Background: Overactive signaling through NLRs is associated with inflammatory disease.
Results: In vivo inhibition of RIPK2 alleviates inflammation in two inflammatory disease models: an acute peritonitis model and a spontaneous CD-like ileitis model.
Conclusion: Inhibition of RIPK2 may be beneficial in certain inflammatory states.
Significance: This work supports further development of RIPK2-targeted therapies as well as proposes biomarkers to guide treatment.
Keywords: Enzyme Inhibitor, Immunology, Innate Immunity, Nod-like Receptor (NLR), Signal Transduction, Dual Specificity Kinase
Abstract
The RIPK2 kinase transduces signaling downstream of the intracellular peptidoglycan sensors NOD1 and NOD2 to promote a productive inflammatory response. However, excessive NOD2 signaling has been associated with numerous diseases, including inflammatory bowel disease (IBD), sarcoidosis and inflammatory arthritis, making pharmacologic inhibition of RIPK2 an appealing strategy. In this work, we report the generation, identification, and evaluation of novel RIPK2 specific inhibitors. These compounds potently inhibit the RIPK2 tyrosine kinase activity in in vitro biochemical assays and cellular assays, as well as effectively reduce RIPK2-mediated effects in an in vivo peritonitis model. In conjunction with the development of these inhibitors, we have also defined a panel of genes whose expression is regulated by RIPK2 kinase activity. Such RIPK2 activation markers may serve as a useful tool for predicting settings likely to benefit from RIPK2 inhibition. Using these markers and the FDA-approved RIPK2 inhibitor Gefitinib, we show that pharmacologic RIPK2 inhibition drastically improves disease in a spontaneous model of Crohn Disease-like ileitis. Furthermore, using novel RIPK2-specific inhibitors, we show that cellular recruitment is inhibited in an in vivo peritonitis model. Altogether, the data presented in this work provides a strong rationale for further development and optimization of RIPK2-targeted pharmaceuticals and diagnostics.
Introduction
NOD2 is a cytosolic sensor for the bacterially-derived peptidoglycan breakdown product muramyl dipeptide (MDP)2 (1, 2). NOD2 originally gained notoriety as the first identified genetic susceptibility gene for Crohn Disease (CD) following studies showing that loss-of-function (LOF) polymorphisms occurring within the MDP sensing region increase the risk for development of CD (3–5). However, such loss-of-function NOD2 polymorphisms are present in ∼7–10% of the general Caucasian population, and the vast majority of people carrying NOD2 loss-of-function alleles do not develop CD (3–5). In fact, there are far greater numbers of CD patients who are WT for NOD2 than those who are polymorphic, and as NOD2 is regulated at the transcriptional level by NF-κB (6), it has been suggested that increased expression of WT NOD2 may represent a feed-forward mechanism by which inflammation is exacerbated. Studies have demonstrated that CD patients with WT NOD2 demonstrate increased expression of both NOD2 and its associated kinase RIPK2 as well as increased RIPK2 kinase activity (7, 8). In addition, compared with WT healthy controls, monocytes from CD patients WT for NOD2 show increased proinflammatory IL-8 secretion in response to MDP stimulation (9). Given the fact that aberrant overactive WT NOD2 signaling has been associated with an increasing number of inflammatory diseases including Early Onset Sarcoidosis (EOS) and Blau Syndrome (10–13), inflammatory arthritis (14, 15), allergic inflammation (16), and multiple sclerosis (17), pharmacologic inhibition of NOD2 signaling may, therefore, be efficacious in certain clinical settings.
The dual-specificity kinase RIPK2 is integral to propagating signals resulting from NOD2 activation, including the initiation of downstream NF-κB, MAPK, and autophagy pathways (18–20). The fact that RIPK2 is also utilized by the closely related NLR NOD1 (21), makes RIPK2 inhibition an attractive option should one wish to suppress an entire arm of innate immune signaling dedicated to sensing cytosolic bacterial peptidoglycan. Protein kinases have been successfully pharmacologically targeted in both cancer and inflammatory diseases and therefore have a history of successful translational intervention. Previous work by our laboratory has reclassified RIPK2 as a dual-specificity kinase (22). This finding not only allowed for the identification of 2 FDA-approved drugs that inhibit RIPK2 activity (Erlotinib and Gefitinib), but also helped rationalize the development of specific RIPK2 inhibitor programs by multiple pharmaceutical companies.
In the present work, we identify and characterize two novel RIPK2 inhibitors. Unlike kinase inhibitors traditionally discovered through high-throughput screening of previously generated type-I inhibitor compound libraries, the RIPK2 inhibitors we describe herein result from screening a novel Nanokinib® library comprised of compounds generated through a small-molecule macrocylization process (Nanocyclix®), which result in a unique binding and mode-of-action compared with currently described kinase inhibitors. Not only do we demonstrate the potency of such compounds against RIPK2 activity in biochemical and cellular assays, we also show that these RIPK2 inhibitors are also very potent in inhibiting RIPK2 activity in vivo using an MDP-induced peritonitis model. Using this assay, these novel compounds were found to significantly inhibit inflammatory cell recruitment compared with vehicle-treated animals. These data support further optimization and larger-scale synthesis of such RIPK2 inhibitors to facilitate longer-term in vivo testing in various disease models in which RIPK2 is thought to play a role. To demonstrate the feasibility of RIPK2 inhibition in inflammatory disease over a longer term, we used the well-studied, widely-available drug, Gefitinib (Iressa™, AstraZeneca). Gefitinib is an ATP-competitive kinase inhibitor designed against the EGF-R and has been shown to be a very effective first-line treatment for non-small cell lung cancer (NSCLC) in patients harboring activating EGF-R mutations (23, 24). We have previously demonstrated that Gefitinib directly inhibits RIPK2 activity with a potency equal to that of the EGF-R (IC50 in the low nanomolar range). Studies that have retested Gefitinib against a panel of more than 300 kinases show that Gefitinib is a highly specific inhibitor, affecting predominantly EGF-R and RIPK2 (25).3 The dosage, pharmacokinetics, absorption, distribution, metabolism, excretion, and toxicology of Gefitinib have all been well studied. Therefore, having all of these parameters defined, enabled us to test the in vivo efficacy of RIPK2 inhibition using Gefitinib in a setting of inflammatory disease.
The use of RIPK2 inhibitors in long-term inflammatory disease treatment will need to be guided by robust and reliable assays to detect RIPK2 activity and inhibition in disease. To this end, we utilized pharmacologic inhibition of RIPK2 in combination with RNA sequencing to define a 9-gene panel that may help predict the efficacy of RIPK2 inhibition. We validate this panel with the development of novel RIPK2 inhibitors that target RIPK2 without targeting EGFR. Using this 9-gene signature, we identify a mouse model of CD in which RIPK2 inhibition is potentially efficacious. We demonstrate that Gefitinib-mediated inhibition of RIPK2 is beneficial in vivo in the SAMP1/YitFc mouse, a spontaneous mouse model of Crohn's Disease in which NOD2 is WT (28, 29). We show that inflammatory cytokine secretion in macrophages from these mice was also markedly reduced upon inhibition of RIPK2 and pharmacologic inhibition of RIPK2 tyrosine phosphorylation correlated with improvement in disease. These results suggest that RIPK2 inhibition might be effective in the treatment of specific settings of inflammatory disease and propose a gene expression profile, which may be useful to predict which patients might be particularly helped by RIPK2 inhibition.
EXPERIMENTAL PROCEDURES
Cell Lines, Plasmids, Transfection, and Western Blotting
Transient transfection assays were performed using calcium phosphate transfection of HEK293 cells (ATCC®, CRL-1573). Omni-tagged RIPK2 was generated by PCR cloning HA-tagged RIPK2 (a gift from V. Dixit, Genentech) into pCDNA4/Hismax (Invitrogen) or into the InterPlay® Mammalian TAP system (Stratagene). pMXp-HA-tagged full length NOD2 or NOD2 lacking the LRR region was a kind gift from C. McDonald (Lerner Research Institute, CCF). For immunoprecipitation (IP), cell lysates were prepared with a buffer containing 50 mm Tris HCL (pH 7.5), 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm EGTA, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 5 mm iodoacetimide, 5 mm N-ethylmaleimide, 1 mm PMSF, 1 μm sodium orthovanadate, and protease inhibitor mixture. Immunoprecipitations were performed using anti-Omni rabbit antibody (M-21) (Santa Cruz Biotechnology) overnight followed by the addition of protein G-Sepharose (Invitrogen) or by addition of streptavidin-agarose (Sigma) for NTAP-tagged RIPK2. Immunoprecipitates were washed 5x in lysis buffer before boiling in an equal volume of 2× Laemmli sample buffer. Western blotting was perfomed as described previously (22, 30). Anti-HA antibody (16B12) was obtained from Covance, anti-phosphotyrosine antibody (P-Tyr-100) was obtained from Cell Signaling and anti-Omni mouse antibody(D-8) was obtained from Santa Cruz Biotechnologies.
