Abstract
The ability to examine all chromatids from a single meiosis in yeast tetrads has been indispensable for defining mechanisms of homologous recombination initiated by DNA double-strand breaks (DSBs). Using a broadly applicable strategy for the analysis of chromatids from a single meiosis at two recombination hotspots in mouse oocytes and spermatocytes, we demonstrate here the unidirectional transfer of information — gene conversion — in both crossovers and noncrossovers. Whereas gene conversion in crossovers is associated with reciprocal exchange, the unbroken chromatid is not altered in noncrossover gene conversions, providing strong evidence that noncrossovers arise from a distinct pathway. Gene conversion frequently spares the binding site of the hotspot-specifying protein PRDM9 with the result that erosion of the hotspot is slowed. Thus, mouse tetrad analysis demonstrates how unique aspects of mammalian recombination mechanisms shape hotspot evolutionary dynamics.
Sexual reproduction requires the formation of haploid gametes from diploid precursors through meiosis, which comprises two divisions following a single round of genome duplication. During the first meiotic prophase, recombination establishes physical connections between homologous chromosomes (homologs), essential for proper chromosome segregation1–3.
Recombination is best understood in yeast, in part because all four chromatids from a single meiosis — a tetrad — can be recovered4. Tetrad analysis demonstrated that recombination can occur with an exchange of chromatid arms, a crossover, or without an exchange, a noncrossover5. Importantly, both crossovers and noncrossovers are often associated with gene conversion, the non-reciprocal transfer of information from a donor chromatid to the recipient. A model to account for this, confirmed later by molecular approaches, is that recombination initiated by DNA double-strand breaks (DSBs) leads to gene conversion at the DSB site using information from the uncut donor chromatid6. This model posited formation of a double-Holliday junction intermediate that is resolved as a crossover or a noncrossover, such that either resolution type can lead to conversion of markers on the donor. Work in budding yeast supports this model for crossovers but demonstrated that most noncrossovers arise by pathways that do not involve resolution of a double-Holliday junction or alteration of the donor7–11.
In mammals, crossovers are detected by genetic mapping in pedigrees and by sperm and oocyte typing, and can be inferred from population diversity analysis12–17. Events involving transfer of short patches of genetic information attributed to noncrossovers have also been detected by sperm and oocyte typing18–22. Gene conversion has been inferred in mammals but not formally proven, because, unlike in fungi, only single chromatids could be analyzed18,21,23. Although many aspects of meiotic recombination are likely conserved with yeast13, mammals differ in key features. For example, the ratio of noncrossovers to crossovers appears to be much higher in mammals, and inferred gene conversion tracts are shorter.
In mammals as in other organisms, recombination is initiated by DSBs generated by the SPO11 transesterase3,24,25. DSBs occur most often at preferred sites, termed DSB hotspots, which are presumed to be recombination hotspots26. In mammals, unlike yeast and other organisms, hotspot location is governed largely by PRDM9, a meiosis-specific histone H3 methyltransferase with a DNA binding specificity determined by a tandem array of C2H2 zinc fingers27–30. PRDM9 binding sites occur within hotspots28,31,32, suggesting that these binding sites are likely to undergo gene conversion during repair. This property raises a conundrum about PRDM9 binding site maintenance and thus about hotspot evolutionary dynamics: because the direction of gene conversion is biased (the cut chromosome copies the uncut donor), PRDM9 binding sites are predicted to be rapidly lost in the absence of additional constraints, as is seen for the PRDM9 motif during human evolution29.
To understand these mechanistic and evolutionary aspects of mammalian recombination, we directly interrogated structures of recombinant molecules by developing strategies to analyze all four chromatids of a single meiosis in the mouse Mus musculus. We performed this equivalent of fungal tetrad analysis in both oocytes and spermatocytes because of fundamental differences in female and male meioses. Two recombination hotspots were analyzed, Psmb9 (ref23) and A3 (ref18), which are representative of the fifty or so mammalian hotspots described thus far for the width and the distribution of exchanges12,14. These analyses demonstrate the occurrence of gene conversion, either associated with reciprocal exchange (crossovers) or not (noncrossovers). Importantly, noncrossovers occur without modification of the donor chromatid. Many gene conversions do not include the PRDM9 binding site, providing a mechanism that lengthens hotspot evolutionary lifespan.
Results
Direct evidence for meiotic gene conversion in mice
Meiotic recombination at the Psmb9 hotspot was previously detected using allele-specific PCR of DNA from pooled ovaries23. We adapted this analysis to single oocytes to examine all four chromatids of a given meiosis – a tetrad. As detailed below, the success rate for recovering reciprocal recombinant (CO) molecules when present was very high (100% in most experiments), indicating that we indeed did usually succeed in analyzing all four chromatids.
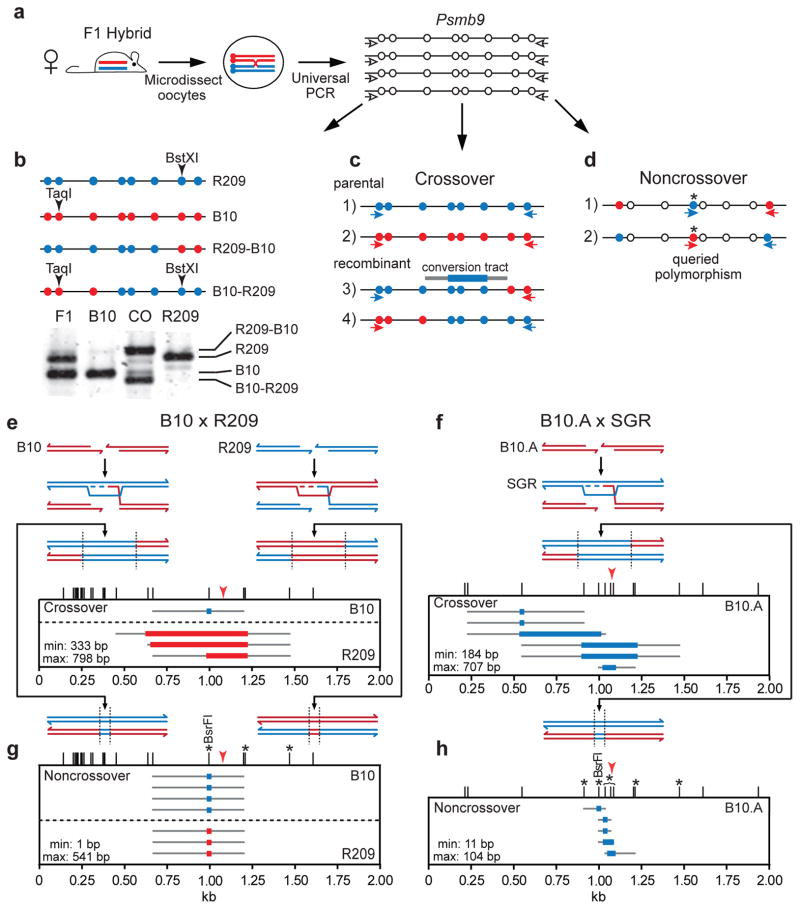
Microdissected dictyate oocytes from 25- to 30-day-old B10 x R209 F1 hybrid mice were individually lysed and the hotspot region was amplified using non-allele-specific primers (universal PCR, Fig. 1a). Crossover and parental chromatids were distinguished by TaqI/BstXI digestion of the PCR product. For example, for the oocyte shown in Figure 1b, four chromatids — two crossover and two parental — were identified. By this approach, 4 of 119 oocytes (3.4%) showed crossover chromatids, translating to a 1.7% per-gamete frequency similar to previous estimates from pooled ovaries (Table 1)23. When allele-specific PCR was used to amplify DNA from the universal PCR and crossover breakpoints were mapped (Fig. 1c), all four oocytes exhibited reciprocal exchange associated with gene conversion, i.e., a 3:1 ratio for one or more polymorphisms between the breakpoints (Fig. 1e). Three crossovers involved conversion to the B10 genotype, indicating a DSB on an R209 chromatid, and one involved conversion to the R209 genotype indicating a DSB on a B10 chromatid (Fig. 1e).
