Background: PINK1 and Parkin promote mitochondrial autophagy upon loss of mitochondrial membrane potential.
Results: smARF induces Parkin/PINK1-mitochondrial autophagy in neurons.
Conclusion: smARF is upstream of Parkin and PINK1 in mitochondrial autophagy.
Significance: Parkin/PINK1-mitochondrial autophagy can be triggered by intrinsic signaling.
Keywords: Mitochondria, Mitophagy, Parkin, Parkinson Disease, PTEN-induced Putative Kinase 1 (PINK1), smARF
Abstract
Parkinson disease (PD) is a complex neurodegenerative disease characterized by the loss of dopaminergic neurons in the substantia nigra. Multiple genes have been associated with PD, including Parkin and PINK1. Recent studies have established that the Parkin and PINK1 proteins function in a common mitochondrial quality control pathway, whereby disruption of the mitochondrial membrane potential leads to PINK1 stabilization at the mitochondrial outer surface. PINK1 accumulation leads to Parkin recruitment from the cytosol, which in turn promotes the degradation of the damaged mitochondria by autophagy (mitophagy). Most studies characterizing PINK1/Parkin mitophagy have relied on high concentrations of chemical uncouplers to trigger mitochondrial depolarization, a stimulus that has been difficult to adapt to neuronal systems and one unlikely to faithfully model the mitochondrial damage that occurs in PD. Here, we report that the short mitochondrial isoform of ARF (smARF), previously identified as an alternate translation product of the tumor suppressor p19ARF, depolarizes mitochondria and promotes mitophagy in a Parkin/PINK1-dependent manner, both in cell lines and in neurons. The work positions smARF upstream of PINK1 and Parkin and demonstrates that mitophagy can be triggered by intrinsic signaling cascades.
Introduction
Elucidating the function of genes responsible for familial forms of Parkinson disease (PD)2 has provided crucial insight into disease mechanisms. For instance, Parkin and PINK1, two genes responsible for autosomal-recessive PD, act together to regulate mitochondrial function (1, 2). In Drosophila, loss of Parkin or PINK1 leads to similar mitochondrial defects, and overexpression of Parkin rescues PINK1 loss of function (3–6). This positions Parkin downstream of PINK1 in a common genetic pathway affecting mitochondrial function. The findings also provide support for the longstanding hypothesis that mitochondrial dysfunction is an early event in PD pathogenesis. Indeed, decreased complex I activity, increased mitochondrial DNA mutations, and exposure to environmental toxins that inhibit complex I of the electron transport chain have all been associated with PD (7–9). However, the mechanisms leading to mitochondrial defects in PD remain elusive. Mitochondria are double-membrane organelles, which carry out oxidative phosphorylation to generate most of the ATP within cells. This process involves several membrane-associated protein complexes that transport electrons from NADH or FADH to oxygen and shuttle protons from the matrix to the intermembrane space. This generates an electro-chemical potential across the inner membrane, which is then used by the ATP synthase to generate ATP. The mitochondrial membrane potential can be dissipated by chemical protonophores, such as carbonyl cyanide m-chlorophenylhydrazone (CCCP), which are also called mitochondrial uncouplers because they uncouple the electron transport chain from the ATP synthesis. Depolarization of mitochondria in mammalian cells using such chemical uncouplers induces the translocation of Parkin, an E3 ubiquitin ligase, from the cytosol to the mitochondria (10). Subsequently, these depolarized mitochondria are targeted for degradation by autophagy (mitophagy). PINK1, a normally short lived mitochondrial protein kinase, needs to accumulate to high levels on the outer surface of such depolarized mitochondria in order to trigger Parkin recruitment and mitophagy (10–14). Recent work resolving the structure of Parkin reveals that Parkin is autoinhibited under basal conditions, and becomes deinhibited in response to mitochondrial depolarization and PINK1 kinase activity (15–21). Parkin then ubiquitinates mitochondrial proteins, such as the mitofusins and voltage-dependent anion channels, which ultimately leads to mitochondrial degradation by autophagy (13, 22–26). Although the downstream events in this pathway have been increasingly characterized, the physiological stimuli and the upstream signaling leading to the activation of PINK1/Parkin mitophagy remain largely unexplored. Interestingly, the short mitochondrial isoform of ARF (smARF), an alternative translation product of the tumor suppressor p19ARF, localizes to mitochondria, where it induces their depolarization and autophagy (27). We report here that the expression of smARF in cell lines and in neurons depolarizes mitochondria and triggers mitophagy in a Parkin- and PINK1-dependent fashion, without the requirement of chemical uncouplers. The findings place smARF upstream of PINK1 and Parkin in a novel signaling pathway, which may shed light on the intrinsic mechanisms of PINK1/Parkin mitophagy in PD.
MATERIALS AND METHODS
Cell Culture and Microscopy
HEK293T and HeLa cells were obtained from ATCC and were grown at 37 °C with 5% CO2 in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. MitoTrackerTM dyes were used at 100 nm in medium for 30 min, followed by a 30-min wash in media without the dye. Cells were fixed with 4% formaldehyde in PBS, permeabilized with 0.25% Triton X-100 in PBS, and blocked in 5% bovine serum albumin before incubation with primary and secondary antibodies. Cortical and hippocampal neurons were isolated from 14- and 15-day-old embryos, respectively, from WT or PINK1−/− mice. Neurons were plated on poly-l-lysine and kept in culture for 5 days in Neurobasal medium (Invitrogen) supplemented with penicillin/streptomycin, N2, B27, and l-glutamine and then transfected overnight with Lipofectamine 2000 in 50% Opti-MEM (Invitrogen), 50% neurobasal medium. For recruitment and mitophagy experiments, HeLa cells were transduced at a multiplicity of infection of 3:1 with lentivirus encoding GFP-FLAG-Parkin or GFP for 24 h, followed by transfections using Lipofectamine 2000TM (Invitrogen) following the manufacturer's instructions. Cells were imaged using a Zeiss LSM710 confocal microscope using a ×63 Plan Apo objective (Zeiss). For Parkin recruitment and mitophagy experiments, images were captured and scored in a blinded fashion, and at least 50 cells were counted for each condition. Experiments were repeated n = 3 times.