Identification and Screening of RIPK2 Inhibitors
RIPK2 compounds were generated through a proprietary small molecule macrocylization process (Nanocyclix®, Oncodesign). Initial screening of compound libraries was performed using in vitro radiometric kinase assays utilizing recombinantly purified RIPK2 expressed in insect cells as kinase and RBER-CHKtide as a substrate (see supplemental Fig. S1A for complete sequence, assays performed by ProQinase). Ten concentrations of inhibitor were tested ranging from 3 × 10−6 m to 9 × 10−11 m using 15.7 nm (50 ng) recombinant RIPK2 and 2 μg of recombinant RBER-CHKtide substrate per 50 μl reaction. Compounds which showed in vitro IC50 values of < 100 nm were tested in a cellular assay where RIPK2 activity (tyrosine autophosphorylation) was induced by co-expression of NOD2 with RIPK2 and inhibition of kinase activity was assessed by loss of tyrosine autophosphorylation upon treatment with RIPK2 inhibitor (2 concentrations tested: 250 nm and 500 nm). The two compounds which maintained potent inhibition of RIPK2 tyrosine phosphorylation in the cellular assay at the lower 250 nm dose were then used for further in vitro and in vivo assays.
Determining Kinase Specificity of RIPK2 Inhibitors
A contract research organization (ProQinase) was utilized to determine the inhibition of 366 kinases (Gefitinib or OD36) or 88 kinases (OD38) in response to 1 μm Gefitinib, 1 μm OD36, 1 μm OD38, 100 nm OD36 and 100 nm OD38. In brief, each recombinant kinase was pre-incubated with the indicated dose of inhibitor for 15 min before an in vitro kinase assay was performed using a known substrate. After 30 min, the reaction was stopped and phosphate incorporation was measured. Dendrograms indicating kinase specificity were generated according to the following characteristics: Inhibition ≥ 90% = green + 300–350 pixel size, Inhibition ≥75% = light green + 200–300 pixel size, Inhibition ≥ 50 = light green + 175–200 pixel size, Inhibition ≥ 25 = yellow + 50–175 pixel size, Inhibition ≥ 10 = red + 20–50 pixel size.
RNA Isolation
Bone marrow-derived macrophages (BMDMs) were generated by culturing bone marrow for 7 days in 10% DMEM with 25% conditioned Ladmac media (gift from Clifford Harding, CWRU). Cells were rested in 10% FBS-DMEM media overnight before use. BMDMs were pretreated with Gefitinib (LC Labs), RIPK2 inhibitor (Oncodesign) or vehicle (DMSO) for 30 min before stimulating with MDP for 4 h. Cells were then harvested and RNA extracted using a Qiagen RNeasy kit using the manufacturer's instructions.
RNA Sequencing
RNA was sent to Oklahoma Medical Research Foundation for RNA sequencing. The 100-bp paired-end Illumina reads were processed to remove the 3′ bases with Phred quality score of lower than 20. Reads that are less than 20 bases after quality trimming were removed from further analysis. The reads from each replicate of each sample were mapped to mouse genome release mm9 using tophat v1.4.1 program before guided assembly using cufflinks v1.3.0 program with mouse genome annotation from RefSeq database and the lincRNA annotation from Ensembl database. Differential expression of transcripts was analyzed using two-tailed Student's t test with Benjamini and Hochberg correction of false discovery rate (FDR). FDR-corrected p value of 0.05 was set as the cutoff of statistical significance.
RT-PCR
RNA was reverse transcribed using a Quantitect reverse transcription kit (Qiagen). The following primer pairs were used for amplification: mCXCL10-F 5′-TCCTTGTCCTCCCTAGCTCA-3′, mCXCL10-R 5′-ATAACCCCTTGGGAAGATGG-3′, mGPR84-F 5′-GGGAACCTCAGTCTCCAT-3′, mGPR84-R 5′-TGCCACGCCCCAGATAATG-3′, mIRG1-F 5′-GTTTGGGGTCGACCAGACTT-3′, mIRG1-R 5′-CAGGTCGAGGCCAGAAAACT-3′, mMARCKSL1-F 5′-TTGTGCTGTGCCTAGTGGC-3′, mMARCKSL1-R 5′-GCTTATCAAGTCAGGGACATGG-3′, mRASGRP1-F 5′-CGCTTCTTGCACTCGAACC-3′, mRASGRP1-R 5′-GGGCCAGCTCCATCTATTC-3′, mSLC2a6-F 5′-CGAGCCGGGGCCTTG-3′, mSLC2a6-R 5′-CACCCAAGGTGAACACGGA-3′, mCLEC4e-F 5′-TCCCACCACACAGAGAGAGG-3′, mCLEC4e-R 5′-CCCGGAAATTTGAGAGCTGC-3′, mICAM1-F 5′-CGTGCAGTCGTCCGCTTCCG-3′, mICAM1-R 5′-TGAGAGCTGGGTCCGCGGTG3-′, mCD40-F 5′-TCTCGCCCTGCGATGGTGTCT-3′, mCD40-R 5′-CGGCTTCCTGGCTGGCACAA-3′, mGAPDH-F 5′-AGGCCGGTGCTGAGTATGTC-3′, mGAPDH-R 5′-TGCCTGCTTCACCACCTTCT-3′. Sybr Green was obtained from Bio-Rad, and the real-time PCR reactions were carried out using a CFX96 C1000 Real-Time Thermal Cycler from Bio-Rad. RT-PCR data is presented as the means ± S.E. RT-PCR experiments were performed in duplicate and repeated twice. A representative experiment is shown. Significance of comparisons shown was assessed by Student's two-tailed t test with the cut-off for significance set at p = 0.05.
Mice and Inhibitor Treatment
C57BL/6 mice were obtained from Jackson Labs and maintained in a rodent barrier facility. SAMP1/YitFc mice were obtained from the IBD Program Project Animal Core directed by Dr. Theresa Pizarro. Gefitinib (LC Laboratories) was incorporated into RMH3000 rodent chow at 0.025% of food weight (Research Diets). This amount of drug was calculated based on previous studies on the typical food intake of various mouse strains (31) and the average weight of our mice, resulting in a dose of 50 mg/kg/day. Diet containing Gefitinib was administered to SAMP1/YitFc mice starting at 5 weeks of age for a duration of 7.5 weeks. Mice were euthanized and tissue was harvested for histology (the IBD Program Project Animal Core). H & E sections were scored by a blinded pathologist using a published and well-established histopathological scoring system for these mice. Histology was scored by 1) active inflammation (infiltration with neutrophils especially at the base of the muscularis; Score = 0–4 × % ileal involvement), 2) chronic inflammation (lymphocytes, plasma cells, and macrophages in the mucosa and submucosa; Score = 0–4 × % ileal involvement), and 3) villus distortion (flattening and/or widening of normal villus architecture; Score = 0–4 × % ileal involvement). The sum of all 3 individual components was then expressed as the total inflammatory score. For the MDP-induced peritonitis model, C57BL/6 mice (Jackson Labs) were given vehicle or 6.25 mg/kg of Gefitinib or OD36 intraperitoneal in a 500 μl volume. Thirty minutes later, mice were injected with 150 μg of MDP intraperitoneal Four hours later, mice were sacrificed and a peritoneal lavage using 5 ml of PBS was performed.
In Situ Proximity Ligation Assay
Anti-phosphotyrosine antibody generated in mouse (P-Tyr-100, Cell Signaling) and anti-RIPK2/RICK antibody generated in rabbit (H-300, Santa Cruz Biotechnologies) were used for primary antibodies. Duolink PLA was performed on paraffin-embedded, formalin-fixed sections using the secondary probes, amplification reagents, and polymerase as directed by the manufacturer's instructions (Sigma).
