the molecular details are still unclear.
Ultimately, our knowledge of RNA virus replication should lead to the discovery and development of new RNA virus inhibitors. A promising new inhibitor that has already been shown to affect the replication of many RNA viruses, including IAV, PA, WNV, YFV, and MACV, is the nucleotide analogue Favipiravir (T-705) [168]. Specific virus-specific inhibitors have also been found based on new polymerase structures. Co-crystallisation of IAV PB1 and PB2, for instance, has let to the discovery of peptides that interfere with RdRp assembly [169, 170]. In addition, screening assays are rapidly becoming invaluable for drug discovery and have led, for instance, to the identification of compounds that act as in vitro inhibitors of the IAV endonuclease activity [171]. Research has also demonstrated that general polymerase principles exist that may inspire strategies for a rapid and standardised design of live, attenuated vaccine strains [44]. In part, this latter insight relies on polymerase dynamics, which is also pointing to new druggable sites elsewhere in the structure [27, 133, 172].
In conclusion, it is clear that the RNA replication and transcription strategies vary greatly among RNA viruses. But many of the biochemical and biophysical properties of the polymerase domain have been uncovered, allowing for the development of novel antiviral strategies. Fundamental research into RNA-dependent polymerases is thus proving to be invaluable, and will likely continue to be, in combating current and emerging RNA virus infections.
Acknowledgments
Apologies to colleagues whose work could not be cited due to length restrictions. This work was supported by Grants 021.001.037 and 825.11.029 from the Netherlands Organization for Scientific Research (NWO), Grant 098721/Z/12/Z from the Wellcome Trust and a Kemp postdoctoral fellowship from Lincoln College Oxford. Dr. Eric Snijder, Dr. Ervin Fodor, and Dr. Frank Vreede are kindly acknowledged for helpful comments and suggestions.
References
- 1.Butel JS. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraaij PL, Bodewes R, Osterhaus AD, Rimmelzwaan GF. The ins and outs of universal childhood influenza vaccination. Future Microbiol. 2011;6:1171–1184. doi: 10.2217/fmb.11.106. [DOI] [PubMed] [Google Scholar]
- 4.Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel MM, Glass R, Desai R, Tate JE, Parashar UD. Fulfilling the promise of rotavirus vaccines: how far have we come since licensure? Lancet Infect Dis. 2012;12:561–570. doi: 10.1016/S1473-3099(12)70029-4. [DOI] [PubMed] [Google Scholar]
- 6.Kyle JL, Harris E. Global spread and persistence of dengue. Ann Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 7.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Micro. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, Alabdullatif ZN, Assad M, Almulhim A, Makhdoom H, Madani H, Alhakeem R, Al-Tawfiq JA, Cotten M, Watson SJ, Kellam P, Zumla AI, Memish ZA, KSA MERS-CoV Investigation Team Hospital outbreak of middle east respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliot RM, Brennan B. Emerging pleboviruses. Curr Opin Virol. 2014;5:50–57. doi: 10.1016/j.coviro.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerase. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruenn JA. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 1991;19:217–226. doi: 10.1093/nar/19.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2:a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya AE, Pringle FM, Zeddam J, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya A, Pringle F, Zeddam J, Luke B, Cameron C, Kalmakoff J, Hanzlik T, Gordon K, Ward V. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang DM, Zemla AT, Zhou CL. Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res. 2013;41:1464–1482. doi: 10.1093/nar/gks1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen JL, Long AM, Schultz SC. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 17.Tao L, Farsetta DL, Nibert ML, Harrison SC. RNA Synthesis in a Cage—structural studies of reovirus polymerase λ3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 18.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 19.Bressanelli S, Tomei L, Rey FA, De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong P, Peersen OB. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J, Das K, Hsiou Y, Sarafianos SG, Clark JA, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with double-stranded DNA and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 22.Pan J, Vakharia VN, Tao YJ. The structure of a birnavirus polymerase reveals a distinct active site topology. Proc Natl Acad Sci USA. 2007;104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrer-Orta C, Arias A, Escarmís C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Choi KH, Rossmann MG. RNA-dependent RNA polymerases from Flaviviridae . Curr Opin Struct Biol. 2009;19:746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Yap TL, Xu T, Chen Y-L, Malet H, Egloff M-P, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright S, Poranen MM, Bamford DH, Stuart DI, Grimes JM. Noncatalytic ions direct the RNA-dependent RNA polymerase of bacterial double-stranded RNA virus ϕ6 from de novo initiation to elongation. J Virol. 