Cloning, Vectors, and RNAi
p19ARF-FLAG and p19ΔNT-FLAG constructs were a generous gift from Adi Kimchi (27). p19M1I-FLAG and p19M45I-FLAG were generated by mutagenesis using the QuikChangeTM mutagenesis kit (Agilent Technologies). Human mCherry-Parkin was obtained from Addgene (catalog no. 23956). PINK1-c-Myc was obtained from Addgene (catalog no. 13314). Rat GFP-Parkin was generated by excising the Parkin coding sequence from pcDNA3.1 Parkin (28) and inserting it into pEGFP-C1. Rat FLAG-Parkin (28) was excised from pcDNA3.1 and inserted into pRRLsinPPTeGFP to generate the rat GFP-FLAG-Parkin lentivirus construct. Human GFP-Parkin was a gift from Michael Schlossmacher (University of Ottawa). siGENOME human PINK1 siRNA (Dharmacon) was used either as a SMARTpool (M-004030-02) containing four different sequences or individually as indicated: PINK1-02, GAAAUCCGACAACAUCCUU; PINK1-04, GGAGCCAUCGCCUAUGAA. Parkin siRNA (Invitrogen) sequences were as follows: Parkin 1989, UUUACAGAGAAACACCUUGUCAAUG; Parkin 2551, CCGACUCUCUCCAUCAGAAGGGUUU.
Flow Cytometry
HEK293T cells were stained with 200 nm tetramethylrhodamine methyl ester (TMRM) in medium (30 min, followed by a 15-min wash), trypsinized, and washed before processing on a FACSCalibur cell analyzer (BD Biosciences) with an argon laser at 488 nm and band pass filters at 530 ± 15 nm for green and 585 ± 21 nm for red. A minimum of 50,000 cells were used per condition per experiment. pcDNA-transfected cells were used as a standard to delimitate quadrants for TMRM-positive and TMRM-negative cells. Experiments were repeated n = 3 times.
Immunoblotting
Cells were lysed using radioimmune precipitation buffer (0.1% SDS, 0.5% Triton X-100, 0.5% sodium deoxycholate, 150 mm Tris, pH 7.5, 300 mm NaCl) with a mixture of protease inhibitors (aprotinin, leupeptin, benzamidine, PMSF). Samples were boiled in a final concentration of 1× Laemmli buffer with 30 mm DTT, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. Membranes were blocked with 5% milk, washed, and incubated overnight at 4 °C with primary antibody in 3% BSA. Membranes were blocked again with 5% milk and washed and then incubated with peroxidase-conjugated secondary antibody (Jackson) at a 1:5000 dilution. Membranes were developed using ECL substrate (PerkinElmer Life Sciences). All washing, blocking, and staining steps were performed with PBS containing 0.1% Tween. Densitometric quantification was performed using ImageJ.
Cell Fractionation
Two confluent 10-cm plates of HEK293 or HeLa cells were harvested in PBS, pelleted, and resuspended in 1.5 ml of mitochondrial isolation buffer, pH 7.4, with 1 mg/ml BSA (0.1%). Mitochondrial isolation buffer was composed of 210 mm mannitol, 70 mm sucrose, 5 mm Tris, pH 7.4, 200 μm EGTA, 100 μm EDTA, and protease inhibitors (aprotinin, leupeptin, benzamidine, PMSF). Cells were homogenized using 15 strokes of a Teflon/glass homogenizer at 900 rpm. Cell debris was pelleted twice by centrifugation at 600 × g for 10 min. The supernatant was then spun at 12000 × g to pellet mitochondria. Mitochondria were washed in 1 ml of mitochondrial isolation buffer, pH 7.4, without BSA, EGTA, or EDTA. Mitochondria were then washed again with the same buffer and repelleted and the resuspended in 50–100 μl of the same buffer. Protein concentration was measured by BCA (Pierce).
MitoTracker Fluorescence Calculation
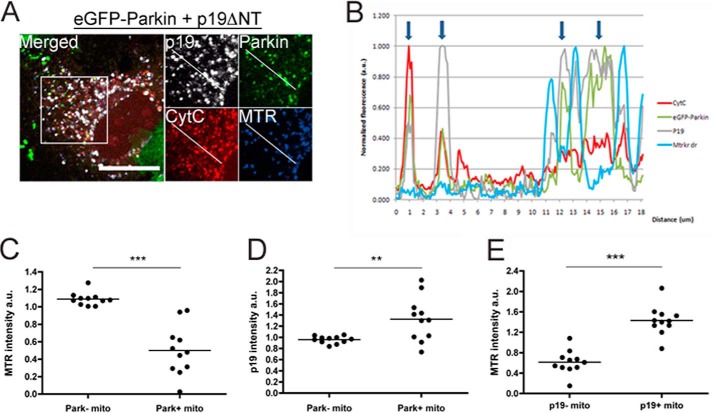
Cells exhibiting incomplete GFP-FLAG-Parkin translocation to mitochondria were selected. Using ImageJ software, cytochrome c staining was used to select mitochondrial objects. GFP and MitoTracker Deep Red fluorescence were measured for every object. Objects with ≤10% relative GFP fluorescence were counted as Parkin-negative, whereas those with >10% were scored as positive. Each point on the scatter plot represents the mean relative intensity of MitoTracker Deep Red for each subset (Parkin-negative (−) or Parkin-positive (+)) for all objects in one cell. Student's paired t test was used for statistics. n = 11.
Antibodies
Immunofluorescence
The antibodies used were as follows: p19ARF (Abcam ab80; diluted 1:500), p14ARF (Abcam ab3212; diluted 1:1000), AP-2 (Sigma A7107; diluted 1:2000), Hsp60 (Sigma H-3524; diluted 1:1000), Tim23 (BD 611222; diluted 1:1000), cytochrome c (BD 556432; diluted 1:500).
Immunoblotting
The antibodies used were as follows: p19ARF (Abcam ab80; diluted 1:1000), PINK1 (Novus BC100-494; diluted 1:3000), Hsp60 (H-3524; diluted 1:10,000), Mitofusin1 (Mfn1) (Novus NB110-58853; diluted 1:2000), Mitofusin 2 (MFN2) (Sigma M6319; diluted 1:2000), Parkin (Santa Cruz Biotechnology, Inc. SC-32282; diluted 1:1000), actin (Millipore MAB1501; diluted 1:200,000) GAPDH (Novus NB300-320; diluted 1:10,000); p32 (Millipore AB2991; diluted 1:3000).
Statistical Tests
All statistical analyses were performed by either one-way or two-way parametric analysis of variance using Bonferroni's t test for post hoc comparison and a requisite p value of 0.05 or less except for in Fig. 3, C–E, where a paired t test was used. All error bars represent S.E.