RESULTS
Transcriptomic Profiling Reveals a Novel Gene Set Specifically Regulated by RIPK2 Kinase Activity
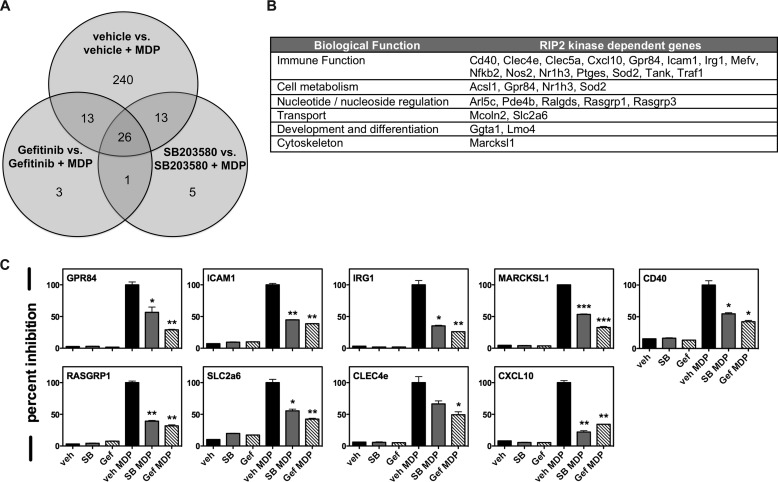
Currently, RIPK2 activity is most accurately measured biochemically, by performing in vitro kinase assays or through Western blotting for tyrosine-phosphorylated RIPK2. However, a simpler, more robust method of detecting RIPK2 activation would be more amenable to potential future clinical applications. With this in mind, we set out to determine a unique set of NOD2-driven genes that were particularly sensitive and specific to RIPK2 kinase inhibition. Previously published studies as well as our own findings indicate that the EGF-R inhibitor Gefitinib and the p38 inhibitor SB203580 showed inhibition of RIPK2 activity with an IC50 equivalent to their intended targets (22, 25, 27). Taking advantage of this, we treated primary bone marrow-derived macrophages with either vehicle, 500 nm SB203580 or 500 nm Gefitinib for 30 min before stimulation with 10 μg/ml MDP for 4 h. We then extracted RNA and performed RNA sequencing for each condition. These 2 compounds, having been designed against distinct targets, possess different binding preferences and off-target profiles (25, 32). The only kinase they both inhibit is RIPK2 (25, 32). Therefore, focusing on transcripts that were differentially affected by MDP treatment and jointly affected by both SB203580 and Gefitinib allowed us to identify genes that most likely reflect the function and activation of RIPK2 (intersection of Venn Diagram, Fig. 1A). While our previously published studies using Affymetrix chips showed only 40–60 genes that were consistently up- or down-regulated 2-fold upon MDP stimulation (33), due to its increased linear range and increased sensitivity RNA-seq transcriptomic analysis revealed 10–15× as many genes whose expression is induced by MDP (30), and this allowed us to select for genes particularly sensitive to RIPK2 kinase inhibition. Further classification of these genes based on biological function showed that a majority of these were involved in immune processes (Fig. 1B). The gene list identified was curated against published TNF, IL-1, and LPS induced expression profiles. While genes such as NOS2, GPR84, ICAM1, and CXCL10 were general inflammatory response genes, others such as SLC26a, MARCKSL1, and RASGRP1 were more specific to a NOD2-driven gene expression profile. The gene set of RIPK2 kinase-driven genes was then narrowed to a set of 9 such that both general inflammatory response genes and NOD2-specific genes were represented. We then independently verified the dependence of these 9 genes on the kinase activity of RIPK2 by qRT-PCR (Fig. 1C).
FIGURE 1.
Transcriptomic profiling reveals a novel gene set specifically regulated by RIPK2 kinase activity. A, primary BMDMs were stimulated with vehicle or with each of the RIPK2 kinase inhibitors Gefitinib or SB203580, in the absence or presence of, the NOD2 agonist MDP. RNA was extracted from each condition and subjected to RNA sequencing. While Gefitinib inhibits both RIPK2 and the EGF-R, and SB203580 inhibits both RIPK2 and p38, the only shared “off-target” kinase they inhibit to the same extent as their intended target is RIPK2. Focusing on transcripts differentially expressed by both RIPK2 inhibitors during NOD2 activation allowed us to determine 26 genes that were specifically regulated by RIPK2 kinase activity. B, of the genes identified, pathway analysis showed that the RIPK2 kinase-dependent genes were preferentially involved in immune functions, cell metabolism and nucleotide/nucleoside regulation. C, of the RIPK2 kinase-dependent genes identified using RNA sequencing, 9 genes were selected and validated using qRT-PCR. Using BMDMs, all 9 genes showed significant up-regulation in the presence of MDP and a corresponding down-regulation when either of the 2 RIPK2 inhibitors, Gefitinib or SB203580, were present. Experiments were performed in duplicate on three separate occasions (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001).
Development of Novel Inhibitors of RIPK2
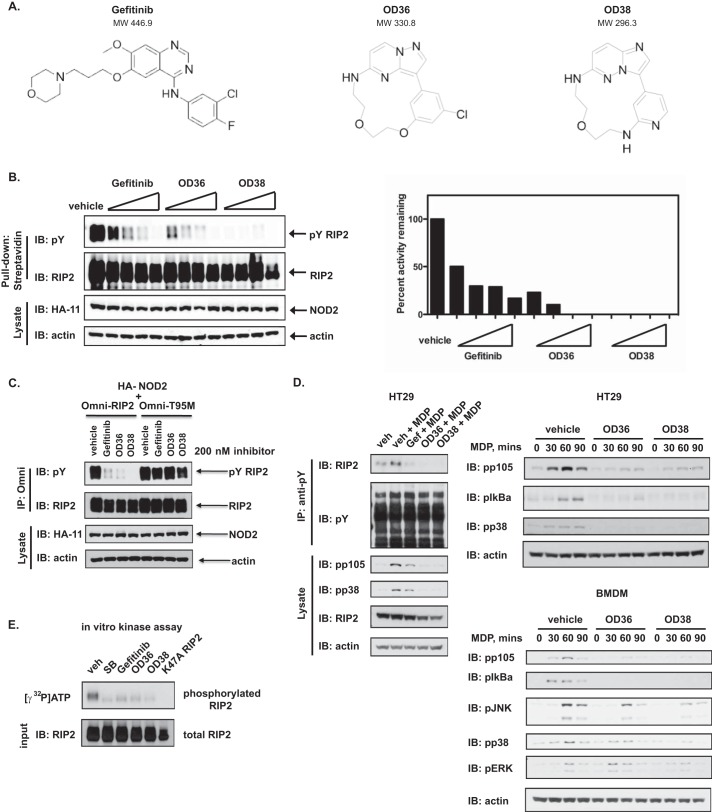
NOD2 hyperactivity has been published to occur in patients with Crohn disease who are WT for NOD2 (7, 8), in patients with Early Onset Sarcoidosis (10–13) and in mouse models of allergic inflammation (16) and multiple sclerosis (17). Additionally, since loss-of-function NOD2 polymorphisms have been shown to be protective against Ulcerative Colitis (34), inhibition of RIPK2 kinase activity might be efficacious in UC as well. For these reasons, we screened a proprietary kinase-directed Nanocyclix® library for novel RIPK2 kinase inhibitors and compared their activity to Gefitinib. These compounds were developed using a novel, small-molecule macrocyclisation platform comprised of an ATP scaffold and functionalized linker. These compounds are of low molecular weight, have predictable structure-activity relationships and drug-like properties. Initial screening was performed using a radiometric kinase assay on a recombinant kinase substrate (supplemental Fig. S1A). Those which showed IC50 < 100 nm were then further subjected to a cellular screen for RIPK2 activity (RIPK2 tyrosine autophosphorylation). Out of hundreds of such compounds, we identified 2 (OD36 and OD38), which inhibited RIPK2 activity at low nanomolar range (supplemental Fig. S1B; Fig. 2A, structures shown side-by-side with Gefitinib for comparison). When dose-response experiments were performed, these novel compounds inhibited RIPK2 activity even more potently than Gefitinib (Fig. 2B). To explore the mechanism of action of these inhibitors, we utilized a Gefitinib-resistant mutant form of RIPK2 in which the gatekeeper residue, threonine 95 (T95), has been mutated to a bulky methionine (RIPK2 T95M). The T95M RIPK2 mutation is homologous to that occurring within the EGF-R in lung cancer patients who develop resistance to Gefitinib (T790M) (22, 35). Such a mutation creates steric hindrance and prevents entry of Gefitinib into the ATP-binding pocket, thereby changing the sensitivity of RIPK2 to this drug. Neither OD36 nor OD38 could inhibit T95M RIPK2 at the concentrations used (Fig. 2C), indicating that Gefitinib, OD36 and OD38 compete with ATP for binding and that all agents were directly capable of inhibiting RIPK2. To prevent any potential artifacts generated by activation of RIPK2 induced by overexpression of NOD2, we also stimulated a colorectal epithelial cell line HT29 with MDP and looked for the effects of Gefitinib or OD36 or OD38 on inducible RIPK2 tyrosine autophosphorylation (Fig. 2D). We additionally looked at the effect of these novel RIPK2 inhibitors on MAPK and NF-κB pathways (Fig. 2D, HT29 and BMDM time course). In both cases, Gefitinib, OD36 and OD38 inhibited both RIPK2 tyrosine autophosphorylation as well as downstream NF-κB and MAPK signaling induced by MDP. As these compounds were found to act as ATP-competitive inhibitors of RIPK2, we would expect that OD36 and OD38 equally affect both the Tyr and Ser/Thr-directed activities of RIPK2. RIPK2 is known to autophosphorylate on Ser, Thr and Tyr residues (22), and indeed, subjecting RIP2 to a radiometric in vitro kinase assay demonstrates loss of γ-32P autophosphorylation in the presence of Gefitinib, SB203580, OD36, and OD38 suggesting that these agents inhibit total kinase activity (kinase activity targeted against Tyr, Ser, and Thr) (Fig. 2E). We additionally tested the effects of SB20350 (a Ser/Thr inhibitor) as well as the new RIPK2 inhibitors on the reported pS176 RIPK2 autophosphorylation site (36)(supplemental Fig. S2). All 3 showed inhibition of MDP-induced RIPK2 phosphorylation suggesting that these inhibitors affect the kinase function of RIPK2 regardless of the amino acid targeted for phosphorylation.