2012;86:2837–2849. doi: 10.1128/JVI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moustafa IM, Shen H, Morton B, Colina CM, Cameron CE. Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J Mol Biol. 2011;410:159–181. doi: 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 29.Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001;20:1153–1163. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak HB, Ghosh SK, Roberts AW, Sharma SD, Yoder JD, Arnold JJ, Gohara DW, Barton DJ, Paul AV, Cameron CE. Structure–function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol): a surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J Biol Chem. 2002;277:31551–31562. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- 31.Johansson M, Brooks AJ, Jans DA, Vasudevan SG. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-β and the viral helicase, NS3. J Gen Virol. 2001;82:735–745. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- 32.Jablonski SA, Morrow CD. Mutation of the aspartic acid residues of the GDD sequence motif of poliovirus RNA-dependent RNA polymerase results in enzymes with altered metal ion requirements for activity. J Virol. 1995;69:1532–1539. doi: 10.1128/jvi.69.3.1532-1539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.te Velthuis AJ, Arnold JJ, Cameron CE, van den Worm SH, Snijder EJ. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogden KM, Ramanathan HN, Patton JT. Mutational analysis of residues involved in nucleotide and divalent cation stabilization in the rotavirus RNA-dependent RNA polymerase catalytic pocket. Virology. 2012;431:12–20. doi: 10.1016/j.virol.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valverde-Garduño V, Gariglio P, Gutiérrez L. Functional analysis of HIV-1 reverse transcriptase motif C: site-directed mutagenesis and metal cation interaction. J Mol Evol. 1998;47:73–80. doi: 10.1007/pl00006364. [DOI] [PubMed] [Google Scholar]
- 36.Vreede FT, Jung TE, Brownlee GG. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J Virol. 2004;78:9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Götte M, Konigsberg W, Cameron CE. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci USA. 2007;104(11):4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas SK, Nayak DP. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu C, Fan S, Li C, Macken C, Kim JH, Hatta M, Neumann G, Kawaoka Y. Functional analysis of conserved motifs in influenza virus PB1 protein. PLoS One. 2012;7:e36113. doi: 10.1371/journal.pone.0036113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai VC, Kao CC, Ferrari E, Park J, Uss AS, Wright-Minogue J, Hong Z, Lau JY. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J Virol. 1999;73:10129–10136. doi: 10.1128/jvi.73.12.10129-10136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-depedent RNA polymerease and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 44.Yang X, Smidansky ED, Maksimchuk KR, Lum D, Welch JL, Arnold JJ, Cameron CE, Boehr DD. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure. 2012;20:1519–1527. doi: 10.1016/j.str.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen H, Sun H, Li G. What is the role of motif D in the nucleotide incorporation catalyzed by the RNA-dependent RNA polymerase from poliovirus? PLoS Comput Biol. 2012;8:e1002851. doi: 10.1371/journal.pcbi.1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Yang X, Lee CA, Moustafa IM, Smidansky ED, Lum D, Arnold JJ, Cameron CE, Boehr DD. Vaccine-derived mutation in motif D of poliovirus RNA-dependent RNA polymerase lowers nucleotide incorporation fidelity. J Biol Chem. 2013;288:32753–32765. doi: 10.1074/jbc.M113.484428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrer-Orta C, Arias A, Pérez-Luque R, Escarmís C, Domingo E, Verdaguer N. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci USA. 2007;104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J Biol Chem. 2000;275:25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 49.Sankar S, Porter AG. Point mutations which drastically affect the polymerization activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase I. J Biol Chem. 1992;267:10168–10176. [PubMed] [Google Scholar]