FIGURE 3.
smARF induces Parkin recruitment to individual mitochondria with low Δψm and high levels of smARF. HeLa cells transduced with eGFP-FLAG-Parkin for 24 h and transfected with p19ΔNT were stained with MitoTracker Deep Red 24 h after transfection followed by immunofluorescence using cytochrome c (CytC) and p19ARF antibodies. Only cells with incomplete eGFP-FLAG-Parkin translocation to the mitochondria were selected. A representative image is shown in A (scale bar, 20 μm). B, fluorescence intensity plot of the line drawn with ImageJ in A. Arrows, mitochondria with eGFP-Parkin fluorescence. C and D, the mean MitoTracker Deep Red fluorescence intensity (C) or p19ARF intensity (D) of Parkin+ (mitochondria with higher than cell average eGFP-Parkin signal) versus Parkin− (mitochondria with lower than cell average eGFP-Parkin signal) mitochondria is plotted. E, point histogram of MitoTracker Deep Red intensity of p19+ (with higher than cell average p19 signal) versus p19− (lower than cell average p19 signal) mitochondria derived from the fluorescence analysis. n = 11. **, p < 0.01; ***, p < 0.001.
RESULTS
smARF Triggers Mitochondrial Depolarization and Stabilizes PINK1
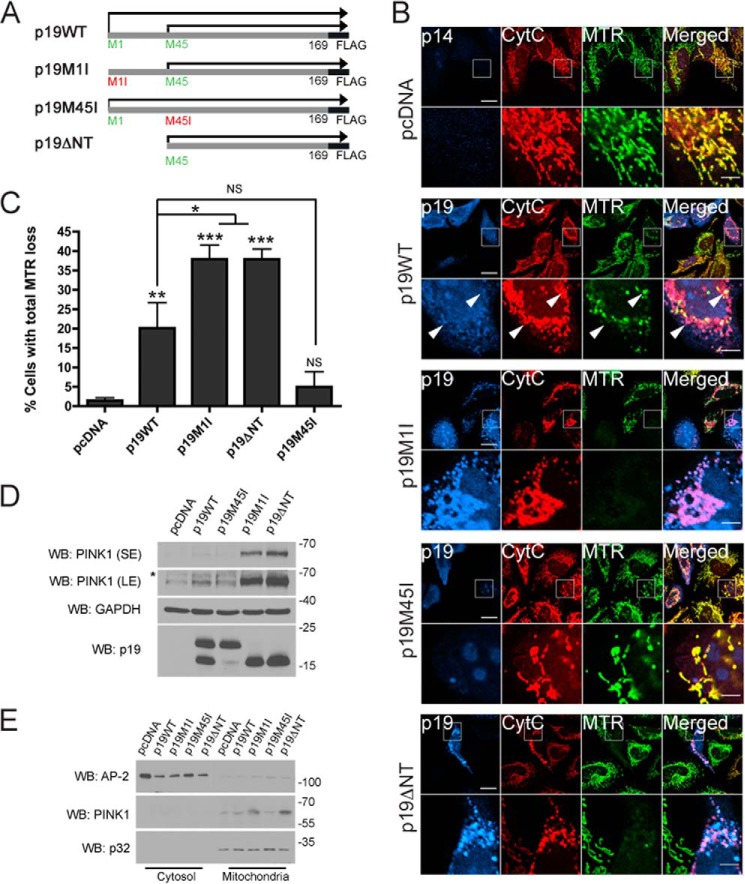
p19ARF, a tumor suppressor gene in the INK4/ARF locus, encodes p19ARF, a 169-amino acid protein predominantly localized to the nucleus. A previous study demonstrated that translation of p19ARF results in two protein products, migrating at ∼19 and 15 kDa (27). The shorter product, which was named smARF for short mitochondrial ARF, was shown to be the result of an alternative translation site at codon 45 of the p19ARF mRNA. In the same study, smARF was shown to localize to mitochondria and induce the loss of mitochondrial membrane potential (27). To characterize further the specificity of smARF-induced mitochondrial depolarization, we expressed either wild-type p19ARF (p19WT-FLAG) or various p19ARF mutants in HeLa cells and surveyed mitochondrial polarization using the potentiometric dye MitoTracker Deep Red (MTR) (Fig. 1, A and B). To express smARF but not full-length p19ARF, we forced translation initiation to begin at methionine 45 by using either a truncated construct lacking the first 44 amino acids of the N terminus (p19ΔNT-FLAG) or a construct in which the first methionine in full-length p19ARF was mutated to isoleucine (p19M1I-FLAG) as described previously (27). Conversely, a p19M45I-FLAG mutant was used as a control to express full-length p19ARF but not smARF. In agreement with Reef et al. (27), we found that smARF (p19M1I and p19ΔNT) predominantly localizes to mitochondria (Fig. 1B). As expected, smARF expression (p19M1I and p19ΔNT) led to a complete loss of mitochondrial membrane potential in a significant fraction of transfected cells (Fig. 1, B and C). In contrast, few cells expressing p19M45I showed depolarized mitochondria, whereas p19WT expression led to an intermediate phenotype, consistent with the predominantly nuclear localization of p19M45I and the localization of p19WT in the nucleus (about 75% of cells) or in the nucleus and at the mitochondria (about 25% of cells) (data not shown). Although it is not known how full-length p19 or smARF can depolarize mitochondria, we observed that not all p19-positive mitochondria are depolarized (Fig. 1B, arrows), suggesting that full-length p19 or smARF does not directly depolarize mitochondria. This also suggests that smARF depolarizes mitochondria locally rather than globally. Mitochondrial depolarization with chemical uncouplers has been shown to increase the levels of full-length PINK1 by stabilizing it at the outer mitochondrial membrane (OMM) (11, 12, 14, 29). We found that smARF expression strongly increased the levels of endogenous full-length 64-kDa PINK1 in HeLa cells (Fig. 1D). PINK1 levels were highest in cells exclusively expressing smARF but were also slightly elevated in p19WT-transfected cells, which expressed both smARF and full-length p19ARF (Fig. 1D). As established previously, 64-kDa PINK1 was found predominantly in mitochondrial fractions and not in the cytosol (Fig. 1E). Taken together, these results indicate that the expression of smARF but not full-length p19ARF in cells depolarizes mitochondria and increases the levels of full-length PINK1.