FIGURE 2.
Development of novel inhibitors of RIPK2. An inhibitor library developed using Nanocyclix technology was screened for activity directed against RIPK2's tyrosine kinase function. Initial screening was performed using in vitro radiometric kinase assays testing recombinantly expressed RIPK2 activity on a synthetic substrate in the presence of the various Nanocyclix compounds. Of over hundreds screened, 2 showed strong activity against RIPK2 (supplemental Fig. S1). A. The structure of these two novel RIPK2 inhibitory compounds, as compared with Gefitinib, are presented here. B, these two compounds, OD36 and OD38, were tested in dose-response assays against Gefitinib to determine their comparable ability to inhibit RIPK2. While all three compounds showed that nanomolar concentrations could inhibit NOD2-mediated RIPK2 tyrosine autophosphorylation in HEK293 cells, both OD36 and OD38 displayed a more potent activity against RIPK2 (quantified data using a chemiluminescent CCD imager are shown to the right). Experiments were performed twice with similar results. C, to further determine the mode of action of OD36 and OD38, the T95M RIPK2 mutant was utilized. This mutation reduces the ATP binding pocket of RIPK2, preventing entry of Gefitinib and conferring a corresponding resistance to the drug. Observing a similar response using the novel RIPK2 inhibitors indicates a similar mode of action. As with Gefitinib, both OD36 and OD38 were unable to inhibit T95M RIPK2, indicating they are specific for the RIPK2 ATP-binding pocket. D, OD36 and OD38 inhibit endogenous MDP-induced RIPK2 tyrosine autophosphorylation as well as MDP-induced MAPK and NF-kB activation in two cell types. HT29 cells or BMDMs were treated with RIPK2 inhibitors overnight (250 nm) prior to stimulation with MDP. Lysates were collected 1 h after stimulation for assessment of endogenous RIPK2 tyrosine autophosphorylation (D, left panels) or at the timepoints indicated for analysis of downstream signaling (D, right panels). Experiments were performed three times with similar results. E, radiometric in vitro kinase assays using tyrosine autophosphorylation of RIPK2 as a readout of RIPK2 kinase activity show that SB203580, Gefitinib, OD36, and OD38 (500 nm) all inhibit the total kinase activity of RIPK2, indicating that as expected from their mechanism of action, both Tyr or Ser/Thr directed-activities are affected.
Specificity of Novel RIPK2 Inhibitors
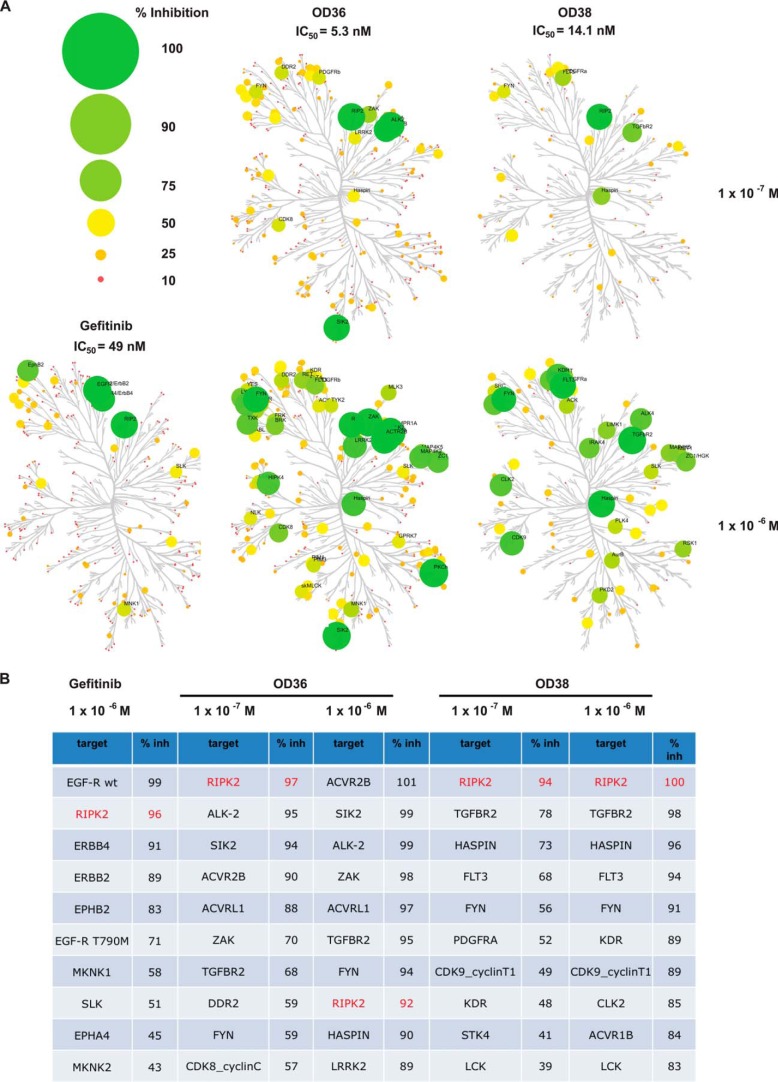
To determine the selectivity profile of these newly discovered RIPK2 inhibitors, biochemical kinase assays were performed using two concentrations of inhibitor, 100 nm and 1uM. The most potent compound (OD36) and Gefitinib were profiled against 366 kinases, whereas OD38 was screened against 88 kinases. The kinase profiles, visualized as dendrograms, and the extent of inhibition are graphically represented in Fig. 3A. At 100 nm, both OD36 and OD38 show very good initial specificity, inhibiting few other kinases to the same extent as RIPK2 and showing very high potency with IC50 values in the lower nanomolar range (OD36 having an IC50 of 5.3 nm and OD38 having an IC50 of 14.1 nm, Fig. 3A). As expected, at higher concentrations, off-target effects are increased while retaining a strong inhibitory activity against RIPK2 (summarized in Fig. 3B). The results of these specificity and dose-response assays are important for a number of reasons. First, as none of the specific RIPK2 inhibitors affect the EGF-R, this fact can be used to further validate our genetic RIPK2 activation panel independently of an effect on the EGF-R. Secondly, while Gefitinib inhibits both the EGF-R and RIPK2, it is remarkably specific for these two kinases (Fig. 3B), suggesting that it may be possible to titrate in vivo doses such that these are the only two kinases inhibited. Finally, given that these inhibitors were developed using a novel medicinal chemistry technology (Nanocyclix®) for which a wealth of data exists with regard to the properties of the proprietary library of linkers and scaffolds used to generate such compounds, knowledge of this drug discovery platform is currently being leveraged to further optimize the overall profile of these through iterative rounds of chemical modification and in vitro/in vivo testing
FIGURE 3.
Specificity of novel RIPK2 inhibitors. A, to determine the specificities of OD36 and OD38, a panel of 366 kinases were subjected to radiometric in vitro kinase assays in the presence of 100 nm or 1 mm of inhibitor and a specificity profile was then generated. Small molecule-kinase inhibition maps are shown for each of the RIPK2 inhibitors at a concentration of either 100 nm or 1 mm with the extent of inhibition indicated by both size and color (refer to key). The kinase dendogram was adapted and used with permission from Science (sciencemag.org) and Cell Signaling Technology (cellsignal.com). As indicated by the circle color and size, at 100 nm, OD36 and OD38 show good potency and selectivity for RIPK2. B, kinases most inhibited by each compound at the specified concentration and the extent of inhibition are listed. RIPK2 (highlighted) is inhibited greater than 90% by all 3 compounds at all doses tested. Furthermore, at a dose of 100 nm, few kinases other than RIPK2 are inhibited to the same extent. Additionally, this Table indicates that the new RIPK2 inhibitors have distinct off-target profiles from Gefitinib, information that is useful in ascribing effects directly to RIPK2 inhibition.