- 50.Morin B, Coutard B, Lelke M, Ferron F, Kerber R, Jamal S, Frangeul A, Baronti C, Charrel R, de Lamballerie X, Vonrhein C, Lescar J, Bricogne G, Günther S, Canard B. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 2010;6:e1001038. doi: 10.1371/journal.ppat.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reguera J, Weber F, Cusack S. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 2010;6:e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SP. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Fontaine-Rodriguez EC, Whelan SP. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol. 2005;79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogino T, Yadav SP, Banerjee AK. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 2010;107:3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kranzusch PJ, Whelan SP. Architecture and regulation of negative-strand viral enzymatic machinery. RNA Biol. 2012;9:941–948. doi: 10.4161/rna.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogino T, Kobayashi M, Iwama M, Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J Biol Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- 59.Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 60.Ruigrok RW, Crepin T, Hart DJ, Cusack S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr Opin Struct Biol. 2010;20:104–113. doi: 10.1016/j.sbi.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 61.Hostomsky Z, Hostomska Z, Fu TB, Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J Virol. 1992;66:3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Smerdon SJ, Jäger J, Kohlstaedt LA, Rice PA, Friedman JM, Steitz TA. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc Natl Acad Sci USA. 1991;91:7242–7246. doi: 10.1073/pnas.91.15.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Telesnitsky A, Goff SP. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J. 1993;12:4433–4438. doi: 10.1002/j.1460-2075.1993.tb06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joyce CM. How DNA travels between the separate polymerase and 3′-5′-exonuclease sites of DNA polymerase I (Klenow fragment) J Biol Chem. 1989;264:10858–10866. [PubMed] [Google Scholar]
- 68.Steitz T, Yin Y. Accuracy, lesion bypass, strand displacement and translocation by DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2004;359:17–23. doi: 10.1098/rstb.2003.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spagnolo JF, Rossignol E, Bullitt E, Kirkegaard K. Enzymatic and nonenzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays. RNA. 2010;16:382–393. doi: 10.1261/rna.1955410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lyle JM, Bullitt E, Bienz K, Kirkegaard K. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science. 2002;296:2218–2222. doi: 10.1126/science.1070585. [DOI] [PubMed] [Google Scholar]
- 71.Högbom M, Jäger K, Robel I, Unge T, Rohayem J. The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J Gen Virol. 2009;90:281–291. doi: 10.1099/vir.0.005629-0. [DOI] [PubMed] [Google Scholar]
- 72.Bentham M, Holmes K, Forrest S, Rowlands DJ, Stonehouse NJ. Formation of higher-order foot-and-mouth disease virus 3Dpol complexes is dependent on elongation activity. J Virol. 2012;86:2371–2374. doi: 10.1128/JVI.05696-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chinnaswamy S, Murali A, Li P, Fujisaki K, Kao CC. Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions. J Virol. 2010;84:5923–5935. doi: 10.1128/JVI.02446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tellez AB, Wang J, Tanner EJ, Spagnolo JF, Kirkegaard K, Bullitt E. Interstitial contacts in an RNA-dependent RNA polymerase lattice. J Mol Biol. 2011;412:737–750. doi: 10.1016/j.jmb.2011.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jorba N, Area E, Ortin J. Oligomerization of the influenza virus polymerase complex in vivo. J Gen Virol. 2007;89:520–524. doi: 10.1099/vir.0.83387-0. [DOI] [PubMed] [Google Scholar]
- 77.York A, Hengrung N, Vreede FT, Huiskonen J, Fodor E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc Natl Acad Sci USA. 2013;110:E4238–E4245. doi: 10.1073/pnas.1315068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jorba N, Coloma R, Ortin J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 2009;5:e1000462. doi: 10.1371/journal.ppat.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Imbert I, Guillemot J, Bourhis J, Bussetta C, Coutard B, Egloff M, Ferron F, Gorbalenya A, Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boursnell MEG, Brown TD, Foulds IJ, Green PF, Tomley FM, Binns MM. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 83.Xu X, Liu Y, Weiss S, Arnold E, Sarafianos SG, Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 2003;31:7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.te Velthuis AJ, van den Worm SH, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiao Y, Ma Q, Restle T, Shang W, Svergun D, Ponnusamy R, Sczakiel G, Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J Virol. 2012;40:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barr JN, Wertz GW. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′- and 5′-terminal sequences. J Virol. 2004;78:1129–1138. doi: 10.1128/JVI.78.3.1129-1138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flick R, Hobom G. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J Gen Virol. 1999;80:2565–2572. doi: 10.1099/0022-1317-80-10-2565. [DOI] [PubMed] [Google Scholar]