FIGURE 1.
smARF triggers loss of mitochondrial membrane potential and stabilizes PINK1. A, diagram of the p19ARF constructs used in this study: p19WT-FLAG, translated both from Met-1 and Met-45; p19M1I-FLAG, translated from Met-45 only; p19M45I-FLAG, translated from Met-1 only; and p19ΔNT-FLAG, which lacks the first 44 amino acids. B, HeLa cells expressing pcDNA, p19WT-FLAG (p19WT), p19M1I-FLAG (p19M1I), p19M45I-FLAG (p19M45I), or p19ΔNT-FLAG (p19ΔNT) for 24 h and stained with the mitochondrial membrane potential indicator MTR were processed for immunofluorescence against cytochrome c (CytC) and p19 or p14 antibodies. Scale bar, 20 μm for low power and 5 μm for zoom. Arrows, polarized and depolarized mitochondria in the same p19WT-transfected cell. C, the percentage of cells in B negative for MitoTracker Deep Red staining while retaining mitochondrial cytochrome c staining was measured. At least 50 cells were counted per condition per experiment. All columns are compared with pcDNA or with p19WT. n = 3. *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, HeLa cells expressing pcDNA, p19WT, p19M1I, p19M45I, or p19ΔNT for 24 h were lysed, and 50 μg of protein lysate were separated by SDS-PAGE followed by immunoblotting against PINK1, GAPDH, and p19ARF. SE, short exposure; LE, long exposure. *, nonspecific band. E, cells treated as in D were fractionated into cytosolic and mitochondria-enriched fractions and processed for immunoblotting against PINK1, AP-2 (cytosolic), and p32 (mitochondrial) antibodies. WB, Western blot. Error bars, S.E.
smARF Triggers Parkin Translocation to Mitochondria and Promotes Mitophagy
Given that smARF depolarized mitochondria and led to the stabilization of mitochondrial PINK1, we tested whether it could promote recruitment of Parkin to mitochondria. HeLa cells, which contain no endogenous Parkin, were transduced with either an eGFP-FLAG-Parkin or eGFP lentivirus, followed 24 h later with the p19ARF constructs described above or with PINK1-Myc. 24 h after the p19ARF constructs were transfected, we observed robust recruitment of Parkin to mitochondria in cells expressing either p19M1I or p19ΔNT, comparable with the recruitment seen with PINK1 overexpression (not shown). In contrast, expression of p19M45I did not lead to Parkin recruitment, whereas p19WT again led to an intermediate phenotype (Fig. 2, A and B). Thus, the expression of smARF but not of full-length p19ARF leads to Parkin recruitment. To better understand the relationship between Parkin recruitment to the mitochondria and smARF expression, we analyzed the fluorescence intensity of eGFP-Parkin, p19, and MTR on mitochondria (marked by cytochrome c staining) in cells transfected with eGFP-Parkin and p19ΔNT showing partial eGFP-Parkin recruitment. A representative cell used for these measurements is shown in Fig. 3A. A line drawn by ImageJ was used to capture the fluorescence intensity of representative mitochondria, and these values are shown in Fig. 3B. Mitochondria were classified as being either Park+ or Park−, depending on whether eGFP-Parkin fluorescence was higher or lower, respectively, than the average eGFP-Parkin fluorescence for all of the labeled mitochondria in the cell. Relative MTR fluorescence (against the average) was measured for each Park+ and Park− mitochondrion, and the average MTR fluorescence for Park+ and Park− groups was calculated for each cell (Fig. 3C). The same method was used to measure the relationship between eGFP-Parkin and p19 fluorescence intensity (Fig. 3D) and between p19 and MTR fluorescence intensity (Fig. 3E). We found that Parkin is predominantly recruited to mitochondria that lack membrane potential (Fig. 3C) and that express high levels of smARF (Fig. 3D). This suggests that smARF locally depolarizes mitochondria, leading to localized recruitment of Parkin. Surprisingly, high levels of smARF are found on mitochondria with high Δψm (Fig. 3E). Therefore, Parkin is recruited to a small subset of p19+ mitochondria that have low Δψm and are not representative of most of p19+-expressing mitochondria in these cells. Given that in most cells expressing smARF, the whole mitochondrial network is depolarized (Fig. 1C) and that smARF levels are not proportional with Δψm reduction (Figs. 1B and 3E), we conclude that smARF levels need to reach a certain threshold before collapsing Δψm and triggering subsequent Parkin recruitment. This also means that smARF is probably not directly depolarizing mitochondria by carrying protons across the inner mitochondrial membrane because p19 levels would be inversely proportional to MTR fluorescence.
FIGURE 2.
smARF expression leads to translocation of Parkin from the cytosol to mitochondria. A, HeLa cells transduced with eGFP-FLAG-Parkin or eGFP lentivirus for 24 h were transfected with PINK1-Myc, pcDNA, p19WT, p19M1I, p19M45I, p19ΔNT, or PINK1-c-Myc and processed for immunofluorescence after 24 h using cytochrome c (CytC) and p19 or p14 antibodies. Micrographs of both eGFP and eGFP-FLAG-Parkin are shown for p19WT, p19M1I, p19M45I, and p19ΔNT. Only the eGFP-Parkin micrograph is shown for the pcDNA condition. Scale bar, 20 μm for low power and 5 μm for zoom. B, histogram showing the proportion of transfected cells with eGFP-FLAG-Parkin co-localizing with cytochrome c. For each construct, at least 100 cells were counted per condition per experiment. All columns are compared with pcDNA. n = 3. **, p < 0.01; ***, p < 0.001. Error bars, S.E.