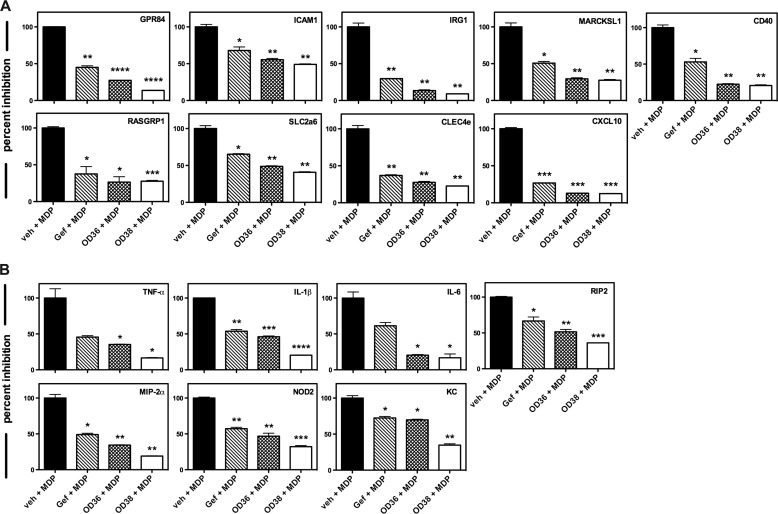
Novel RIPK2 Inhibitors Down-regulate our Defined Panel of RIPK2 Kinase-dependent Genes
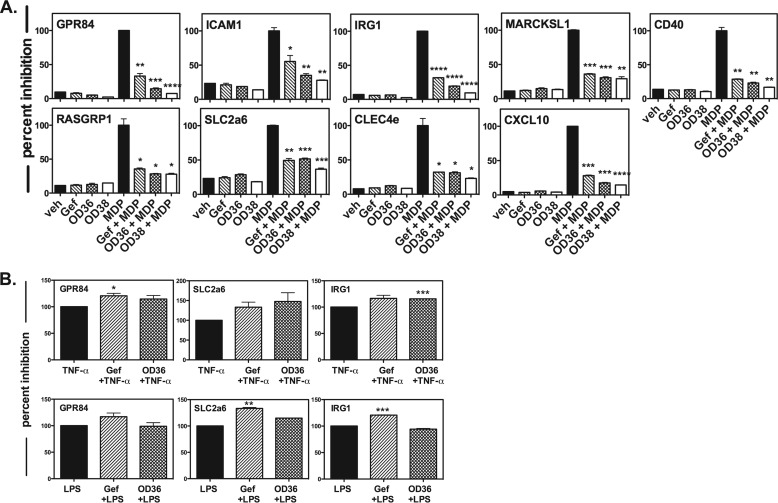
Given that the OD36 and OD38 RIPK2 inhibitors were developed using an independent medicinal chemistry technology and show different off-target effects compared with Gefitinib, down-regulation of our genetic RIPK2 activation panel would be an additional verification that this 9 gene biomarker set is suitable to confirm activation status of RIPK2. As such, we stimulated primary BMDMs with the NOD2 agonist MDP in the absence or in the presence of either Gefitinib or our novel RIPK2 inhibitors. In all cases, this 9 gene set was inhibited by OD36 and OD38 to a similar or better extent than Gefitinib (Fig. 4A). These effects were not reflective of a global down-regulation of inflammation-induced genes by these inhibitors as both Gefitinib and OD36 did not show a decrease in the 3 selected genes tested when TNF-α or LPS were used as stimuli (Fig. 4B). These finding suggest that this genetic RIPK2 activation panel may be a useful tool in addition to biochemical assays to monitor RIPK2 function as influenced by either a disease state or pharmacologic inhibition. Second, these data also support further optimization of lead compounds such as OD36 and OD38, which display a very strong inhibitory activity against RIPK2.
FIGURE 4.
Novel RIPK2 inhibitors downregulate our defined panel of RIPK2 kinase-dependent genes. A, RIPK2 kinase-dependent gene panel established in Fig. 1 was tested against the novel RIPK2 inhibitors OD36 and OD38. Primary BMDMs were incubated with vehicle or with 500 nm OD36, OD38, or Gefitinib in the absence or in the presence of 10 μg/ml MDP. OD36 and OD38 inhibited the panel of 9 MDP-induced RIPK2 kinase-dependent genes, indicating a specificity for RIPK2 kinase activity and providing additional validation for our genetic RIPK2 activation panel (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). Experiments were performed in duplicate on three separate occasions. B, to confirm that both Gefitinib and our novel RIPK2 inhibitors down-regulated the selected genes as a result of RIPK2 activation, as opposed to in response to other inflammatory stimuli, BMDMs were treated with either 10 ng/ml TNF-a (top row) or 10 ng/ml LPS (bottom row) in the absence or in the presence of either Gefitinib or OD36. Of the three genes selected (GPR84, SLC2a6 and IRG1), both inhibitors failed to down-regulate these genes in response to TNF-a or LPS stimulation, confirming their specificity for RIPK2 and RIPK2-mediated gene expression.
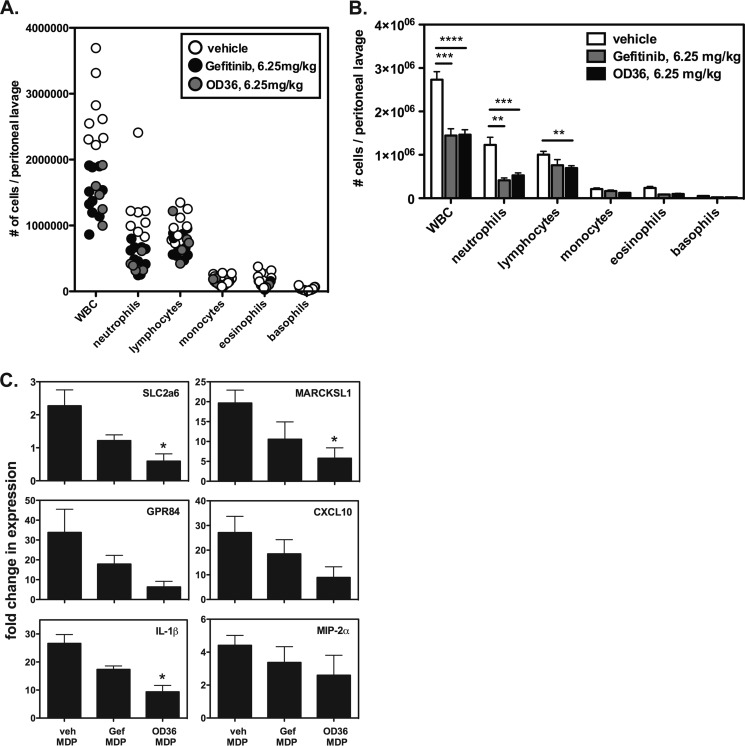
The Novel RIPK2 Inhibitor OD36 Reduces Cellular Infiltration in An in Vivo MDP-induced Peritonitis Model
To demonstrate that these novel RIPK2 inhibitors, which show potent in vitro inhibitory activity, are also functional in vivo, we subjected mice to an MDP-induced model of peritonitis. Prior to administration of MDP, we intraperitoneally (intraperitoneal) administered vehicle, 6.25 mg/kg Gefitinib, or 6.25 mg/kg OD36 30 min prior to intraperitoneal delivery of 150 μg of MDP for an additional 4 h. As shown in Fig. 5A, both Gefitinib and OD36 inhibited the recruitment of inflammatory cells to the peritoneum, specifically that of neutrophils, and, to a lesser extent, lymphocytes. Statistical analysis of peritoneal lavage cell subsets shows a significant effect of both Gefitinib and OD36 in inhibiting MDP-induced peritonitis (Fig. 5B). When we isolated RNA from the infiltrating cells and subjected these to qRT-PCR, we observed a decrease in both expression of RIPK2-specific genes as well as a decrease in inflammatory cytokine and chemokine gene expression (Fig. 5C). We also took supernatants from the peritoneal lavage and performed multiplex analysis for various chemokines. Although there was quite some variability between animals, for a number of analytes tested, there was also an observable trend in decrease of secreted chemokines in either the Gefitinib or in the OD36-treated mice (supplemental Fig. S3). These very promising in vivo results for an early-stage RIPK2 inhibitor such as OD36, which also shows potent in vitro activity and good initial selectivity, encourages continued optimization of such RIPK2 inhibitors that will retain potency but in addition display enhanced specificity and an improved half-life in vivo.
FIGURE 5.
The novel RIPK2 inhibitor OD36 reduces cellular infiltration in an in vivo MDP-induced peritonitis model. A, mice were administered vehicle, Gefitinib or the novel RIPK2 inhibitor OD36 at 6.25 mg/kg intraperitoneal 30 min before intraperitoneal delivery of 150 μg of MDP for an additional 4 h. Cellular differentials of peritoneal lavages from individual mice are shown. The data are represented as a bar graph in B for clearer presentation of statistical analysis. Both Gefitinib and OD36 show reduction of the MDP-induced recruitment of WBC, particularly that of neutrophils and lymphocytes (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). C, RNA was extracted from cellular infiltrate obtained from each experimental group and qRT-PCR was performed. Expression of both genetic RIPK2 activation markers as well as cytokines and chemokines were reduced in the Gefitinib-treated animals but more so in the animals receiving the same dose of OD36.