- 89.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baudin F, Bach C, Cusack S, Ruigrok R. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deng T, Vreede F, Brownlee GG. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol. 2006;80:2337–2348. doi: 10.1128/JVI.80.5.2337-2348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leahy MB, Zecchin G, Brownlee GG. Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters. J Virol. 2002;76:2019–2023. doi: 10.1128/JVI.76.4.2019-2023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.te Velthuis AJ, Turrell L, Vreede FT, Fodor E. Uncoupling of influenza A virus transcription and replication through mutation of the unpaired adenosine in the viral RNA promoter. J Virol. 2013;87:10381–10384. doi: 10.1128/JVI.00636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tchatalbachev S, Flick R, Hobom G. The packaging signal of influenza viral RNA molecules. RNA. 2001;7:979–989. doi: 10.1017/s1355838201002424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paterson D, te Velthuis AJ, Vreede FT, Fodor E. Host restriction of influenza virus polymerase activity by PB2 627E is diminished on short viral templates in a nucleoprotein-independent manner. J Virol. 2014;88:339–344. doi: 10.1128/JVI.02022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5’ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem. 2011;286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brinton MA, Fernandez AV, Dispoto JH. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- 99.Proutski V, Gould EA, Holmes EC. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acid Res. 1997;25:1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khromykh AA, Kondratieva N, Sgro Y, Palmenberg A, Westaway EG. Significance in replication of the terminal nucleotides of the flavivirus genome. J Virol. 2003;77:10623–10629. doi: 10.1128/JVI.77.19.10623-10629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R. De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J Virol. 2003;77:8831–8842. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen J, Noueiry A, Ahlquist P. An alternate pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J Virol. 2003;77:2568–2577. doi: 10.1128/JVI.77.4.2568-2577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Subba-Reddy CV, Tragesser B, Xu Z, Stein B, Ranjith-Kumar CT, Kao CC. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase in human cells reveals requirements for de novo initiation and protein–protein interaction. J Virol. 2012;86:4317–4327. doi: 10.1128/JVI.00069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hema M, Kao CC. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J Virol. 2004;78:1169–1180. doi: 10.1128/JVI.78.3.1169-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beerens N, Selisko B, Ricagno S, Imbert I, van der Zanden L, Snijder EJ, Canard B. De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J Virol. 2007;81:8384–8395. doi: 10.1128/JVI.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kranzusch PJ, Schenk AD, Rahmeh AA, Radoshitzky SR, Bavari S, Walz T, Whelan SP. Assembly of a functional machupo virus polymerase complex. Proc Natl Acad Sci USA. 2010;107:20069–20074. doi: 10.1073/pnas.1007152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vogt DA, Andino R. An RNA element at the 5′-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 2010;6:e1000936. doi: 10.1371/journal.ppat.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gamarnik AV, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 109.Andino R, Rieckhof GE, Achacoso PL, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 111.Herold J, Andino R. Poliovirus RNA replication requires genome circulization through a protein bridge. Mol Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Paul AV, Yin J, Mugavero J, Rieder E, Liu Y, Wimmer E. A “slide-back” mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J Biol Chem. 2003;278:43951–43960. doi: 10.1074/jbc.M307441200. [DOI] [PubMed] [Google Scholar]
- 113.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Steil BP, Barton DJ. Conversion of VPg into VPgpUpUOH before and during poliovirus negative-strand RNA synthesis. J Virol. 2009;83:12660–12670. doi: 10.1128/JVI.01676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. The 5′ ends of hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chinnaswamy S, Yarbrough I, Palaninathan S, Kumar CT, Vijayaraghavan V, Demeler B, Lemon SM, Sacchettini JC, Kao CC. A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis C virus polymerase. J Biol Chem. 2008;283:20535–20546. doi: 10.1074/jbc.M801490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Surana P, Satchidanandam V, Nair DT. RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res. 2014;42:2758–2773. doi: 10.1093/nar/gkt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Poranen M, Salgado P, Koivunen M, Wright S, Bamford D, Stuart D, Grimes J. Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase. Nucleic Acids Res. 2008;36:6633–6644. doi: 10.1093/nar/gkn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gong P, Kortus MG, Nix JC, Davis RE, Peersen OB. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS One. 2013;8:e60272. doi: 10.1371/journal.pone.0060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J Biol Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- 121.Hong Z, Cameron CE, Walker MP, Castro C, Yao N, Lau JY, Zhong W. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology. 2001;285:6–11. doi: 10.1006/viro.2001.0948. [DOI] [PubMed] [Google Scholar]