To test whether smARF can also promote mitophagy, cells were examined at 48 h after transfection with p19ARF constructs for the loss of mitochondria. We observed that a large fraction of cells expressing Parkin and either p19M1I or p19ΔNT lacked mitochondria (Fig. 4, A–C). In contrast, neither p19M45I- nor p19WT-expressing cells lacked mitochondria. Thus, the expression of Parkin with smARF but not with full-length p19ARF leads to mitophagy. Indeed, the lower levels of depolarization (Fig. 1, B and C), PINK1 stabilization (Fig. 1, D and E), and Parkin recruitment (Fig. 2, A and B) with the p19WT construct appeared insufficient to induce mitophagy. To confirm that all suborganellar compartments of the mitochondria are being degraded following 48 h of smARF and eGFP-Parkin expression, we used the additional mitochondrial markers Hsp60, a mitochondrial matrix protein, and Tim23, a mitochondrial intermembrane protein (Fig. 4B). Like cytochrome c (Fig. 4A), these markers also disappeared in cells expressing both smARF and GFP-Parkin, consistent with previous work (10, 11, 14). Indeed, both Parkin and smARF (p19M1I) expression were required to promote the turnover of mitochondrial proteins Hsp60 and MFN2 in HeLa cells (Fig. 5A). Moreover, in eGFP-Parkin-expressing cells, smARF (p19M1I) led to a reduction of NDUFA10 levels compared with cells expressing pcDNA (Fig. 5A). Although non-significant, there was also a trend toward eGFP-Parkin leading to a greater reduction in NDUFA10 levels compared with eGFP in smARF-expressing cells (Fig. 5A). Interestingly, overexpression of eGFP-Parkin seems to increase full-length p19ARF levels without affecting the ratio of smARF/full-length p19 (Fig. 5B), suggesting that cells expressing high levels of Parkin might also contain higher levels of p19ARF. Although data show that smARF levels are not significantly altered by Parkin (Fig. 5B), it is likely that they are also increased and are being degraded during mitophagy, masking this increase. smARF was initially shown to induce autophagy in HEK293 cells (27), which, in contrast to HeLa cells, express low levels of endogenous Parkin. To test whether endogenous Parkin is activated downstream of smARF, we transfected HEK293 cells with either non-targeting (NT) siRNA or one of two different Parkin siRNAs (park1989 or park2551). At 48 h, we transfected pcDNA, p19WT, p19M1I, or p19M45I and, 48 h later, surveyed ubiquitination of MFN2 (Fig. 5C), a well characterized downstream substrate in the Parkin/PINK1 mitophagy pathway (23, 25, 26). We found that only in the presence of endogenous Parkin and smARF, but not in the presence of full-length p19ARF, did MFN2 become ubiquitinated, despite similar levels of PINK1 (Fig. 5, C and D). Because high levels of Parkin have been shown to be required for MFN2 ubiquitination (23, 25), the findings fit well with the observation that only p19M1I resulted in significantly higher levels of endogenous Parkin in the mitochondria-enriched fraction (Fig. 5D). We also confirmed that smARF (p19M1I and p19ΔNT) led to loss of mitochondrial membrane potential in HEK293T cells by using TMRM staining followed by flow cytometry (Fig. 5E). Intriguingly, expression of p19M45I also led to slight mitochondrial depolarization in these cells (Fig. 5E) and, albeit non-significant, to a small increase in PINK1 levels (Fig. 5D). This suggests that full-length p19 might also be able to depolarize mitochondria, depending on the cell type in which it is expressed (i.e. Fig. 5E (HEK293) versus Fig. 1C (HeLa)). Regardless of these cell type differences, taken together, our findings suggest that smARF acts upstream of Parkin in mitophagy.
FIGURE 4.
smARF triggers Parkin-dependent mitophagy. A, HeLa cells transduced with eGFP-FLAG-Parkin or eGFP for 24 h were transfected with pcDNA, p19WT, p19M1I, p19M45I, or p19ΔNT (not shown) and processed for immunofluorescence after 48 h using cytochrome c (CytC) and p19 or p14 antibodies. Scale bar, 20 μm. B, antibodies against the mitochondrial matrix marker Hsp60 and the inner mitochondrial membrane protein Tim23 were also used to monitor the disappearance of mitochondria in p19M1I-transfected cells. For Tim23, mCherry and mCherry-Parkin transfection was used instead of eGFP and eGFP-FLAG-Parkin lentiviral infection. Scale bar, 20 μm. C, histogram showing the proportion of transfected cells that lacked staining for cytochrome c. At least 50 cells were counted per condition per experiment. All GFP-Parkin conditions are compared with GFP conditions for each construct. All columns are also compared with pcDNA (separately for GFP and GFP-Parkin series). n = 3. ***, p < 0.001. Error bars, S.E.
FIGURE 5.
Parkin is essential for smARF-induced mitophagy. A, HeLa cells were transfected with eGFP or eGFP-Parkin along with pcDNA, p19WT, p19M1I, or p19M45I constructs for 48 h. Cells were lysed and processed for immunoblotting against indicated proteins. Levels of mitochondrial proteins NDUFA10, Hsp60, MFN2, and PINK1 are shown in histograms. eGFP conditions are compared with eGFP-Parkin conditions for all constructs, and eGFP-Parkin conditions for each construct are compared with eGFP-Parkin + pcDNA. n = 3. *, p < 0.05; **, p < 0.01; ***, p < 0.001. B, levels of full-length p19ARF, smARF, and the ratio between the two from immunoblots shown in A are plotted in histograms. eGFP conditions are compared with eGFP-Parkin conditions for all constructs, and eGFP-Parkin conditions for each construct are compared with eGFP-Parkin + pcDNA. n = 3. **, p < 0.01; ***, p < 0.001. C, HEK293 cells were treated with NT or Parkin (1989 and 2551) siRNA. pcDNA, p19WT, p19M45I, or p19M1I was transfected 48 h after siRNA transfection, and the cells were harvested 48 h later. 20 μg of mitochondria-enriched fractions were immunoblotted for the indicated proteins. A shift in MFN2 migration (arrow), consistent with ubiquitination is only observed in the presence of endogenous Parkin and smARF and is quantified in a histogram (D). PINK1 and Parkin levels are also quantified in histograms (D). NT, park1989, and park2551 conditions are compared for all constructs, and the NT siRNA + pcDNA condition is compared with each other NT siRNA + (p19 construct) condition. n = 3. **, p < 0.01; ***, p < 0.001. E, HEK293T cells expressing pcDNA, p19WT, p19M1I, p19ΔNT, or p19M45I for 24 h were stained with the mitochondrial membrane potential indicator TMRM, and the cells were sorted by FACS. TMRM-negative cells were plotted. At least 50,000 cells were sorted for each condition per experiment. All columns are compared with pcDNA or with p19WT. n = 3. ***, p < 0.001. Error bars, S.E.