Figure 1. Mouse oocyte tetrad analysis provides direct evidence for meiotic gene conversion.
a. Individual germinal vesicle stage oocytes from F1 hybrid mice were microdissected for PCR to amplify the Psmb9 hotspot (chromosome 17) from all 4 chromatids using primers that recognize both parental chromosomes (white arrows). These universal PCRs provided DNA for tetrad analysis of recombinants.
b. Identification of crossovers at the Psmb9 hotspot from B10 x R209 hybrids by restriction fragment length polymorphisms. DNA from universal PCR of each oocyte (a) was digested with TaqI and BstXI and electrophoresed to differentiate PCR products from parental and crossover chromatids. Polymorphisms are schematically represented by blue and red circles. CO, crossover.
c. Tetrad amplification strategy for crossovers. DNA from universal PCR of each oocyte (a) was used to seed 4 separate PCRs, each with a primer set to amplify the Psmb9 hotspot from either a parental or recombinant chromatid (red and blue arrows). For B10 x R209 hybrids, any oocyte suspected of containing a crossover (b) was tested by this assay; for the B10.A x SGR hybrids, all oocytes were tested this way.
d. Tetrad amplification strategy for noncrossovers. Universally amplified DNA from (a) was used to seed 2 separate PCRs, each with one primer directed to a polymorphism being queried for conversion (asterisk) and a second primer directed to a polymorphism at one end of the Psmb9 hotspot.
e. Reciprocal crossovers with associated gene conversion in the B10 x R209 hybrid. Meiotic DSBs can form at the Psmb9 hotspot on the B10 (red) or R209 (blue) chromosome, leading to strand invasion into the intact donor homolog, which serves as a template for repair synthesis (dotted line). Thus, gene conversion will occur in favor of the donor homolog, as delineated by dashed vertical lines. Only the 2 chromatids engaged in homologous recombination are shown for simplicity. Tetrad analysis (n=119 oocytes, from 3 mice) identified reciprocal crossovers with associated gene conversion in which Psmb9 sequences on the B10 chromosome (1 crossover) or R209 chromosome (3 crossovers) were converted to those of the homolog. Thus, on a per-meiosis basis, the crossover frequency was 3.4% (95% CI = 1.1–8.9%), translating to a per-gamete frequency of 1.7% (8 crossover chromatids per 476 haploid genome equivalents; Table 1). Red arrowhead indicates the position of the PRDM9wm7 binding site32. The mean minimal and maximal gene conversion tracts are as indicated. Colored bar, minimal gene conversion tract; gray lines, maximal gene conversion tract; ticks, polymorphisms.
f. Reciprocal crossovers with associated gene conversion in the B10.A x SGR hybrid. DSBs form preferentially at the Psmb9 hotspot on the B10.A chromosome (red), such that repair is templated by the R209 (blue) chromosome. Tetrad analysis (n=205 oocytes, from 3 mice) identified reciprocal crossovers in which the Psmb9 sequences on the B10.A chromosome were converted to those of the SGR chromosome. The 2.9% (95% CI = 1.2–6.6%) per-meiosis crossover frequency agrees with previous estimates from ovaries (Table 1)23. Red arrowhead indicates the 2 polymorphisms that confer preferential PRDM9wm7 binding to the hotspot center from the B10.A chromosome32.
g. Noncrossovers in the B10 x R209 hybrid. Tetrad analysis identified noncrossovers in which Psmb9 sequences on the B10 chromosome (4 noncrossovers) or R209 chromosome (3 noncrossovers) were converted to those of the homolog, while the homolog was unaltered in each case. The noncrossover frequency was 5.9% (95% CI = 2.6–12.2%) per meiosis. Asterisks, the four polymorphisms queried for noncrossovers, using the approach shown in (d). Monitoring the same 4 polymorphisms and 3 others in either pooled sperm or oocytes, >95% of detected noncrossovers were restricted to the 4 polymorphisms (Supplementary Fig. 1; Supplementary Table 1).
h. Noncrossovers in the B10.A x SGR hybrid. All five noncrossovers identified by tetrad analysis involved conversion of Psmb9 hotspot sequences on the B10.A chromosome to those of the SGR chromosome. The noncrossover frequency was 2.4% (95% CI = 0.9–5.9%) per meiosis. Asterisks, polymorphisms queried for noncrossovers, as in (g).
Table 1.
Summary of recombination frequencies from tetrads and pooled gametes.
| Hotspot | F1 Hybrid | Frequency of recombinant chromatids per gametea
|
|||
|---|---|---|---|---|---|
| Crossovers (95% CI, %)b | Noncrossovers (95% CI, %)b | ||||
|
| |||||
| Oocyte | Ovary | Oocyte | Ovary | ||
| Psmb9 | B10 x R209 | 1.7% (0.54–4.4) | 2.0%c (1.6–2.5) | 1.5% (0.65–3.0) | 0.77%c (0.54–1.1) |
| B10.A x SGR | 1.5% (0.60–3.3) | 0.79%c (0.50–1.2) | 0.6% (0.22–1.5) | 1.0%c,d (0.75–1.4) | |
|
| |||||
| Spermatocyte | Sperm | Spermatocyte | Sperm | ||
|
| |||||
| A3 | A/J x DBA/2J | 0.04% (0.02–0.06) | 0.02%e (0.01–0.07) | 0.31% (0.26–0.37) | 0.29%f (0.23–0.36) |
To calculate recombination events on a per meiosis basis from a per gamete frequency, the crossover frequency is multiplied by 2 and the noncrossover is multiplied by 4, given that crossovers modify 2 of 4 chromatids while noncrossovers modify 1 of 4 chromatids.
Confidence interval (CI) calculations here and in the text use the Wilson procedure with continuity correction.
Crossover and noncrossover frequency calculations are restricted to those samples used for noncrossover measurements, as shown in Supplementary Figure 1 and Supplementary Table 1, accounting for the small differences from that previously reported23.
Noncrossover frequency is derived from the B10.A chromosome only, which represents >95% of events23.
The crossover frequency is lower than that previously determined18, largely attributable to results from one of the animals in the current study.
Noncrossover frequency is derived from the DBA/2J chromosome only, which represents >80% of events as in Supplementary Fig. 2.
For some hotspots, preferential recombination initiation on one homolog has been proposed because crossover breakpoints cluster asymmetrically depending upon which crossover chromatid is amplified33. In a few cases, such as Psmb9, this property is directly correlated with PRDM9 binding and H3K4me3 enrichment on the homolog with predicted high initiation activity32,34. Tetrad experiments were thus performed at Psmb9 with a second F1 hybrid, B10.A x SGR, in which recombination is thought to initiate preferentially on the B10.A chromosome23. Moreover, the polymorphism density in this hybrid is higher in the center of Psmb9, allowing finer scale recombination maps.
Six oocytes out of 205 (2.9%) displayed crossovers, all reciprocal (Fig. 1f), for a 1.5% per-gamete frequency (Table 1). All six displayed gene conversion to the SGR genotype, providing direct evidence for preferential initiation on the B10.A chromosome and substantiating the model that asymmetric crossover breakpoint distributions result from biased initiation. Overall, analysis in both hybrids provides formal proof for gene conversion and for generation of reciprocal crossover molecules within a single meiotic cell.
The mean gene conversion tract length for B10.A x SGR was 446 bp (Fig. 1f), similar to inference from the asymmetric crossover breakpoint distribution23. Because tetrad analysis captures both recombinant chromatids, gene conversion tract length could also be determined from the B10 x R209 hybrid which does not show biased initiation. The mean tract length was 566 bp (Fig. 1e), similar to B10.A x SGR. Thus, mean tract lengths from hotspots with unbiased initiation are similar to those with biased initiation.