- 122.Laurila MR, Makeyev EV, Bamford DH. Bacteriophage π6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J Biol Chem. 2001;277:17117–17124. doi: 10.1074/jbc.M111220200. [DOI] [PubMed] [Google Scholar]
- 123.Mosley RT, Edwards TE, Murakami E, Lam AM, Grice RL, Du J, Sofia MJ, Furman PA, Otto MJ. tructure of hepatitis C virus polymerase in complex with primer-template RNA. J Virol. 2012;86:6503–6511. doi: 10.1128/JVI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ferrer-Orta C, Arias A, Agudo R, Pérez-Luque R, Escarmís C, Domingo E, Verdaguer N. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 2006;25:880–888. doi: 10.1038/sj.emboj.7600971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sharma N, O’Donnell BJ, Flanegan JB. 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J Virol. 2005;79:3565–3577. doi: 10.1128/JVI.79.6.3565-3577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Morasco BJ, Sharma N, Parilla J, Flanegan JB. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J Virol. 2003;77:5136–5144. doi: 10.1128/JVI.77.9.5136-5144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol. 2008;15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 129.Li ML, Rao P, Krug RM. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 2001;20:2078–2086. doi: 10.1093/emboj/20.8.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu Y, Qin K, Meng G, Zhang J, Zhou J, Zhao G, Luo M, Zheng X. Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus. J Biol Chem. 2013;288:11013–11023. doi: 10.1074/jbc.M112.392878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Datta K, Wolkerstorfer A, Szolar OH, Cusack S, Klumpp K. Characterization of PA-N terminal domain of Influenza A polymerase reveals sequence specific RNA cleavage. Nucl Acid Res. 2013;41:8289–8299. doi: 10.1093/nar/gkt603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 133.Davis BC, Thorpe IF. Molecular simulations illuminate the role of regulatory components of the RNA polymerase from the hepatitis C virus in influencing protein structure and dynamics. Biochemistry. 2013;52:4541–4552. doi: 10.1021/bi400251g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thompson AA, Albertini RA, Peersen OB. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J Mol Biol. 2007;366:1459–1474. doi: 10.1016/j.jmb.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ren Z, Wang H, Ghose R. Dynamics on multiple timescales in the RNA-directed RNA polymerase from the cystovirus ϕ6. Nucleic Acids Res. 2010;38:5105–5118. doi: 10.1093/nar/gkq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ . Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vilfan ID, Candelli A, Hage S, Aalto AP, Poranen MM, Bamford DH, Dekker NH. Reinitiated viral RNA-dependent RNA polymerase resumes replication at a reduced rate. Nucleic Acid Res. 2008;36:7059–7067. doi: 10.1093/nar/gkn836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Aggarwal S, Bradel-Tretheway B, Takimoto T, Dewhurst S, Kim B. Biochemical characterization of enzyme fidelity of influenza A virus RNA polymerase complex. PLoS One. 2010;5:e10372. doi: 10.1371/journal.pone.0010372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Freistadt MS, Vaccaro JA, Eberle KE. Biochemical characterization of the fidelity of poliovirus RNA-dependent RNA polymerase. Virol J. 2007;4:44. doi: 10.1186/1743-422X-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Domingo E, Holland J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 142.Drake J, Holland J. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Perrino FW, Preston BD, Sandell LL, Loeb LA. Extension of mismatched 3′ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1989;86:8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Eckert KA, Kunkel TA. Fidelity of DNA synthesis catalyzed by human DNA polymerase alpha and HIV-1 reverse transcriptase: effect of reaction pH. Nucleic Acids Res. 1993;21(22):5212–5220. doi: 10.1093/nar/21.22.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 146.Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 147.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci USA. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sadeghipour S, Bek EJ, McMinn PC. Ribavirin-resistant mutants of human enterovirus 71 express a high replication fidelity phenotype during growth in cell culture. J Virol. 2013;87:1759–1769. doi: 10.1128/JVI.02139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Coffey LL, Beeharry Y, Bordería AV, Blanc H, Vignuzzi M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci USA. 2011;108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sierra M, Airaksinen A, González-López C, Agudo R, Arias A, Domingo E. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J Virol. 2007;81:2012–2024. doi: 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Binh N, Wakai C, Kawaguchi A, Nagata K. Involvement of the N-terminal portion of influenza virus RNA polymerase subunit PB1 in nucleotide recognition. Biochem Biophys Res Commun. 2014;443:975–979. doi: 10.1016/j.bbrc.2013.12.071. [DOI] [PubMed] [Google Scholar]