PINK1 Is Required for Parkin- and smARF-induced Mitophagy in Neurons
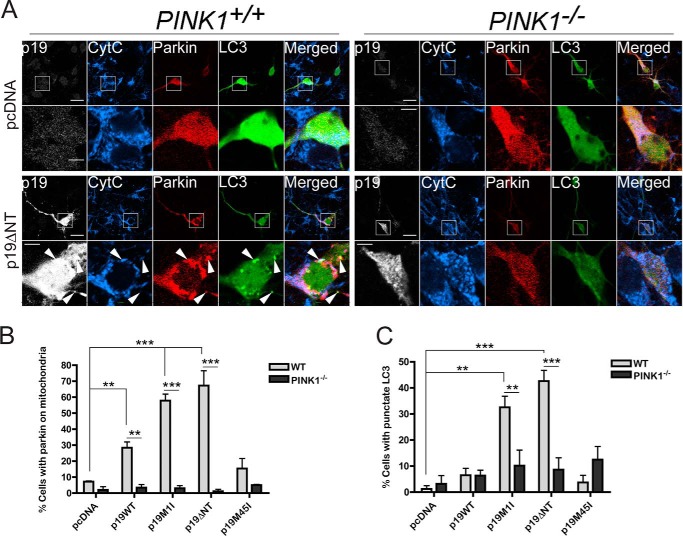
PINK1 is necessary for the recruitment of Parkin to mitochondria and for subsequent Parkin-dependent mitophagy upon treatment with chemical uncouplers (11–14). However, it is not known whether PINK1 is also required for smARF-induced Parkin recruitment and mitophagy. Silencing of PINK1 using siRNA (Fig. 6A) significantly reduced smARF-induced Parkin translocation (Fig. 6B) and mitophagy (Fig. 6, C and D) in HeLa cells, confirming that smARF, PINK1, and Parkin function in a common mitophagy pathway. To investigate whether smARF can trigger Parkin recruitment in neurons, we co-transfected cortical (Fig. 7A) and hippocampal (data not shown) primary neurons with both mCherry-Parkin and p19ARF constructs. Strikingly, p19M1I and p19ΔNT induced Parkin translocation to mitochondria, whereas p19M45I did not (Fig. 7B). Similar to what had been observed in immortalized cells, p19WT expression resulted in an intermediate phenotype. Moreover, in neurons transfected with GFP-LC3, a punctate GFP pattern was apparent upon co-expression of mCherry-Parkin with p19M1I or p19ΔNT but not with p19M45I or p19WT (Fig. 7C). This indicates that co-expression of smARF, but not full-length p19ARF, with Parkin leads to the induction of autophagy. As indicated with arrows in Fig. 7A, GFP-LC3 puncta either co-localize with mitochondrial staining and Parkin or only with Parkin, which suggests ongoing mitophagy in these neurons. Perhaps most importantly, Parkin translocation to mitochondria and LC3 puncta could not be induced by smARF in neurons isolated from PINK1−/− mice (Fig. 7, A–C). Together, these findings demonstrate that the smARF/PINK1/Parkin mitophagy pathway operates in neurons.
FIGURE 6.
PINK1 is required for smARF-induced Parkin-dependent mitophagy. A, immunoblot showing PINK1 levels after transfection with PINK1-targeted SMARTpool siRNA (24-h treatment) or with individual siRNAs (PINK1-02, -03, and -04) from the SMARTpool (48-h treatment). B–D, HeLa cells were transfected with NT or PINK1 siRNA (SMARTpool for the recruitment experiment and individual PINK1 siRNAs for the mitophagy experiment). After 48 h, the cells were transduced with eGFP-FLAG-Parkin, and 24 h later, they were transduced with pcDNA, p19WT, p19M1I, and p19ΔNT. After a further 24 h (recruitment experiment) or 48 h (mitophagy experiment), the cells were processed for immunofluorescence using Hsp60 and p19 or p14 antibodies. As a further positive control, cells were treated with 20 μm CCCP for either 3 h (recruitment) or 24 h (mitophagy) at 48 h post-transfection. B, histogram showing the proportion of transfected cells with eGFP-Parkin co-localizing with the mitochondrial marker Hsp60. At least 100 cells were counted per condition per experiment. The PINK1 siRNA condition was compared with NT siRNA for each construct. n = 3. ***, p < 0.001. C, micrographs of NT, PINK1-02 (P-02), and PINK1-04 (P-04) siRNA conditions processed for immunofluorescence after 48 h of p19ΔNT expression. Scale bar, 20 μm. Arrows, smARF- and eGFP-Parkin-transfected cells that have either lost (NT condition) or retained (PINK1-02 and PINK1-04 conditions) their mitochondrial staining. D, histogram showing the proportion of transfected cells that lacked staining for Hsp60 for each of the indicated siRNA conditions. At least 50 cells were counted per condition per experiment. PINK1-02 and PINK-04 siRNA conditions are compared with the NT siRNA condition for each construct and for the 24-h CCCP treatment. n = 3. ***, p < 0.001. Error bars, S.E.
FIGURE 7.
smARF induces Parkin/PINK1-dependent mitophagy in neurons. A, cortical neurons generated from embryonic day 14 WT and PINK1−/− embryos were transfected with mCherry-Parkin, GFP-LC3, and either pcDNA, p19WT, p19M1I, p19M45I, or p19ΔNT constructs and processed 24 h later for immunofluorescence using cytochrome c (CytC) and p19 antibodies (only pcDNA and p19ΔNT shown). Scale bar, 20 μm for low power and 5 μm for zoom. Arrows, co-localization of p19ARF, mCherry-Parkin, cytochrome c, and GFP-LC3 (two top arrows) and co-localization of p19ARF, mCherry-Parkin, and GFP-LC3 without cytochrome c (bottom arrow). B, histogram showing the proportion of transfected cells with mCherry-Parkin co-localizing with the mitochondrial marker cytochrome c in WT versus PINK1−/− neurons. At least 25 neurons were counted per condition per experiment. All WT conditions are compared with PINK1−/− for each construct. All columns are also compared with pcDNA (separately for WT and PINK1−/− series) n = 3. **, p < 0.01; ***, p < 0.001. C, histogram showing the proportion of transfected cells with punctate GFP-LC3 staining in WT versus PINK1−/− neurons. At least 25 neurons were counted per condition per experiment. n = 3. All WT conditions are compared with PINK1−/− for each construct. All columns are also compared with pcDNA (separately for WT and PINK1−/− series). **, p < 0.01; ***, = p < 0.001. Error bars, S.E.