Noncrossovers identified by oocyte tetrad analysis
Noncrossovers are predicted to be the major outcome of mammalian meiotic recombination and to arise by a distinct mechanism from that of crossovers13. To identify noncrossovers, PCR was performed with DNA from the universal PCR of individual oocytes using allele-specific primers directed to specific polymorphisms (asterisks, Fig. 1d). DNA sequencing then verified the presence and extent of the noncrossover.
In the B10 x R209 hybrid, three polymorphisms were tested (Fig. 1g). Seven oocytes had detectable noncrossovers (5.9%), all of which converted only the BsrFI polymorphism in the hotspot center. Four noncrossovers derived from initiation on the B10 chromosome and three on R209. The noncrossover frequency was 1.5% per gamete, similar to pooled ovaries (Table 1) 23.
In the B10.A x SGR hybrid, seven polymorphisms were queried (Fig. 1h). Five oocytes had detectable noncrossovers (2.4%), all incorporating polymorphisms within the central ~200 bp of the hotspot. The per-gamete noncrossover frequency was 0.6%, similar to pooled ovaries (Table 1; Supplementary Table 1). As with oocytes, noncrossovers from pooled ovaries or sperm typing were also concentrated near the hotspot center (Supplementary Fig. 1; Supplementary Table 1). Noncrossovers incorporating more than one polymorphism (co-conversion) were also detected at similar proportions in oocytes (2 of 5 noncrossovers) and pooled ovaries (21 of 42), although, interestingly, sperm typing showed fewer co-conversions (16 of 62, p=0.0134, Fisher’s exact test, two-tailed). Overall, taking into account the distance to the adjacent polymorphisms, the mean noncrossover gene conversion tract length was 86 bp (minimum: 23 bp; maximum: 148 bp) in pooled ovaries and 68 bp (minimum: 15 bp; maximum: 124 bp) in sperm (Supplementary Fig. 1). All noncrossovers from tetrads were conversions to SGR genotype on a B10.A chromosome (Fig. 1h), implying preferential initiation on B10.A as with pooled ovaries (Supplementary Table 1). Importantly, the donor chromosome was unaffected in all noncrossovers, excluding the possibility that they arose from two nearby crossovers.
Unidirectional transfer of genetic information in noncrossovers
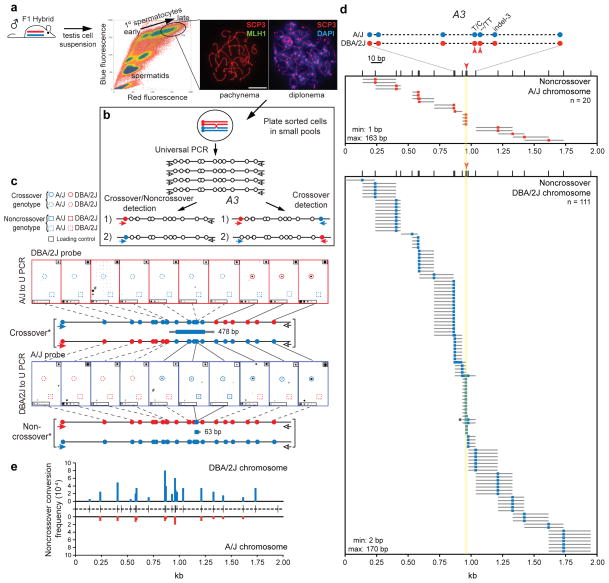
Tetrad analysis was also applied to spermatocytes. Flow cytometry was used to isolate late prophase I primary spermatocytes35 which have completed meiotic recombination19 (Fig. 2a). To maximize noncrossover recovery, we analyzed the A3 hotspot, which has a high ratio of detectable noncrossovers to crossovers (~10:1)18. Fine-scale recombination analysis is possible because of the high density and relatively even distribution of polymorphisms, mostly single nucleotide polymorphisms. In total, 22 polymorphisms were queried in A/J x DBA/2J F1 hybrids, averaging ~100 bp apart and with a higher density at the hotspot center. In this hybrid, A3 is inferred to have preferential initiation on the DBA/2J chromosome.
Figure 2. Mouse spermatocyte tetrads demonstrate that noncrossovers result from unidirectional transfer of information.
a. Single cell suspensions from the testes of adult F1 hybrid mice were stained with Hoechst 33342 and propidium iodide. Cells with the highest blue and red fluorescence were sorted using the indicated gate (oval). In three independent experiments, all sorted cells were primary spermatocytes, with ~98% in diplonema and MLH1-positive pachynema, based on staining with the axial element marker SYCP3 and the crossover marker MLH137,58,59, as shown in the representative images.
b. A3 hotspot amplification strategy for spermatocytes. To amplify both noncrossovers and crossovers (left), cells were plated in pools of ~20 cells per well and universally amplified across the A3 hotspot on chromosome 1. The amplified DNA was used to seed 2 separate PCRs using an allele-specific forward primer and a universal reverse primer. To amplify crossovers only (right), cells were plated in larger pools of ~100 cells per well for universal amplification. The universally amplified DNA was used to seed 2 separate PCRs using primer sets to detect both recombinant chromatids.
c. Representative crossover and noncrossover from spermatocyte analysis. Replicate blots were made from PCRs in the A/J to universal (U) orientation (top) and the DBA/2J to U orientation (bottom) and probed with allele-specific oligonucleotides to genotype polymorphisms across the A3 hotspot. The genotype of a representative crossover with the length of the gene conversion tract is shown between the blots and that of a representative noncrossover is shown below the blots. Dot blot legend: solid colored circles/squares, genotype determined by blotting; dashed colored circles/squares, inferred genotype; black squares upper right corner, loading control of amplified DBA/2J or A/J DNA; black rectangles lower left corner, dilutions of loading control; #, well containing noncrossovers on both the A/J and DBA/2J chromosomes.
d. Spermatocyte noncrossovers at the A3 hotspot. Because recombination initiates preferentially on the DBA/2J chromosome (red), the majority of noncrossovers involve conversion of DBA/2J polymorphisms to the A/J genotype (blue). The noncrossover frequency was 1.25% (95% CI = 1.0–1.5%) per meiosis. Schematic of the central polymorphisms is shown at the top. Red arrowheads indicate polymorphisms implicated in differential PRDM9 binding between A/J and DBA/2J; yellow shading is the predicted PRDM9 binding site (Fig. 3b). Indel-3 is located within a direct repeat such that only the A/J polymorphism can be genotyped; <4% of noncrossovers on the DBA/2J chromosome were converted only at this polymorphism. Asterisk, noncrossover highlighted in c.
e.Total Poisson adjusted noncrossover frequencies on the DBA/2J (top) and A/J (bottom) chromosomes. Noncrossover frequencies at each tested polymorphism are normalized for co-conversions. Ticks in the center represent the 22 polymorphisms tested.
Pools of spermatocytes were analysed rather than single cells to increase recovery of recombination events. Pools were small so that most recombinants detected would derive from a single spermatocyte. Sorted spermatocytes were lysed in 522 pools of ~20 cells each, and the A3 hotspot was amplified by universal PCR then nested PCRs with allele-specific forward primers and universal reverse primers (Fig. 2b, left). Amplified DNA was blotted onto replicate filters and probed with allele-specific oligonucleotides to identify and map recombinants (Fig. 2c)36. From ~10,440 analyzed spermatocytes, 111 noncrossovers converted a DBA/2J chromosome segment to the A/J genotype, versus 20 in the opposite orientation (Fig. 2d). Thus, the noncrossover frequency was 1.25% per meiosis (0.31% per gamete), with most initiation on the DBA/2J chromosome, similar to pooled sperm18. Even with the closely spaced polymorphisms, noncrossover gene conversion tracts most often included only a single polymorphism and averaged 86 ± 49 bp. While noncrossovers clustered in the central 200 bp of A3, a significant fraction was distributed ~1 kb to either side (Fig. 2d, e and Supplementary Fig. 2). All noncrossovers were non-reciprocal. Thus, nonselectively typing many polymorphisms allowed high-resolution noncrossover mapping throughout the hotspot. For comparison, sperm from the same mice used to sort spermatocytes were also assessed for noncrossovers on the DBA/2J chromosome. A nearly identical frequency (0.29% per gamete; Table 1) and similar distribution (Supplementary Fig. 2) of noncrossovers were obtained.