Genetic RIPK2 Activation Markers Predict the Efficacy of RIPK2 Inhibition in a Spontaneous Model of CD-like Ileitis
Because loss-of-function NOD2 polymorphisms cause Crohn disease in an appreciable subset of CD patients, and because hyperactive NOD2 function has also been shown to cause inflammatory disease, when thinking about RIPK2 inhibition and disease treatment, it is necessary to be able to predict which patients are likely to respond to RIPK2 inhibition. To this end, we tested the 9-gene biomarker panel in a well-established mouse model of CD. The SAMP1/YitFc mouse develops severe, spontaneous ileitis that recapitulates human Crohn's disease in a number of ways. This mouse develops discontinuous transmural inflammation, crypt abscesses, granulomas, and ileal structures (29). Additionally, like human Crohn disease, SAMP1/YitFc disease is not limited to the ileum as a significant percentage of these mice develop Crohn-like gastritis. Importantly, unlike DSS or TNBS colitis models which require disruption of the epithelium requiring subsequent EGF-R activity to re-epithelialize the ulcer bed, the SAMP1/YitFc mouse only rarely develops ulcerations (29). The NOD2 and RIPK2 genes of this strain have been sequenced and found to be wild-type. Lastly but most importantly, treatments that have shown to be efficacious in this mouse strain have been predictive of efficacy in humans (29). Given all these factors, the SAMP1/YitFc mouse is therefore a good model to determine whether RIPK2 kinase inhibition using Gefitinib would be efficacious. In order to ascertain whether the 9-gene RIPK2 activation signature we have established may be useful in predicting responsiveness of the SAMP1/YitFc mice to RIPK2 inhibition, primary bone marrow-derived macrophages (BMDMs) were generated from these mice and treated with MDP in the absence or in the presence of either Gefitinib, OD36, or OD38. Quantitative RT-PCR of the isolated RNA indicated that expression of MDP-induced genes was downregulated by pharmacologic RIPK2 inhibition (Fig. 6A). Additionally, all three RIPK2 inhibitors caused down-regulation of various inflammatory cytokines and chemokines, as well as expression of NOD2 and RIPK2 themselves (both NF-kB-driven genes) (Fig. 6B). These findings suggest that RIPK2 inhibition might be efficacious in this spontaneous ileitis-prone mouse strain.
FIGURE 6.
Pharmacologic inhibition of RIPK2 in macrophages from SAMP1/YitFc mice show down-regulation of both MDP-induced genetic RIPK2 activation markers and pro-inflammatory gene expression. A, primary BMDMs were generated from SAMP1/YitFc mice and were stimulated with 10 mg/ml MDP for 4 h either in the presence or absence of 500 nm Gefitinib, OD36 or OD38. RNA was then harvested and qRT-PCR was performed. Gefitinib, OD36, and OD38 all significantly inhibited MDP-induced, RIPK2 kinase-dependent gene expression (*, p ≤ 0.05; **, p ≤ 0.01; *** p ≤ 0.001). Experiments were performed in duplicate on three separate occasions. B, primary BMDMs were generated from SAMP1/YitFc mice and were stimulated with 10 μg/ml MDP for 4 h either in the presence or absence of 500 nm Gefitinib, OD36, or OD38. RNA was then harvested, and qRT-PCR was performed. The RIPK2 inhibitors significantly inhibited MDP-induced pro-inflammatory cytokine production in the SAMP1/YitFc macrophages (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). Experiments were performed in duplicate on three separate occasions.
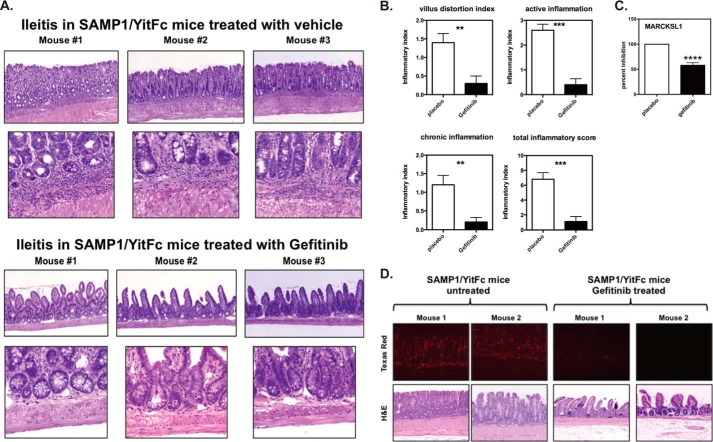
To determine if this was indeed the case in vivo, Gefitinib was incorporated into mouse food to deliver a dose each of 50 mg/kg/day. Based on the FDA Center for Drug Evaluation and Research guidelines (37), when converted, this value roughly corresponds to the lowest clinical dosage recommended in humans (250 mg daily). After 7.5 weeks of treatment, SAMP1/YitFc mice given Gefitinib or vehicle control were sacrificed, and their entire ileums and jejunums were evaluated by two blinded board-certified pathologists (Wei Xin, M.D. and Derek Abbott, M.D, Ph.D., University Hospitals of Cleveland). Mice treated with vehicle showed blunted villi and severe acute inflammation at the border of the lamina propria and muscularis mucosa (top panels, Fig. 7A). In contrast, mice treated with low dose Gefitinib showed increased villous height and no acute inflammation at the border of the muscularis mucosa and lamina propria (bottom panels, Fig. 7A). Blinded histopathology scoring using well-established criteria (29, 38) showed significant decreased villous distortion and statistically significantly decreased acute and chronic inflammation (Fig. 7B).
FIGURE 7.
Inhibition of RIPK2's kinase activity significantly ameliorates ileitis in SAMP1/YitFc mice. A, given that RIPK2 kinase inhibition decreases MDP-induced pro-inflammatory gene expression in SAMP1/YitFc macrophages, SAMP1/YitFc mice were treated with either vehicle control or Gefitinib (50 mg/kg/daily for 7.5 weeks). While the SAMP1/YitFc mice treated with vehicle control showed severe villous blunting, crypt abscesses, mucosal, and transmural inflammation and fibrosis (top panels), these features were significantly diminished in mice treated with Gefitinib (lower panels). The experiment was repeated three times with 5 mice per experimental group. Representative histology is shown. B, blinded histopathological scoring using published criteria for ileal inflammation in the SAMP1/YitFc mouse was performed. There was significantly less acute inflammation, villous distortion, and chronic inflammation in SAMP1/YitFc mice treated with Gefitinib (**, p ≤ 0.01; ***, p ≤ 0.001). C, RNA was extracted from ileal tissue obtained from placebo or Gefitinib-treated SAMP1/YitFc and subjected to qRT-PCR. SAMP1/YitFc mice treated with Gefitinib show decreased expression of MARCKSL1. D, SAMP1/YitFc mice were treated with Gefitinib (50 mg/kg/daily for 7.5 weeks) or with vehicle control. Formalin-fixed tissue was immunostained with both an anti-phospho tyrosine antibody (mouse) and an anti-RIPK2 antibody (rabbit) and subjected to a proximity ligation assay to detect tyrosine phosphorylated RIPK2 (DuoLink, Sigma). Mice treated with Gefitinib showed significantly less tyrosine phosphorylated RIPK2 staining, indicating that Gefitinib inhibits RIPK2 autophosphorylation in vivo in this ileitis-prone mouse model.
Gefitinib is well-known to inhibit EGF-R in vivo at the doses used, and, despite equal potency in in vitro kinase assays and tissue culture systems, Gefitinib has never been shown to inhibit RIPK2 in in vivo disease systems. To confirm the effect of Gefitinib in affecting RIPK2 activity in vivo, we extracted RNA from ileum obtained from either placebo or Gefitinib-treated SAMP/YitFc mice, and subjected the RNA to qRT-PCR for our genetic RIPK2 activation markers. However, as this panel was initially established using BMDMs, few showed expression within the disease tissue (ileum). However, one marker that was reproducibly down-regulated upon treatment with Gefitinib was MARCKSL1 (Macrophage Myristoylated Alanine-Rich C Kinase Substrate), which has also been reported to be expressed in tissues such as the small intestine and colon (Fig. 7C). In addition, given that Gefitinib potently inhibits RIPK2's tyrosine autophosphorylation, we were able to utilize the DuoLink proximity ligation assay (Sigma Aldrich) to show that Gefitinib inhibits RIPK2 tyrosine autophosphorylation in situ in the Samp/YitFc mouse. In this system, a signal is generated only if two events (in this case, tyrosine phosphorylation and the presence of RIPK2) occur in very close proximity. Fluorescence is then indicative of RIPK2 tyrosine phosphorylation. Using this assay, RIPK2 was found to be heavily tyrosine phosphorylated in the vehicle-treated SAMP/YitFc mice but this tyrosine phosphorylation (and inflammation) was greatly decreased when Gefitinib was present (Fig. 7D). This data suggest that RIPK2 is, in fact, inhibited by Gefitinib in vivo and that such inhibition correlates with a decrease in disease severity in this mouse model of ileitis.