DISCUSSION
Parkin and PINK1 have been shown to mediate the degradation of mitochondria with compromised membrane potential in mammalian cells (10–14). However, the high doses of chemical uncouplers used to trigger mitochondrial depolarization in these studies are unlikely to accurately reflect the natural stimuli encountered during disease progression in vivo. This highlights our lack of understanding of the upstream signaling pathways that trigger Parkin/PINK1 mitophagy endogenously. Moreover, it may underpin the difficulty in inducing Parkin/PINK1-mediated mitophagy in neurons. Indeed, whereas some studies have shown that Parkin can translocate to mitochondria depolarized by chemical uncouplers in neurons (30–35), others have called into question both the existence of the pathway and the relevance of treating neurons with high doses of chemical uncouplers (36, 37). Parkin translocation to mitochondria in response to CCCP was observed in neurons preincubated with inhibitors of apoptosis (30). This suggests that, in neurons, chemical uncouplers activate apoptotic pathways, making it difficult to use these compounds. Another obstacle is that the presence of antioxidants in neuronal medium tends to counteract the depolarizing effects of uncouplers (31). Accordingly, whereas certain studies have shown Parkin translocation to mitochondria, no study to date using chemical uncouplers has been able to reliably quantify Parkin-dependent mitophagy in neurons (1).
We postulated that proteins known to depolarize mitochondria might represent an alternative and perhaps more physiological trigger of Parkin/PINK1 mitophagy. Mitochondrial membrane potential fluctuates in healthy cells and can be modulated by the expression of a number of nuclear encoded gene products (38). smARF, an alternative translation product of p19ARF mRNA, was demonstrated to induce mitochondrial depolarization and autophagy in HEK293 cells (27). However, mitophagy per se was not examined in this study. We hypothesized that Parkin and PINK1 function downstream of smARF-induced mitochondrial depolarization in a common mitophagy pathway. Here, we showed that Parkin is recruited to depolarized mitochondria following smARF expression, which eventually leads to the autophagy of the mitochondrial network in a PINK1-dependent manner. Thus, smARF overexpression recapitulates Parkin/PINK1 mitophagy without the need for high doses of chemical uncouplers. Experimentally, the work may provide a useful system to better control mitochondrial depolarization and PINK1/Parkin mitophagy by inducing the graded expression smARF in cells. Full-length p19ARF, which is predominantly localized to the nucleus, was also shown to partially localize to mitochondria and promote autophagy (39–41). However, the ability of full-length p19ARF to induce mitochondrial depolarization was not assessed. This is of importance because there are reports in the literature of both full-length p19ARF and smARF being able to induce autophagic cell death (27, 39, 40). Our results demonstrate that smARF but not full-length p19ARF localizes to mitochondria and induces mitophagy. Moreover, although WT p19ARF induced some Parkin recruitment, it did not lead to complete mitophagy. We speculate that cells transfected with WT p19ARF activated apoptosis in addition to mitophagy, resulting in cell death before mitochondrial network removal by autophagy. Indeed, p19ARF was previously shown to induce apoptosis (39, 42), and although we did not quantify it, we often observed cell death induced by p19ARF transfection. Intriguingly, it is possible that certain stimuli favor the production of smARF over full-length p19ARF in cells through leaky scanning or internal ribosome entry site regulation, providing a transcription-independent mechanism to rapidly trigger mitophagy. Interestingly, Reef et al. (27) demonstrated in the original smARF report that both viral and cellular oncogene expression increases p19ARF and smARF levels in cells. This raises the possibility that viral infections, which can induce oncogene expression (43), or increased oncogenic signaling itself might represent a natural trigger of smARF expression.
We show here that Parkin and PINK1 are essential for smARF to promote mitochondrial autophagy, suggesting that smARF may play a role in PD. Interestingly, p19ARF induces cellular senescence, and its levels are known to increase with age (44, 45). It would be interesting to determine whether smARF levels also increase with age in healthy and PD brains. Intriguingly, we also showed that exogenous levels of Parkin seem to increase p19ARF and smARF levels, suggesting that neurons expressing high levels of Parkin might also co-express smARF. Conversely, neurons lacking Parkin might lack smARF and display an even greater impairment in their ability to trigger mitophagy to eliminate damaged mitochondria. Impairment of the Parkin/PINK1 mitophagy pathway was previously hypothesized to lead to the accumulation of damaged mitochondria in PD. This might occur through various mechanisms during normal aging, such as exposure to mitochondrial toxins, exposure to reactive oxygen species, and accumulation of mitochondrial DNA mutations. Based on our results, we suggest that damaged mitochondria might also accumulate through defects in the smARF mitophagy pathway. Strikingly, we demonstrate that Parkin and PINK1 are downstream actors in the smARF autophagy pathway in neurons. However, whether or not this pathway operates endogenously in neurons and how it might contribute to neurodegeneration remain to be determined. Indeed, to our knowledge, very few studies have explored the biochemical role p19ARF in neurons or in the brain (46–48), and virtually nothing is known about the mechanisms and the endogenous pathways that trigger smARF expression from the alternative translational start site on the p19ARF mRNA. Thus, although much work remains to be done, our findings demonstrating that Parkin and PINK1 are essential for smARF-mediated autophagy provide a novel experimental system to assay PINK1/Parkin mitophagy in neurons and implicate the p19ARF pathway for the first time in the pathogenesis of PD.
This work was supported by a Graduate Student Award from Parkinson Society Canada (to K. G.) and an operating grant (MOP-62714) from the Canadian Institutes of Health Research (to E. A. F.).
- PD
- Parkinson disease
- smARF
- short mitochondrial isoform of ARF
- TMRM
- tetramethylrhodamine methyl ester
- MTR
- MitoTracker Deep Red
- OMM
- outer mitochondrial membrane
- eGFP
- enhanced green fluorescent protein
- NT
- non-targeting
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone.