Overall, a single noncrossover was detected in each of 118 pools, while multiple noncrossovers on the DBA/2J chromosome were detected in 10 pools, close to the number predicted assuming recombinant spermatocytes were Poisson-distributed among pools (12.5). Only 3 pools showed noncrossovers on both the DBA/2J and A/J chromosomes, matching expectation for noncrossovers from independent spermatocytes (3.9). These results indicate that gene conversion on the A/J chromosome results from infrequent recombination initiation on that chromosome, not initiation on DBA/2J. Thus, the large number of analyzed spermatocytes and high polymorphism density provided compelling evidence that noncrossovers arise from a unidirectional transfer of genetic information.
Crossover gene conversion tracts often encompass the hotspot center
Crossovers at A3 were detected in spermatocytes but in much smaller numbers than noncrossovers: Two reciprocal crossovers in the same experiments as noncrossovers (Fig. 2b, left) and 13 more from larger pools of spermatocytes (90 pools of ~100 spermatocytes) using two allele-specific primers (Fig. 2b, right), 10 of which were reciprocal. Consistent with preferential initiation on the DBA/2J chromosome, twice as many crossovers showed gene conversion to the A/J genotype (Fig. 3a). Recovery of three crossovers without the reciprocal product may reflect less efficient amplification from larger pools (thus, a 77% success rate for both products). This 0.08% crossover frequency per meiosis translates to 0.04% per gamete, similar to sperm typing (Table 1). The net noncrossover to crossover ratio at A3 was therefore ~15 to 1 (noncrossover: 1.25%; crossover: 0.08%).
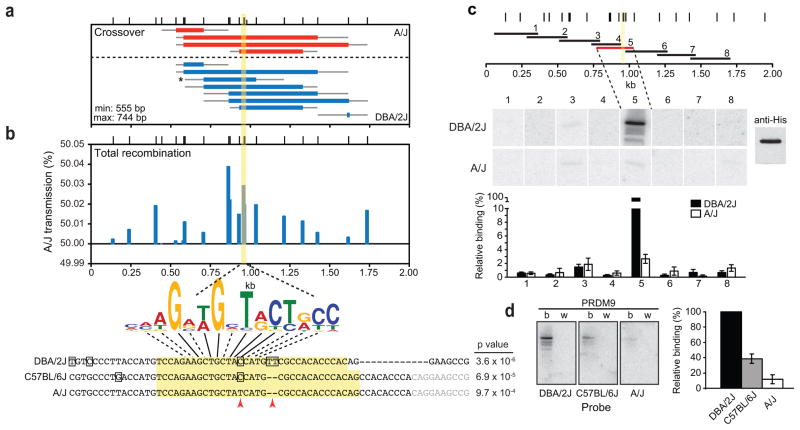
Figure 3. PRDM9 binds to the center of the A3 hotspot.
a. Spermatocyte crossovers with associated gene conversion at the A3 hotspot. Tetrad analysis identified 4 reciprocal crossovers in which A3 sequences on the A/J chromosome were converted to those of the DBA/2J chromosome and 8 reciprocal crossovers in which A3 sequences on the DBA/2J chromosome were converted to those of the A/J chromosome. Crossovers were isolated from experiments with 20 cells (2 crossovers) or 100 cells (10 crossovers) per well. The crossover frequency per meiosis was 0.08% (95% CI = 0.05–0.13). Asterisk, crossover highlighted in Figure 2c; yellow bar, predicted PRDM9 binding.
b. Deviation from the Mendelian 50% transmission frequency due to crossover and noncrossover gene conversion. The A/J transmission frequency per gamete at each indicated polymorphism is shown. Anything over 50% represents the non-Mendelian transmission of A/J sequences, i.e., transmission distortion. The consensus PRDM9b binding motif derived by Brick et al28 is indicated below the graph. The sequence of the 57-bp probes used in d is shown at the bottom. A match with indicated p value to the consensus binding motif for the PRDM9b allele is shown. Yellow bar and yellow shading predicted PRDM9 binding site; red arrowheads, polymorphisms implicated in differential PRDM9 binding. Noncrossover gene conversion tracts incorporated the PRDM9 binding site polymorphisms in 26 of 131 events (20%), 22 of which were conversions to A/J (Fig. 2d). Among these events, the C/T transition (~4-fold) and the 2-bp insertion/deletion (≥7-fold) were much more likely to be converted in favor of A/J sequences (Fig. 2d).
c. Mapping the PRDM9 binding site at the A3 hotspot. DBA/2J and A/J mice carry the Mus musculus domesticus Prdm9b allele from, which is identical to that in C57BL/6J mice. The horizontal bars represent the positions of the 8 overlapping ~250 bp DNA probes generated for the A/J and DBA/2J genotype. Representative southwesterns for each probe against His-tagged PRDM9b are shown along with a loading control (probed with anti-His antibody) on the far right. The lower molecular weight bands correspond to PRDM9 degradation products. Quantification of the relative binding of the PRDM9b protein is shown at the bottom (± standard error of the mean). PRDM9b showed substantial binding to DBA/2J probe 5 from the hotspot center but much less efficiently to the A/J probe 5. Weak binding was also detected for probe 3 from both DBA/2J and A/J. The relevance of this binding is uncertain, however, as recombination in this region of the hotspot is considerably higher on the DBA/2J chromosome than the A/J chromosome (Fig. 2e). PRDM9wm7 also bound weakly to probe 3 from A/J, but did not bind any of the DBA/2J fragments (Supplementary Fig. 3d).
d. Southwestern analysis localizing PRDM9b, but not PRDM9wm7, binding to the center of the A3 hotspot. The 57-bp central probe (sequence shown in b) was used together against His-tagged PRDM9b (b) and PRDM9wm7 (w) with quantification of relative binding on the right (± standard error of the mean). PRDM9b bound to the all three probes, but best to DBA/2J. None of the probes were bound by PRDM9wm7. Note: PRDM9wm7, but not PRDM9b, binds to the center of Psmb9 in strains where this hotspot is active (red arrowhead, Fig. 1)32.
The mean crossover gene conversion tract length at A3 was 626 ± 319 bp, similar to Psmb9 but substantially longer than for noncrossovers at A3 (86 ± 49 bp). Unlike noncrossovers, most crossover gene conversion tracts overlapped each other and the hotspot center, such that 5 polymorphisms spanning ~200 bp of the center were converted in 75% of crossovers (Fig. 3a). Of note, however, three crossovers had short, off-center gene conversion tracts (Fig. 3a), which could not have been inferred from sperm typing (Supplementary Fig. 3).
We determined A/J transmission relative to DBA/2J at each polymorphism (Fig. 3b). Gene conversion, primarily from noncrossovers, resulted in transmission distortion (deviation from the Mendelian ratio) in favor of A/J sequences for nearly all polymorphisms, especially those in the central ~200 bp, agreeing with sperm typing18. The polymorphism that showed the greatest distortion was 8.2-fold more likely to convert to the A/J genotype, resulting in A/J transmission to 50.04% of gametes. Both crossover and noncrossover gene conversion contributed to transmission distortion of this polymorphism: 5 of 7 crossovers (71%) and 17 of 19 noncrossovers (89%) involved conversion to the A/J genotype. However, because noncrossovers greatly outnumber crossovers, the net impact of noncrossovers is greater.
PRDM9 binds to the center of the A3 hotspot
PRDM9 specifies the location of recombination hotspots in mice and humans. DBA/2J and A/J mice express PRDM9b from Mus musculus domesticus27,30. We assayed PRDM9b binding to A3 sequences in vitro to determine if preferential recombination initiation on the DBA/2J chromosome is associated with higher binding affinity. M. m. molossinus PRDM9wm7 (which has different DNA contact residues27) was used as a control.