DISCUSSION
IBD is a chronic, relapsing inflammatory disorder of the gastrointestinal tract whose treatment and management is less than ideal (39). Evidence of increased NOD2 activity in IBD patients lacking the common loss of function NOD2 variants combined with our discovery of RIPK2 as a dual-specificity kinase, has encouraged the development of specific RIPK2 inhibitors by multiple companies with the goal of introducing a new therapy for this as well as other diseases showing excessive NOD2/RIPK2 activity (40, 41). In this work, we report the development of such compounds for inhibition of RIPK2 kinase function that display potent activity against RIPK2 both in vitro and in vivo. The RIPK2 inhibitors we use in this study have been developed using a proprietary novel small molecule macrocyclization platform (Nanocyclix®, Oncodesign) comprised of an ATP scaffold and functionalized linker. Most current kinase inhibitors are ATP competitive and mimic the interactions between the adenine ring and the hinge region of the target kinase. Aside from possessing a strong interaction with the hinge region, Nanoclyclix® compounds additionally utilize interactions with the hydrophobic back pocket as well interactions with the ribose binding region, thereby, more fully occupying the ATP-binding site. The beauty of such a technology is that by combining readouts of RIPK2 kinase activity (in vitro kinase assays, RIPK2 tyrosine autophosphorylation, MDP-driven cytokine release) and the in-depth knowledge of this platform by the company, Oncodesign, lead compound optimization can be iteratively improved until desired properties of selectivity and metabolic half-life are achieved. The early RIPK2 specific compounds resulting from such technology (OD36 and OD38) already display very potent inhibitory activity against RIPK2 in multiple in vitro assays, as well as an off-target profile distinct from that of Gefitinib. Importantly, we additionally demonstrate that these novel RIPK2 inhibitors are potent in vivo using an MDP-induced peritonitis model. These findings encourage the continued optimization of these lead compounds such that they may be amenable to prolonged administration in vivo, as will be the need in chronic inflammatory conditions. Because of the fact that optimization of such compounds for preclinical in vivo use requires enormous resources, repurposing already approved therapies to demonstrate in vivo feasibility of RIPK2 inhibition would provide the rationale needed to further invest in improving the in vivo half-life and selectivity of such lead compounds. In the current work, we show that inhibition of RIPK2 by repurposing the EGF-R inhibitor Gefitinib resulted in a striking reduction in inflammation in a spontaneous model of Crohn Disease ileitis. Using a proximity ligation assay (PLA), we also demonstrate that RIPK2 tyrosine phosphorylation is reduced within the ileal tissue in Gefitinib-treated animals concurrent with a reduction of inflammation. Collectively, these results support further optimization and pre-clinical testing of specific RIPK2 inhibitors.
Given that targeting RIPK2 in inflammatory disease requires a functional and potentially hyperactive NOD2 response, we also introduce and utilize additional assays to detect RIPK2 activity and inhibition. We utilized Next-Gen sequencing to establish a genetic RIPK2 activation panel, which may be useful in determining whether a particular setting may be amenable to RIPK2 inhibition or if a patient is likely to be responsive to RIPK2 treatment. This technology may be broadly applicable to inflammatory disease as both NOD2 and RIPK2 are NF-κB regulated genes, and their expression is up-regulated upon exposure to TNF, IL-1, and IFN (6) as well as in acute ileitis and colitis (7, 8). Studies have shown that NOD1, NOD2, or RIPK2 loss causes decreased inflammation in a number of mouse models of inflammatory disease, including EAE (17), inflammatory arthritis (14, 15), allergic inflammation (16), sarcoidosis (42), and ulcerative colitis (43). Studies in human pediatric Crohn's disease in which patients are WT for NOD2 has shown heightened NOD2 and RIPK2 activity (7, 8), and agents targeting both RIPK2 and p38 have been efficacious in mouse models of inflammatory bowel disease (26, 44). Genotyping IBD patients with the aim of personalized treatment for those harboring the commonly occurring, CD-predisposing, loss-of-function NOD2 alleles has, so far, not altered the course of clinical treatment. This is, in part, due to the poor predictive ability of having a single loss-of-function allele on the disease phenotype. Although compound heterozygosity and homozygosity for these variants appears to correlate with more aggressive disease, there is still no proven treatment that will be beneficial for this subset. However, genotyping IBD patients for NOD2 status may be useful for the remaining CD as well as UC patients who lack such polymorphisms and are, therefore, candidates for RIPK2 inhibition. Preselecting for such individuals will allow us to utilize the genetic RIPK2 activation panel we describe herein to determine which patients show heightened RIPK2 activity compared with controls as well as display down-regulation of RIPK2 induced genes upon exposure to inhibitor in vitro, ultimately defining a subset of individuals who are likely to benefit from in vivo RIPK2 inhibition. Taken together, in this work we have identified novel RIPK2 inhibitors, we demonstrate the feasibility and efficacy of in vivo pharmacologic inhibition of RIPK2 in a mouse model of IBD, and we establish genetic and immunohistochemical methods of monitoring RIPK2 activation with the ultimate goal of moving this potential therapy closer to a personalized treatment for inflammatory disease.
Supplementary Material
Acknowledgments
We thank Drs. XiaoXia Li and Christine McDonald (CCF, Cleveland OH), Klaus Ley, Mitchell Kronenberg, Hilde Cheroutre (LIAI, La Jolla, CA), and Jerrold Turner (Univ. Chicago) for helpful comments and critiques on the manuscript.
This work was supported by a Burroughs Wellcome Career Award (to D. W. A.), an R01 GM086550 (DWA) and a P01 DK091222 (to F. C., T. T. P., and D. W. A.) and an American Cancer Society Postdoctoral Fellowship Award PF-1 1-058-01-MPC and K99 HL122365-01 award (to J. T. T-A). Financial Conflict of Interest: Drs. Benderitter, Hoflack, and Abbott have financial interests in the development of novel RIP2 inhibitors.

This article contains supplemental Figs. S1–S3.
IUPHAR database (2013).
- MDP
- muramyl dipeptide
- IBD
- inflammatory bowel disease
- NLR
- Nod-like receptor
- CD
- Crohn Disease
- LOF
- loss-of-function
- FDR
- false discovery rate.
REFERENCES
- 1. Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 [DOI] [PubMed] [Google Scholar]
- 2. Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S. J., Moran A. P., Fernandez-Luna J. L., Nuñez G. (2003) Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278, 5509–5512 [DOI] [PubMed] [Google Scholar]
- 3. Hampe J., Cuthbert A., Croucher P. J., Mirza M. M., Mascheretti S., Fisher S., Frenzel H., King K., Hasselmeyer A., MacPherson A. J., Bridger S., van Deventer S., Forbes A., Nikolaus S., Lennard-Jones J. E., Foelsch U. R., Krawczak M., Lewis C., Schreiber S., Mathew C. G. (2001) Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet 357, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 4. Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 5. Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 6. Gutierrez O., Pipaon C., Inohara N., Fontalba A., Ogura Y., Prosper F., Nunez G., Fernandez-Luna J. L. (2002) Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-κB activation. J. Biol. Chem. 277, 41701–41705 [DOI] [PubMed] [Google Scholar]
- 7. Negroni A., Stronati L., Pierdomenico M., Tirindelli D., Di Nardo G., Mancini V., Maiella G., Cucchiara S. (2009) Activation of NOD2-mediated intestinal pathway in a pediatric population with Crohn's disease. Inflammatory Bowel Diseases 15, 1145–1154 [DOI] [PubMed] [Google Scholar]
- 8. Stronati L., Negroni A., Merola P., Pannone V., Borrelli O., Cirulli M., Annese V., Cucchiara S. (2008) Mucosal NOD2 expression and NF-κB activation in pediatric Crohn's disease. Inflammatory Bowel Diseases 14, 295–302 [DOI] [PubMed] [Google Scholar]
- 9. Li J., Moran T., Swanson E., Julian C., Harris J., Bonen D. K., Hedl M., Nicolae D. L., Abraham C., Cho J. H. (2004) Regulation of IL-8 and IL-1β expression in Crohn's disease associated NOD2/CARD15 mutations. Human Molecular Genetics 13, 1715–1725 [DOI] [PubMed] [Google Scholar]
- 10. Henckaerts L., Vermeire S. (2007) NOD2/CARD15 disease associations other than Crohn's disease. Inflammatory Bowel Diseases 13, 235–241 [DOI] [PubMed] [Google Scholar]
- 11. Kanazawa N., Okafuji I., Kambe N., Nishikomori R., Nakata-Hizume M., Nagai S., Fuji A., Yuasa T., Manki A., Sakurai Y., Nakajima M., Kobayashi H., Fujiwara I., Tsutsumi H., Utani A., Nishigori C., Heike T., Nakahata T., Miyachi Y. (2005) Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-κB activation: common genetic etiology with Blau syndrome. Blood 105, 1195–1197 [DOI] [PubMed] [Google Scholar]
- 12. Okafuji I., Nishikomori R., Kanazawa N., Kambe N., Fujisawa A., Yamazaki S., Saito M., Yoshioka T., Kawai T., Sakai H., Tanizaki H., Heike T., Miyachi Y., Nakahata T. (2009) Role of the NOD2 genotype in the clinical phenotype of Blau syndrome and early-onset sarcoidosis. Arthritis Rheumatism 60, 242–250 [DOI] [PubMed] [Google Scholar]