REFERENCES
- 1. Grenier K., McLelland G. L., Fon E. A. (2013) Parkin- and PINK1-dependent Mitophagy in Neurons: Will the Real Pathway Please Stand Up? Front. Neurol. 4, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corti O., Brice A. (2013) Mitochondrial quality control turns out to be the principal suspect in parkin and PINK1-related autosomal recessive Parkinson's disease. Curr. Opin. Neurobiol. 23, 100–108 [DOI] [PubMed] [Google Scholar]
- 3. Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J. (2003) Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U.S.A. 100, 4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 5. Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 6. Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., Lu B. (2006) Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U.S.A. 103, 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schapira A. H., Cooper J. M., Dexter D., Jenner P., Clark J. B., Marsden C. D. (1989) Mitochondrial complex I deficiency in Parkinson's disease. Lancet 1, 1269. [DOI] [PubMed] [Google Scholar]
- 8. Langston J. W., Ballard P., Tetrud J. W., Irwin I. (1983) Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 219, 979–980 [DOI] [PubMed] [Google Scholar]
- 9. Bender A., Krishnan K. J., Morris C. M., Taylor G. A., Reeve A. K., Perry R. H., Jaros E., Hersheson J. S., Betts J., Klopstock T., Taylor R. W., Turnbull D. M. (2006) High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 38, 515–517 [DOI] [PubMed] [Google Scholar]
- 10. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 14. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J, Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trempe J. F., Sauvé V., Grenier K., Seirafi M., Tang M. Y., Ménade M., Al-Abdul-Wahid S., Krett J., Wong K., Kozlov G., Nagar B., Fon E. A., Gehring K. (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 [DOI] [PubMed] [Google Scholar]
- 16. Wauer T., Komander D. (2013) Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Riley B. E., Lougheed J. C., Callaway K., Velasquez M., Brecht E., Nguyen L., Shaler T., Walker D., Yang Y., Regnstrom K., Diep L., Zhang Z., Chiou S., Bova M., Artis D. R., Yao N., Baker J., Yednock T., Johnston J. A. (2013) Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spratt D. E., Martinez-Torres R. J., Noh Y. J., Mercier P., Manczyk N., Barber K. R., Aguirre J. D., Burchell L., Purkiss A., Walden H., Shaw G. S. (2013) A molecular explanation for the recessive nature of parkin-linked Parkinson's disease. Nat. Commun. 4, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., Endo T., Fon E. A., Trempe J. F., Saeki Y., Tanaka K., Matsuda N. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 [DOI] [PubMed] [Google Scholar]
- 20. Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D. G., Ritorto M. S., Hofmann K., Alessi D. R., Knebel A., Trost M., Muqit M. M. (2014) Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kane L. A., Lazarou M., Fogel A. I., Li Y., Yamano K., Sarraf S. A., Banerjee S., Youle R. J. (2014) PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glauser L., Sonnay S., Stafa K., Moore D. J. (2011) Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118, 636–645 [DOI] [PubMed] [Google Scholar]
- 23. Chan N. C., Salazar A. M., Pham A. H., Sweredoski M. J., Kolawa N. J., Graham R. L., Hess S., Chan D. C. (2011) Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20, 1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ziviani E., Tao R. N., Whitworth A. J. (2010) Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. U.S.A. 107, 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D. F., Karbowski M., Youle R. J. (2010) Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., Taanman J. W. (2010) Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19, 4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reef S., Zalckvar E., Shifman O., Bialik S., Sabanay H., Oren M., Kimchi A. (2006) A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol. Cell 22, 463–475 [DOI] [PubMed] [Google Scholar]
- 28. Fallon L., Bélanger C. M., Corera A. T., Kontogiannea M., Regan-Klapisz E., Moreau F., Voortman J., Haber M., Rouleau G., Thorarinsdottir T., Brice A., van Bergen En Henegouwen P. M., Fon E. A. (2006) A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 8, 834–842 [DOI] [PubMed] [Google Scholar]
- 29. Greene A. W., Grenier K., Aguileta M. A., Muise S., Farazifard R., Haque M. E., McBride H. M., Park D. S., Fon E. A. (2012) Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cai Q., Zakaria H. M., Simone A., Sheng Z. H. (2012) Spatial Parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 22, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joselin A. P., Hewitt S. J., Callaghan S. M., Kim R. H., Chung Y. H., Mak T. W., Shen J., Slack R. S., Park D. S. (2012) ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum. Mol. Genet. 21, 4888–4903 [DOI] [PubMed] [Google Scholar]
- 32. Seibler P., Graziotto J., Jeong H., Simunovic F., Klein C., Krainc D. (2011) Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J. Neurosci. 31, 5970–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koyano F., Okatsu K., Ishigaki S., Fujioka Y., Kimura M., Sobue G., Tanaka K., Matsuda N. (2013) The principal PINK1 and Parkin cellular events triggered in response to dissipation of mitochondrial membrane potential occur in primary neurons. Genes Cells 18, 672–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y. L., Selkoe D., Rice S., Steen J., LaVoie M. J., Schwarz T. L. (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bingol B., Tea J. S., Phu L., Reichelt M., Bakalarski C. E., Song Q., Foreman O., Kirkpatrick D. S., Sheng M. (2014) The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375 [DOI] [PubMed] [Google Scholar]
- 36. Van Laar V. S., Arnold B., Cassady S. J., Chu C. T., Burton E. A., Berman S. B. (2011) Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum. Mol. Genet. 20, 927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rakovic A., Shurkewitsch K., Seibler P., Grünewald A., Zanon A., Hagenah J., Krainc D., Klein C. (2013) Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J. Biol. Chem. 288, 2223–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Andrews Z. B., Diano S., Horvath T. L. (2005) Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat. Rev. Neurosci. 6, 829–840 [DOI] [PubMed] [Google Scholar]
- 39. Abida W. M., Gu W. (2008) p53-Dependent and p53-independent activation of autophagy by ARF. Cancer Res. 68, 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pimkina J., Humbey O., Zilfou J. T., Jarnik M., Murphy M. E. (2009) ARF induces autophagy by virtue of interaction with Bcl-xl. J. Biol. Chem. 284, 2803–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reef S., Kimchi A. (2008) Nucleolar p19ARF, unlike mitochondrial smARF, is incapable of inducing p53-independent autophagy. Autophagy 4, 866–869 [DOI] [PubMed] [Google Scholar]
- 42. Qi Y., Gregory M. A., Li Z., Brousal J. P., West K., Hann S. R. (2004) p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431, 712–717 [DOI] [PubMed] [Google Scholar]
- 43. Moore P. S., Chang Y. (2010) Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 10, 878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baker D. J., Perez-Terzic C., Jin F., Pitel K. S., Pitel K., Niederländer N. J., Jeganathan K., Yamada S., Reyes S., Rowe L., Hiddinga H. J., Eberhardt N. L., Terzic A., van Deursen J. M. (2008) Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat. Cell Biol. 10, 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krishnamurthy J., Torrice C., Ramsey M. R., Kovalev G. I., Al-Regaiey K., Su L., Sharpless N. E. (2004) Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 114, 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abdouh M., Chatoo W., El Hajjar J., David J., Ferreira J., Bernier G. (2012) Bmi1 is down-regulated in the aging brain and displays antioxidant and protective activities in neurons. PloS One 7, e31870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bruggeman S. W., Valk-Lingbeek M. E., van der Stoop P. P., Jacobs J. J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., van Lohuizen M. (2005) Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 19, 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hulleman E., Helin K. (2005) Molecular mechanisms in gliomagenesis. Adv. Cancer Res. 94, 1–27 [DOI] [PubMed] [Google Scholar]