Overlapping fragments spanning ~1.7 kb of the A3 alleles from DBA/2J and A/J were used as probes in southwestern analysis (Fig. 3c). PRDM9b showed substantial binding to a DBA/2J fragment from the hotspot center (probe 5) but not to other DBA/2J fragments. PRDM9b also bound the A/J fragment from the hotspot center but much less efficiently (Fig. 3c), consistent with less frequent DSB formation on the A/J chromosome. PRDM9wm7 bound poorly or not at all to DBA/2J or A/J fragments (Supplementary Fig. 3d), confirming the specificity of PRDM9b binding.
To further localize binding, we tested a 57-bp fragment (Fig. 3b) spanning sequences unique to probe 5. This hotspot-center fragment contains a partial match to the site predicted from PRDM9b zinc finger composition, and a nearly exact match to the consensus from whole-genome DSB analysis (Fig. 3b)28. Accordingly, PRDM9b bound to both the DBA/2J and A/J probes, but ~8-fold better to DBA/2J (Fig. 3d).
The C57BL/6J A3 allele has substantially more activity than A/J, although not as high as DBA/2J18. Likewise, the corresponding 57-bp fragment from C57BL/6J (Fig. 3b) had an intermediate level of PRDM9b binding (Fig. 3d). The predicted PRDM9 binding site contains two sequence differences between these strains: a C/T base substitution and a 2-bp insertion/deletion (red arrowheads, Fig. 3b). The hierarchy of PRDM9b binding (DBA/2J>C57BL/6J≫A/J) suggests that the 2-bp deletion common to C57BL/6J and A/J reduces binding efficiency by about half and the A/J-specific base substitution reduces it even further. Thus, despite the long PRDM9 recognition site owing to its zinc finger array, one or a few residues can significantly affect PRDM9 binding, reinforcing the idea that PRDM9 binding efficiency modulates recombination hotspot activity31,32.
We investigated how recombination affects transmission of the PRDM9 binding site, considering transmission from either crossover and noncrossover gene conversion of the polymorphisms inferred to have the greatest impact on PRDM9 binding. The C/T transition and the 2-bp insertion/deletion were much more likely to convert to A/J sequences, resulting in significant transmission distortion (Fig. 3b). However, 80% of noncrossovers converted polymorphisms flanking the center of A3 without converting those affecting PRDM9 binding. Thus, most recombination events are predicted not to affect A3 hotspot activity.
Discussion
In this study, we sought to test long-standing assumptions about the mechanism of meiotic recombination in mammals by developing mouse tetrad analysis, a strategy to assess multiple chromatids from a single meiosis at recombination hotspots. Our experiments demonstrated non-Mendelian transfer of information — gene conversion — during recombination. Crossing-over in both oocytes and spermatocytes was associated with gene conversion. Noncrossovers were non-reciprocal exchanges of genetic information without observable modification of the donor locus. Importantly, gene conversion frequently occurred away from the PRDM9 binding site, with implications for understanding recombination mechanisms and hotspot evolution.
Gene conversion tracts were observed in each of 22 crossovers recovered from mouse tetrads. The strong association of gene conversion with crossovers together with relatively uniform conversion tract lengths (average 566 ± 277 bp) suggest that most crossovers arise from a common mechanism. Consistent with this, ~90% of crossovers in mouse are dependent on the MLH1 protein37,38. Gene conversion associated with crossing-over is compatible with double-Holliday junction resolution, as predicted by the original DSB repair model from yeast6. However, in mammals the two Holliday junctions may be closer at the time of resolution given that the mean gene conversion tract length is 3 to 4-fold shorter than in yeast10,39. Consistent with a common mechanism driving crossover formation, all of the characterized human and mouse hotspots have a relatively uniform inferred mean conversion tract length of ~500 bp18,23,33,40–44, similar to what we report here. However, it remains possible that hotspot-specific, regional, chromosomal, or polymorphism density-dependent effects may exist and cause variations, which could be uncovered by genome-wide deep sequencing15.
Double-Holliday junction resolution can in principle also give rise to noncrossovers6. However, this model predicts the presence of heteroduplex DNA on both the recipient and donor chromatids such that the donor can become modified. The absence of donor modification in the >140 noncrossovers we examined is more consistent with an alternative pathway(s) involving a unidirectional transfer of information from the donor to the recipient, for example, synthesis-dependent strand annealing. Dissolution of a double-Holliday junction by branch migration can also lead to noncrossover formation without donor modification45. In yeast, most noncrossovers are thought to derive from either of these latter mechanisms rather than double-Holliday junction resolution7,10.
This study found short noncrossover gene conversion tract lengths (94 ± 62 bp), matching previous reports in mouse and human18,21–23,43. As with crossovers, noncrossover gene conversion tract lengths are much shorter than in yeast (≥20-fold shorter)10,39,46, suggesting there are significant differences in either the proteins involved or the chromatin organization of the recombining region, which could affect DNA end processing, heteroduplex formation/extension, or mismatch correction. Mechanistic differences between mammals and yeast may reflect differences in the biological processes in which meiotic recombination participates. For example, strand invasion involves a search for sequence homology and is thought to contribute to stabilization of interactions between homologs47. Limiting the extent of repair synthesis in mammals as compared to yeast until multiple interhomolog recombination interactions occur along the chromosome may reduce opportunities to involve repetitive, non-allelic DNA, in turn reducing the potential for ectopic exchanges. Moreover, shorter strand extension could produce a structure favorable for strand displacement, and therefore participate in controlling the noncrossover pathway.
An unanticipated and novel finding that affects our understanding of meiotic DSB repair mechanisms is the broad distribution of gene conversion at the A3 hotspot despite highly localized PRDM9 binding. From ssDNA mapping at resected DSBs, it has been proposed that DSBs arise most frequently near PRDM9 binding sites28,48. This hypothesis is supported by the mapping of meiotic DSBs by SPO11-oligonucleotide sequencing (ref49; J. Lange, M.J., and S.K., unpublished data): At the A3 hotspot in C57BL/6 mice, 6-fold more DSBs (SPO11 oligonucleotides) occurred in the central 200 bp compared to the flanking 1.8 kb (900 bp on each side; J. Lange, M.J., and S.K., unpublished data). By contrast, only half of the detected noncrossovers mapped to this central region (57 of 111 noncrossovers). Thus, it seems likely that gene conversion distribution reflects a feature(s) of the recombination mechanism rather than DSB distribution alone.
Crossover gene conversion tracts include central polymorphisms more often than noncrossovers; however, some crossover gene conversion tracts do not (25–50%), which would not have been evident by single chromatid analysis (Supplementary Fig. 3). Hotspots with biased initiation show crossover breakpoint asymmetry, such that most breakpoints from one orientation are offset from those of the other orientation33. The subset of breakpoints from both orientations that overlap could have been attributed to rare initiation on the other chromosome. Instead, our results reveal these to be crossovers with both breakpoints (and the intervening gene conversion tract) offset to one side of the hotspot center. These tracts are typically shorter, suggesting that they have other mechanistic differences as well.