- 152.Guan H, Simon AE. Polymerization of nontemplate bases before transcription initiation at the 3’ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′ end repair of viral RNAs. Proc Natl Acad Sci USA. 2000;97:12451–12456. doi: 10.1073/pnas.97.23.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Teramoto T, Kohno Y, Mattoo P, Markoff L, Falgout B, Padmanabhan R. Genome 3′-end repair in dengue virus type 2. RNA. 2008;14:2645–2656. doi: 10.1261/rna.1051208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol. 2002;76:8989–9001. doi: 10.1128/JVI.76.18.8989-9001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Neufeld KL, Galarza JM, Richards OC, Summers DF, Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol . J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Poranen MM, Koivunen MR, Bamford DH. Nontemplated terminal nucleotidyltransferase activity of double-stranded RNA bacteriophage phi6 RNA-dependent RNA polymerase. J Virol. 2008;82:9254–9264. doi: 10.1128/JVI.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Graham SC, Sarin LP, Bahar MW, Myers RA, Stuart DI, Bamford DH, Grimes JM. The N-terminus of the RNA polymerase from infectious pancreatic necrosis virus is the determinant of genome attachment. PLoS Pathog. 2011;7:e1002085. doi: 10.1371/journal.ppat.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Urban S, Urban S, Fischer KP, Tyrell DL. Efficient pyrophosphorolysis by a hepatitis B virus polymerase may be a primer-unblocking mechanism. Proc Natl Acad Sci USA. 2001;98:4984–4989. doi: 10.1073/pnas.091324398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tu X, Das K, Han Q, Bauman JD, Clark ADJ, Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E. Structural basis of HIV-1 resistance to AZT by excision. Nat Struct Mol Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.D’Abramo CM, Cellai L, Götte M. Excision of incorporated nucleotide analogue chain-terminators can diminish their inhibitory effects on viral RNA-dependent RNA polymerases. J Mol Biol. 2004;337:1–14. doi: 10.1016/j.jmb.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 161.Das K, Bandwar RP, White KL, Feng JY, Sarafianos SG, Tuske S, Tu X, Clark ADJ, Boyer PL, Hou X, Gaffney BL, Jones RA, Miller MD, Hughes SH, Arnold E. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J Biol Chem. 2009;284:35092–35100. doi: 10.1074/jbc.M109.022525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lemm JA, Rumenapf T, Strauss EG, Rice CM, Strauss JH. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fodor E. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol. 2013;57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 164.Paterson D, Fodor E. Emerging roles for the influenza A virus nuclear export protein (NEP) PLoS Pathog. 2012;8:e1003019. doi: 10.1371/journal.ppat.1003019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, García-Sastre A, tenOever BR. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci USA. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nedialkova DD, Gorbalenya AE, Snijder EJ. Arterivirus nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 2010;6:e1000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Furuta Y, Gowen B, Takahashi K, Shiraki K, Smee D, Barnard D. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Muratore G, Goracci L, Mercorelli B, Foeglein Á, Digard P, Cruciani G, Palù G, Loregian A. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc Natl Acad Sci USA. 2012;109:6247–6252. doi: 10.1073/pnas.1119817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mänz B, Götz V, Wunderlich K, Eisel J, Kirchmair J, Stech J, Stech O, Chase G, Frank R, Schwemmle M. Disruption of the viral polymerase complex assembly as a novel approach to attenuate influenza A virus. J Biol Chem. 2011;286:8414–8424. doi: 10.1074/jbc.M110.205534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kowalinski E, Zubieta C, Wolkerstorfer A, Szolar O, Ruigrok R, Cusack S. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 2012;8:e1002831. doi: 10.1371/journal.ppat.1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Garriga D, Ferrer-Orta C, Querol-Audí J, Oliva B, Verdaguer N. Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J Mol Biol. 2013;425:2279–2287. doi: 10.1016/j.jmb.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 173.Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 174.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 175.Trask SD, McDonald SM, Patton JT. Structural insights into the coupling of virion assembly and rotavirus replication. Nat Rev Microbiol. 2012;10:165–177. doi: 10.1038/nrmicro2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mertens P. The dsRNA viruses. Virus Res. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 177.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymearse encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vasquez-del Carpio R, Morales JL, Barro M, Ricardo A, Spencer E. Bioinformatic prediction of polymerase elements in the rotavirus VP1 protein. Biol Res. 2006;39:649–659. doi: 10.4067/s0716-97602006000500008. [DOI] [PubMed] [Google Scholar]