- 13. Schürmann M., Valentonyte R., Hampe J., Müller-Quernheim J., Schwinger E., Schreiber S. (2003) CARD15 gene mutations in sarcoidosis. European Respiratory Journal 22, 748–754 [DOI] [PubMed] [Google Scholar]
- 14. Rosenzweig H. L., Clowers J. S., Nunez G., Rosenbaum J. T., Davey M. P. (2011) Dectin-1 and NOD2 mediate cathepsin activation in zymosan-induced arthritis in mice. Inflammation Research 60, 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosenzweig H. L., Jann M. J., Vance E. E., Planck S. R., Rosenbaum J. T., Davey M. P. (2010) Nucleotide-binding oligomerization domain 2 and Toll-like receptor 2 function independently in a murine model of arthritis triggered by intraarticular peptidoglycan. Arthritis Rheumatism 62, 1051–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duan W., Mehta A. K., Magalhaes J. G., Ziegler S. F., Dong C., Philpott D. J., Croft M. (2010) Innate signals from Nod2 block respiratory tolerance and program T(H)2-driven allergic inflammation. Journal of Allergy and Clinical Immunology 126, 1284–1293 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shaw P. J., Barr M. J., Lukens J. R., McGargill M. A., Chi H., Mak T. W., Kanneganti T. D. (2011) Signaling via the RIP2 adaptor protein in central nervous system-infiltrating dendritic cells promotes inflammation and autoimmunity. Immunity 34, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kobayashi K., Inohara N., Hernandez L. D., Galán J. E., Nuñez G., Janeway C. A., Medzhitov R., Flavell R. A. (2002) RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199 [DOI] [PubMed] [Google Scholar]
- 19. Park J. H., Kim Y. G., McDonald C., Kanneganti T. D., Hasegawa M., Body-Malapel M., Inohara N., Núñez G. (2007) RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 178, 2380–2386 [DOI] [PubMed] [Google Scholar]
- 20. Homer C. R., Kabi A., Marina-García N., Sreekumar A., Nesvizhskii A. I., Nickerson K. P., Chinnaiyan A. M., Nuñez G., McDonald C. (2012) A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J. Biol. Chem. 287, 25565–25576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 22. Tigno-Aranjuez J. T., Asara J. M., Abbott D. W. (2010) Inhibition of RIP2's tyrosine kinase activity limits NOD2-driven cytokine responses. Genes Dev. 24, 2666–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mok T. S., Wu Y. L., Thongprasert S., Yang C. H., Chu D. T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., Nishiwaki Y., Ohe Y., Yang J. J., Chewaskulyong B., Jiang H., Duffield E. L., Watkins C. L., Armour A. A., Fukuoka M. (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. New England Journal of Medicine 361, 947–957 [DOI] [PubMed] [Google Scholar]
- 24. Thongprasert S., Duffield E., Saijo N., Wu Y. L., Yang J. C., Chu D. T., Liao M., Chen Y. M., Kuo H. P., Negoro S., Lam K. C., Armour A., Magill P., Fukuoka M. (2011) Health-related quality-of-life in a randomized phase III first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients from Asia with advanced NSCLC (IPASS). J. Thorac. Oncol. 6, 1872–1880 [DOI] [PubMed] [Google Scholar]
- 25. Brehmer D., Greff Z., Godl K., Blencke S., Kurtenbach A., Weber M., Müller S., Klebl B., Cotten M., Kéri G., Wissing J., Daub H. (2005) Cellular targets of gefitinib. Cancer Res. 65, 379–382 [PubMed] [Google Scholar]
- 26. Hollenbach E., Vieth M., Roessner A., Neumann M., Malfertheiner P., Naumann M. (2005) Inhibition of RICK/nuclear factor-κB and p38 signaling attenuates the inflammatory response in a murine model of Crohn disease. J. Biol. Chem. 280, 14981–14988 [DOI] [PubMed] [Google Scholar]
- 27. Godl K., Wissing J., Kurtenbach A., Habenberger P., Blencke S., Gutbrod H., Salassidis K., Stein-Gerlach M., Missio A., Cotten M., Daub H. (2003) An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 100, 15434–15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reuter B. K., Pastorelli L., Brogi M., Garg R. R., McBride J. A., Rowlett R. M., Arrieta M. C., Wang X. M., Keller E. J., Feldman S. H., Mize J. R., Cominelli F., Meddings J. B., Pizarro T. T. (2011) Spontaneous, immune-mediated gastric inflammation in SAMP1/YitFc mice, a model of Crohn's-like gastritis. Gastroenterology 141, 1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pizarro T. T., Pastorelli L., Bamias G., Garg R. R., Reuter B. K., Mercado J. R., Chieppa M., Arseneau K. O., Ley K., Cominelli F. (2011) SAMP1/YitFc mouse strain: a spontaneous model of Crohn's disease-like ileitis. Inflammatory Bowel Diseases 17, 2566–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tigno-Aranjuez J. T., Bai X., Abbott D. W. (2013) A Discrete Ubiquitin-Mediated Network Regulates the Strength of NOD2 Signaling. Mol. Cell. Biol. 33, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bachmanov A. A., Reed D. R., Beauchamp G. K., Tordoff M. G. (2002) Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behavior Genetics 32, 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tao M., Scacheri P. C., Marinis J. M., Harhaj E. W., Matesic L. E., Abbott D. W. (2009) ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr. Biol. 19, 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jostins L., Ripke S., Weersma R. K., Duerr R. H., McGovern D. P., Hui K. Y., Lee J. C., Schumm L. P., Sharma Y., Anderson C. A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S. L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J. P., Ahmad T., Amininejad L., Ananthakrishnan A. N., Andersen V., Andrews J. M., Baidoo L., Balschun T., Bampton P. A., Bitton A., Boucher G., Brand S., Büning C., Cohain A., Cichon S., D'Amato M., De Jong D., Devaney K. L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L. R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T. H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I. C., Lees C. W., Louis E., Mahy G., Mansfield J., Morgan A. R., Mowat C., Newman W., Palmieri O., Ponsioen C. Y., Potocnik U., Prescott N. J., Regueiro M., Rotter J. I., Russell R. K., Sanderson J. D., Sans M., Satsangi J., Schreiber S., Simms L. A., Sventoraityte J., Targan S. R., Taylor K. D., Tremelling M., Verspaget H. W., De Vos M., Wijmenga C., Wilson D. C., Winkelmann J., Xavier R. J., Zeissig S., Zhang B., Zhang C. K., Zhao H., International I. B. D. G. C., Silverberg M. S., Annese V., Hakonarson H., Brant S. R., Radford-Smith G., Mathew C. G., Rioux J. D., Schadt E. E., Daly M. J., Franke A., Parkes M., Vermeire S., Barrett J. C., Cho J. H. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobayashi S., Boggon T. J., Dayaram T., Jänne P. A., Kocher O., Meyerson M., Johnson B. E., Eck M. J., Tenen D. G., Halmos B. (2005) EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. New England Journal of Medicine 352, 786–792 [DOI] [PubMed] [Google Scholar]
- 36. Dorsch M., Wang A., Cheng H., Lu C., Bielecki A., Charron K., Clauser K., Ren H., Polakiewicz R. D., Parsons T., Li P., Ocain T., Xu Y. (2006) Identification of a regulatory autophosphorylation site in the serine-threonine kinase RIP2. Cellular Signalling 18, 2223–2229 [DOI] [PubMed] [Google Scholar]
- 37. U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drugs Evaluation Research (2005) Guidance for Industry: Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers. U.S. Food and Drug Administration, Bethesda, MD [Google Scholar]
- 38. Gorfu G., Rivera-Nieves J., Hoang S., Abbott D. W., Arbenz-Smith K., Azar D. W., Pizarro T. T., Cominelli F., McDuffie M., Ley K. (2010) β7 integrin deficiency suppresses B cell homing and attenuates chronic ileitis in SAMP1/YitFc mice. J. Immunol. 185, 5561–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Girardin M., Manz M., Manser C., Biedermann L., Wanner R., Frei P., Safroneeva E., Mottet C., Rogler G., Schoepfer A. M. (2012) First-line therapies in inflammatory bowel disease. Digestion 86, 6–10 [DOI] [PubMed] [Google Scholar]
- 40. Zhao Y., Song J. H., Ajami A. M., Reinecker H-C. (2012) A new Rip2 kinase inhibitor for the treatment of intestinal inflammation. Abstract 169.5. J. Immunol. 188, 169.5 [Google Scholar]
- 41. Desai B., Haile P., Casillas L., Gough P., Bertin J., Votta B. (2012) Use of novel small molecule inhibitors to investigate the role of endogenous RIP2 kinase activity in modulating NOD2/RIP2-dependent cytokine signaling. Abstract 172.4. J. Immunol. 188, 172.4 [Google Scholar]
- 42. Tanabe T., Ishige I., Suzuki Y., Aita Y., Furukawa A., Ishige Y., Uchida K., Suzuki T., Takemura T., Ikushima S., Oritsu M., Yokoyama T., Fujimoto Y., Fukase K., Inohara N., Nunez G., Eishi Y. (2006) Sarcoidosis and NOD1 variation with impaired recognition of intracellular Propionibacterium acnes. Biochim. Biophys. Acta 1762, 794–801 [DOI] [PubMed] [Google Scholar]
- 43. Amendola A., Butera A., Sanchez M., Strober W., Boirivant M. (2013) Nod2 deficiency is associated with an increased mucosal immunoregulatory response to commensal microorganisms. Mucosal Immunol. 7, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hollenbach E., Neumann M., Vieth M., Roessner A., Malfertheiner P., Naumann M. (2004) Inhibition of p38 MAP kinase- and RICK/NF-κB-signaling suppresses inflammatory bowel disease. FASEB J. 18, 1550–1552 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.