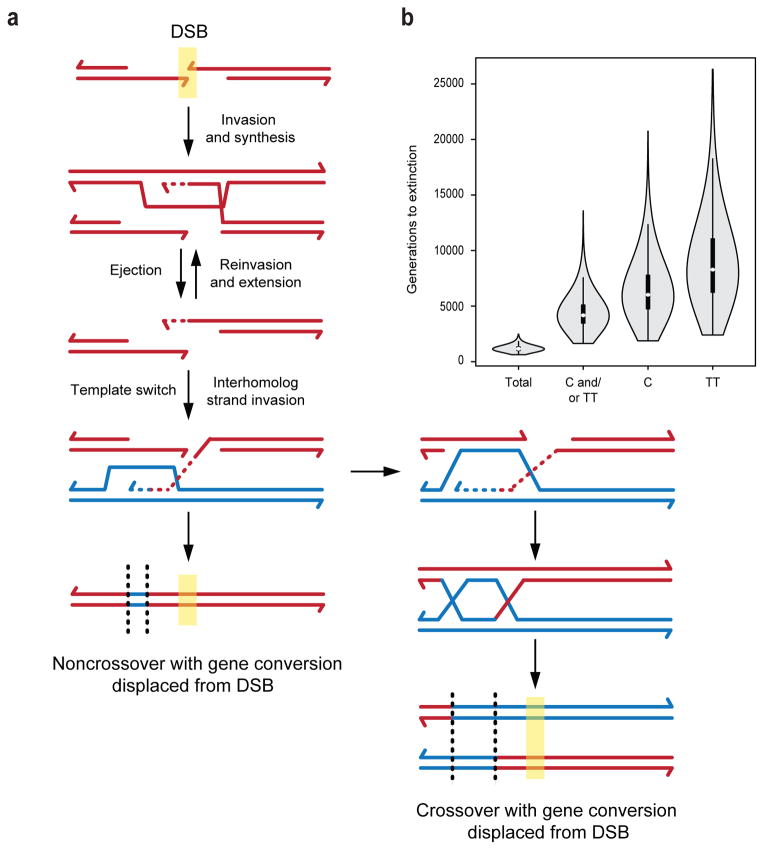
In principle, gene conversion at the hotspot center could be reduced relative to DSB frequency by mismatch repair of heteroduplex intermediates biased toward restoration of parental sequences at the initial site of strand invasion10,50. Alternatively, parental sequences could be restored by initial strand invasion into the sister chromatid followed by polymerization1 (Fig. 4a). Ejection of the newly synthesized DNA from the sister would free it to switch templates and prime further DNA synthesis from the homolog, leading to gene conversion at a distance from the DSB site. In support of this model, intermediates involving invasion into the sister have been detected physically in budding yeast51,52 as have intersister repair events53. Our data suggest that these template switches may be prevalent in mouse, and one could speculate that sister invasion may be favored in situations where heterologies occur near DSB sites, as in A3. A variation of this model is that both ends initiate strand invasion, one into the sister and the other into the homolog1,10, such that asymmetry in intermediates leads to off-center gene conversion. Such multi-template engagement may enhance the efficiency of recombination1, as well as reducing gene conversion at the hotspot center.
Figure 4. Models of recombination and hotspot loss.
a. A model for how intersister recombination can result in displacement of gene conversion away from DSB sites.
b. Modelling the speed of hotspot loss at A3. Assuming that every recombination event results in conversion to the hotspot-disrupting allele, the polymorphisms inferred to make the hotspot more active are extinguished in 1160 generations on average in Monte Carlo simulations (Total). However, given that 80% of the gene conversions detected at A3 by tetrad analysis (crossovers and noncrossovers combined) preserved the PRDM9 binding site, extinction of either one of the hotspot-active polymorphisms is predicted to take a much more substantial 4176 generations (C and/or TT). Even a more modest frequency of PRDM9 binding site retention (i.e., in 40% of gene conversions) would substantially extend the time to extinction (3665 generations). Not shown are similar simulations for the Psmb9 hotspot; however, recombination would lead to its extinction in 113 generations if the hotspot was extinguished in every event, whereas the observed retention of the critical PRDM9 binding polymorphism in a substantial fraction of gene conversions (6/11) predicts a much longer time to extinction (536 generations). Transmission frequencies were used to estimate the number of generations until loss of the indicated PRDM9 binding site polymorphisms using Wright-Fischer modeling. A violin plot is used to show the median number of generations (white dot), the first to third quartiles (black bar), and standard deviation (whiskers) and is framed by the probability density at each point.
Gene conversion presents a significant dilemma for understanding how hotspot locations might be maintained in organisms in which the recombination landscape is determined by the sequence-specific DNA binding protein, PRDM9. In individuals heterozygous for hotspot alleles that display differential PRDM9 binding, gene conversion will tend to favor the transmission of the hotspot-disrupting polymorphisms to offspring, leading to hotspot erosion over time. That hotspots exist despite this notion that they will rapidly extinguish themselves has been termed the “hotspot paradox”54. Indeed, signatures of PRDM9 motif decay are readily apparent in the human genome compared to chimpanzee29. Further, some human recombination hotspots show transmission distortion of hotspot-disrupting polymorphisms that has been modelled to lead to rapid hotspot extinction33,41. Yet, hotspots are long-lived enough to cause linkage disequilibrium, implying that they can be maintained over tens of thousands of generations55.
Given that PRDM9 binds to hotspot centers, as shown here for A3 and previously for Psmb9 and other hotspots31,32, how can hotspots persist over evolutionary time scales? Examining a large number of gene conversions at A3, we found that polymorphisms favoring PRDM9 binding are frequently preserved in recombinant molecules. This would be predicted to promote hotspot longevity. Multiple factors contribute to preservation of the PRDM9 binding site. Noncrossover gene conversion tracts are short and are distributed such that only a fraction includes the hotspot-disrupting polymorphisms (20%). While crossover gene conversion tracts are longer, they also do not always incorporate the hotspot-disrupting polymorphisms (75%). Further, noncrossovers are much more frequent (10 to 20-fold) than crossovers, such that the more frequent conversion at the PRDM9 site in crossovers is offset by their lower numbers.
Thus, the polymorphisms inferred to make the A3 hotspot highly active would be expected to be much more slowly extinguished when considering transmission data from tetrad analysis than assuming that every recombination event results in conversion to the hotspot-disrupting allele, as is usually modelled56,57 (C and/or TT versus Total, Fig. 4b). Even at Psmb9, where the numbers of crossovers and detectable noncrossovers are more similar, hotspot-disrupting polymorphisms are included in only ~60% of gene conversions. Thus, gene conversion frequently spares the PRDM9 binding site with the result that erosion of hotspots is slowed.
Methods
Animal husbandry
In this study F1 hybrids are identified as haplotype 1 x haplotype 2 regardless of the parent of origin. A/J x DBA/2J F1 hybrids were derived from parental strains A/J and DBA/2J obtained from The Jackson Laboratory. B10 x R209 and B10.A x SGR F1 hybrids were bred from parental strains C57BL/10JCrl (B10), B10.A-H2a H2-T18a/SgSnJ (B10.A), B10.A(R209) (R209), and B10.MOL-SGR (SGR)60 at the Institute of Human Genetics, Montpellier, France. All experiments were performed in accordance with relevant regulatory standards and were approved by the MSKCC Institutional Animal Care and Use Committee or carried out according to CNRS (Centre National de la Recherche Scientifique) guidelines.
Detection of recombinant molecules in isolated oocytes
Mammalian female meiosis initiates during embryogenesis and then arrests in the dictyate state, between diplonema and diakinesis of meiotic prophase I, until ovulation; dictyate oocytes have completed meiotic recombination but have yet to undergo metaphase I, such that they contain all four chromatids from a single meiosis. Ovaries from 25 to 30-day-old mice were dissected and germinal vesicle stage oocytes were isolated in M9 medium supplemented with 0.1% BSA. Single oocytes were transferred to 5 μl of PBND lysis solution (10 mM Tris-HCl pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.1 mg/mL gelatin, 0.45% NP40, 0.45% Tween-2061), incubated at 50 °C for 30 min followed by 95 °C for 10 min. A first round of PCR was performed in a volume of 50 μl with 3.125 U Taq DNA polymerase (MP Biochemicals) and 0.315 U Pfu DNA polymerase (Promega), with primers 881U21 and 10225L21 (Supplementary Table 2a), in the buffer described in62. One μl of PCR product was incubated with 3.5 U S1 Nuclease (Invitrogen) in a volume of 10 μl at room temperature for 30 min, followed by the addition of 90 μl of 5 mM Tris-HCl, pH 8.0. One μl of the S1 nuclease-digested PCR product was used for a second round of nested PCR, in a volume of 10 μl. For detecting parental alleles and crossover products in B10 x R209 F1 hybrid, the second PCR was performed with primers 7716U21 and 3457L19. The PCR products were then digested with TaqI and BstXI, generating fragments of allele-specific length (B10, 2,099 bp; R209, 2,274 bp; B10-R209, 2,020 bp; R209-B10, 2,355 bp) (Fig. 1b). Crossover products in the B10.A x SGR F1 hybrid, as well as noncrossovers in both hybrids were detected by performing the second PCR reaction with allele-specific primers (Fig. 1c,d and Supplementary Table 2a,b).
From 128 and 210 oocytes isolated from 3 B10xR209 F1 mice and 3 B10.AxSGR F1 mice, detectable PCR products were obtained from 119 and 205 oocytes, respectively. Amplification efficiency was further estimated by assessing the amplification of each parental allele in every oocyte in universal PCR reactions. B10 and R209 were distinguished following TaqI/BstXI digestion as described above. The B10.A allele is distinguished from SGR by the presence of a 57-bp insertion, which was detected by agarose gel electrophoresis following universal PCR amplification. Under the assumption that the 4 chromatids present in one oocyte are amplified independently from each other and therefore that the number of amplified chromatids per oocyte follows a binomial distribution, this estimate predicted the probability of detecting either a crossover or noncrossover (Supplementary Table 3). All four possible types of molecules (both parental alleles and both reciprocal crossover products) were amplified independently with allele-specific primers from oocytes in which a crossover was detected (Fig. 1c). The positions of the exchanges were determined by restriction digestion and sequencing of the PCR products. Products that displayed either a mix of both alleles along their entire length or a distal exchange point were discarded, as they resulted from PCR artefacts. Noncrossovers were confirmed by amplifying the recipient parental allele with flanking allele-specific primers and sequencing. Only samples in which the converted allele could be detected on sequence profiles were retained as bona fide noncrossover events. Seven of 9 PCR products from B10xR209 F1 hybrid and 5 of 14 PCR products from B10.AxSGR F1 hybrid were confirmed by sequencing.
Isolation of 4C spermatocytes
Testes from 2 to 4-month old A/J x DBA/2J F1 hybrids were decapsulated and seminiferous tubules were incubated at 33°C in Gey’s balanced salt solution (GBSS) with 0.5 mg/ml collagenase (Worthington CLS4) for 15 min shaking at 500 rpm. The tubules were rinsed and treated with 0.5 mg/ml trypsin (Sigma T9935) supplemented with 1 μg/ml DNase I (Sigma DNEP) at 33 °C shaking at 500 rpm for 15 min. Trypsin was inactivated by addition of 5% fetal calf serum. Cells were individualized by repeated pipetting and filtration through a 70-μM cell strainer (BD Falcon 352350). Cells were washed several times in GBSS with 5% FCS and 1 μg/ml DNase I and stained with 5 μg/ml Hoechst 33342 (resuspended in DMSO) for 2 hr at 33°C shaking at 500 rpm. Just prior to fluorescent activated cell sorting, cells were stained with 0.2 μg/ml propidium iodide (PI) and filtered through a 40-μm cell strainer. Cells were sorted on a MoFlo cytometer (Dako) with a 350 nm argon laser35. Late-stage primary spermatocytes exhibit the highest blue and red fluorescence due to their DNA content and chromatin structure63, respectively, allowing them to be readily sorted from cells that do not contain a full complement of chromatids, such as secondary spermatocytes and spermatids. Dead cells were gated by PI staining. Live cells with the highest blue and red fluorescence intensity were sorted, repeatedly washed with TBS, and counted with a hemocytometer. Cells were diluted and plated at a density of 20 (noncrossover/crossover assay) or 100 (crossover assay) cells in 5 μl TBS per well in 96-well plates and frozen at −80 °C for storage. A portion of sorted cells were surface spread64 and stained59 with antibodies that recognize the axis component, SYCP3, the crossover marker, MLH1, and DAPI to allow staging and purity assessment. Splenic cells from the same mice were used as a somatic control. They were separated by macerating through a 70-μm cell strainer, repeatedly washed in GBSS with 5% FCS and 1 μg/ml DNase I and stained with Hoechst and PI for cell sorting. Live splenic cells were plated at the same cell numbers as spermatocytes and no recombinants were detected (16,200 tested cells or 64,800 molecules). Epididymides (sperm) and liver (somatic control) from the same mice were isolated for DNA extraction and recombination analysis36. No recombinants were detected in liver controls (14,400 molecules).
Amplification and detection of recombinants in isolated spermatocytes
Cells were lysed and extracted in 0.38% Igepal CA-630 (Sigma), 0.38% Tween-20, and 0.3 μg/μl Proteinase K (Qiagen) at 55°C for 2 hr. Proteinase K was inactivated at 96 °C for 15 min prior to PCR. Extracted wells were diluted 6-fold into PCR buffer containing the universal primers A3f600 and A3r6000 (5.4 kb, Supplementary Table 2c,d) that amplify across the entire A3 hotspot. PCR conditions were 96 °C for 1 min followed by 6 cycles at 96 °C for 20 sec, 60 °C for 30 sec, and 65 °C for 7 min and 19 cycles at 96 °C for 20 sec, 58 °C for 30 sec, and 65 °C for 7 min. 1/30th of the reaction was treated with 7.0 U S1 nuclease for 20 min at room temperature in a volume of 10 μl and then diluted 10-fold in 5 mM Tris buffer (pH 7.4) containing 2 μg/μl of sonicated salmon sperm DNA as carrier. 1.6 μl of the diluted S1 nuclease reaction was seeded into separate primary allele-specific PCR for each crossover configuration or using allele-specific primers on one side of the hotspot and universal primers on the other to amplify noncrossovers and crossovers (Fig. 2b and Supplementary Table 2c,d)36. Recombinants were genotyped by dot blotting (Supplementary Table 2e) and several were confirmed by cloning and restriction digest and/or genotyping36. Spermatocytes were analyzed from three mice.
Southwestern blotting assays
Southwestern blotting assays were performed as described previously32, using full-length His-tagged mouse PRDM9wm7 and PRDM9b. The 200 to 300-bp probes covering the A3 hotspot were generated by PCR amplification of genomic DNA from either strain (A/J or DBA/2J), with XbaI or NheI site-tailed primers (Supplementary Table 2f). PCR fragments were digested with XbaI or NheI to generate a CTAG 5′-overhang at each end. The 57-bp probes containing the PRDM9-binding motif at the center of the A3 hotspot were made by annealing complementary oligonucleotides leaving a 3-bp 5′-overhang with a G in second position at each end (Supplementary Table 2g). DNA fragments were then labeled by end-filling with alpha-32P dCTP as described previously32. His-tagged PRDM9 was detected with a monoclonal mouse anti-polyhistidine primary antibody (Sigma, H1029, clone HIS-1).
Transmission distortion calculations
To estimate the number of generations required for polymorphisms that are causative for transmission distortion to become fixed in a population, Monte Carlo simulations were performed with 1,000 samples using a Wright-Fisher model. For each generation starting from an initial population of 10,000 heterozygous individuals, 20,000 gametes were chosen at random, with alleles transmitted at the frequency experimentally determined by mouse tetrad analysis (A3: TT = 0.4998040, C = 0.4997082, both sites = 0.4995121; Psmb9: polymorphism 70 = 0.4939024) or at the frequency predicted from the model that all DSBs lead to loss of PRDM9 binding (Total recombination for A3 = 0.4975647 and for Psmb9 = 0.4646341). Transmission frequencies were determined at A3 using the formula:
where Ncell is the number of cells tested and Aconv and Dconv are the number of conversions to the A/J or DBA2/J genotype at a particular polymorphism, respectively. The same approach was used to determine transmission frequencies at Psmb9. Each simulation ended when the allele was fixed or it reached 50,000 generations. Simulations were performed in R version 2.15.3.
URLs
R version 2.15.3, http://cran.r-project.org; R script for the transmission distortion simulation algorithm, http://cbio.mskcc.org/public/Cole_Mouse_Tetrads/; Confidence Intervals (CI) were calculated at http://vassarstats.net/.
Supplementary Material
Acknowledgments
We thank members of the Jasin, Keeney, and de Massy laboratories, especially Julian Lange, Esther de Boer, Sam Tischfield, Shintaro Yamada, and Shaun Peterson. We also thank Pat Hunt (WSU) for encouraging the oocyte experiments and Aaron Gabow (MSKCC) and Yue Lu (MDACC) for fixation calculations, and the Réseau des Animaleries de Montpellier (RAM) and Dominique Haddou for management of mice. This work was supported by Ruth L. Kirschstein NRSA F32HD51392 (F.C.) and Cancer Prevention and Research Institute of Texas award R1213 (F.C.), Centre National de la Recherche Scientifique (F.B., C.G., and B.dM), Agence Nationale pour la Recherche (program ANR-09-BLAN-0269-01 to B.dM), Association pour la Recherche contre le Cancer (B.dM), and NIH grants R01GM105421 (M.J., S.K.) and R01HD53855 (S.K., M.J.). This paper is dedicated to the memory of our colleague Jérôme Buard.
Footnotes
AUTHOR CONTRIBUTIONS
F.C., F.B., S.K., B.dM, and M.J. conceived the study, interpreted the data and wrote the manuscript. F.C. and F.B. performed the spermatocyte and oocyte (and associated) recombination experiments, respectively. F.B and C.G. performed the Southwestern blotting. F.C., M.J. and S.K. estimated the fixation rates.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hunter N. Meiotic Recombination. In: Aguilera A, Rothstein R, editors. Topics in Current Genetics, Molecular Genetics of Recombination. 17/2007. Springer-Verlag; Heidelberg: 2007. pp. 381–442. [Google Scholar]
- 2.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 3.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–99. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 4.Winge O, Laustsen O. On two types of spore germination, and on genetic segregations in Saccharomycetes demonstrated through single-spore culture. CR Trav Lab Carlsberg, Ser Physiol. 1937;22:99–117. [Google Scholar]
- 5.Hurst DD, Fogel S, Mortimer RK. Conversion-associated recombination in yeast (hybrids-meiosis-tetrads-marker loci-models) Proc Natl Acad Sci USA. 1972;69:101–5. doi: 10.1073/pnas.69.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 7.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 8.Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 9.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 10.Martini E, et al. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 2011;7:e1002305. doi: 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–99. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]
- 13.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 14.Paigen K, Petkov P. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet. 2010;11:221–33. doi: 10.1038/nrg2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 16.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–30. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–12. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole F, Keeney S, Jasin M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell. 2010;39:700–10. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol Cell. 2005;20:563–73. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 20.Guillon H, de Massy B. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat Genet. 2002;32:296–9. doi: 10.1038/ng990. [DOI] [PubMed] [Google Scholar]
- 21.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–6. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 22.Ng SH, Parvanov E, Petkov PM, Paigen K. A quantitative assay for crossover and noncrossover molecular events at individual recombination hotspots in both male and female gametes. Genomics. 2008;92:204–9. doi: 10.1016/j.ygeno.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baudat F, de Massy B. Cis- and trans-acting elements regulate the mouse Psmb9 meiotic recombination hotspot. PLoS Genet. 2007;3:e100. doi: 10.1371/journal.pgen.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Egel R, Lankenau D-H, editors. Recombination and Meiosis. Springer-Verlag; Berlin Heidelberg: 2007. pp. 81–123. [Google Scholar]
- 25.Cole F, Keeney S, Jasin M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 2010;24:1201–7. doi: 10.1101/gad.1944710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–24. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 27.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–5. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers S, et al. Drive Against Hotspot Motifs in Primates Implicates the PRDM9 Gene in Meiotic Recombination. Science. 2010;327:876–9. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of Mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billings T, et al. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol. 2013;14:R35. doi: 10.1186/gb-2013-14-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grey C, et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31:267–71. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 34.Buard J, Barthes P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–24. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastos H, et al. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–9. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- 36.Cole F, Jasin M. Isolation of meiotic recombinants from mouse sperm. Methods Mol Biol. 2011;745:251–82. doi: 10.1007/978-1-61779-129-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker SM, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–42. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 38.Woods LM, et al. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J Cell Biol. 1999;145:1395–406. doi: 10.1083/jcb.145.7.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–85. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bois PR. A highly polymorphic meiotic recombination mouse hot spot exhibits incomplete repair. Mol Cell Biol. 2007;27:7053–62. doi: 10.1128/MCB.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–87. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- 42.Odenthal-Hesse L, Berg IL, Veselis A, Jeffreys AJ, May CA. Transmission distortion affecting human noncrossover but not crossover recombination: a hidden source of meiotic drive. PLoS Genet. 2014;10:e1004106. doi: 10.1371/journal.pgen.1004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarbajna S, et al. A major recombination hotspot in the XqYq pseudoautosomal region gives new insight into processing of human gene conversion events. Hum Mol Genet. 2012;21:2029–38. doi: 10.1093/hmg/dds019. [DOI] [PubMed] [Google Scholar]
- 44.Yauk CL, Bois PR, Jeffreys AJ. High-resolution sperm typing of meiotic recombination in the mouse MHC Ebeta gene. EMBO J. 2003;22:1389–97. doi: 10.1093/emboj/cdg136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 46.Chen SY, et al. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15:401–15. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kauppi L, et al. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013;27:873–86. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker CL, Walker M, Kajita S, Petkov PM, Paigen K. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 2014;24:724–32. doi: 10.1101/gr.170167.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–31. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radford SJ, Sabourin MM, McMahan S, Sekelsky J. Meiotic recombination in Drosophila Msh6 mutants yields discontinuous gene conversion tracts. Genetics. 2007;176:53–62. doi: 10.1534/genetics.107.070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh SD, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–72. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31:324–36. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boulton A, Myers RS, Redfield RJ. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci USA. 1997;94:8058–63. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coop G, Myers SR. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007;3:e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calabrese P. A population genetics model with recombination hotspots that are heterogeneous across the population. Proc Natl Acad Sci USA. 2007;104:4748–52. doi: 10.1073/pnas.0610195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pineda-Krch M, Redfield RJ. Persistence and loss of meiotic recombination hotspots. Genetics. 2005;169:2319–33. doi: 10.1534/genetics.104.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–79. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cole F, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nature Cell Biol. 2012;14:424–30. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shiroishi T, Sagai T, Hanzawa N, Gotoh H, Moriwaki K. Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 1991;10:681–6. doi: 10.1002/j.1460-2075.1991.tb07997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanneman WH, Schimenti KJ, Schimenti JC. Molecular analysis of gene conversion in spermatids from transgenic mice. Gene. 1997;200:185–92. doi: 10.1016/s0378-1119(97)00416-2. [DOI] [PubMed] [Google Scholar]
- 62.Jeffreys AJ, Neumann R, Wilson V. Repeat unit sequence variation in minisatellites: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell. 1990;60:473–85. doi: 10.1016/0092-8674(90)90598-9. [DOI] [PubMed] [Google Scholar]
- 63.Belloc F, et al. A flow cytometric method using Hoechst 33342 and propidium iodide for simultaneous cell cycle analysis and apoptosis determination in unfixed cells. Cytometry. 1994;17:59–65. doi: 10.1002/cyto.990170108. [DOI] [PubMed] [Google Scholar]
- 64.Barchi M, et al. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 2008;4:e1000076. doi: 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baudat F, de Massy B. Parallel detection of crossovers and noncrossovers in mouse germ cells. Methods Mol Biol. 2009;557:305–22. doi: 10.1007/978-1-59745-527-5_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.